Abstract
Nonconservative substitutions for Tyr-115 in the reverse transcriptase (RT) of human immunodeficiency virus type 1 (HIV-1) lead to enzymes displaying lower affinity for deoxynucleoside triphosphates (dNTPs) (A. M. Martín-Hernández, E. Domingo, and L. Menéndez-Arias, EMBO J. 15:4434–4442, 1996). Several mutations at this position (Y115W, Y115L, Y115A, and Y115D) were introduced in an infectious HIV-1 clone, and the replicative capacity of the mutant viruses was monitored. Y115W was the only mutant able to replicate in MT-4 cells, albeit very poorly. Nucleotide sequence analysis of the progeny virus recovered from supernatants of four independent transfection experiments showed that the Y115W mutation was maintained. However, in all cases an additional substitution in the primer grip of the RT (M230I) emerged when the virus increased its replication capacity. Using recombinant HIV-1 RT, we demonstrate that M230I mitigates the polymerase activity defect of the Y115W mutant, by increasing the dNTP binding affinity of the enzyme. The second-site suppressor effects observed were mediated by mutations in the 66-kDa subunit of the RT, as demonstrated with chimeric heterodimers. Examination of available crystal structures of HIV-1 RT suggests a possible mechanism for restoration of enzyme activity by the second-site revertant.
Human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) plays an essential role in the replication of the single-stranded genomic RNA of the virus. Reverse transcription involves the synthesis of a double-stranded proviral DNA which integrates in the host genome, in a process mediated by the RNA-dependent and DNA-dependent polymerase and the endonuclease (RNase H) activities of the enzyme (for recent reviews, see references 1, 10, and 31). HIV-1 RT is also a target for chemotherapeutic intervention in the control of AIDS. The crystallographic structure of HIV-1 RT has been determined in the absence of ligands (28), complexed with nonnucleoside RT inhibitors (15), and complexed with double-stranded DNA (13). In addition, a crystal structure of a covalently trapped catalytic complex of HIV-1 RT containing a DNA template-primer and a deoxynucleoside triphosphate (dNTP) has been recently reported (11). HIV-1 RT is a heterodimer composed of two subunits of 66 and 51 kDa, with subdomains termed fingers, thumb, palm, and connection in both subunits and an RNase H domain in the larger subunit only. The polymerase active site resides within the palm subdomain of the 66-kDa subunit, which contains the catalytic aspartic acid residues 110, 185, and 186. Other residues in their vicinity, such as Lys-65, Arg-72, Asp-113, Ala-114, Tyr-115, and Gln-151, are involved in the interaction with the incoming dNTP (11). The role of these amino acids in polymerase function has been investigated by site-directed mutagenesis (3, 33, 36). Nonconservative substitutions at several of these positions often lead to the loss of RT function and virus viability (16).
In previous studies, we used site-directed mutagenesis to produce HIV-1 RT variants with substitutions at Tyr-115. This amino acid was systematically replaced by Phe, Trp, Val, Ile, Met, Leu, Ala, Ser, Cys, Asn, His, Gly, Asp, Lys, and Pro. While the substitution of Phe for Tyr-115 rendered RT fully active, other replacements affected the polymerase activity of the enzyme, by increasing the Km values for the incorporation of dNTPs (17, 18), suggesting an altered dNTP binding function. This effect was more pronounced in those variant RTs with smaller and less hydrophobic side chains at position 115. To study the viability of virus harboring mutations at position 115, we introduced the substitutions Y115W, Y115L, Y115A, and Y115D in the RT of an infectious HIV-1 clone by site-directed mutagenesis. In transfection experiments with one of the variant RTs (Y115W mutant), viable virus was recovered, but in all cases, the genotypic analysis of the progeny revealed that it contained an additional substitution at position 230 (Ile instead of Met). This second-site reversion appears to compensate for the dNTP binding defect shown by the Y115W mutant.
MATERIALS AND METHODS
Cell lines and molecular clones.
COS-1 cells and MT-4 cells were maintained as monolayer and suspension cultures, respectively, in RPMI 1640 medium supplemented with 10% fetal bovine serum and 2 mM glutamine. Virus used in this study was an infectious molecular clone of HIV-1 isolate 89ES061 previously described (21, 22). The 5′ end of the proviral DNA, including the 5′ long terminal repeat region and the viral genes gag, pol, and vif, was cloned in the XbaI and EcoRI sites of plasmid pBSK (Stratagene) to obtain plasmid p61F A. The 3′ end of the proviral DNA, including the env gene and the 3′ long terminal repeat, was cloned in pBSK at the EcoRI and XbaI sites to generate plasmid p61F B. Cotransfection with plasmids p61F A and p61F B renders virus infectious after in vivo ligation (21). MT-4 cells were a gift of D. D. Richman (University of California at San Diego). Mutant HIV clones were obtained by site-directed mutagenesis. A 4,385-bp PstI-SalI fragment derived from p61F A, which contains the entire pol gene, was cloned in the mutagenesis vector pALTER-1 (Promega). In vitro mutagenesis reactions were carried out by following the manufacturer’s instructions, and mutations Y115W, Y115L, Y115A, and Y115D were introduced with the mutagenic oligonucleotides previously described (17, 18). The mutagenized PstI-SalI insert was then excised and cloned back into p61F A for its use in the transfection experiments.
DNA transfection experiments.
Transfections were performed as previously reported (21). Briefly, 3 × 106 COS-1 cells were electroporated with 10 μg of subgenomic clones p61F A and p61F B. Forty-eight hours after transfection, 4 × 106 MT-4 cells were added to the culture. Viral replication was monitored by RT activity and p24 antigen detection in culture supernatants. Virion-associated RT activity was determined as described by Willey et al. (34), after 10 μl of transfection supernatant was mixed with 50 μl of an RT reaction mixture which contained a template-primer of poly(rA) (5 μg/ml) and oligo(dT)12–18 (1.57 μg/ml) in 50 mM Tris (pH 7.8)–75 mM KCl–2 mM dithiothreitol–5 mM MgCl2–0.05% Nonidet P-40–0.5 μCi of [32P]dTTP (800 Ci/mmol). In some assays, cold dTTP was added to the reaction mixture to achieve a final nucleotide concentration of 10 μM. Production of p24 antigen in cell-free supernatants was measured with an antigen capture kit (SAIC Frederick, AIDS Vaccine Program).
Infections.
Supernatants from transfection experiments collected on days when maximum levels of p24 antigen were detected were used to infect 5 × 106 fresh MT-4 cells. Viruses were grown to obtain a stock, which was then titrated in an MT-4 plaque assay (9, 29). Infections were carried out at a multiplicity of infection of 0.01 PFU per cell, and viral replication was monitored by measuring RT activity in culture supernatants and determination of cell viability by the trypan blue staining method.
RNA isolation, amplification, and nucleotide sequence analysis.
Culture supernatants were passed through 0.45-μm-pore-size filters and treated with DNase A for 30 min to eliminate input DNA. Total RNA was obtained from 20 μl of treated transfection supernatants (2). RNA amplification was done with the one-tube RT-PCR PCR system (Titan; Boehringer Mannheim) with primers 47RU (5′ GTATTAGTAGGACCTACACCT 3′, positions 2055 to 2075) and 59RD (5′ ATGATTCCTAATGCATATTGTGAGT 3′, complementary to positions 3622 to 3646). Primers are numbered according to the sequence of Ratner et al. (27). These primers amplify a 1,591-bp fragment. After reverse transcription at 50°C for 30 min and a 5-min incubation at 94°C, samples were subjected to 35 rounds of amplification. Each cycle comprised a 1-min denaturation step at 94°C, a 1-min annealing step at 55°C, and a 2-min extension step at 72°C. Nucleotide sequence analysis of the amplified product was performed with the fmol DNA sequencing system (Promega, Madison, Wis.) with the following primers: 14RD (5′ GCACGATATCTAATCCTGGTGTCTCA 3′, complementary to positions 2540 to 2561), 3′RU (5′ GCGGGATCCTGAAAATCCATACAATACTC 3′, positions 2278 to 2304), 15′RU (5′ TAGATATCAGTACAATGTGCTTCCAC 3′, positions 2555 to 2580), 58RU (5′ GCCAGAAAAAGACAGCTGGACTGT 3′, positions 2867 to 2890), and 59RD (5′ ATGATTCCTAATGCATATTGTGAGT 3′, complementary to positions 3622 to 3646). When sequences revealed a heterogeneity at a given position, the relative proportion of each mutant was estimated by densitometry of the specific bands in the sequencing gel, with a PhosphorImager apparatus and the PCBAS program.
Expression and purification of recombinant HIV-1 RTs.
Recombinant HIV-1 RT mutants used in this study were previously described (17, 18), except for those having Ile-230 instead of Met. In this case, the mutation was introduced with the Altered Sites in vitro mutagenesis system kit from Promega, following the manufacturer’s instructions. The mutagenic oligonucleotide used was 5′ GGAGTTCATAACCGATCCAAAGGAATGG 3′, and the introduced mutation was confirmed by DNA sequencing. Purification of mutant and wild-type (WT) HIV-1 RTs was carried out after independent expression of their subunits (p66 and p51), by following a previously described procedure (17). The 51-kDa subunit was obtained with an extension of 14 amino acid residues at its N-terminal end, including six consecutive histidines to facilitate its purification by metal chelate affinity chromatography. All RTs were purified as p66-p51 heterodimers. Briefly, RTs were prepared by mixing the cell pellets of p66 and p51 clones at a 10:1-to-15:1 ratio. The bacteria were lysed, sonicated, and centrifuged, and supernatants were applied to a nickel-nitriloacetic acid-agarose column (Invitrogen Corporation, Carlsbad, Calif.). The column was washed, and histidine-tagged RT was eluted with an imidazole gradient. RT-containing fractions were pooled; dialyzed against 50 mM Tris-HCl (pH 7.0) containing 25 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, and 10% glycerol (buffer A); and then passed through Q Sepharose (Amersham Pharmacia, Uppsala, Sweden). The RT was recovered in the nonbinding fraction and loaded onto Bio-Rex 70 (Bio-Rad Laboratories, Hercules, Calif.). RT was eluted with an NaCl gradient, and the RT-containing fractions were dialyzed against buffer A, concentrated in Centriprep-30 and Centricon-30 (Amicon, Beverly, Mass.) to less than 0.5 ml, and stored at −20°C. RT preparations were up to 90 to 95% pure, as judged by examination of Coomassie blue-stained gels.
Single nucleotide extension assays.
Oligonucleotides PG5 (5′ TGGTAGGGCTATACAT 3′) and pT (5′ GGATTTTAGACAGGAACGGT 3′) were labeled at their 5′ termini with [γ-32P]ATP with T4 polynucleotide kinase, as previously described (18). The phosphorylated primers were then annealed to templates. In the case of PG5, the template used was D2 (5′ GGGATTAAATAAAATAGTAAGAATGTATAGCCCTACCA 3′), a 38-mer synthetic oligonucleotide representing the HIV-1 gag sequence that extends from nucleotides 1137 (5′ end) to 1174 (3′ end) according to the sequence numbering of Ratner et al. (27). M13mp2 single-stranded DNA was the template used with oligonucleotide pT. The templates and their corresponding primers were annealed in 150 mM NaCl–150 mM magnesium aspartate as described previously (18). Nucleotide incorporation assays were performed in 20 μl of 50 mM HEPES (pH 7.0)–15 mM NaCl–15 mM magnesium aspartate–130 mM KCH3COO–1 mM dithiothreitol–5% polyethylene glycol 6000 (18). The template-primer concentration was 30 nM for D2/PG5 and 20 nM for M13mp2 single-stranded DNA/pT. The active enzyme concentration in these assays was around 6 nM. Reactions were initiated by incubating the enzyme with the corresponding annealed template-primer in the absence of dNTP (10 min at 37°C), followed by the addition of appropriate dNTPs at various concentrations. The reaction mixtures were incubated for 30 s at 37°C, and then the reactions were stopped by adding 8 μl of 10 mM EDTA in 90% formamide containing 3 mg of xylene cyanol FF per ml and 3 mg of bromophenol blue per ml. The extension products were resolved by electrophoresis in 20% polyacrylamide–urea gels and quantitated with a BAS 1500 scanner. Elongation measurements were fitted to the Michaelis-Menten equation, and the kcat and Km values were determined as previously described (17).
RESULTS
In vitro replication of HIV-1 with mutations affecting Tyr-115 of RT.
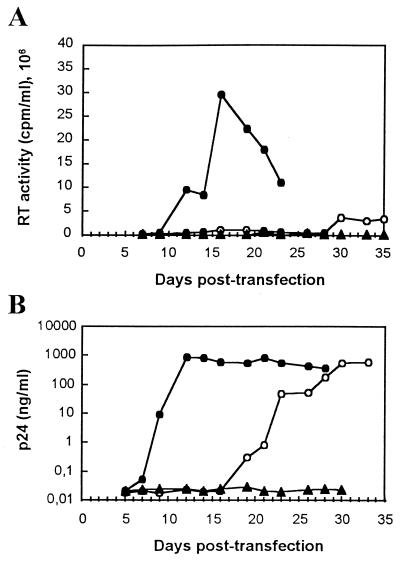
To examine the effect on virus replication of several mutations affecting Tyr-115 of HIV-1 RT, we introduced the mutations Y115W, Y115L, Y115A, and Y115D into the proviral genome. COS-1 cells were transfected with WT or mutated proviral DNA, and after addition of MT-4 cells, coculture supernatants were monitored at various times for RT activity and p24 antigen level. When WT DNA was used, RT activity was detected around 12 days after transfection, reaching a maximum level between days 14 and 18 (Fig. 1A), and p24 was detected in the culture supernatants from day 9 (Fig. 1B). In contrast, cultures transfected with mutants Y115L, Y115A, and Y115D failed to produce any detectable RT activity even after 52 days of culture and were found to be negative for the presence of p24 (values indistinguishable from those of the mock-transfection assays in Fig. 1). Mutant Y115W displayed significant levels of RT activity from day 30 after transfection (Fig. 1A) and significant amounts of p24 in culture supernatants from day 19 after transfection (Fig. 1B). Supernatants of these cultures were able to infect MT-4 cells. The data shown in Fig. 1 correspond to a single transfection experiment, but similar results were obtained in three additional transfection experiments. The WT virus and the virus recovered from transfections with Y115W were the only ones that produced infectious progeny, although the detection of RT activity in the supernatant was significantly delayed in the case of Y115W, emerging more than 30 days after transfection in the three other experiments (Table 1).
FIG. 1.
Replication kinetics of WT HIV-1 and Tyr-115 mutant virus in MT-4 cells. COS-1 cells were transfected by electroporation with 10 μg of each proviral DNA, as indicated in Materials and Methods. Forty-eight hours after transfection, MT-4 cells were added. Samples were withdrawn from cultures transfected with WT provirus (●), mutant Y115W (○), and mock (no DNA) (▴) at various days after transfection and then assessed for RT activity (A) and for the presence of p24 antigen (B).
TABLE 1.
Cell transfections with HIV-1 cDNA clones
| Expt no. and clone | Maximum level in culture supernatantsa
|
Nucleotide sequence of recovered virusb
|
||||
|---|---|---|---|---|---|---|
| RT activity
|
p24 antigen
|
|||||
| Day | 106 cpm/ml | Day | ng/ml | Codon 115 | Codon 230 | |
| 1c | ||||||
| WT | 16 | 29.5 | 12 | 900 | TAT (Tyr) | ATG (Met) |
| Y115W | 30 | 3.8 | 30 | 600 | TGG (Trp) | ATA (Ile) |
| Y115L | − | <0.5 | ND | |||
| Y115A | − | <0.5 | ND | |||
| Y115D | − | <0.5 | ND | |||
| 2 | ||||||
| WT | 14 | 19 | 12 | 600 | TAT (Tyr) | ATG (Met) |
| Y115W | 44 | 5.0 | 40 | 1,500 | TGG (Trp) | ATA/ATT (Ile)d |
| Y115L | − | <0.5 | − | <0.05 | ||
| Y115A | − | <0.5 | − | <0.05 | ||
| Y115D | − | <0.5 | − | <0.05 | ||
| 3 | ||||||
| WT | 12 | 22.0 | ND | TAT (Tyr) | ATG (Met) | |
| Y115W | 35 | 3.15 | ND | TGG (Trp) | ATA (Ile) | |
| Y115L | − | <0.5 | ND | |||
| Y115A | − | <0.5 | ND | |||
| Y115D | − | <0.5 | ND | |||
| 4 | ||||||
| WT | 12 | 22.0 | ND | TAT (Tyr) | ATG (Met) | |
| Y115W | 45 | 2.0 | ND | TGG (Trp) | ATA/ATT (Ile)d | |
RT activity and p24 antigen level were determined periodically in the cell culture medium of all transfections. Days and values reported were those obtained at the peak of RT activity or p24 antigen. The minus sign indicates that levels were not distinguishable from background (mock-infected cells), at all times tested (up to 30 days after transfection in experiments 1 and 3 and up to 45 days in experiments 2 and 4). ND, not determined.
The sequences reported were obtained from viral RNA amplified from supernatants collected when maximum RT activity was detected. Procedures are described in Materials and Methods.
The variation in RT activity and p24 antigen level following transfection and infection by WT and mutant Y115W in this experiment is depicted in Fig. 1.
ATA/ATT means that a mixture of the two nucleotides at the third base of the codon was present, according to the band pattern observed in sequencing gels.
Mutation M230I restores the replication capacity of the Y115W mutant.
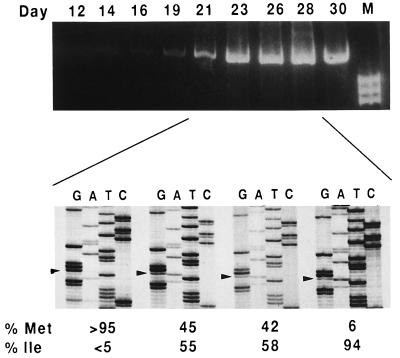
The complete RT coding region of viruses recovered from transfections with the Y115W mutant was determined, after amplification of viral RNA from culture supernatants obtained at the peak of RT activity. In all four transfection experiments, only one mutation was observed in the RT coding region, which resulted in the substitution of Ile for Met-230 (Table 1). This amino acid change was produced by a G-to-A transition at the third base of codon 230. In two transfections, a mixture of A and T was detected at this position (Table 1). In all cases, Trp-115 was maintained. To monitor the kinetics of appearance of the Ile-230 variant, we carried out RT-PCR and nucleotide sequence analysis with transfection supernatants withdrawn on various days after transfection (Fig. 2). A weak band of amplification was obtained from supernatants collected on days 16 and 19 after transfection, indicating a very low level of viral replication. By day 23, a significant increase in the amplified band was observed. Nucleotide sequencing of the amplified products and a quantitation of the relative proportions of Met and Ile at position 230 were also carried out. In the supernatants of day 23, Ile-230 appeared instead of Met, and this was coincident with a sharp increase in the amount of RT-PCR-amplified material. The appearance of substitution M230I correlated with an increase in p24 antigen production (compare Fig. 1B and 2). In the peak of RT activity (day 30 posttransfection of transfection experiment 1), more than 95% of the viral population contained Ile at position 230. These data reveal the rapid imposition of a mutant HIV-1 with Ile at position 230. Mutations Y115W and M230I were the only ones found in the RT-encoding region of viruses recovered in any of the four experiments in which cells were transfected with mutant Y115W. Thus, substitution M230I appears to restore the replication capacity of mutant Y115W, as a compensatory mutation for the presence of Trp at RT position 115.
FIG. 2.
Kinetics of imposition of Ile over Met-230 in viruses evolving after transfection with a clone harboring replacement Y115W. RT-PCR was carried out with transfection supernatants obtained at various days after transfection. Bands shown in lane M derive from digestion of ΦX174 DNA with HaeIII and correspond to fragments of 1,353, 1,078, and 872 bp, top to bottom, respectively. The relative amounts of Met and Ile at position 230 are shown below and were estimated by densitometry of the specific bands in the sequencing gels of the PCR products corresponding to days 21, 23, 26, and 28. Arrowheads indicate the position of the third base of codon 230 (G for Met-230 and A for Ile-230).
Enzymatic characterization of RT mutants.
To study whether M230I affected RT activity, steady-state kinetic parameters for the incorporation of nucleotides at the 3′ end of the primer were obtained for the WT RT, the single mutants Y115W and M230I, and the double mutant Y115W/M230I (Table 2). All of them displayed roughly similar kcat values. However, the Km values for the incorporation of dNTP were 75- to 130-fold higher for the single mutant Y115W than for the WT enzyme, resulting in a reduced catalytic efficiency, which is the likely cause of the replication defect observed in viruses harboring this mutation. On the other hand, WT RT and single mutant M230I had similar affinities for the incoming nucleotide. Interestingly, the presence of M230I together with Y115W resulted in an enzyme with an increased catalytic efficiency, which displayed a 9- to 65-fold-higher affinity for dNTP, relative to the single mutant Y115W. This effect can be attributed to the presence of both amino acid substitutions in the 66-kDa subunit of HIV-1 RT, as demonstrated by using chimeric heterodimers where substitutions were introduced in p66, p51, or both subunits (Table 2).
TABLE 2.
Kinetic parameters for dNTP incorporation of WT and mutant RTsa
| Enzyme | Value for template-primer:
|
|||
|---|---|---|---|---|
| D2/PG5
|
M13mp2 ssDNA/pT
|
|||
| Km (μM) | kcat (min−1) | Km (μM) | kcat (min−1) | |
| WT | 0.173 ± 0.027b | 3.07 ± 0.54b | 0.082 ± 0.032 | 2.54 ± 0.11 |
| Y115W | 13.4 ± 1.6 | 2.92 ± 0.12 | 10.6 ± 1.6 | 4.12 ± 0.19 |
| M230I | 0.266 ± 0.031 | 1.97 ± 0.53 | 0.045 ± 0.009 | 6.73 ± 0.48 |
| Y115W/M230I | 1.49 ± 0.09 | 3.87 ± 0.35 | 0.162 ± 0.021 | 3.06 ± 0.86 |
| p66Y115W/p51WT | 11.5 ± 1.1 | 3.76 ± 0.11 | 7.5 ± 0.6 | 3.14 ± 0.06 |
| p66Y115W/p51M230I | 13.6 ± 1.1 | 3.60 ± 0.09 | 12.5 ± 1.2 | 4.65 ± 0.51 |
| p66Y115W/M230I/p51WT | 3.09 ± 0.45 | 7.52 ± 3.50 | 0.226 ± 0.013 | 7.48 ± 0.12 |
The incorporated nucleotides at the 3′ end of the primer were dTTP for template-primer D2/PG5 and dATP for M13mp2 ssDNA/pT. Data shown are the mean values ± standard deviations, obtained from a nonlinear least-squares fit of the kinetics data to the Michaelis-Menten equation. Each of the experiments was performed independently at least twice. The substitutions present in each subunit of the heterodimer are shown by a superscript.
Reported data for this enzyme with template-primer D2/PG5 were taken from reference 19.
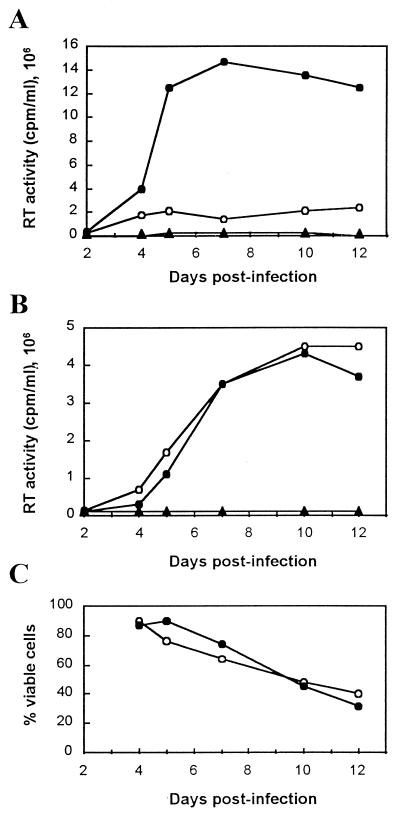
The lower levels of RT activity obtained from transfection supernatants with viruses having both mutations, Y115W and M230I (Fig. 1A), could be explained by the lower affinity for dNTP of the double mutant than of the WT enzyme, since those RT activity assays were carried out in the presence of very low concentrations of dTTP. However, the high levels of p24 antigen detected in transfection supernatants of WT and Y115W (Fig. 1) suggested that viruses recovered from both transfection experiments were viable and replicated normally. The growth curves obtained upon infection of MT-4 cells with WT HIV-1 and the double mutant Y115W/M230I show that the two viruses had similar replication kinetics (Fig. 3). Interestingly, the double-mutant virus was more sensitive to the concentration of dTTP in the RT activity assay. In agreement with the enzymological data, the differences in RT activity between the WT HIV-1 and the double-mutant virus were minimized when the concentration of dTTP in the assay was increased.
FIG. 3.
Replication kinetics of WT HIV-1 and the double mutant Y115W/M230I. MT-4 cells (106) were infected at a multiplicity of infection of 0.01 PFU per cell. Infections were monitored by measuring RT activity (A and B) and cell viability (C). RT activity was determined either in the presence of 12.5 nM dTTP (A) or in the presence of 10 μM dTTP (B). ●, WT HIV-1; ○, double mutant Y115W/M230I; ▴, mock.
DISCUSSION
Several lines of evidence support the role of Tyr-115 and Met-230 in RT function. We have found that viruses harboring the mutation Y115L, Y115A, or Y115D showed no signs of virus replication (Table 1). Similar results were previously reported for mutant Y115N (16). In the present study, we have also shown that the Y115W variant replicates very poorly until the emergence of mutation M230I. Tyr-115 is part of a conserved motif in many polymerases (25). Tyr and Phe are the only amino acids found at this position in retroviral RTs, and Phe is the only residue which can replace Tyr-115 of HIV-1 RT without causing a detrimental effect in its DNA polymerase activity (17, 18). The Y115F appears in vitro after passage of the virus in the presence of abacavir and confers low-level resistance to this carbocyclic nucleoside inhibitor (32). In Moloney murine leukemia virus RT, an aromatic amino acid residue at this position is required for infectivity, since only the WT Phe-155 or Tyr can support virus replication (7). Met-230 is part of a conserved motif found in many RNA-dependent DNA polymerases, including RTs and telomerases (20, 25, 37). All retroviral RTs have Met or Leu at this position. In the case of HIV-1, the substitution of Leu for Met-230 has been detected in virus that was passaged in cell culture, in the presence of the nonnucleoside RT inhibitor delavirdine (23). Ile-230 has been found in one nonproductive HIV-1 clone (5).
In this report, we demonstrate that the substitution of Ile for Met-230 restores the replication capacity of viruses having the Y115W mutation in the RT to WT levels. To our knowledge, this is the most dramatic compensatory effect described so far for HIV-1 clones with a defect in polymerase function. Compensatory mutations play a role in the acquisition of drug resistance, by increasing viral fitness. However, they usually appear in viruses which already display a significant replicative capacity. Examples for RT are the substitution of Ile for Val-75 that improves the replication capacity of multidrug-resistant viruses having the mutations Q151M and F77L (12) and the substitution L74V, which, besides V75I, can stimulate viral growth in quinoxaline-resistant viruses having substitution G190E (4, 14). In both cases, the compensatory mutation appears in the FINGERS subdomain and could affect the positioning of the template in the binding cleft of HIV-1 RT. Our results show that the substitution Y115W impairs RT activity by decreasing the dNTP binding affinity of the polymerase, but the acquisition of the M230I mutation compensates for the dNTP binding defect. The crystallographic structure of HIV-1 RT reveals that Tyr-115 is located close to the polymerase active site, while Met-230 is part of the primer grip which forms the β12-β13 hairpin, including residues 227 to 235. The ribose moiety of the incoming dNTP projects into a small pocket lined by the side chains of Asp-113, Tyr-115, Phe-116, and Gln-151 (11). Several amino acid substitutions associated with resistance to nucleoside analogue inhibitors lie within the dNTP binding site (e.g., K65R, L74V, M184V, F116Y, and Q151M) (1, 6, 30). Met-230 interacts with the 3′-terminal phosphate of the primer, making extensive contacts with nucleotides of the 3′ primer terminus and with the side chain of Tyr-183 (6, 11, 13). It has been suggested that binding of nonnucleoside RT inhibitors could induce a conformational change leading to repositioning of the primer grip (6), but none of the amino acids surrounding Met-230 appears to be involved in major drug resistance mutations. Met-230 influences the proper positioning of the RT on the template-primer. Its replacement by Ala led to alterations of both polymerase and RNase H activities (8, 24) and affected RNA primer selection, a step which is important at the initiation of minus-strand DNA synthesis (26). Not surprisingly, the mutation M230A caused severe defects in proviral DNA synthesis and rendered virus noninfectious (38). Crystallographic data have implicated the primer grip in orienting the primer terminus for nucleophilic attack on an incoming dNTP. In agreement with this proposal, it has been shown that substitution of Ala for Met-230 produces a significant decrease in the dNTP binding affinity of the enzyme (35), an effect which is also observed with mutant HIV-1 RTs having nonconservative substitutions at position 115 (17, 18). Therefore, it is likely that Y115W leads to a less favorable positioning of the α-phosphate dNTP for attack by the 3′ OH of the primer terminus, and mutation M230I restores, at least in part, the proper alignment required for catalysis.
Restoration of RT function by direct reversion of Trp-115 to Tyr was probably limited by the two mutations needed to convert the triplet for Trp (UGG) into a triplet for Tyr (UAU or UAC). The two required mutations had to occur simultaneously in the same genomic RNA molecule since G→A alone would lead to the termination codon UAG, and G→U (or G→C) alone would produce an RT with Cys at position 115, an enzyme with a very high Km for dNTP binding (18). Therefore, the genetic makeup of the virus (its initial position in sequence space) may have prompted replacement M230I, thereby revealing the implication of this residue in RT catalysis. The results described in this paper illustrate the enormous adaptative potential of HIV-1 and should guide future mutagenesis experiments with the aim of gaining a better knowledge of the DNA polymerization mechanism catalyzed by RTs, by combining a biochemical with an evolutionary approach.
ACKNOWLEDGMENTS
We thank Gema Gómez Mariano for help with DNA sequencing.
This work was supported by Fondo de Investigación Sanitaria grants 98/0054-01, -02, and -03 and by an institutional grant of Fundación Ramón Areces to Centro de Biología Molecular “Severo Ochoa.”
REFERENCES
- 1.Arts E J, Wainberg M A. Human immunodeficiency virus type 1 reverse transcriptase and early events in reverse transcription. Adv Virus Res. 1996;46:97–163. doi: 10.1016/s0065-3527(08)60071-8. [DOI] [PubMed] [Google Scholar]
- 2.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer P L, Ferris A L, Clark P, Whitmer J, Frank P, Tantillo C, Arnold E, Hughes S H. Mutational analysis of the fingers and palm subdomains of human immunodeficiency virus type-1 (HIV-1) reverse transcriptase. J Mol Biol. 1994;243:472–483. doi: 10.1006/jmbi.1994.1673. [DOI] [PubMed] [Google Scholar]
- 4.Boyer P L, Gao H-Q, Hughes S H. A mutation at position 190 of human immunodeficiency virus type 1 reverse transcriptase interacts with mutations at positions 74 and 75 via the template-primer. Antimicrob Agents Chemother. 1998;42:447–452. doi: 10.1128/aac.42.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlini F, Federic M, Equestre M, Ricci S, Ratti G, Zibai Q, Verani P, Rossi G B. Sequence analysis of HIV-1 proviral DNA from a non producer chronically infected HUT-78 cellular clone. J Viral Dis. 1992;1:40–55. [Google Scholar]
- 6.Ding J, Das K, Hsiou Y, Sarafianos S G, Clark A D, Jr, Jacobo-Molina A, Tantillo C, Hughes S H, Arnold E. Structural and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 Å resolution. J Mol Biol. 1998;284:1095–1111. doi: 10.1006/jmbi.1998.2208. [DOI] [PubMed] [Google Scholar]
- 7.Gao G, Goff S P. Replication defect of Moloney murine leukemia virus with a mutant reverse transcriptase that can incorporate ribonucleotides and deoxyribonucleotides. J Virol. 1998;72:5905–5911. doi: 10.1128/jvi.72.7.5905-5911.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh M, Jacques P S, Rodgers D W, Ottmann M, Darlix J-L, Le Grice S F J. Alterations to the primer grip of p66 HIV-1 reverse transcriptase and their consequences for template-primer utilization. Biochemistry. 1996;35:8553–8562. doi: 10.1021/bi952773j. [DOI] [PubMed] [Google Scholar]
- 9.Harada S, Koyanagi Y, Yamamoto N. Infection of HTLV-III/LAV in HTLV-I-carrying cells MT-2 and MT-4 and application in a plaque assay. Science. 1985;229:563–566. doi: 10.1126/science.2992081. [DOI] [PubMed] [Google Scholar]
- 10.Hottiger M, Hübscher U. Human immunodeficiency virus type 1 reverse transcriptase. Biol Chem Hoppe-Seyler. 1996;377:97–120. [PubMed] [Google Scholar]
- 11.Huang H, Chopra R, Verdine G L, Harrison S C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 12.Iversen A K N, Shafer R W, Wehrly K, Winters M A, Mullins J I, Chesebro B, Merigan T C. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral therapy. J Virol. 1996;70:1086–1090. doi: 10.1128/jvi.70.2.1086-1090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, Hizi A, Hughes S H, Arnold E. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleim J-P, Rösner M, Winkler I, Paessens A, Kirsch R, Hsiou Y, Arnold E, Riess G. Selective pressure of a quinoxaline nonnucleoside inhibitor of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) on HIV-1 replication results in the emergence of nucleoside RT-inhibitor-specific (RT Leu-74→Val or Ile and Val-75→Leu or Ile) HIV-1 mutants. Proc Natl Acad Sci USA. 1996;93:34–38. doi: 10.1073/pnas.93.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 16.Larder B A, Kemp S D, Purifoy D J M. Infectious potential of human immunodeficiency virus type 1 reverse transcriptase mutants with altered inhibitor sensitivity. Proc Natl Acad Sci USA. 1989;86:4803–4807. doi: 10.1073/pnas.86.13.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martín-Hernández A M, Domingo E, Menéndez-Arias L. Human immunodeficiency virus type 1 reverse transcriptase: role of Tyr115 in deoxynucleotide binding and misinsertion fidelity of DNA synthesis. EMBO J. 1996;15:4434–4442. [PMC free article] [PubMed] [Google Scholar]
- 18.Martín-Hernández A M, Gutiérrez-Rivas M, Domingo E, Menéndez-Arias L. Mispair extension fidelity of human immunodeficiency virus type 1 reverse transcriptases with amino acid substitutions affecting Tyr115. Nucleic Acids Res. 1997;25:1383–1389. doi: 10.1093/nar/25.7.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menéndez-Arias L. Studies on the effects of truncating α-helix E′ of p66 human immunodeficiency virus type 1 reverse transcriptase on template-primer binding and fidelity of DNA synthesis. Biochemistry. 1998;37:16636–16644. doi: 10.1021/bi981830g. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 21.Olivares I, Shaw G, López-Galíndez C. Phenotypic switch in a Spanish HIV type 1 isolate on serial passage on MT-4 cells. AIDS Res Hum Retroviruses. 1997;13:979–984. doi: 10.1089/aid.1997.13.979. [DOI] [PubMed] [Google Scholar]
- 22.Olivares I, Casado Herrero C, Iglesias-Ussel M D, Dietrich U, López-Galíndez C. Complete sequence of an infectious molecular clone derived from a Spanish HIV type I isolate. AIDS Res Hum Retroviruses. 1998;14:1649–1651. doi: 10.1089/aid.1998.14.1649. [DOI] [PubMed] [Google Scholar]
- 23.Olmsted R A, Slade D E, Kopta L A, Poppe S M, Poel T J, Newport S W, Rank K B, Biles C, Morge R A, Dueweke T J, Yagi Y, Romero D L, Thomas R C, Sharma S K, Tarpley W G. (Alkylamino)piperidine bis(heteroaryl)piperizine analogs are potent, broad-spectrum nonnucleoside reverse transcriptase inhibitors of drug-resistant isolates of human immunodeficiency virus type 1 (HIV-1) and select for drug-resistant variants of HIV-1IIIB with reduced replication phenotypes. J Virol. 1996;70:3698–3705. doi: 10.1128/jvi.70.6.3698-3705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palaniappan C, Wisniewski M, Jacques P S, Le Grice S F J, Fay P J, Bambara R A. Mutations within the primer grip region of HIV-1 reverse transcriptase result in loss of RNase H function. J Biol Chem. 1997;272:11157–11164. doi: 10.1074/jbc.272.17.11157. [DOI] [PubMed] [Google Scholar]
- 25.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powell M D, Ghosh M, Jacques P S, Howard K J, Le Grice S F J, Levin J G. Alanine-scanning mutations in the “primer grip” of p66 HIV-1 reverse transcriptase result in selective loss of RNA priming activity. J Biol Chem. 1997;272:13262–13269. doi: 10.1074/jbc.272.20.13262. [DOI] [PubMed] [Google Scholar]
- 27.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E, Baumeister K, Ivanoff L, Petteway S R, Jr, Pearson M L, Lautenberger J A, Papas T S, Ghrayeb J, Chang N T, Gallo R C, Wong-Staal F. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 28.Rodgers D W, Gamblin S J, Harris B A, Ray S, Culp J S, Hellmig B, Woolf D J, Debouck C, Harrison S C. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sánchez-Palomino S, Rojas J M, Martínez M A, Fenyö E M, Nájera R, Domingo E, López-Galíndez C. Dilute passage promotes expression of genetic and phenotypic variants of human immunodeficiency virus type 1 in cell culture. J Virol. 1993;67:2938–2943. doi: 10.1128/jvi.67.5.2938-2943.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tantillo C, Ding J, Jacobo-Molina A, Nanni R G, Boyer P L, Hughes S H, Pauwels R, Andries K, Janssen P A J, Arnold E. Location of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase. Implications for mechanisms of drug inhibition and resistance. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 31.Telesnitsky A, Goff S P. Reverse transcriptase and the generation of retroviral DNA. In: Coffin J W, Hughes S H, Varmus H E, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 121–160. [PubMed] [Google Scholar]
- 32.Tisdale M, Alnadaf T, Cousens D. Combination of mutations in human immunodeficiency virus type 1 reverse transcriptase required for resistance to the carbocyclic nucleoside 1592U89. Antimicrob Agents Chemother. 1997;41:1094–1098. doi: 10.1128/aac.41.5.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wakefield J K, Jablonski S A, Morrow C D. In vitro enzymatic activity of human immunodeficiency virus type 1 reverse transcriptase mutants in the highly conserved YMDD amino acid motif correlates with the infectious potential of the proviral genome. J Virol. 1992;66:6806–6812. doi: 10.1128/jvi.66.11.6806-6812.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willey R L, Smith D H, Lasky L A, Theodore T S, Earl P L, Moss D, Capon D J, Martin M A. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wöhrl B M, Krebs R, Thrall S H, Le Grice S F J, Scheidig A J, Goody R S. Kinetic analysis of four HIV-1 reverse transcriptase enzymes mutated in the primer grip region of p66. Implications for DNA synthesis and dimerization. J Biol Chem. 1997;272:17581–17587. doi: 10.1074/jbc.272.28.17581. [DOI] [PubMed] [Google Scholar]
- 36.Wrobel J A, Chao S-F, Conrad M J, Merker J D, Swanstrom R, Pielak G J, Hutchison C A., III A genetic approach for identifying critical residues in the fingers and palm subdomains of HIV-1 reverse transcriptase. Proc Natl Acad Sci USA. 1998;95:638–645. doi: 10.1073/pnas.95.2.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiong Y, Eickbush T H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu Q, Ottmann M, Pechoux C, Le Grice S, Darlix J-L. Mutations in the primer grip of human immunodeficiency virus type 1 reverse transcriptase impair proviral DNA synthesis and virion maturation. J Virol. 1998;72:7676–7680. doi: 10.1128/jvi.72.9.7676-7680.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]