Abstract
Artificial intelligence (AI) based predictive models for early detection of cardiovascular disease (CVD) risk are increasingly being utilised. However, AI based risk prediction models that account for right-censored data have been overlooked. This systematic review (PROSPERO protocol CRD42023492655) includes 33 studies that utilised machine learning (ML) and deep learning (DL) models for survival outcome in CVD prediction. We provided details on the employed ML and DL models, eXplainable AI (XAI) techniques, and type of included variables, with a focus on social determinants of health (SDoH) and gender-stratification. Approximately half of the studies were published in 2023 with the majority from the United States. Random Survival Forest (RSF), Survival Gradient Boosting models, and Penalised Cox models were the most frequently employed ML models. DeepSurv was the most frequently employed DL model. DL models were better at predicting CVD outcomes than ML models. Permutation-based feature importance and Shapley values were the most utilised XAI methods for explaining AI models. Moreover, only one in five studies performed gender-stratification analysis and very few incorporate the wide range of SDoH factors in their prediction model. In conclusion, the evidence indicates that RSF and DeepSurv models are currently the optimal models for predicting CVD outcomes. This study also highlights the better predictive ability of DL survival models, compared to ML models. Future research should ensure the appropriate interpretation of AI models, accounting for SDoH, and gender stratification, as gender plays a significant role in CVD occurrence.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10916-024-02087-7.
Keywords: Cardiovascular Disease, Prediction, Machine Learning, Deep Learning, Artificial Intelligence
Introduction
Cardiovascular diseases (CVD) cause 32% of all global deaths [1]. A confluence of environmental, genetic, social, and physiological factors leads to the development of CVD incidence, hospitalisation, and mortality. Early detection of these factors and appropriate interventions are recommended approaches for decreasing CVD burden and impact. Various multivariable prediction models exist for early detection of CVD risk [2], including the latest risk estimation tools like pooled cohort equations (PCE) [3], Systematic COronary Risk Evaluations (SCORE, SCORE2, and SCORE2 for older people) [4, 5], and Predicting Risk of cardiovascular disease EVENTs (PREVENT) [6].
However, due to the availability of big data and advancing technology, machine learning (ML) and deep learning (DL) prediction models, the two subfields of artificial intelligence (AI), have increasingly being utilised. Several studies have indicated that ML and DL models surpass traditional multivariable models in predicting CVD risk and specific events such as stroke [7]. These AI models exhibit enhanced discrimination and risk stratification abilities.
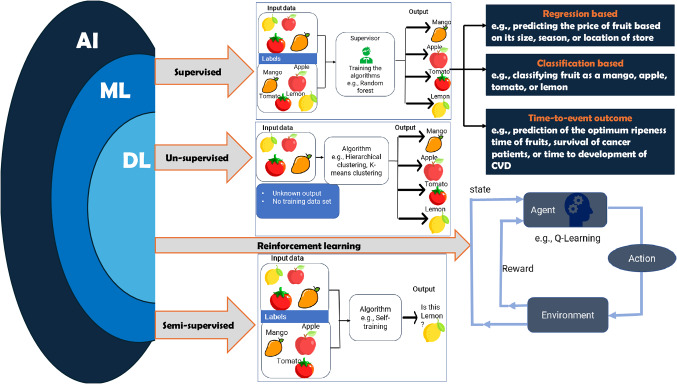
ML uses algorithms and statistical models to analyse and draw inferences from patterns in the data. DL is a subset of ML that uses artificial neural networks. As illustrated in Fig. 1, there are primarily four types of ML and DL [8, 9]; supervised, unsupervised, semi-supervised, and reinforcement learning (RL). Supervised ML algorithms have been utilised for future risk prediction. They require training using labelled data, that is data that contains inputs and correct corresponding outputs. Depending on the type of the outcome variable that is available for the learning phase, regression and classification algorithms (including binary, multi-class, multi-level, and imbalanced classification) are commonly employed. Regression algorithms predict continuous variables while classification algorithms determine the likelihood that a certain event will occur. Unsupervised ML algorithms (such as anomaly detection, clustering) use unlabelled data and are intended to find groups/clusters of similar characteristics without human supervision. Semi-supervised ML combines the features of supervised and unsupervised ML approaches, i.e., utilises both labelled and unlabelled data. RL algorithms interact with an environment to learn the optimal behaviour to maximise the overall reward.
Fig. 1.
Overview of machine learning and deep learning models
ML models have been in existence since 1957. The perceptron, which laid the foundation for supervised ML models and artificial neural networks, was one of the earliest neural network models. Since then, ML has passed several important milestones: the development of decision trees in the 1960s, support vector machines (SVM) in the 1990s, random forest in 2001, DL models in the 2010s, large language models such as ChatGPT in 2022, and many others recently [10]. These models are supervised ML algorithms for classification and regression, and are applied for predicting or forecasting chronic diseases, including CVD risk [11]. However, ML and DL algorithms for survival prediction were not widely used until Random Survival Forest (RSF) was developed by Ishwaran et al. in 2008 [12]. In particular, survival AI prediction algorithms, which estimate the time until a health outcome occurs, have not received as much attention as classification and regression ML algorithms [13]. Currently, various AI models for right-censored data are gaining popularity, even though they are predominantly used for predicting cancer patient survival outcomes [14–17]. Survival forest models [16, 17], NonLinear Cox proportional hazard (Cox PH) model (also known as DeepSurv model), and Neural Multi-Task Logistic Regression (NMTLR) [14–16] are among the commonly utilised models. Some studies have also employed algorithms such as CoxTime and Cox-CC [14].
Numerous systematic reviews on AI-based CVD prediction have been conducted [18–21]; yet they primarily focus on classification-based models. For instance, Baashar et al.‘s research assessed the effectiveness of ML and DL in CVD prediction through network meta-analysis [20], covering 17 studies from 2016 to 2021 and suggesting that DL might yield better results than ML. Nonetheless, a systematic review that succinctly summarise ML and DL models for right-censored data is still lacking. The justification for exploring AI models for right-censored data stems from the unique nature of survival outcomes. Unlike regression and classification problems, survival outcome must account for two components during model training: the follow-up time, which is continuous, and the event status, indicating whether a specific event has occurred, such as CVD, represented as a binary outcome.
Additionally, previous risk prediction models, including the latest multivariable prediction models and AI-based models mentioned above, primarily focus on standard modifiable risk factors of CVD, demographics (age and sex/gender), and lifestyle factors (particularly smoking). This means that social determinants of health (SDoH), defined as the social and environmental circumstances in which people grow, live, work, worship, and age, have been overlooked in disease prediction models [22], including CVD [18, 23]. For example, only race in the PCE [3] and social deprivation in the PREVENT tool [6] are incorporated when predicting CVD. Similarly, AI-based risk prediction models consider only a limited number of SDoH variables, like race, income, and occupation [18]. SDoH are detailed in the Healthy People 2030 framework using five domains, namely, economic stability, education quality and access, social and community context, neighborhood and built environment, and healthcare access and quality [24]. Using the Healthy People framework as a foundation, our umbrella review [25] demonstrated that SDoH have a major role in development of CVD. In general, disparities in SDoH give rise to health inequalities, which are systematic discrepancies in the opportunities people need to attain optimal health.
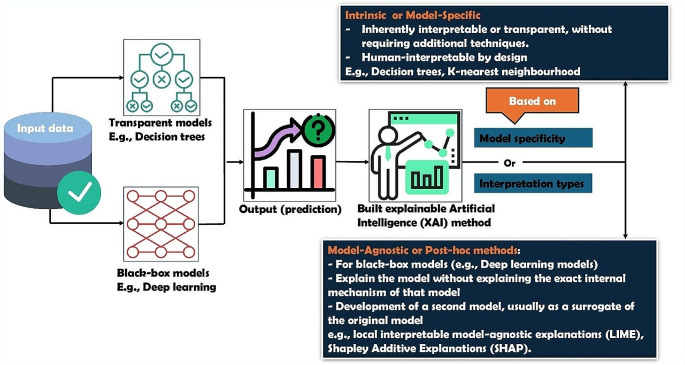
It is also important to focus on the explainability of the AI models to improve confidence in their application. These are called eXplainable AI (XAI) techniques and, as shown in Fig. 2, these can be model-specific (use the structure of the model itself, e.g., built in feature importance measures in ensemble models) or model-agnostic (provide post-hoc explanations e.g., Local Interpretable Model-agnostic Explanations (LIME), Shapley Additive exPlanations (SHAP)) [26]. However, these techniques have limited application for survival ML and DL methods. New XAI techniques for survival models such as Survival SHAP (SurvSHAP), survival neural additive model (SurvNAM), and survival LIME (SurvLIME) are currently gaining attention but are only used for explaining some algorithms [27, 28]. XAI in general, aims to increase user trust in a model to different stakeholders: (1) those with model expertise (e.g., ML experts, researchers); and (2) those without (clinicians, patients). However, the explainability of time to event AI models has been less explored.
Fig. 2.
Overview of eXplainable AI approaches
Moreover, various systematic reviews have been conducted on AI-based CVD prediction [18–21]; however, they have all focused-on classification-based models (models designed for classification problems). For example, research conducted by Baashar et al. evaluated the efficacy of ML and DL in the prediction of CVD through network meta-analysis [20]. The study encompassed 17 studies spanning from 2016 to 2021, concluding that DL may offer more favorable outcomes than ML in predicting CVD.
Therefore, this systematic review aims to (1) investigate AI models for survival prediction employed in predicting CVD; (2) indicate whether XAI is applied for interpreting the models; and (3) examine whether the identified AI models account for SDoH as well as gender stratification.
Methods
Registration and Reporting
The study protocol was registered in the International Prospective Register of Systematic Reviews (PROSPERO CRD42023492655). The Preferred Reporting Items for a Systematic Review and Meta-analysis (PRISMA) statement is used for reporting [29] (Table S1).
Eligibility Criteria
Studies were deemed eligible if they intended to predict CVD outcomes using AI methods for survival prediction. There were no restrictions based on country, study design, language, and study period (Table 1). Grey literature (including conference abstracts), case reports, letters, editorials, and reviews were not eligible. AI models based on simulation and imaging/text data are ineligible as they do not use structured population-level data.
Table 1.
Key items for framing the aim, search strategy, and study inclusion and exclusion criteria
| Domain | Description |
|---|---|
| Population | Adult population (age ≥ 18 years) |
| Intervention (Models) | Machine learning and deep learning predictive models for time to event outcome |
| Comparator | Not applicable |
| Outcomes | One or more of the following cardiovascular disease outcomes; (1) cardiovascular disease or major adverse cardiovascular event (MACE); (2) cardiovascular disease subtypes such as coronary heart disease, angina, myocardial infarction, heart failure, cerebrovascular disease (hemorrhagic and ischemic stroke), heart disease; or (3) hospitalisation or mortality due to cardiovascular disease or subtypes of cardiovascular diseases. |
| Time | No restriction |
| Setting | Both community and institution (e.g., hospital). |
Information Sources and Search Strategy
We carried out a comprehensive search using five electronic databases, from their inception to December 21, 2023: Embase via Ovid, Scopus, Web of Science, IEEE Xplore, and Ovid Medline. Further studies were identified by a manual search using Google Scholar, and through backward and forward reference searching using Web of Science. Various terms related to CVD, AI methods, and risk prediction were utilised (Table 2), linked through Boolean and adjacency (or proximity) operators. The comprehensive search terms used in Ovid Medline are available in the supplementary file (Table S2).
Table 2.
Summary of keywords/search terms per each concept
| No. | Categories | Keyword |
|---|---|---|
| 1. | Cardiovascular disease (including mortality or hospitalisation) | Cardiovascular disease, coronary heart disease, ischemic heart disease, angina, atrial fibrillation, major cardiovascular event, myocardial infarction, heart failure, congestive heart failure, heart disease, cerebrovascular disease/event, stroke, ischemic stroke, hemorrhagic stroke, and peripheral arterial disease |
| 2. | Artificial intelligence | Artificial intelligence, machine learning, deep learning, random survival forest, Extra Survival Trees, survival ensembles, survival support vector machine, Multi-Task Logistic Regression, DeepSurv, Non-Linear Cox proportional hazard model, CoxTime, CoxCC, probability mass function, Nnet-survival, DeepHit, DeepHitSingle, Piecewise Constant Hazard model, Discrete-Time Models, Continuous-Time Models, Neural network, survival neural network, deep neural survival networks |
| 3. | Risk prediction | Prediction, risk assessment, prognosis, predict, predictive modeling, detect, identify, identification, detection, risk stratification |
| 4. | Right censored time to event out come | Time to event, right censored, survival analysis, survival data, censoring |
Study Selection and Data Extraction
Identified records from databases were exported to Endnote Version 20 and then to ASReview [30] and Covidence [31]. Following deduplication, eligible articles at the title and abstract stage were selected using ASReview. Full text screening was done using Covidence. Using the data extraction sheet prepared based on the 11 CHecklist for critical Appraisal and data extraction for systematic Reviews of prediction Modelling Studies (CHARMS) domains [32], data were extracted. Two reviewers (ABT and HLH) selected eligible studies and undertook data extraction, resolving conflicts through discussion (full-text review: proportionate agreement = 96%, Cohen’s κ = 0.92).
Data Synthesis
Characteristics of studies were summarised based on items from the CHARMS statement and our specific aims. If a study used more than one ML or DL algorithm, we reported the prediction performance measure for the best performing algorithm. ML and DL models were compared with each other and with the standard Cox PH model. Utilised SDoH variables, based on the Healthy People 2030 framework, were reported. XAI methods employed were described. However, due to variations in the study population, the endpoint description, the different ML and DL algorithms utilised, and the variety in the types and numbers of variables, the prediction performance of the models was not pooled (i.e., meta-analysis was not conducted).
Assessment of Risk of Bias
To evaluate the risk of bias (RoB), we used the Prediction Model Risk of Bias Assessment Tool (PROBAST) [32] with four domains: participant selection; predictors; outcome; and analysis, and different signaling questions per each domain. Using the PROBAST, we also assessed applicability using three domains: participant, predictors, and outcome. Two authors (ABT and HLH) assessed RoB independently and any disagreements were resolved by discussion.
Results
Screening Result
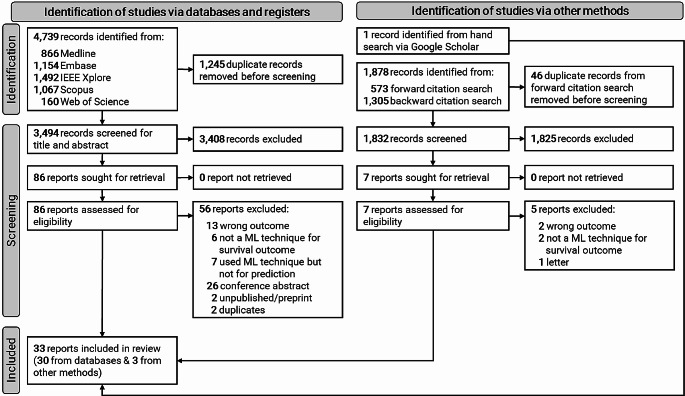
Out of a total of 4,739 studies retrieved through database searching, 86 were eligible for a full-text review. Thirty-three studies in total, 30 studies [33–62] from database searching and three studies [63–65] from other sources, qualified for inclusion in this study (Fig. 3). The studies that were excluded during the full-text review are provided in the supplementary file (Table S3).
Fig. 3.
PRISMA flow diagram showing the study selection process
Characteristics of the Included Studies
The majority of studies were published in 2023 (n = 16/33; 48.5%) and originated from the United States (n = 13/32; 40.6%; one study did not report the country). Approximately, 39.4% (n = 13/33) of the studies used a sample size of 10,000 or more. Two studies focused exclusively on one gender (one on men and the other on women), whilst the majority of studies analysing both genders had a higher percentage of women (50% and above) (Table S4).
Follow-up Time and Incidence of Cardiovascular Diseases
The mean or median follow-up time ranges from 4.4 years to 25.03 years in studies of community-dwelling people, and 4.3 months to 8.05 years in studies of institutionalised-people (Table S4). Various CVD outcomes, with their definitions and corresponding ICD codes detailed in Table S4, were identified (Fig. 4). The incidence of CVD outcomes ranged from 2.1% (CVD-related mortality) to 43.7% (MACE) (Table S4).
Fig. 4.
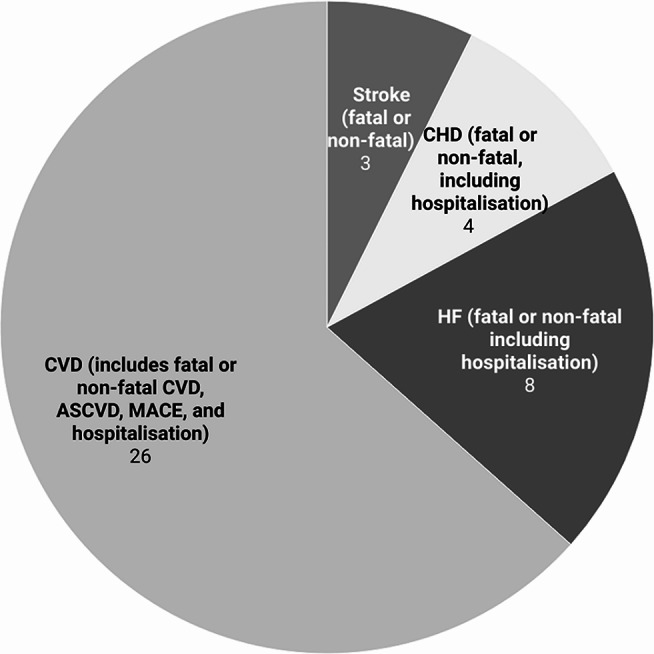
Number of studies based on predicted cardiovascular diseases outcomes. ASCVD: Atherosclerotic cardiovascular disease; CHD: Coronary heart disease; CVD: Cardiovascular disease; HF: Heart failure; and MACE; Major adverse cardiovascular events. Note: Since one study can incorporate more than one outcome, the sum total reported here exceeds the total number of included studies
Model Related Characteristics
Employed ML and DL Models
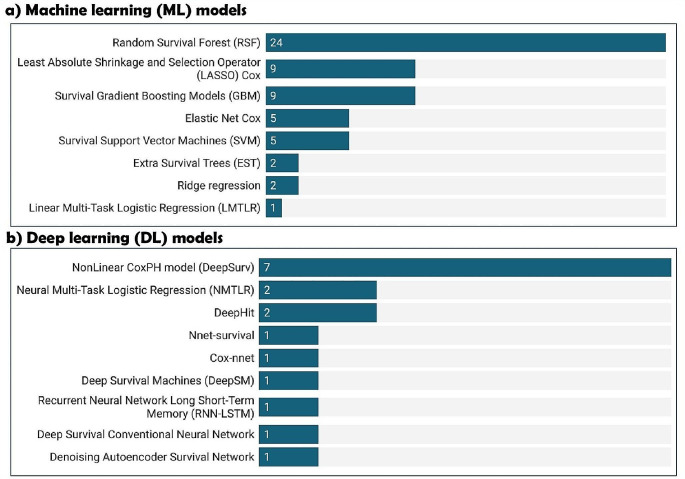
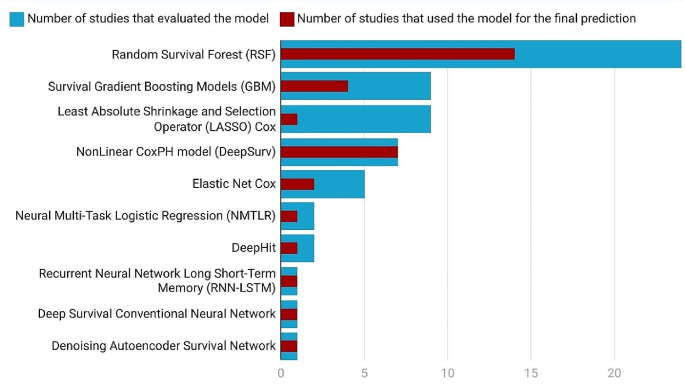
Eight ML and nine DL models were utilised, with Fig. 5 presenting the names of each model and the number of studies that employed them, and Table S5 providing further details.
Fig. 5.
Identified ML and DL models. Since a single study could utilise multiple ML and/or DL models, the total number of studies presented here exceeds 33 (the total number of studies included)
Best Performing ML and DL Models
To evaluate the predictive performance, studies utilised C-index, area under the curve (AUC), and Brier score or calibration plot. In addition, some studies also explored other measures such as decision curve analysis. These performance evaluation metrics are presented in Table S5. The mean C-index (standard deviation) was 0.79 (0.069) for ML models, 0.82 (0.061) for DL models, 0.81 (0.144) for Penalised Cox, 0.80 (0.058) for RSF, 0.79 (0.032) for DeepSurv, and 0.77 (0.055) for survival Gradient Boosting Models (GBM) (Table 3).
Table 3.
Descriptive statistics of predictive performance (C-index/area under the curve) by ML and DL algorithms
| Machine learning and deep learning models (number of studiesf) | Mean C-index (Standard deviation)g | Median C-indexg | Min | Max | Inter Quartile Range |
|---|---|---|---|---|---|
| Random survival forest (n = 23) | 0.80 (0.058) | 0.80 | 0.65 | 0.92 | 0.77-0.0.83 |
| DeepSurv (n = 10) | 0.79 (0.032) | 0.79 | 0.74 | 0.85 | 0.77–0.81 |
| Survival Gradient Boosting Models (n = 7) | 0.77 (0.055) | 0.75 | 0.72 | 0.87 | 0.72–0.81 |
| Other deep learning modelsa (n = 6) | 0.85 (0.081) | 0.83 | 0.76 | 0.96 | 0.79–0.94 |
| Penalised Coxb (n = 4) | 0.81 (0.144) | 0.81 | 0.66 | 0.93 | 0.69–0.93 |
| Machine learning modelsc (n = 34) | 0.79 (0.069) | 0.79 | 0.65 | 0.96 | 0.75–0.83 |
| Deep learning modelsd (n = 16) | 0.82 (0.061) | 0.80 | 0.74 | 0.96 | 0.78–0.84 |
aIncludes DeepHit, Neural Multi-Task Logistic Regression, Recurrent Neural Network Long Short-Term Memory, and deep survival conventional neural network
bIncludes LASSO and Elastic Net Cox models
cIncludes Random Survival Forest, survival Gradient Boosting Models, and Penalised Cox models
dIncludes Deepsurv, Neural Multi-Task Logistic Regression, Recurrent Neural Network Long Short-Term Memory, and Deep Survival Conventional Neural Network
fTotal number of models may differ from total number of included studies, because some studies reported for men and women separately or fitted a model based on race and studies may not report the C-index or area under the curve quantitatively
gArea under the curve if the study did not report C-index
Eight studies only used one ML [35, 37, 39, 41, 47, 50, 57, 58] and four studies only used one DL [34, 36, 43, 59] algorithm to predict CVD. The 26 studies [33–37, 39–46, 48, 49, 52–57, 59, 62–65] that compared ML and DL models with the Cox PH model revealed that ML and DL models were better in predicting CVD. Nine studies [33, 38, 51–53, 55, 60, 62, 65] among 18 studies [33, 38, 42, 44, 46, 48, 49, 51–55, 60–65] that compared the RSF with other models, selected RSF as the best performing model. Six studies [35, 37, 39, 41, 50, 57] used RSF without comparison with other ML or DL models. Among the seven studies [38, 44, 55, 56, 63–65] that compared the survival GBM with other models, the boosting models were better in two studies [56, 64]. In another two studies [47, 58], the boosting method was used to predict CVD without comparing with other models. All of the five studies [38, 44, 45, 56, 63] that evaluated Elastic Net Cox compared the model with other models, and in the two studies [45, 63] Elastic Net Cox was the best performing model for CVD prediction. In the nine studies [33, 45, 49, 52, 53, 60–62, 65] that evaluated LASSO-Cox, all compared the model with other models, it was in only one study [61] that LASSO-Cox was selected as the best performing model. As for the DL models, the DeepSurv model was the best performing model in all the five studies [34, 40, 43, 46, 48, 49, 54] that compared the model with other models. In the other two studies [34, 40], DeepSurv was not compared with other ML or DL models. Some studies also selected NMTLR [42, 46], DeepHit [51, 52], and denoising autoencoder survival network [44] as best performing models in predicting CVD. Others [36, 59] examined Recurrent Neural Network Long Short-Term Memory and Deep Survival Conventional Neural Network, however, without comparing with any other ML or DL models (Fig. 6 and Table S5).
Fig. 6.
Number of studies that evaluated the prediction model and used it for their final prediction
Comparison of ML and DL Models
Among the eight studies [42, 44, 46, 48, 49, 51, 52, 54] that compared ML and DL models together, DL models were better in predicting CVD risk in seven studies [42, 44, 46, 48, 49, 51, 54]. Four studies [46, 48, 49, 54] compared the DeepSurv model with other ML and DL models and all four found this model to be the best for predicting CVD outcomes (Table 4 and Table S5).
Table 4.
Selected models, based on their performance, among studies that compared machine learning and deep learning models together
| Author and year | Selected best model | Deep Learning models | Machine learning models |
|---|---|---|---|
| Feng 2022 [42] | Neural Multi-Task Logistic Regression | Neural Multi-Task Logistic Regression | Random survival forest and Linear Multi-Task Logistic Regression |
| Gao 2023 [44] | Denoising autoencoder Survival network | Denoising autoencoder Survival network | Elastic Net Cox, Gradient Boosted Survival, Support Vector Machine, and Random survival forest |
| Hathaway 2021 [46] | DeepSurv/NonLinear CoxPH model | Deepsurv/NonLinear CoxPH model and Neural Multi-Task Logistic Regression | Random survival forest and Support Vector Machine |
| Kim 2023 [48] | DeepSurv/NonLinear CoxPH model | Deepsurv/NonLinear CoxPH model and Deep Survival Machines (DeepSM) | Random survival forest |
| Lin 2023 [49] | DeepSurv/NonLinear CoxPH model | Deepsurv/NonLinear CoxPH model | LASSO-Cox and Random survival forest |
| Ren 2022 [54] | DeepSurv/NonLinear CoxPH model | Deepsurv/NonLinear CoxPH model | Random survival forest |
| Morris 2023 [51] | DeepHit | DeepHit | Random survival forest and Penalised cox proportional hazards |
| Nguyen 2023 [52] | Random Survival Forest | DeepHit | Random survival forest and LASSO-Cox |
Utilised XAI Techniques
The 25 studies interpreted their model using different approaches (Table 5 and Table S5). Out of those studies, four [49, 52, 63, 64] used more than one method. Eight studies [33, 34, 37, 40, 42, 44, 58, 60] provided no model interpretation.
Table 5.
Model interpretation methods
| Model interpretation technique utilised | Number of studies |
|---|---|
| Feature importance (e.g., permutation (majority), Mean Decrease Gini, mean of the minimal depth of the maximal subtree) | 14 [35, 39, 41, 46, 48–50, 52–54, 56, 62, 65] |
| Shapley Additive exPlanations (SHAP) | 6 [47, 51, 52, 61, 63, 64] |
| Partial dependence plots (PDPs) | 4 [43, 55, 63, 64] |
| Machine learning derived/simplified risk score (including nomogram) | 3 [45, 49, 57] |
| Temporal Importance Model Explanation (TIME) | 1 [52] |
| Layer-wise Relevance Propagation (LRP) | 1 [59] |
| Component wise gradient boosting coefficients | 1 [38] |
| Contribution of features using weighted ratio (WR) | 1 [36] |
Model Validation
All studies internally validated their prediction model using either train-test splitting or using resampling methods such as k-fold cross-validation and Bootstrapping. However, only six studies [40, 46, 56, 57, 59, 65] externally validated their prediction model. The commonly employed models were RSF and DeepSurv (Table S5).
Gender Stratification
Of 31 studies that used gender in their prediction models, only six studies [34, 36, 39, 40, 53, 59] performed gender-stratified prediction. RSF and DL models such as DeepSurv were most popular (Table S5).
Number of Predictors and Feature Selection Methods
The number of candidate predictors ranged from 7 to 950. Mostly the number of candidate predictors was greater than 50 (n = 13/33; 39.4%). The number of final predictors used ranged from 3 to 613, with the majority (n = 13/33; 39.4%) incorporating 21 to 50 variables (Table S5). In 22 studies, variable selection was not performed. Of those studies that performed variable selection prior to training, two used RSF, two used LASSO-Cox, one used stepwise forward selection, and one used Elastic Net Cox. Additionally, five studies used more than one variable selection method (Table S5).
Types of Predictors
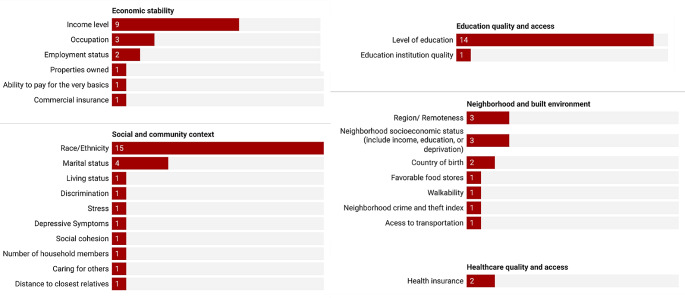
Included studies used a wide range of risk factors such as standard modifiable cardiovascular risk factors, demographic factors, imaging features, biomarkers, variables related to sleep and diet, environmental chemicals, and SDoH variables (Table S5). Regarding our study’s interest in SDoH, at least one SDoH was included by 20 studies [33, 34, 38–40, 42, 46, 47, 50–53, 55, 57, 58, 60–63, 65] as a candidate variable. Of these 20 studies, all except for two [53, 57] incorporated at least one SDoH as a final predictor to train the model. However, only two studies [51, 58] employed a wide range of SDoH variables from the Healthy People 2030 framework. The most frequently considered SDoH variables were race/ethnicity, level of education, and income (Fig. 7 and Table S5).
Fig. 7.
Number of studies that incorporated social determinants of health variables for predicting cardiovascular disease
Models Based on Population
Eighteen studies [33, 34, 36, 37, 39, 40, 46, 50–53, 55, 56, 59, 61–63, 65] were conducted among the general population (relatively healthy adults) (Table S4). Half of the studies [33, 37, 39, 50, 52, 53, 55, 62, 65] employed RSF, after comparison [33, 52, 53, 55, 62, 65] or used RSF as the only model [37, 39, 50]. Six studies [34, 36, 40, 46, 51, 59] used DL models in which three used DeepSurv [34, 40, 46], three did comparison with other models [40, 46, 51], and three did not do comparison [34, 36, 59]. In three studies, Elastic Net Cox [63], LASSO-Cox [61], and Survival GBM [56] were the best performing models (Table S5).
The remaining 15 studies [35, 38, 41–45, 47–49, 54, 57, 58, 60, 64] were conducted among institutionalised populations or individuals with specific medical conditions (Table S4). Nine studies [35, 38, 41, 45, 47, 57, 58, 60, 64] used ML models, five studies [35, 38, 41, 57, 60] utilised RSF (after comparison [38, 60] or as the only model [35, 41, 57]), three studies [47, 58, 64] survival GBM (after comparison [64] or as the only model [47, 58]), and one study [45] utilised Elastic Net Cox (after comparison with other models) for their final prediction. Six studies [42–44, 48, 49, 54], all after comparison with other models, used DL models. Four of the studies [43, 48, 49, 54] used the DeepSurv model and the other two used NMTLR [42] and denoising autoencoder survival network [44] (Table S5).
Models Based on Types of Variables
Models Incorporated Imaging Features
11 studies [33, 35, 38, 44, 46, 48, 50, 56, 57, 62, 65] included image features, with standard modifiable risk factors, SDoH factors, or other factors, in their prediction models (Table S5). Seven of them [33, 38, 50, 57, 62, 65] utilised RSF, after comparing with other models [33, 38, 62, 65] or as the only model [35, 50, 65]. No studies compared RSF with DL models; instead, they compared RSF with other ML models. One study [56] selected survival GBM, after comparing with other ML models. Three studies utilised DL models, two DeepSurv [46, 48] and one denoising autoencoder survival network [44]. These three models were compared with ML learning models (such as RSF) and DL models (such as NMTLR).
Models Accounted for SDoH
Among the 18 studies that included at least one SDoH (detailed above) as their final predictor (Table S5), nine studies utilised RSF, because RSF was the best performing model in seven studies [33, 38, 52, 55, 60, 62, 65] or used as the only model [39, 50]. Two studies employed Elastic Net Cox [63] and LASSO-Cox [61] since they were the best performing models as compared to other models. Two studies [47, 58] utilised survival GBM without comparing the model with other ML or DL models. Five models utilised DL models; three studies used DeepSurv [34, 40, 46], two studies NMTLR [42], and one study DeepHit [51]. All these studies compared their DL model with other ML models (mostly with RSF) and/or other DL models.
Software and Related Information
Twelve studies [33, 41, 44, 45, 50, 53, 55, 57, 58, 60, 62, 65] used R software (packages: glmnet, ranger, randomForestSRC, mlr3, mlr3proba). Meanwhile, nine studies [34, 36, 38, 47, 48, 51, 56, 61, 64] utilised Python software (packages: scikit-survival, PySurvival, pycox). Studies also used more than one software, for instance, 11 studies [35, 37, 39, 40, 42, 43, 46, 49, 52, 54, 63] used R and Python. The specifics of the missing data management and hyperparameter tuning methods, along with the libraries and packages (including source codes) utilised in the studies are presented in Table S6.
Risk of Bias
Among all 33 studies, 15 [33, 38, 45, 49–51, 53, 54, 57–59, 61, 62, 64, 65] had low (RoB); 14 [34–37, 39–42, 44, 46, 52, 55, 56, 63] had high RoB; and four [43, 47, 48, 60] had uncertain RoB. As for applicability, 19 studies [34, 38–40, 42, 43, 45, 47, 49, 53, 55–58, 60–62, 64, 65] had low applicability concerns and 14 [33, 35–37, 41, 44, 46, 48, 50–52, 54, 59, 63] had high applicability concerns (Table 6).
Table 6.
Risk of bias and applicability assessment
| Author, Year | Risk of Bias | Applicability | Overall | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1. Participants | 2. Predictors | 3. Outcome | 4. Analysis | 1. Participants | 2. Predictors | 3. Outcome | Risk of Bias | Applicability | |
| Ambale-Venkatesh, 2017 | + | + | + | + | + | - | + | + | - |
| Barbieri, 2022 | + | + | + | - | + | + | + | - | + |
| Bauer, 2023 | + | + | + | - | + | - | + | - | - |
| Blanchard, 2022 | + | + | + | - | + | - | + | - | - |
| Brester, 2023 | + | + | + | - | + | - | + | - | - |
| Chhoa, 2023 | + | + | + | + | + | + | + | + | + |
| Chun, 2021 | + | + | + | - | + | + | + | - | + |
| Deng, 2023 | + | + | + | - | + | + | + | - | + |
| Duan, 2024* | - | + | + | + | - | + | + | - | - |
| Farhadian, 2021 | ? | + | + | - | + | + | - | - | - |
| Feng, 2022 | + | + | + | - | + | + | + | - | + |
| Gandin, 2023 | ? | + | + | ? | + | + | + | ? | + |
| Gao, 2023 | + | + | + | - | + | + | - | - | - |
| Garcia-Carretero, 2019 | + | + | + | + | + | + | + | + | + |
| Hathaway, 2021 | + | + | + | + | + | - | + | - | - |
| Jain, 2021 | + | + | + | ? | + | + | + | ? | + |
| Kim, 2023 | + | + | + | ? | + | + | - | ? | - |
| Lin, 2023 | + | + | + | + | + | + | + | + | + |
| Mauger, 2023 | + | + | + | + | + | + | - | + | - |
| Moreno-Sánchez, 2023 | + | + | + | + | + | + | + | + | + |
| Morris, 2023 | + | + | + | + | + | + | - | + | - |
| Nguyen, 2023 | + | + | + | - | + | + | - | - | - |
| Qian, 2023 | + | + | + | + | + | + | + | + | + |
| Ren, 2022 | + | + | + | + | + | + | - | + | - |
| Rigdon, 2019 | - | + | + | - | + | + | + | - | + |
| Sabovcik, 2022 | + | + | + | - | + | + | + | - | + |
| Segar, 2019 | + | + | + | + | + | + | + | + | + |
| Segar, 2021 | + | + | + | + | + | + | + | + | + |
| Stabellini, 2023 | + | + | + | + | + | + | + | + | + |
| Sung, 2019 | + | + | + | + | + | + | - | + | - |
| Turchin, 2023 | + | + | ? | + | + | + | + | ? | + |
| Wang, 2023 | + | + | + | + | + | + | + | + | + |
| Zhuang, 2022 | + | + | + | + | + | + | + | + | + |
* The accepted manuscript (pre-proof) was found during the search and was published in January 2024
+ indicates low risk of bias/low concern regarding applicability
- indicates high risk of bias/high concern regarding applicability
? indicates unclear risk of bias/unclear concern regarding applicability
Discussion
Main Findings
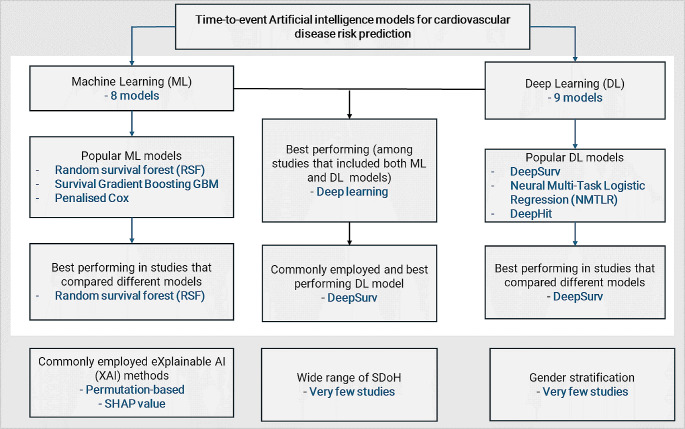
To enhance clarity, our principal findings are depicted in Fig. 8. A variety of ML and DL models for survival prediction in CVD were identified. The popular ML methods were RSF, survival GBM, and Penalised Cox models. These three ML models also performed best at predicting time to CVD occurrence, when compared to numerous other ML models considered in the included studies. Regarding DL models, the models that were utilised most frequently were DeepSurv, NMTLR, and DeepHit. These three DL models had better performance, compared with other DL or ML models. Permutation based feature importance and SHAP values were the predominant XAI methods utilised for explaining the models. While a variety of variables were incorporated to predict CVD, there was a noticeable lack of consideration for a wide range of SDoH variables. Additionally, prediction modeling with gender stratification was rarely explored.
Fig. 8.
Summary of the principal findings
AI Models for Survival Prediction and Year of Publication
Our systematic review revealed ML and DL models for survival prediction are increasingly gaining attention, while nearly all studies were published in 2019 or afterwards, half were published in 2023. This is not surprising since the packages in R and Python (e.g., mlr3proba package in R, and pycox, scikit-survival, PySurvival Python packages) for survival prediction using AI models became available in 2019 or later [66–69].
ML Models for Survival Outcome in CVD Prediction
The most utilised ML model was RSF. Our finding corroborate a scoping review on applications of ML in predicting survival outcomes, which identified RSF as the most frequently utilised model [13]. RSF has become a well-developed and user-friendly model, since its introduction by Ishwaran et al. in 2008 [12]. RSF is effective at handling complex interactions, has built-in variable importance measures, and is robust to overfitting [70]. The other plausible explanation is due to the availability of numerous open-source packages in standard software such as R and Python for appropriate training of RSF [66–69].
The next commonly applied ML models were survival GBM and Penalised Cox models. This may be at least partly explained by survival GBM considers non-linear interactions, has a high reported prediction accuracy, has greater ease of interpretability, and has automatic variable selection [71]. The availability of packages in R and Python (effectively open-source) makes the model easily trainable and accessible. For instance, survival GBM can be efficiently trained using the newly developed Python package, scikit-survival [69]. Both the RSF model and survival GBM are ensemble models that combine the decisions from several baseline models to improve the overall performance and robustness [72]. The Penalised Cox models are commonly used because they are important for penalising and provide a parsimonious model [73]. They are easier to apply (having a few, maximum two, parameters to tune). Finally, a few studies also utilised survival SVM, Linear Multi-Task Logistic Regression (LMTLR), and Extra survival trees in predicting CVD.
DL Models for Survival Outcome in CVD Prediction
Different DL models were also utilised to predict CVD. The DeepSurv model was mostly utilised. The other models were the NMTLR and DeepHit. These models are also commonly applied in oncologic studies [15, 74–76]. Most of these models can be well-trained using the two important Python packages, PySurvival and Pycox, which have been popular since 2019 [66–68]. Denoising autoencoder survival network, Recurrent Neural Network Long Short-Term Memory, and Deep Survival Conventional Neural Network [66, 68] were also utilised by some of the studies.
Best Performing ML and DL Models for Survival Outcome in CVD Prediction
Compared to the standard Cox proportional hazards model, both ML and DL models have demonstrated superior performance, in terms of discriminative ability and calibration. Another review has also shown that ML models outperform conventional methods in predicting health outcomes [13]. This may be due to the limited capability of the standard Cox model to handle high-dimensional datasets and its reliance on a linear relationship assumption, which are often not met.
The most frequently selected ML models (based on their prediction performance) were ensemble methods (RSF and survival GBM). RSF and survival GBM are ensemble models that are known to have superior prediction performance because they are drawn from several baseline learners [72]. However, this finding might also be a result of RSF and survival GBM models being considered in many of the included studies. In three studies, Penalised Cox-models were also selected as the best preforming. Penalised Cox models reduce overfitting, handle multicollinearity (particularly the Elastic Net Cox), enhance interpretability, and automate variable selection by shrinking less important predictors’ coefficients to zero [77].
As for DL models, in almost all studies, DeepSurv was selected as the best performing model. Our finding corroborates multiple individual studies on the survival prediction of cancer patients, demonstrating that DeepSurv surpasses alternative methods in predictive accuracy [14, 15, 76]. DeepSurv computes complex and non-linear features without a priori selection or domain expertise and is helpful for personalised risk prediction, even better than other linear and non-linear survival methods [78]. Notably, DeepSurv was also popular and, therefore, available for comparison.
Comparison of ML and DL Models for Survival Outcome in CVD Prediction
Consistent with studies that have examined both ML and DL models in the context of predicting the survival of cancer patients [76, 79], our study found that DL models surpass ML models in predicting time to CVD occurrence. That is, among the eight studies that compared ML and DL models together, DL models outperformed in seven, whereas ML models excelled in only one study. This is because DL models can improve prediction by (1) enhancing discrimination and calibration, (2) leveraging large datasets effectively, and (3) autonomously learning complex representations for better risk stratification [80].
XAI Techniques Utilised
All studies included black-box ML models (except Penalised Cox models) and DL models. Black-box models are not explainable unless XAIs are utilised, which means that humans cannot understand how predictions are made [81]. Despite the included studies considering the black-box models, not all studies interpreted their models. Studies that interpreted their models mostly used permutation-based feature importance followed by SHAP value. Using XAIs, studies identified key factors driving predictions and provide transparency in model decision-making. However, feature importance alone cannot ensure a responsible and effective translation of the model into clinical practice.
SDoH Variables Accounted for in ML and DL Models for Survival Outcome in CVD Prediction
All studies evaluated the standard modifiable cardiovascular risk factors. Biomarkers, imaging features, and variables related to sleep and diet were also considered. However, despite recent studies revealing the major role of SDoH in CVD [25, 82], only a handful of prediction models incorporated a wide range of SDoH variables. Our findings expand on another systematic review aimed at identifying SDoH in ML based CVD prediction models, which also reported that included models did not give much emphasis to SDoH [83]. In this systematic review, most studies considered certain SDoH variables, such as race, education level, and income. However, the use of specific SDoH variables such as race in deploying ML models is controversial [84, 85]. For example, there is a notion that race is a biological construct, rather than a social one, and the race-aware ML model deployment could perpetuate existing biases and discrimination [86, 87]. While we agree that poorly implemented race-conscious models might perpetuate existing biases, including race in ML models’ deployment is helpful for accurate predictions and addressing racial disparities in health outcomes [87, 88]. Additionally, by incorporating race, models can help tailor interventions and allocate resources more effectively to communities in need. Therefore, rather than simply omitting race in the deployment of ML models, it is essential to implement race-aware models with nuanced considerations tailored to the specific context, purpose, and application of the model [88].
Gender Stratification in CVD Prediction
In 80% of the studies, gender-stratified prediction was overlooked despite gender playing a role in CVD presentation, diagnosis, and survival [89, 90]. Moreover, the role of gender is a critical determinant of CVD as it shapes one’s norms, roles, social relations, and behaviors [91]. Due to the challenges in distinguishing gender and sex from the studies, we used the general term “gender”. Additionally, it is important to acknowledge the following when considering gender versus sex in the deployment of ML models [92, 93]: (1) Viewing gender strictly as a binary biological construct fails to account for the intricate social factors that shape gender identity and expression, (2) Inferring gender solely based on biological sex characteristics can lead to discrimination against transgender and non-binary individuals. Generally, gender-stratified prediction models are beneficial for pinpointing gender-specific predictive factors for tailored and potentially more effective interventions [94]. However, we recommend that gender-stratified prediction models be undertaken after meticulous attention to the representativeness of data, potential biases, and the fundamental factors driving gender disparities in health outcomes.
Model Validation
Almost all studies internally validated their models. However, a few studies did external validation. Another review also highlighted that most studies did not perform external validation of their ML models [13]. Although external validation is commonly viewed as a critical step in transitioning clinical prediction models from development to implementation, it should not be seen as an automatic green light for model deployment. Moreover, there is no single recommended validation design, external validation is not always essential, and at times, multiple external validations may be required. Generally, the necessity and scope of external validations are contingent upon the intended application of the model and the justification for conducting an external validation study [95].
Implications for Clinical Practice and Recommendations
AI-based risk prediction models have an increased discrimination ability and accuracy as compared to the conventional multivariable models [96]. However, there are misconceptions that ML requires large amounts of data [97]. Despite ML models often benefiting from large datasets, they can still be effectively applied to smaller health-related datasets as long as the right balance between data quantity and quality is ensured and interpretability is prioritised [97]. It is also imperative to consider the nature of the dataset. For example, when considering longitudinal data with available follow-up time classification-based ML methods should not be used. Right-censoring should be accounted for, since excluding those who lost to follow-up, may result in a biased estimate. ML models for right-censored data have been utilised since 2008 and have recently flourished. Since 2018/19 numerous new models (particularly DL models) for right-censored data with their respective open-source coding packages have become available [66–69]. While it is encouraging that survival ML and DL models are gaining more focus and the development of cutting-edge models is accelerating, their interpretability still poses a challenge. There are open-source XAI methods such as SurvSHAP and SurvLIME for interpreting ML and DL models for right-censored data [27, 28]. However, it is noted that models trained using the PySurvival package, for instance, are not yet supported. Therefore, it is crucial to also focus on their XAI, whether it is model-agnostic or model-specific. In this systematic review, the quality assessment tool PROBAST, typically used for standard prediction models, was employed. However, its application to AI-based prediction models was not direct, leading to the omission or alteration of some signaling questions to evaluate the studies’ quality. Notably, PROBAST + AI tools are currently in development [98, 99], but at this stage, they remain as protocols and should be made available to researchers and decision-makers soon.
Additionally, a standardised measuring tool for most SDoH variables is lacking. SDoH are complex and specific to context and setting, necessitating tailored approaches. Taking these factors into account when measuring SDoH could aid in the creation of effective, context-specific strategies that precisely reflect the impact of SDoH on health outcomes. Inadequately designed SDoH (e.g., race)-sensitive models have the potential to exacerbate existing biases and discrimination within healthcare systems [86, 87]. Consequently, it is imperative to apply nuanced considerations that are specific to the context, purpose, and application of the predictive model. In this systematic review, despite having not differentiated between gender and sex, we found that a common limitation in CVD risk prediction studies is the rarity of gender-specific analysis. Future prediction studies should focus on gender-stratification while incorporating a range of SDoH in the AI prediction models for enhanced prediction and wise decision making.
Strengths and Limitations
The strengths of this systematic review are its novelty in concisely summarising the ML and DL models utilised for time to CVD outcomes, the applied interpretation techniques, and the assessment of whether SDoH variables or gender-stratification were accounted for. However, despite this systematic review had compared ML and DL in the context of CVD and found that DL is more effective for predicting incident CVD, due to the heterogeneity of studies (e.g., in terms of population, type and number of variables incorporated), we did not do direct comparison through meta-analysis. Additionally, we note that that the most commonly used models also had the best performance. Therefore, our findings may be biased due to the availability of these models for comparison.
Conclusion
This review identified and compared the different ML and DL models for survival outcomes in CVD prediction. RSF, survival GBM, and Penalised Cox models were the most popular and optimal predicting ML methods. Among DL models, DeepSurv was the most popular and optimal predicting model. Compared to ML models, DL models had better prediction performance. In general, RSF and DeepSurv models were the most popular and better performing models, regardless of the types of variables included (e.g., SDoH) or the population (e.g., community-based, institutionalised). Permutation-based feature importance and SHAP value were the commonly utilised XAI methods for interpretating the AI models. Despite the evidence for SDoH as predictors of CVD and gender-desegregated findings, they were considered by only a few of the included studies. To improve CVD risk prediction and inform clinicians decision-making future studies need to assess SDoH, in addition to the traditional factors and other emerging risk factors. While men and women share many traditional risk factors for CVD, additional gender-specific risk factors and mechanisms are at play. Therefore, it is crucial to consider gender differences when it comes to predicting and managing CVD risks. Moreover, more methodological work is still required to improve ease of interpretability of deep survival learning models, particularly as they have no built-in feature importance methods.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
For her crucial assistance in the development of our search strategy, we extend our sincere gratitude to Lorena Romero, the senior librarian of the Ian Potter Library at the Alfred Hospital in Melbourne, Victoria, Australia. We acknowledge the Wurundjeri People who are the Traditional Custodians of the lands on which the first and senior authors predominately work and live, and we pay our respects to their Elders, past and present.
Author Contributions
A.B.T: Conceptualisation, Design and Search Strategy, Literature Search and Screening, Data Extraction, Data Synthesis and Interpretation, Writing - Original Draft, Finalising the Manuscript. H.L.H: Literature Search and Screening, Data Extraction, Data Synthesis and Interpretation, Editing, Finalising the Manuscript. M.V: Conceptualisation, Data Synthesis and Interpretation, Editing, Finalising the Manuscript, Supervision. A.J.O: Data Synthesis and Interpretation, Editing, Finalising the Manuscript, Supervision. R.F.P: Conceptualisation, Data Synthesis and Interpretation, Editing, Finalising the Manuscript, Supervision.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. ABT and HLH are supported by Monash International Tuition Scholarship and Monash Graduate Scholarship. Funders played no role in the design of the study, data collection, analysis and interpretation of data, the decision to publish, and in the writing of the manuscript.
Open Access funding enabled and organized by CAUL and its Member Institutions
Data Availability
Data is provided within the manuscript or supplementary information files.
Declarations
Ethical Approval
Not applicable.
Competing Interests
The authors declare no competing interests.
Disclosures
None.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organisation. Noncommunicable diseases [updated 16 September, 2023; cited 2023 16 November,]. Available from: https://www.who.int/news-room/fact-sheets/detail/noncommunicable-diseases.
- 2.Sofogianni A, Stalikas N, Antza C, Tziomalos K. Cardiovascular Risk Prediction Models and Scores in the Era of Personalized Medicine. J Pers Med. 2022;12(7).
- 3.Yadlowsky S, Hayward RA, Sussman JB, McClelland RL, Min Y-I, Basu S. Clinical Implications of Revised Pooled Cohort Equations for Estimating Atherosclerotic Cardiovascular Disease Risk. Annals of Internal Medicine. 2018;169(1):20–9. [DOI] [PubMed] [Google Scholar]
- 4.SCORE2 working group and ESC Cardiovascular risk collaboration. SCORE2-OP risk prediction algorithms: estimating incident cardiovascular event risk in older persons in four geographical risk regions. European Heart Journal. 2021;42(25):2455–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.SCORE2 working group and ESC Cardiovascular risk collaboration. SCORE2 risk prediction algorithms: new models to estimate 10-year risk of cardiovascular disease in Europe. European Heart Journal. 2021;42(25):2439–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan SS, Matsushita K, Sang Y, Ballew SH, Grams ME, Surapaneni A, et al. Development and Validation of the American Heart Association’s PREVENT Equations. Circulation. 2024;149(6):430–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jalepalli SK, Gupta P, Dekker A, Bermejo I, Kar S. Development and validation of multicentre study on novel Artificial Intelligence-based Cardiovascular Risk Score (AICVD). Fam Med Community Health. 2024;12(Suppl 1).
- 8.Mohammed M, Khan MB, Bashier EBM. Machine learning: algorithms and applications: Crc Press; 2016.
- 9.Ayodele TO. Machine learning overview. New Advances in Machine Learning. 2010;2(9–18):16. [Google Scholar]
- 10.Klokova M. Important dates in history of Machine Learning 2023 [Available from: https://medium.com/mlearning-ai/important-dates-in-history-of-machine-learning-d52d7f24b797.
- 11.Delpino FM, Costa ÂK, Farias SR, Chiavegatto Filho ADP, Arcêncio RA, Nunes BP. Machine learning for predicting chronic diseases: a systematic review. Public Health. 2022;205:14–25. [DOI] [PubMed] [Google Scholar]
- 12.Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random survival forests. 2008.
- 13.Huang Y, Li J, Li M, Aparasu RR. Application of machine learning in predicting survival outcomes involving real-world data: a scoping review. BMC Medical Research Methodology. 2023;23(1):268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Lin S, He Y, Wang J, Pan Y. Deep learning survival model for colorectal cancer patients (DeepCRC) with Asian clinical data compared with different theories. Arch Med Sci. 2023;19(1):264–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Bao H, Shi Y, Zhu W, Peng Z, Yan L, et al. Machine learning methods for accurately predicting survival and guiding treatment in stage I and II hepatocellular carcinoma. Medicine (Baltimore). 2023;102(45):e35892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung J-O, Crnovrsanin N, Wirsik NM, Nienhüser H, Peters L, Popp F, et al. Machine learning for optimized individual survival prediction in resectable upper gastrointestinal cancer. Journal of Cancer Research and Clinical Oncology. 2023;149(5):1691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tizi W, Berrado A. Machine learning for survival analysis in cancer research: A comparative study. Scientific African. 2023;21:e01880. [Google Scholar]
- 18.Zhao Y, Wood EP, Mirin N, Cook SH, Chunara R. Social Determinants in Machine Learning Cardiovascular Disease Prediction Models: A Systematic Review. Am J Prev Med. 2021;61(4):596–605. [DOI] [PubMed] [Google Scholar]
- 19.Krittanawong C, Virk HUH, Bangalore S, Wang Z, Johnson KW, Pinotti R, et al. Machine learning prediction in cardiovascular diseases: a meta-analysis. Scientific Reports. 2020;10(1):16057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baashar Y, Alkawsi G, Alhussian H, Capretz LF, Alwadain A, Alkahtani AA, et al. Effectiveness of Artificial Intelligence Models for Cardiovascular Disease Prediction: Network Meta-Analysis. Comput Intell Neurosci. 2022;2022:5849995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai Y, Cai Y-Q, Tang L-Y, Wang Y-H, Gong M, Jing T-C, et al. Artificial intelligence in the risk prediction models of cardiovascular disease and development of an independent validation screening tool: a systematic review. BMC Medicine. 2024;22(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lans A, Kanbier LN, Bernstein DN, Groot OQ, Ogink PT, Tobert DG, et al. Social determinants of health in prognostic machine learning models for orthopaedic outcomes: A systematic review. J Eval Clin Pract. 2023;29(2):292–9. [DOI] [PubMed] [Google Scholar]
- 23.Segar MW, Hall JL, Jhund PS, Powell-Wiley TM, Morris AA, Kao D, et al. Machine Learning-Based Models Incorporating Social Determinants of Health vs Traditional Models for Predicting In-Hospital Mortality in Patients With Heart Failure. JAMA Cardiol. 2022;7(8):844–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Healthy People 2030, U.S. Department of Health and Human Services, Office of Disease Prevention and Health Promotion.
- 25.Teshale AB, Htun HL, Owen A, Gasevic D, Phyo AZZ, Fancourt D, et al. The Role of Social Determinants of Health in Cardiovascular Diseases: An Umbrella Review. J Am Heart Assoc. 2023;12(13):e029765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linardatos P, Papastefanopoulos V, Kotsiantis S. Explainable AI: A Review of Machine Learning Interpretability Methods. Entropy (Basel). 2020;23(1).
- 27.Krzyziński M, Spytek M, Baniecki H, Biecek P. SurvSHAP(t): Time-dependent explanations of machine learning survival models. Knowledge-Based Systems. 2023;262:110234. [Google Scholar]
- 28.Chaddad A, Peng J, Xu J, Bouridane A. Survey of Explainable AI Techniques in Healthcare. Sensors. 2023;23(2):634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salameh J-P, Bossuyt PM, McGrath TA, Thombs BD, Hyde CJ, Macaskill P, et al. Preferred reporting items for systematic review and meta-analysis of diagnostic test accuracy studies (PRISMA-DTA): explanation, elaboration, and checklist. BMJ. 2020;370:m2632. [DOI] [PubMed] [Google Scholar]
- 30.van de Schoot R, de Bruin J, Schram R, Zahedi P, de Boer J, Weijdema F, et al. An open source machine learning framework for efficient and transparent systematic reviews. Nature Machine Intelligence. 2021;3(2):125–33. [Google Scholar]
- 31.Babineau J. Product review: Covidence (systematic review software). Journal of the Canadian Health Libraries Association/Journal de l’Association des bibliothèques de la santé du Canada. 2014;35(2):68–71. [Google Scholar]
- 32.Fernandez-Felix BM, López-Alcalde J, Roqué M, Muriel A, Zamora J. CHARMS and PROBAST at your fingertips: a template for data extraction and risk of bias assessment in systematic reviews of predictive models. BMC Med Res Methodol. 2023;23(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ambale-Venkatesh B, Yang X, Wu CO, Liu K, Gregory Hundley W, McClelland R, et al. Cardiovascular Event Prediction by Machine Learning: The Multi-Ethnic Study of Atherosclerosis. CIRCULATION RESEARCH. 2017;121(9):1092–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barbieri S, Mehta S, Wu B, Bharat C, Poppe K, Jorm L, et al. Predicting cardiovascular risk from national administrative databases using a combined survival analysis and deep learning approach. International Journal of Epidemiology. 2022;51(3):931–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bauer MJ, Nano N, Adolf R, Will A, Hendrich E, Martinoff SA, et al. Prognostic Value of Machine Learning-based Time-toEvent Analysis Using Coronary CT Angiography in Patients with Suspected Coronary Artery Disease. Radiology: Cardiothoracic Imaging. 2023;5(2):e220107.
- 36.Blanchard M, Feuilloy M, Sabil A, Gerves-Pinquie C, Gagnadoux F, Girault JM. A Deep Survival Learning Approach for Cardiovascular Risk Estimation in Patients With Sleep Apnea. IEEE Access. 2022;10:133468–78. [Google Scholar]
- 37.Brester C, Tuomainen TP, Voutilainen A, Kauhanen J, Kolehmainen M. Predicting risk of cardiovascular death in the high-dimensional cohort follow-up data in the presence of competing events: a guide for building a modeling pipeline. Biostatistics and Epidemiology. 2023;7(1):e2084704. [Google Scholar]
- 38.Chhoa H, Chabriat H, Chevret S, Biard L. Comparison of models for stroke-free survival prediction in patients with CADASIL. SCIENTIFIC REPORTS. 2023;13(1):22443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chun M, Clarke R, Cairns BJ, Clifton D, Bennett D, Chen Y, et al. Stroke risk prediction using machine learning: a prospective cohort study of 0.5 million Chinese adults. Journal of the American Medical Informatics Association: JAMIA. 2021;28(8):1719–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng Y, Liu L, Jiang H, Peng Y, Wei Y, Zhou Z, et al. Comparison of State-of-the-Art Neural Network Survival Models with the Pooled Cohort Equations for Cardiovascular Disease Risk Prediction. BMC medical research methodology. 2023;23(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farhadian M, Dehdar Karsidani S, Mozayanimonfared A, Mahjub H. Risk factors associated with major adverse cardiac and cerebrovascular events following percutaneous coronary intervention: a 10-year follow-up comparing random survival forest and Cox proportional-hazards model. BMC Cardiovascular Disorders. 2021;21(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng Y, Leung AA, Lu X, Liang Z, Quan H, Walker RL. Personalized prediction of incident hospitalization for cardiovascular disease in patients with hypertension using machine learning. BMC medical research methodology. 2022;22(1):325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gandin I, Saccani S, Coser A, Scagnetto A, Cappelletto C, Candido R, et al. Deep-learning-based prognostic modeling for incident heart failure in patients with diabetes using electronic health records: A retrospective cohort study. PLoS ONE. 2023;18(2):e0281878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao Y, Zhou Z, Zhang B, Guo S, Bo K, Li S, et al. Deep learning-based prognostic model using non-enhanced cardiac cine MRI for outcome prediction in patients with heart failure. European radiology. 2023;33(11):8203–13. [DOI] [PubMed] [Google Scholar]
- 45.Garcia-Carretero R, Barquero-Perez O, Mora-Jimenez I, Soguero-Ruiz C, Goya-Esteban R, Ramos-Lopez J. Identification of clinically relevant features in hypertensive patients using penalized regression: a case study of cardiovascular events. Medical and Biological Engineering and Computing. 2019;57(9):2011–26. [DOI] [PubMed] [Google Scholar]
- 46.Hathaway QA, Yanamala N, Budoff MJ, Sengupta PP, Zeb I. Deep neural survival networks for cardiovascular risk prediction: The Multi-Ethnic Study of Atherosclerosis (MESA). Computers in Biology and Medicine. 2021;139:104983. [DOI] [PubMed] [Google Scholar]
- 47.Jain V, Bansal A, Radakovich N, Sharma V, Khan MZ, Harris K, et al. Machine Learning Models to Predict Major Adverse Cardiovascular Events After Orthotopic Liver Transplantation: A Cohort Study. Journal of Cardiothoracic and Vascular Anesthesia. 2021;35(7):2063–9. [DOI] [PubMed] [Google Scholar]
- 48.Kim D-Y, Choi K-H, Kim J-H, Hong J, Choi S-M, Park M-S, et al. Deep learning-based personalised outcome prediction after acute ischaemic stroke. Journal of neurology, neurosurgery, and psychiatry. 2023;94(5):369–78. [DOI] [PubMed] [Google Scholar]
- 49.Lin C-H, Kuo Y-W, Huang Y-C, Lee M, Huang Y-W, Kuo C-F, et al. Development and Validation of a Novel Score for Predicting Long-Term Mortality after an Acute Ischemic Stroke. INTERNATIONAL JOURNAL OF ENVIRONMENTAL RESEARCH AND PUBLIC HEALTH. 2023;20(4).
- 50.Mauger CA, Gilbert K, Suinesiaputra A, Bluemke DA, Wu CO, Lima JAC, et al. Multi-Ethnic Study of Atherosclerosis: Relationship between Left Ventricular Shape at Cardiac MRI and 10-year Outcomes. Radiology. 2023;306(2):e220122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morris MC, Moradi H, Aslani M, Sims M, Schlundt D, Kouros CD, et al. Predicting incident cardiovascular disease among African-American adults: A deep learning approach to evaluate social determinants of health in the Jackson heart study. PLoS ONE. 2023;18(11):e0294050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen HT, Vasconcellos HD, Keck K, Reis JP, Lewis CE, Sidney S, et al. Multivariate longitudinal data for survival analysis of cardiovascular event prediction in young adults: insights from a comparative explainable study. BMC medical research methodology. 2023;23(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qian X, Keerman M, Zhang X, Guo H, He J, Maimaitijiang R, et al. Study on the prediction model of atherosclerotic cardiovascular disease in the rural Xinjiang population based on survival analysis. BMC public health. 2023;23(1):1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ren J, Liu D, Li G, Duan J, Dong J, Liu Z. Prediction and Risk Stratification of Cardiovascular Disease in Diabetic Kidney Disease Patients. FRONTIERS IN CARDIOVASCULAR MEDICINE. 2022;9:923549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rigdon J, Basu S. Machine learning with sparse nutrition data to improve cardiovascular mortality risk prediction in the USA using nationally randomly sampled data. BMJ OPEN. 2019;9(11):e032703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sabovcik F, Ntalianis E, Cauwenberghs N, Kuznetsova T. Improving predictive performance in incident heart failure using machine learning and multi-center data. FRONTIERS IN CARDIOVASCULAR MEDICINE. 2022;9:1011071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Segar MW, Vaduganathan M, Patel KV, McGuire DK, Butler J, Fonarow GC, et al. Machine Learning to Predict the Risk of Incident Heart Failure Hospitalization Among Patients With Diabetes: The WATCH-DM Risk Score. Diabetes care. 2019;42(12):2298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stabellini N, Cullen J, Moore JX, Dent S, Sutton AL, Shanahan J, et al. Social Determinants of Health Data Improve the Prediction of Cardiac Outcomes in Females with Breast Cancer. Cancers. 2023;15(18):4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sung JM, Cho I-J, Sung D, Kim S, Kim HC, Chae M-H, et al. Development and verification of prediction models for preventing cardiovascular diseases. PLoS ONE. 2019;14(9):e0222809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turchin A, Morrison FJ, Shubina M, Lipkovich I, Shinde S, Ahmad NN, et al. EXIST: EXamining rIsk of excesS adiposiTy-Machine learning to predict obesity-related complications. Obesity Science and Practice. 2023.
- 61.Wang H, Tucker WJ, Jonnagaddala J, Schutte AE, Jalaludin B, Liaw S-T, et al. Using machine learning to predict cardiovascular risk using self-reported questionnaires: Findings from the 45 and Up Study. INTERNATIONAL JOURNAL OF CARDIOLOGY. 2023;386:149–56. [DOI] [PubMed] [Google Scholar]
- 62.Zhuang X-D, Tian T, Liao L-Z, Dong Y-H, Zhou H-J, Zhang S-Z, et al. Deep Phenotyping and Prediction of Long-term Cardiovascular Disease: Optimized by Machine Learning. The Canadian journal of cardiology. 2022;38(6):774–82. [DOI] [PubMed] [Google Scholar]
- 63.Duan S, Wu Y, Zhu J, Wang X, Zhang Y, Gu C, et al. Development of interpretable machine learning models associated with environmental chemicals to predict all-cause and specific-cause mortality: A longitudinal study based on NHANES. Ecotoxicol Environ Saf. 2024;270:115864. [DOI] [PubMed] [Google Scholar]
- 64.Moreno-Sánchez PA. Improvement of a prediction model for heart failure survival through explainable artificial intelligence. Front Cardiovasc Med. 2023;10:1219586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Segar MW, Jaeger BC, Patel KV, Nambi V, Ndumele CE, Correa A, et al. Development and Validation of Machine Learning-Based Race-Specific Models to Predict 10-Year Risk of Heart Failure: A Multicohort Analysis. Circulation. 2021;143(24):2370–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kvamme H, Borgan Ø, Scheel I. Time-to-event prediction with neural networks and Cox regression. arXiv preprint arXiv:190700825. 2019.
- 67.Sonabend R, Király FJ, Bender A, Bischl B, Lang M. mlr3proba: an R package for machine learning in survival analysis. Bioinformatics. 2021;37(17):2789–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fotso S. PySurvival: open source package for survival analysis modeling 2019 [Available from: https://www.pysurvival.io/.
- 69.Pölsterl S. scikit-survival: A Library for Time-to-Event Analysis Built on Top of scikit-learn. Journal of Machine Learning Research. 2020;21(212):1–6. [PMC free article] [PubMed] [Google Scholar]
- 70.Wang H, Li G. A Selective Review on Random Survival Forests for High Dimensional Data. Quant Biosci. 2017;36(2):85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li K, Yao S, Zhang Z, Cao B, Wilson CM, Kalos D, et al. Efficient gradient boosting for prognostic biomarker discovery. Bioinformatics. 2022;38(6):1631–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Şentürk Acar A, Ata Tutkun N. USE OF ENSEMBLE METHODS FOR SURVIVAL PREDICTION. Mugla Journal of Science and Technology. 2020;6(2):158–64. [Google Scholar]
- 73.Ambler G, Seaman S, Omar R. An evaluation of penalised survival methods for developing prognostic models with rare events. Statistics in medicine. 2012;31(11–12):1150–61. [DOI] [PubMed] [Google Scholar]
- 74.Yan L, Gao N, Ai F, Zhao Y, Kang Y, Chen J, et al. Deep learning models for predicting the survival of patients with chondrosarcoma based on a surveillance, epidemiology, and end results analysis. Front Oncol. 2022;12:967758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bao L, Wang YT, Zhuang JL, Liu AJ, Dong YJ, Chu B, et al. Machine Learning-Based Overall Survival Prediction of Elderly Patients With Multiple Myeloma From Multicentre Real-Life Data. Front Oncol. 2022;12:922039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jiang C, Wang K, Yan L, Yao H, Shi H, Lin R. Predicting the survival of patients with pancreatic neuroendocrine neoplasms using deep learning: A study based on Surveillance, Epidemiology, and End Results database. Cancer Med. 2023;12(11):12413–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tay K, Simon N, Friedman J, Hastie T, Tibshirani R, Narasimhan B. Regularized Cox Regression. 2023.
- 78.Katzman JL, Shaham U, Cloninger A, Bates J, Jiang T, Kluger Y. DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC medical research methodology. 2018;18(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jin L, Zhao Q, Fu S, Cao F, Hou B, Ma J. Development and validation of machine learning models to predict survival of patients with resected stage-III NSCLC. Front Oncol. 2023;13:1092478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sarker IH. Deep Learning: A Comprehensive Overview on Techniques, Taxonomy, Applications and Research Directions. SN Computer Science. 2021;2(6):420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ali S, Abuhmed T, El-Sappagh S, Muhammad K, Alonso-Moral JM, Confalonieri R, et al. Explainable Artificial Intelligence (XAI): What we know and what is left to attain Trustworthy Artificial Intelligence. Information Fusion. 2023;99:101805. [Google Scholar]
- 82.Teshale AB, Htun HL, Hu J, Dalli LL, Lim MH, Neves BB, et al. The relationship between social isolation, social support, and loneliness with cardiovascular disease and shared risk factors: A narrative review. Archives of Gerontology and Geriatrics. 2023;111:105008. [DOI] [PubMed] [Google Scholar]
- 83.Zhao Y, Wood EP, Mirin N, Cook SH, Chunara R. Social Determinants in Machine Learning Cardiovascular Disease Prediction Models: A Systematic Review. American Journal of Preventive Medicine. 2021;61(4):596–605. [DOI] [PubMed] [Google Scholar]
- 84.Basu A. Use of race in clinical algorithms. Sci Adv. 2023;9(21):eadd2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Manski CF, Mullahy J, Venkataramani AS. Using measures of race to make clinical predictions: Decision making, patient health, and fairness. Proc Natl Acad Sci U S A. 2023;120(35):e2303370120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kwabi-Addo B. Race: a Biological or Social Concept. In: Kwabi-Addo B, editor. Health Outcomes in a Foreign Land: A Role for Epigenomic and Environmental Interaction. Cham: Springer International Publishing; 2017. p. 279–97. [Google Scholar]
- 87.Kim PT. Race-aware algorithms: Fairness, nondiscrimination and affirmative action. Cal L Rev. 2022;110:1539. [Google Scholar]
- 88.Khor S, Haupt EC, Hahn EE, Lyons LJL, Shankaran V, Bansal A. Racial and Ethnic Bias in Risk Prediction Models for Colorectal Cancer Recurrence When Race and Ethnicity Are Omitted as Predictors. JAMA Network Open. 2023;6(6):e2318495-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leuzzi C, Sangiorgi GM, Modena MG. Gender-specific aspects in the clinical presentation of cardiovascular disease. Fundamental & clinical pharmacology. 2010;24(6):711–7. [DOI] [PubMed] [Google Scholar]
- 90.Teshale AB, Htun HL, Owen AJ, et al Gender-specific aspects of socialisation and risk of cardiovascular disease among community-dwelling older adults: a prospective cohort study using machine learning algorithms and a conventional method J Epidemiol Community Health Epub ahead of print: [June 1, 2024] doi:10.1136/jech-2023-221860.
- 91.O’Neil A, Scovelle AJ, Milner AJ, Kavanagh A. Gender/Sex as a Social Determinant of Cardiovascular Risk. Circulation. 2018;137(8):854–64. [DOI] [PubMed] [Google Scholar]
- 92.Shrestha S, Das S. Exploring gender biases in ML and AI academic research through systematic literature review. Front Artif Intell. 2022;5:976838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cirillo D, Catuara-Solarz S, Morey C, Guney E, Subirats L, Mellino S, et al. Sex and gender differences and biases in artificial intelligence for biomedicine and healthcare. npj Digital Medicine. 2020;3(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang P, Yao J, Xie Y, Luo M. Gender-Specific Predictive Markers of Poor Prognosis for Patients with Acute Myocardial Infarction During a 6-Month Follow-up. J Cardiovasc Transl Res. 2020;13(1):27–38. [DOI] [PubMed] [Google Scholar]
- 95.la Roi-Teeuw HM, van Royen FS, de Hond A, Zahra A, de Vries S, Bartels R, et al. Don’t be misled: Three misconceptions about external validation of clinical prediction models. Journal of Clinical Epidemiology. 2024:111387.
- 96.Liu R, Wang M, Zheng T, Zhang R, Li N, Chen Z, et al. An artificial intelligence-based risk prediction model of myocardial infarction. BMC Bioinformatics. 2022;23(1):217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wilson A. Why ML Is Used. 2018 [updated August 14, 2018. Available from: https://trymachinelearning.com/why-ml-is-used/#:~:text=Another%20misconception%20is%20that%20machine%20learning%20requires%20massive,ML%20algorithms%20perform%20well%20even%20with%20small%20datasets.
- 98.Collins GS, Dhiman P, Andaur Navarro CL, Ma J, Hooft L, Reitsma JB, et al. Protocol for development of a reporting guideline (TRIPOD-AI) and risk of bias tool (PROBAST-AI) for diagnostic and prognostic prediction model studies based on artificial intelligence. BMJ Open. 2021;11(7):e048008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Moons KG, Kaul T, Damen JA, Andaur Navarro C, Hooft L, Reitsma JB, et al. PROBAST + AI: assessing quality, risk of bias and applicability of diagnostic and prognostic prediction models based on AI or ML techniques. Abstracts accepted for the 27th Cochrane Colloquium, London, UK. Cochrane Database of Systematic Reviews 2023; (1 Supp 1). 10.1002/14651858.CD202301.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is provided within the manuscript or supplementary information files.