Abstract
We characterized the peach latent mosaic viroid (PLMVd) replication intermediates that accumulate in infected peach leaves and determined the tissue and subcellular localization of the RNA species. Using in situ hybridization, we showed that PLMVd strands of both plus and minus polarities concentrate in the cells forming the palisade parenchyma. At the cellular level, PLMVd was found to accumulate predominantly in chloroplasts. Northern blot analyses demonstrated that PLMVd replicates via a symmetric mode involving the accumulation of both circular and linear monomeric strands of both polarities. No multimeric conformer was detected, indicating that both strands self-cleave efficiently via their hammerhead sequences. Dot blot hybridizations revealed that PLMVd strands of both polarities accumulate equally but that the relative concentrations vary by more than 50-fold between peach cultivars. Taken together these results establish two hallmarks for the classification of viroids. Group A viroids (e.g., PLMVd), which possess hammerhead structures, replicate in the chloroplasts via the symmetric mode. By contrast, group B viroids, which share a conserved central region, replicate in the nucleus via an asymmetric mechanism. This is an important difference between self-cleaving and non-self-cleaving viroids, and the implications for the evolutionary origin and replication are discussed.
Viroids are small (∼300-nucleotide [nt]), single-stranded, circular RNAs that infect higher plants, causing significant losses in the agricultural industry (see references 15 and 33 for reviews). The 26 known viroid species have been classified in two groups, A and B (see references 6 and 15 for reviews). This classification is based primarily on whether a viroid possesses the five structural domains characteristic of a group B viroid. The group B viroids are further subdivided on the basis of both the sequence and the length of a highly conserved central region. Three viroids possess no sequence or structural similarity with the group B viroids and have been classified in group A. These three viroids possess self-cleaving hammerhead motifs that are essential for their replication (see below). This classification is supported by phylogenetic reconstructions in which a group A viroid (avocado sunblotch viroid [ASBVd]) has been proposed as an evolutionary link between the classical group B viroids and the plant viroid-like satellite RNAs (14).
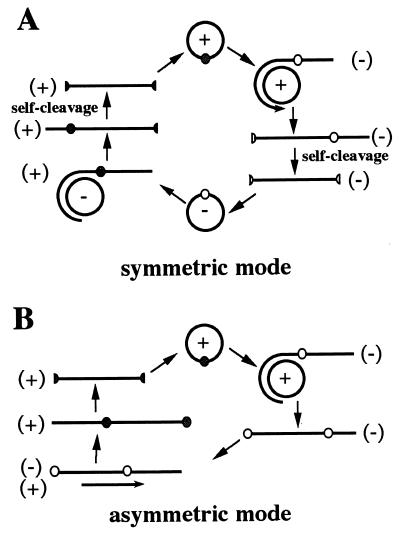
In infected cells, viroids replicate in a DNA-independent manner via a rolling circle mechanism that follows either a symmetric or an asymmetric mode (15) (Fig. 1). In the symmetric mode, the infecting circular monomer (which is assigned plus polarity by convention) is replicated into linear multimeric minus strands, which are then spliced and ligated, yielding minus circular monomers. By using the latter RNA as template, the same three steps are repeated to produce the progeny. In contrast, in the asymmetric mode, the linear multimeric strands serve directly as the template for the synthesis of linear multimeric plus strands. Therefore, both the linear and circular minus monomers are produced only in the symmetric mode. For example, the fact that minus circular monomeric strands of ASBVd are present in RNA isolated from infected avocado plants is taken as evidence that ASBVd replicates via the symmetric mode (10, 20). Similarly, the fact that the circular minus monomer of potato spindle tuber viroid (PSTVd) has not been found in plants infected by this viroid has been taken as evidence that it replicates via the asymmetric mode (5).
FIG. 1.
Schematic representation of the mechanism of viroid rolling circle replication by either the symmetric (A) or the asymmetric (B) mode. The polarity of the strands is indicated in parentheses, and the small circle on the strands denotes the cleavage site. The process begins with the infecting circular (+) strand.
To date most of our knowledge of viroid biology comes from studies of group B viroids (e.g., PSTVd). Three group B viroids have been shown to accumulate in the nuclei of infected cells (4, 18, 29), where they are believed to replicate via the asymmetric mode with host RNA polymerase II as the replicase enzyme. The actual mechanism of processing of the resulting multimeric strands is still a matter of debate. It was proposed that a host endoribonuclease catalyzes the cleavage of multimeric strands into monomers (15); however, a recent report has shown that coconut cadang cadang viroid processing could be mediated by a new self-cleaving motif under specific conditions (25). In contrast, studies on ASBVd have shown that this group A viroid is located mainly in the host chloroplasts (3, 23). In this system, the processing of the multimeric intermediates is mediated by self-catalytic hammerhead motifs. Therefore, it has been proposed that the replication mode and the viroid subcellular localization may be linked and may potentially constitute a fundamental difference between the two major groups of viroids (15). To confirm this analysis, it is essential to determine whether the ASBVd features are common to other group A viroids, more specifically to a member of the peach latent mosaic viroid (PLMVd) subgroup.
PLMVd is an RNA species of 335–338 nucleotides (nt) which causes a latent mosaic of peach trees (19). Both the plus and minus multimeric PLMVd strands efficiently self-cleave in vitro by using hammerhead structures (1, 19). Due to the presence of self-cleavage properties and the absence of a known conserved central region, PLMVd was proposed to be a group A viroid (19). PLMVd, ASBVd, and chrysanthemum chlorotic mottle viroid (CChMVd) are the only hammerhead-containing species for which the group A classification is confirmed. Unlike other viroids, which fold into rod-like or quasi-rod-like structures, CChMVd and PLMVd adopt an unusual branched secondary structure which makes them the only viroids that are insoluble in 2 M lithium chloride (27). These results prompted the classification of the self-cleaving group A viroids into two subgroups, one formed by ASBVd and one formed by PLMVd and CChMVd (27). In this report, we characterize the PLMVd replication intermediates which accumulate in infected peach leaves and determine the tissue and subcellular localization of this RNA species to strengthen the existing criteria for the classification of viroids.
MATERIALS AND METHODS
RNA isolation.
RNA samples were isolated from leaves harvested from a total of nine peach cultivars, six that had been infected by PLMVd and three healthy ones (see Table 1). Four different procedures were used to prepare the RNA samples. The first procedure is a modified version of the procedure involving a polyethylene glycol precipitation (PEG precipitation) (28, 32). Frozen leaf petioles (0.6 g) were ground to a fine powder in liquid nitrogen. The resulting powdered tissue was transferred to a 50-ml centrifuge tube, and 5 ml of phenol mix (phenol-saturated Tris [pH 8.0], chloroform, octanol [100:100:4]), 4 ml of LG buffer (3.5 M LiCl, 0.3 M glycine [pH 9.5]), 80 μl of bentonite solution, 40 μl of 2-mercaptoethanol, and 40 μl of 20% lithium dodecyl sulfate in water) were added. The mixture was vortexed for 30 to 60 s and then centrifuged at 10,000 × g for 20 min. The resulting supernatant was transferred to a fresh tube, and powdered PEG 6000 was added to a final concentration of 20% (wt/vol). After being vortexed for 15 to 30 s, the tubes were successively incubated at 37°C for 5 min, at room temperature for 10 min, and on ice for 20 min and were then vortexed again for 15 s. The mixture was centrifuged at 10,000 × g for 20 min, and the pellet was recovered, washed with 70% ethanol, and air dried. The pellet was dissolved in 400 μl of nuclease-free water, transferred to a sterile Eppendorf tube, and centrifuged at 12,000 × g for 5 min to remove any insoluble material. Finally, the RNA was ethanol precipitated. The second procedure used the RNeasy Plant mini kit (Qiagen) to prepare RNA from ∼150 mg of tissue as specified by the manufacturer. The third procedure was identical to the second, except that 5 mM EDTA was added to all buffers. The fourth procedure was the Tris-EDTA extraction method as previously described (17). All RNA samples were quantified by UV spectrophotometry and electrophoresed on 1.3% agarose gels to assess the quality of the preparation. Dried RNA samples were stored at −70°C.
TABLE 1.
Quantitative analysis of PLMVd in peach tissue
| Peach cultivar | Infected plants | Ratio of linear to circular strandsa
|
Quantity per mg of tissuesb
|
||
|---|---|---|---|---|---|
| Plus | Minus | Plus | Minus | ||
| Hardired | + | 13:1 | 13:1 | 0.4 | 0.3 |
| Siberian C | + | 12:1 | 16:1 | 2.2 | 2.0 |
| Redhaven | + | 9:1 | 14:1 | 5.5 | 3.2 |
| Redgold | + | 16:1 | 14:1 | 0.1 | 0.2 |
| Agua | + | 8:1 | 12:1 | 0.4 | 0.2 |
| Harrow Beauty | + | 8:1 | 15:1 | 1.5 | 1.2 |
| Bailey | − | ||||
| Siberian C | − | ||||
| GF-305 | − | ||||
The proportions of linear versus circular PLMVd strands of both polarities were estimated by the quantification of Northern blot hybridizations.
The quantity of PLMVd (in nanograms per milligram of tissue) was determined by dot blot hybridization.
Preparation of RNA probes.
Both the plus and minus strand-specific riboprobes were synthesized and purified with plasmid pPD1 as the template (1). Briefly, this construction possesses two tandemly repeated PLMVd sequences cloned into the PstI restriction site of pBluescript II KS. The insert is flanked by T3 and T7 promoters for the production of plus- and minus-polarity transcripts, respectively. For Northern blot hybridization, we used the StripEZ transcription kit (Ambion) to obtain probes that can be stripped under mild washing conditions to permit multiple probings of the same membrane. The transcription reaction was performed in the presence of 50 μCi of [α-32P]UTP (or [α-32P]GTP) (3,000 Ci/mmol; Amersham Life Science). For in situ hybridizations, in vitro transcription was performed in the presence of 3.5 mM digoxigenin-11-UTP (DIG-UTP; Boehringer Mannheim) under the conditions recommended by the manufacturer. During transcription, RNAs of both polarities possessing hammerhead sequences are produced and self-cleave efficiently, producing 338-nt linear monomeric transcripts. After transcription, treatment with DNase (RNase free; Pharmacia) was performed and the nucleic acids were ethanol precipitated. In general, the transcripts were resuspended in 20 μl water, 10 μl of stop buffer (0.3% [wt/vol] each bromophenol blue and xylene cyanol, 10 mM EDTA [pH 7.5], 97.5% [vol/vol] deionized formamide) was added, and the resulting mixture was denatured for 2 min at 65°C prior to polyacrylamide gel electrophoresis (PAGE) through a 5% (wt/vol) polyacrylamide gel in 100 mM Tris-borate (pH 8.3)–1 mM EDTA–7 M urea buffer. Transcripts were detected by UV shadowing, excised, eluted, precipitated, purified by passage through Sephadex G-50 spun columns (Pharmacia), lyophilized, quantitated by absorbance spectrophotometry at 260 nm, and stored dry at −70°C. Incorporation of the DIG label into the transcripts was monitored by using dot blots probed with alkaline phosphatase-conjugated anti-DIG antibody (Boehringer Mannheim) as specified by the manufacturer for DIG-labelled transcripts, while Cerenkov counting was used for the 32P-labeled transcripts.
PSTVd probes labeled with DIG were prepared by in vitro transcription from plasmid pHa106 (kindly provided by Martin Tabler [34]). EcoRI-linearized pHa106 was transcribed with SP6 RNA polymerase, producing a plus-polarity 406-nt PSTVd riboprobe, which is slightly longer than the PSTVd monomer (i.e., 359 nt).
In situ hybridization for electron and light microscopy.
Fixation and embedding of peach samples were performed as previously described (23) with minimal modifications. Briefly, leaf pieces from both healthy and PLMVd-infected peach plants (Siberian C cultivar) were fixed with a modified Karnovsky fixation (1% glutaraldehyde and 2% paraformaldehyde in 100 mM cacodylate buffer [pH 7.2] containing 5 mM CaCl2) for 2 to 48 h, dehydrated through an ascending ethanol series at room temperature, and embedded in LR Gold (Electron Microscopy Science) at −20°C under UV light.
In situ hybridizations were performed as previously described (13). For light microscopy, 0.5-μm sections were collected on poly-l-lysine-coated Superfrost glass slides (dried for 3 h at 42°C) and acetylated. Prehybridization (1 h) and hybridization (overnight) were performed as described for Northern blots, except that the salmon sperm DNA was replaced by 0.5 mg of yeast tRNA per ml, the temperature was held constant at 55°C, and both steps were performed in a petri dish humidified with 50% formamide–4× SSC (20× SSC is 3 M NaCl plus 0.3 M sodium citrate [pH 7.0]). After hybridization, the slides were washed three times in 2× SSC at 37°C for 15 min and once in 1× SSC for 15 min at 37°C. Identical results were obtained when the slides were washed after the first 2× SSC wash with 2× SSC plus 1 μg of RNase per ml for 10 min at room temperature. DIG-labeled hybrids were detected as recommended by the manufacturer (Boehringer Mannheim) by using the reaction between alkaline phosphatase-coupled anti-DIG antibody and indoxyl-nitroblue tetrazolium in the presence of polyvinyl alcohol (11). For electron microscopy, ultrathin sections were mounted on 150-mesh nickel grids covered with Parlodion-carbon coating and were then acetylated and washed twice with 2× SSC for 5 min. The grids were placed, section down, on 200 to 400 μl of hybridization buffer and incubated overnight at 60°C as described for light microscopy. After hybridization, the sections were washed at 37°C successively once for 20 min and four times for 5 min each with 2× SSC solution and finally twice for 5 min each with phosphate-buffered saline (0.34 M NaCl, 0.007 M KCl, 0.019 M KH2PO4, 0.004 M Na2HPO4) plus 0.1% Tween 20. For indirect immunodetection, the sections were incubated for 60 min at 37°C with mouse anti-DIG gold-labeled antibodies (Boehringer Mannheim) diluted 1:25 in phosphate-buffered saline–0.1% Tween 20–1% casein. After two washes in PBS buffer, sections were incubated for 60 min at 37°C on gold-labeled mouse anti-DIG (10-nm gold particles [Cedar Lane]) and then briefly rinsed with distilled water. The grids were examined in a Philips 300 electron microscope.
Northern and dot blot hybridizations.
Northern blot hybridizations were performed as described previously (16) with probes radiolabeled with [α-32P]UTP. Isolated RNA (2 μg) or PLMVd transcript (0.5 ng) was either resuspended in formaldehyde gel running buffer, denatured for 3 min at 80°C, and fractionated on 1.3% agarose gels containing formaldehyde and ethidium bromide or resuspended in formamide-dye mixture (95% formamide, 10 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol), denatured for 2 min at 70°C, and subjected to PAGE (5% polyacrylamide) on gels in 50 mM Tris-borate (pH 8.3)–1 mM EDTA–7 M urea running buffer as previously described (1). Nucleic acids were transferred by capillary overnight to nylon filters (Hybond N+; Amersham Life Science) in either 10× SSC (acrylamide gels) or 20× SSC (agarose gels). The filters were then UV cross-linked for 5 min and baked at 80°C for 90 min before being prehybridized for 4 h at 60°C in a solution containing 50% formamide, 5× SSC, 1% sodium dodecyl sulfate (SDS), 5% Denhardt’s solution, and 100 μg of salmon sperm DNA per ml. Hybridizations were performed overnight (∼16 h) at 60°C with fresh prehybridization buffer containing the probe. After hybridization, the filters were successively washed three times for 15 min each in 1× SSC–0.1% SDS at 65°C and once for 15 min in 0.1× SSC–0.1% SDS at 60°C (or 65°C). The filters were analyzed by either autoradiography or with a PhosphorImager (Molecular Dynamics). All Northern blot hybridizations were performed as described above, with the exception of a few controls, which are indicated Results. All blots were successively hybridized with the plus- and the minus-strand probes. Linear PLMVd transcripts used as controls were synthesized as described previously (1). Circular PLMVd transcripts having only 3′,5′-phosphodiester bonds were synthesized as reported previously (2).
Dot blot hybridizations were performed as described for Northern blots, except that serial dilutions of the RNA samples were applied to the filter under vacuum. In addition, quantities ranging from 0.01 to 32 ng of gel-purified monomeric PLMVd transcripts of both polarities (synthesized by runoff transcription) were used as standards. Gel-purified monomeric (338-nt) PLMVd transcripts of both polarities were used as probes, and the filters were quantified with a PhosphorImager.
RESULTS
In situ hybridization of PLMVd strands.
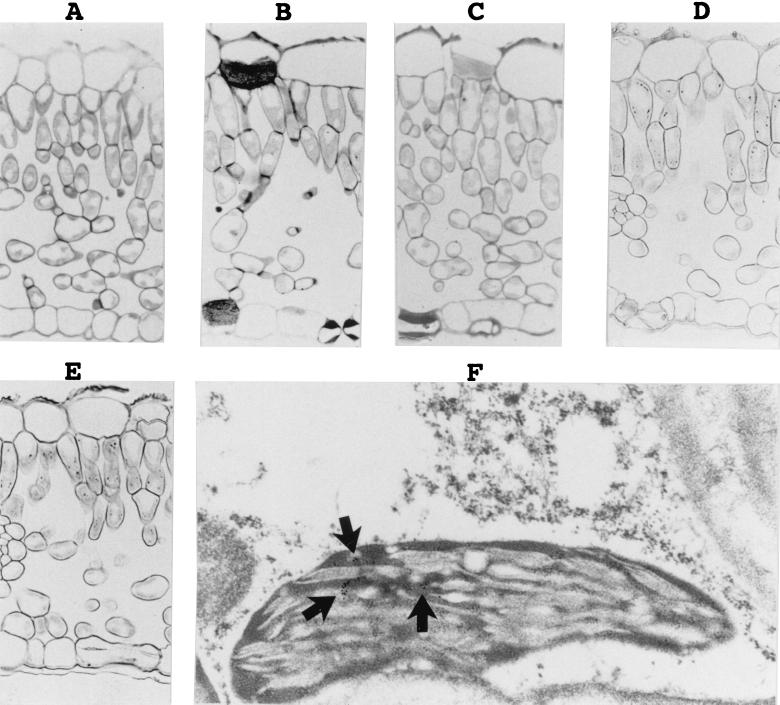
To determine the localization of PLMVd in tissue, leaf pieces from both healthy and PLMVd-infected Siberian C peach plants were harvested, fixed, and studied by in situ hybridization with DIG-UTP-labeled riboprobes (Fig. 2). The PLMVd infection was confirmed by Northern blot hybridization (see below). In the first set of experiments, the DIG-labeled hybrids were detected with alkaline phosphatase-conjugated anti-DIG antibodies (e.g., Fig. 2A to E). No hybridization signal was detected when PLMVd-infected samples were incubated in the absence of a probe or with a DIG-PSTVd probe (Fig. 2A and B). Similarly, no DIG-labeled hybrids were detected in healthy peach leaves (Fig. 2C). In contrast, riboprobes of both polarities of PLMVd reacted with PLMVd-infected samples (Fig. 2D and E). These results were consistent within the various cultivars examined, with the hybridization signals being limited to PLMVd-infected peach leaves (data not shown). At the tissue level, the DIG-labeled hybrids were observed to concentrate in the cells forming the palisade parenchyma. Several DIG-labeled hybrids appeared in each cell; unfortunately, the low resolution of light microscopy could not discern the specific subcellular localization. Variations in the hybridization conditions, washings, and DIG detection revealed different amounts of DIG-labeled hybrids, but in all cases the proportion of the plus and minus strands remained identical.
FIG. 2.
Detection of PLMVd by in situ hybridizations with DIG-labeled riboprobes. (A to E) Observations of the DIG-labeled hybrids by light microscopy. The micrographs show sections of PLMVd-infected leaves hybridized in either the absence (A) or the presence (B) of DIG-PSTVd riboprobe, a section of a healthy peach leaf probed with a minus-polarity PLMVd riboprobe (C), and sections of PLMVd-infected peach leaves probed with either the plus (D) or minus (E) PLMVd riboprobes. Control panels (A to C) were overstained to ensure the detection of trace amounts of PLMVd strands. (F) Typical electron micrograph of the hybridization of PLMVd-infected peach leaves with the minus-strand PLMVd riboprobe. The arrows point to clusters of grains representing PLMVd accumulated strands.
In the second set of experiments, the target-probe hybrids were revealed by using anti-DIG antibodies coupled to gold particles and were visualized by electron microscopy to determine the subcellular localization of PLMVd. Figure 2F shows a typical result when probing with the PLMVd riboprobe of minus polarity. In situ hybridization revealed clusters of PLMVd strands of both polarities in the chloroplasts of infected leaves. In contrast, only negligible numbers of gold particles were found in healthy peach leaves due to the nonspecific binding of the labeled antibody (data not shown). Statistical analysis involved counting the colloidal gold particles in 15 to 20 cells per experiment. In some experiments more than 103 particles were counted in PLMVd-infected samples. These analyses showed an increase of at least 8-fold (with an average of 11.3-fold) in the number of gold particles detected in the chloroplasts of PLMVd-infected peach leaves as compared to the chloroplasts of healthy ones. No corresponding increase was observed in other organelles. Similar increases in chloroplast labeling was observed with both plus and minus PLMVd riboprobes on PLMVd-infected samples but not with PSTVd riboprobes (data not shown). These investigations revealed that more than 80% (81 to 96%) of the total grains observed in the cell were located in the chloroplasts. The chloroplasts in PLMVd-infected tissues contained an average of 10 to 14 grains depending on the experiment. The remaining grains appeared in the cytoplasm, the vacuoles, and the nuclei, while other cellular structures such as the mitochondria and the cell wall had negligible counts. Finally, approximately the same numbers of grains were detected irrespective of the polarity of the PLMVd riboprobe used (i.e. ∼10 to 14 grains per chloroplast), suggesting that both PLMVd strands accumulate to the same degree. These results clearly show that PLMVd is located predominantly in the chloroplasts.
Detection of PLMVd replication intermediates.
To identify the PLMVd replication intermediates which accumulate in cells, RNA samples from both healthy and PLMVd-infected leaves were isolated and analyzed by Northern blot hybridization. Because PLMVd strands of both polarities have the ability to self-cleave in the presence of magnesium, RNA samples were isolated by various procedures (see Materials and Methods). Two of the four methods used included 5 mM EDTA in their extraction buffers to chelate any cations and thereby prevent any self-cleavage of the multimeric strands during purification. All RNA samples were electrophoresed through both agarose and polyacrylamide gels, transferred to filters and successively probed with plus and minus PLMVd riboprobes (Fig. 3).
FIG. 3.
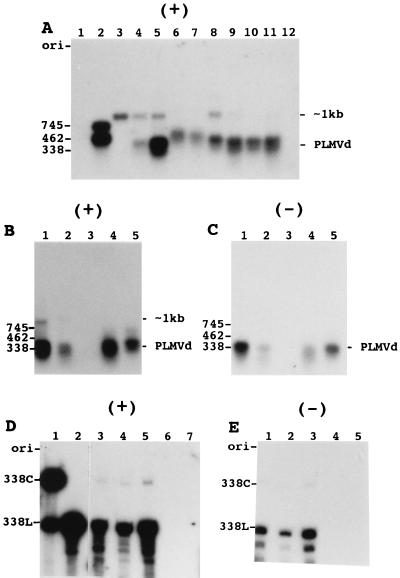
Autoradiograms of Northern blot hybridizations of RNA samples isolated from both healthy and PLMVd-infected peach leaves. RNA samples were fractionated on either agarose (A to C) or polyacrylamide (D and E) gels and then blotted onto nylon filters. The polarity of the PLMVd riboprobe is indicated by the symbol (+) and (−) at the top of the panel. (A) RNA samples were isolated by various extraction procedures: RNeasy Plant mini kit (Qiagen) in the presence of EDTA (lanes 3 to 5); Tris-EDTA isolation (lanes 6 and 7); RNeasy Plant mini kit in the absence of additional EDTA (lane 8); and the PEG precipitation procedure (lanes 9 to 12). In lanes 1 and 2, nonradioactive synthetic PLMVd transcripts of plus (761 and 588 nt) and minus (745 and 462 nt) polarity, respectively, were loaded as controls. Lane 3 contains a sample of healthy GF-305 peach cultivar; lanes 4, 6, 8, 9, 11, and 12 contain RNA samples isolated from a PLMVd-infected RedGold peach cultivar. The samples in lanes 11 and 12 were subjected to either DNase treatment or alkaline hydrolysis prior electrophoresis. Lanes 5 and 7 contain to samples from a PLMVd-infected Redhaven cultivar, while lane 10 contains a sample from the leaves of the Agua cultivar. (B and C) The same filter following hybridization with either the plus- or minus-polarity riboprobe. All samples were isolated by the PEG precipitation procedure. Lanes 1 and 2 contain samples isolated from different leaves of a Redhaven peach. Lane 3 contains an RNA sample from a healthy Bailey cultivar. Lanes 4 and 5 contain RNA samples of PLMVd-infected Redhaven and Siberian C cultivars. (D and E) Filter following PAGE and blotting that was probed with both the plus- and minus-polarity riboprobes, respectively. In panel D, lanes 1 and 2 contain synthetic circular and linear PLMVd controls, while lanes 3 to 5 (which correspond to lanes 1 to 3 of panel E) contain samples from PLMVd-infected Redgold, Hardired, and Siberian C cultivars and lanes 6 and 7 (which correspond to lanes 4 and 5 of panel E) contain samples from healthy GF-305 and Bailey cultivars. Adjacent to the gel, the positions of several PLMVd transcripts are indicated as size references including both the circular (338C) and linear (338L) strands and the mixture of circular and linear molecules (PLMVd). The position of the unknown ∼1-kb species is also indicated in panels A and B.
Initially, the RNA samples were analyzed on 1.3% agarose gels to verify whether PLMVd multimeric strands were accumulated. Nucleic acids were hybridized with radioactive PLMVd-riboprobes synthesized by runoff transcription (see Materials and Methods). Figure 3A shows a typical autoradiogram of a hybridization performed with the plus riboprobe to detect minus-polarity PLMVd strands. Lanes 1 and 2 are controls containing synthetic PLMVd transcripts of plus and minus polarities, respectively. Cross-hybridization between the probe and RNA strands of the same polarity was detectable only after overexposure of the gel (i.e., <2% cross-hybridization). Irrespective of the RNA isolation procedure used, a band corresponding to the PLMVd monomers was consistently detected in the infected samples but not in the healthy ones (compare lanes 4 to 10 with lane 3). The position of this band varied slightly depending on the RNA isolation procedure, most probably because the various RNA isolation methods yielded RNA samples which contained different salts in different concentrations. In addition, the intensity of the band varied depending the extraction procedure used and on the particular cultivar from which the sample was isolated. The RNA nature of the species was confirmed by its resistance to DNase treatment (lane 11) and susceptibility to alkaline hydrolysis (lane 12). An additional nonspecific RNA band of ∼1 kb was detected in both infected and healthy samples and appeared to be more abundant when the Qiagen RNA extraction procedure was used. Regardless of the RNA extraction method used, overexposure of the Northern blots resulted in the appearance of this band (data not shown). However, since the greatest amount was observed in samples from healthy leaves (lane 3), we believe that it is not a PLMVd replication intermediate.
Both the plus and minus PLMVd riboprobes were used to analyze the PLMVd replication intermediates in several peach cultivars. The plus PLMVd-riboprobe (Fig. 3B) detected a band corresponding to monomer minus-strand PLMVd (the nonspecific band of ∼1 kb was detected in one sample), while probing of the same filter (after being stripped) with the minus PLMVd riboprobe detected only monomeric PLMVd of plus polarity (Fig. 3C). These results suggests that both strands of PLMVd accumulate as monomers.
Regardless of the extraction method used, no multimeric strands of either polarity were detected except after overexposure. The presence of 5 mM EDTA in the buffers, which chelates the divalent cations necessary for self-cleavage, did not increase the concentration of multimeric strands. To rule out the possibility that both the linear multimers and circular monomers self-cleaved to yield mainly linear monomers, we performed a set of control extractions. Synthetic radiolabeled PLMVd transcripts were synthesized as described previously (i.e., linear dimers and circular monomers [8]) and added to the ground leaf powder. RNAs of these mixtures were extracted with the RNeasy plant mini kit (Qiagen), either with or without 5 mM EDTA, and were subsequently fractionated on polyacrylamide gels. Radiolabeled transcripts were revealed by autoradiography (data not shown). Independent of the presence of EDTA, only trace amounts of linear monomer strands were detected, suggesting that self-cleavage was limited during these extractions. Therefore, self-cleavage during extraction is not responsible for either the absence of multimeric strands or the low concentration of circular conformers observed.
PAGE followed by Northern blot hybridization was used to resolve circular and linear PLMVd conformers (Fig. 3D and E). Only the two bands corresponding to the linear and circular PLMVd monomers were detected in RNA samples isolated from infected peach leaves. Multimeric strands of either polarity were not observed. The proportion of linear versus circular monomeric molecules was estimated for both the plus and minus strands of several PLMVd cultivars (Table 1). This proportion varied from 8 to 16 (average, 11) linear conformers per circular molecule for the plus strands and from 12 to 16 (average, 14) linear conformers per circular molecule for the minus strand. These results show that PLMVd strands of both polarities accumulate predominantly as linear conformers.
Quantity of PLMVd strands.
To establish the quantity of PLMVd strands (expressed as nanograms of PLMVd per milligram of tissue), we performed dot blot hybridizations. This technique does not differentiate between the linear and circular conformers; however, since we know from the Northern blot hybridizations that the linear conformer predominates, this is not a concern. Only RNA samples isolated by PEG precipitation including the 3.5 M LiCl, known to favor PLMVd extraction (27), were used. Several known concentrations of linear monomeric (338-nt) PLMVd transcripts of both polarities were used as standards. Serial dilutions of RNA from both healthy and infected leaves were spotted onto filters and analyzed with PLMVd riboprobes of both polarities. The results are reported in Table 1. PLMVd was detected only in RNA samples from infected leaves and not in those from healthy ones (<0.01 ng/mg of tissue). The concentrations of both the plus and minus PLMVd strands were relatively constant within each cultivar except the Redgold cultivar, in which slightly more minus strands were detected. In contrast, the concentrations varied significantly between cultivars, ranging from 0.1 to 5.5 ng of PLMVd strands per mg of tissue. PLMVd was more concentrated in Siberian C, Redhaven, and Harrow Beauty and less concentrated in Hardired, Redgold, and Agua peach trees. The observation that the strands of both polarities accumulate at approximately similar concentrations within a cultivar but that these concentrations varied between the cultivars correlates with the results of the Northern blot analyses. Total PLMVd strands accumulate to levels comparable to those PSTVd, which accumulates to a concentration of 1 ng per mg of tissue (7).
DISCUSSION
Subcellular localization is a trademark of viroid groups.
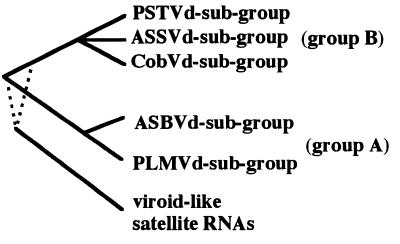
Using in situ hybridization, we have shown that PLMVd replicates predominantly in the palisade parenchyma cells of infected peach leaves (Fig. 2). However, it remains unknown if this cellular localization is a result of the high metabolic activity occurring in these cells or of a PLMVd molecular determinant to either enter or replicate primarily in these cells. Within these cells, PLMVd strands of both polarities accumulate in the chloroplasts (>80% of the strands are found in this organelle). The similar localization of PLMVd and ASBVd, even though they are members of different subgroups (the PLMVd and ASBVd subgroups, respectively), suggests that the subcellular location of a viroid is a fundamental characteristic of their group classification (Fig. 4). Group A viroids are found in chloroplasts, while group B viroids are found in nuclei (see the introduction). Clearly, this fundamental difference has important implications in the molecular biology and evolutionary origin of both viroid groups.
FIG. 4.
Schematic phylogenetic reconstruction of viroids and viroid-like satellite RNAs based on physical characteristics. The dotted lines for the satellite RNAs indicate their precise relationship to viroid group A and B.
Because of the difference in subcellular location, nuclear and chloroplast viroids are likely to use different mechanisms to ensure their replication. For example, host nuclear RNA polymerase II has been proposed to replicate group B viroids (15) but has been shown to be unable to replicate both ASBVd (26) and PLMVd (22). In fact, the host polymerase supporting the replication of group A viroids has yet to be identified. Indeed, two different RNA polymerases transcribe chloroplastic genes: one is a bacteriophage-like RNA polymerase, while the other appears to be related to Escherichia coli RNA polymerase (see reference 24 for a review). One of these RNA polymerases probably replicates group A viroids.
Viroids of both groups also use different strategies in the processing of their multimeric strands. This step is mediated by the hammerhead self-catalytic motif in group A viroids, while group B members are thought to use either a host endoribonuclease or some other catalytic motif. The use of a self-cleavage motif by the chloroplastic viroids might be the result of the absence of an appropriate endoribonuclease in the chloroplast. In contrast, plant viroid-like satellite RNAs are most probably replicated in the cytoplasm by their helper virus replicase (15). The cytoplasm may also lack an appropriate specific endoribonuclease; hence, small satellite RNAs, which are believed to replicate by a rolling circle mechanism, either use hammerhead self-cleavage or have evolved other catalytic motifs (e.g., hairpin self-catalytic motif) for the cleavage of their multimeric strands. If we agree with the hypothesis of a monophyletic origin for both the group A and group B viroids and for the related satellite RNAs (14) (Fig. 4), we conclude that an essential factor for their divergence over time is their cellular localization. The recent observation that the group B viroids may use a new self-cleavage motif (25) supports the belief that the ancestor of today’s viroids possessed a catalytic motif which diverged depending on the cellular location. If satellite RNA and viroids have a common ancestor, the ancestral catalytic motif of both cytoplasmic and chloroplastic plant small RNA pathogens could be a hammerhead.
The PLMVd rolling circle mechanism.
The recent discovery of retroviroid-like elements such as carnation stunt-associated small circular RNA (9) prompted us to investigate whether PLMVd was integrated into the host genome. Southern blot hybridization and PCR amplification with various primers and DNA from infected peach leaves as the template indicated that there is no PLMVd counterpart in either the host genome or the extrachromosomal DNA (22a). Thus, the replication cycle of PLMVd depends solely on RNA intermediates, as is observed for all other viroids. Northern blot hybridizations demonstrated that PLMVd replication follows a symmetric pathway involving predominantly, if not exclusively, accumulation of circular and linear monomeric RNAs of both polarities (summarized in Fig. 1A). Clearly, the linear conformation was the most abundant form, accumulating to levels at least eight times higher than those of circular molecules (Table 1). PLMVd is the first viroid for which replicational intermediates of both polarities are found to be identical and are produced at the same level.
Apart from the type of rolling circle (symmetric versus asymmetric), two features seem to differ significantly between the replication of PLMVd and other viroids. First, group B viroids, as well as ASBVd, accumulate the minus-polarity strand in very low abundance compared to the plus counterpart (15, 20). By contrast, PLMVd accumulate both polarities to the same levels. Therefore, the attribution of a given polarity for PLMVd is not based on biological observation, since both strands are identical with respect to their nature (e.g., linear and circular), concentration, and infectivity. Second, group B viroids accumulate a series of various multimeric linear RNAs (i.e. 1 to 8 units in length), probably as a consequence of a poor cleavage efficiency (15, 20). By contrast, multimeric forms of PLMVd are cleaved to completion and then circularized. Accumulation of a high level of monomeric linear RNA might be the result of a rather inefficient ligation step. The synthesis of multimeric PLMVd strands, which is a prerequisite for a rolling circle mechanism, is supported by reverse transcription experiments allowing the detection of products longer than 1 unit (data not shown). Interestingly, the aforementioned peculiarities of the PLMVd rolling circle are also shared by the newly discovered group A viroid CChMVd (27) and by a small circular viroid-like RNA (csc RNA 1) associated with a cherry disease (12). More importantly, the PLMVd-like rolling circle mechanism implies efficient self-cleavage reactions, rather inefficient ligation steps, and a good stability of the accumulated monomeric RNAs (discussed below).
(i) Self-cleavage.
PLMVd, CChMVd, and csc RNA 1 appear to self-cleave to completion in vivo, although their in vitro self-cleavage efficiency ranges from 10 to 60% (1, 12, 27). However, we have previously shown that the self-cleavage accuracy of PLMVd can reach almost 100% in vitro (1). When PLMVd was transcribed under conditions of slow polymerase activity, which favors sequential production and folding of the hammerhead sequences, self-cleavage of the resulting transcripts was greatly increased (>95%) compared to that under the standard conditions (50 to 60%) (1). Most probably, polymerase processivity or an unknown molecular adaptation (e.g., a cofactor or an interaction with a cellular component mediating structural changes) will explain the high level of self-cleavage of the PLMVd, CChMVd, and csc RNA 1 RNAs in vivo. Interestingly these three RNA species have a common organization of their plus and minus hammerhead sequences that are included within a long hairpin. Therefore, we can expect a similar mode of regulation of self-cleavage for these RNAs that could be responsible for the efficient self-cleavage activity in vivo. Other hammerhead self-cleaving viroids (e.g., ASBVd) and viroid-like RNAs differ by the accumulation of a set of plus multimeric strands (1- to 11-mer) and smaller minus multimeric strands (1- to 3-mer) (20, 30, 31). It has been proposed that these RNA species have reduced self-cleavage activity. For example, the catalytic sequences in ASBVd have been proposed to fold into double-hammerhead structures, which is less favored than the single-hammerhead structures (1).
(ii) Ligation.
The apparently efficient in vivo self-cleavage of multimeric PLMVd strands of both polarities leads exclusively to the accumulation of monomeric conformers. However, it remains unknown why the monomeric linear conformers are the most abundant RNA intermediates. One possible explanation is an inefficient circularization step. Self-ligation has been demonstrated for the circularization of PLMVd strands in vitro (8, 21). In vitro self-ligation is relatively inefficient (i.e., ∼10%), and thus the accumulation of linear monomers is favored. This correlates with the ratio of linear to circular PLMVd strands estimated in this report (11:1 and 14:1 for the plus and minus polarities, respectively). However, the existence of the self-ligation activity in vivo remains to be proved. Whatever the mechanism responsible for the accumulation of monomeric linear PLMVd strands, it is not known if there is only a pool of RNA molecules to be ligated or an RNA population playing a direct role in the life cycle of the viroid.
(iii) Monomeric linear-RNA stability.
The high level of PLMVd linear monomers suggests that this conformation is as stable as the circular conformers, although the circular conformers are proposed to be stabilized by the absence of termini. To prevent degradation by host exonucleases, both the 5′ and 3′ extremities of the linear conformers must be embedded within the RNA structure. This hypothesis is supported by the observation that PLMVd linear monomeric strands are difficult to end label at both termini (28a). Thus, it seems possible that PLMVd linear monomers can accumulate because their termini are located within the RNA structure and are protected from the host exonuclease.
In conclusion, the three group A viroids (PLMVd, ASBVd, and CChMVd) replicate via the symmetric mode, involving self-cleavage activity, and at least two of them are located in the chloroplasts of infected cells. These two characteristics appear to be hallmarks of viroids belonging to this group. However, this does not completely exclude the possibility of finding in the future a group A viroid that will replicate by the asymmetric mechanism. Moreover, the presented data strengthen the relationship between PLMVd and CChMVd regarding their rolling circle mechanism and accumulated intermediates. These two viroids share LiCl solubility (27), a similar hammerhead organization, and surprising structural features (6a). Thus, PLMVd and CChMVd appear to be closely related even though no obvious sequence similarity is observed beside their hammerhead motifs. These observations support the proposed subdivision of group A into ASBVd and PLMVd subgroups (27). Furthermore, PLMVd subgroup viroids show some interesting similarities to the newly discovered satellite RNA csc RNA 1 that extend beyond the presence of a hammerhead sequence on strands of both polarities (see above). These results emphasize the possible evolutionary link between viroids and plant viroid-like satellite RNA.
ACKNOWLEDGMENTS
This work was sponsored by a grant from Natural Sciences and Engineering Research Council (NSERC) of Canada to J.-P.P. F.B. was the recipient of an NSERC studentship. J.-P.P. is a Medical Research Council (MRC, Canada) scholar.
REFERENCES
- 1.Beaudry D, Bussière F, Lareau F, Lessard C, Perreault J P. The RNA of both polarities of the peach latent mosaic viroid self-cleaves in vitro solely by single hammerhead structures. Nucleic Acids Res. 1995;23:745–752. doi: 10.1093/nar/23.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaudry D, Perreault J P. An efficient strategy for the synthesis of circular RNA molecules. Nucleic Acids Res. 1995;23:3064–3066. doi: 10.1093/nar/23.15.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonfiglioli R G, McFadden G I, Symons R H. In situ hybridization localizes avocado sunblotch viroid on chloroplast thylakoid membranes and coconut cadang cadang viroid in the nucleus. Plant J. 1994;6:99–103. [Google Scholar]
- 4.Bonfiglioli R G, Webb D R, Symons R H. Tissue and intra-cellular distribution of coconut cadang cadang viroid and citrus exocortis viroid determined by in situ hybridization and confocal laser scanning and transmission electron microscopy. Plant J. 1996;9:457–465. [Google Scholar]
- 5.Branch A D, Benenfeld B J, Robertson H D. Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc Natl Acad Sci USA. 1988;85:9128–9132. doi: 10.1073/pnas.85.23.9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bussière F, Lafontaine D, Perreault J P. Compilation and analysis of viroid and viroid-like RNA sequences. Nucleic Acids Res. 1995;24:1793–1798. doi: 10.1093/nar/24.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Bussière, F., and J.-P. Perreault. Unpublished data.
- 7.Colpan M, Schumacher J, Brüggemann W, Sänger H L, Reisner D. Large-scale purification of viroid RNA using Cs2SO4 gradient centrifugation and high-performance liquid chromatography. Anal Biochem. 1983;131:257–265. doi: 10.1016/0003-2697(83)90164-1. [DOI] [PubMed] [Google Scholar]
- 8.Côté F, Perreault J P. Peach latent mosaic viroid is locked by a 2′,5′-phosphodiester bond produced by in vitro self-ligation. J Mol Biol. 1997;273:533–543. doi: 10.1006/jmbi.1997.1355. [DOI] [PubMed] [Google Scholar]
- 9.Daros J-A, Flores R. Identification of a retroviroid-like element from plants. Proc Natl Acad Sci USA. 1995;92:6856–6860. doi: 10.1073/pnas.92.15.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daros J-A, Marcos J F, Hernandez C, Flores R. Replication of avocado sunblotch viroid: Evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc Natl Acad Sci USA. 1994;91:12813–12817. doi: 10.1073/pnas.91.26.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeBlock M, Debrouwer D. Nonradioactive in situ hybridization application manual. 2nd ed. Indianapolis, Ind: Boehringer Mannheim Biochemicals; 1996. RNA-RNA in situ hybridization using DIG-labeled probes: the effect of high molecular weight polyvinyl alcohol on the alkaline phosphatase indoxyl-nitroblue tetrazolium reaction; pp. 141–145. [Google Scholar]
- 12.Di Serio F, Daros J A, Ragozzino A, Flores R. A 451-nucleotide circular RNA from cherry with hammerhead ribozymes in its strands of both polarities. J Virol. 1997;79:6603–6610. doi: 10.1128/jvi.71.9.6603-6610.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egger D, Troxler M, Bienz K. Light and electron microscopic in situ hybridization: non-radioactive labeling and detection, double hybridization, and combined hybridization-immunocytochemistry. J Histochem Cytochem. 1994;42:815–822. doi: 10.1177/42.6.8189042. [DOI] [PubMed] [Google Scholar]
- 14.Elena S F, Dopazo J, Flores R, Diener T O, Moya A. Phylogeny of viroids, viroidlike satellite RNAs, and the viroidlike domain of hepatitis δ virus RNA. Proc Natl Acad Sci USA. 1991;88:5631–5634. doi: 10.1073/pnas.88.13.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flores R, Di Serio F, Hernandez C. Viroids: the noncoding genomes. Semin Virol. 1997;8:65–73. [Google Scholar]
- 16.Fourney R M, Miyakoshi J, Day III R S, Paterson M C. Northern blotting: efficient RNA staining and transfer. Focus. 1988;10:5–6. [Google Scholar]
- 17.Hadidi A, Giunchedi L, Schamboul A M, Poggi-Pollini C, Amer M A. Occurrence of peach latent mosaic viroid in stone fruits and its transmission with contaminated blades. Plant Dis. 1997;81:154–158. doi: 10.1094/PDIS.1997.81.2.154. [DOI] [PubMed] [Google Scholar]
- 18.Harders J, Lukacs N, Robert-Nicoud M, Jovin J M, Riesner D. Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridization and confocal laser scanning microscopy. EMBO J. 1989;8:3941–3949. doi: 10.1002/j.1460-2075.1989.tb08577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernandez C, Flores R. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc Natl Acad Sci USA. 1992;89:3711–3715. doi: 10.1073/pnas.89.9.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutchins C J, Keese P, Visvader J E, Rathjen P D, McInnes J L, Symons R H. Comparison of multimeric plus and minus strands of viroids and virusoids. Plant Mol Biol. 1985;4:293–304. doi: 10.1007/BF02418248. [DOI] [PubMed] [Google Scholar]
- 21.Lafontaine D, Beaudry D, Marquis P, Perreault J P. Intra- and intermolecular nonenzymatic ligations occur within transcripts derived from the peach latent mosaic viroid. Virology. 1995;212:705–709. doi: 10.1006/viro.1995.1528. [DOI] [PubMed] [Google Scholar]
- 22.Lareau F. Étude d’éléments de la réplication du viroïde de la mosaique latente du pêcher. M.S. thesis. Sherbrooke, Québec, Canada: Université de Sherbrooke; 1996. [Google Scholar]
- 22a.Laurendeau, S., F. Bussière, and J.-P. Perreault. Unpublished data.
- 23.Lima M I, Fonseca M E N, Flores R, Kitajima E W. Detection of avocado sunblotch viroid in chloroplasts of avocado leaves by in situ hybridization. Arch Virol. 1994;13:385–390. doi: 10.1007/BF01379142. [DOI] [PubMed] [Google Scholar]
- 24.Link G. Green life: control of chloroplast gene transcription. Bioessays. 1996;18:465–471. [Google Scholar]
- 25.Liu Y-H, Symons R H. Specific RNA self-cleavage in coconut cadang cadang viroid: potential for a role in rolling circle replication. RNA. 1998;4:418–429. [PMC free article] [PubMed] [Google Scholar]
- 26.Marcos J F, Flores R. Characterization of RNAs specific to avocado sunblotch viroid synthesized in vitro by a cell-free system from infected avocado leaves. Virology. 1992;186:481–488. doi: 10.1016/0042-6822(92)90013-f. [DOI] [PubMed] [Google Scholar]
- 27.Navarro B, Flores R. Chrysanthemum chlorotic mottle viroid: unusual structural properties of a subgroup of self-cleaving viroids with hammerhead ribozymes. Proc Natl Acad Sci USA. 1997;94:11262–11267. doi: 10.1073/pnas.94.21.11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pallas V, Navarro A, Flores R. Isolation of a viroid-like RNA from hop different from hop stunt viroid. J Gen Virol. 1987;68:3201–3205. [Google Scholar]
- 28a.Perreault, J.-P. Unpublished data.
- 29.Schumacher J, Sänger H L, Riesner D. Subcellular localization of viroids in highly purified nuclei from tomato leaf tissue. EMBO J. 1983;2:1549–1555. doi: 10.1002/j.1460-2075.1983.tb01622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheldon C C, Symons R H. Is hammerhead self-cleavage involved in the replication of a virusoid in vivo? Virology. 1993;194:463–474. doi: 10.1006/viro.1993.1285. [DOI] [PubMed] [Google Scholar]
- 31.Silver S L, Rasochova L, Dineshkumar S P, Miller W A. Replication of barley yellow dwarf virus satellite RNA transcripts in oat protoplasts. Virology. 1994;198:331–335. doi: 10.1006/viro.1994.1036. [DOI] [PubMed] [Google Scholar]
- 32.Skrzeckowski L J, Okely E, Mackiewicz M. AAB Virology Group Meeting. Wellesbourne, United Kingdom: Association of Applied Biologists; 1985. Improved PAGE and cDNA diagnosis of potato spindle tuber viroid; p. 7. [Google Scholar]
- 33.Symons R H. Plant pathogenic RNAs and RNA catalysis. Nucleic Acids Res. 1997;25:2683–2689. doi: 10.1093/nar/25.14.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tabler M, Sänger H L. Infectivity studies on different potato spindle tuber viroid (PSTV) RNAs synthesized in vitro with the SP6 transcription system. EMBO J. 1985;4:2191–2199. doi: 10.1002/j.1460-2075.1985.tb03914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]