Abstract
Background
The oceanic whitetip shark Carcharhinus longimanus (family Carcharhinidae) is one of the largest sharks inhabiting all tropical and subtropical oceanic regions. Due to their life history traits and mortality attributed to pelagic longline fishing practices, this species is experiencing substantial population decline. Currently, C. longimanus is considered by the IUCN Red List of Threatened Species as “vulnerable” throughout its range and “critically endangered” in the western north Atlantic. This study sequences and describes the complete mitochondrial genome of C. longimanus in detail.
Methods and results
The mitochondrial genome of C. longimanus was assembled through next-generation sequencing and then analyzed using specialized bioinformatics tools. The circular, double-stranded AT-rich mitogenome of C. longimanus is 16,704 bp long and contains 22 tRNA genes, 2 rRNA genes, 13 protein coding genes and a 1,065 bp long control region (CR). Out of the 22 tRNA genes, only one (tRNA-Ser1) lacked a typical ‘cloverleaf’ secondary structure. The prevalence of TTA (Leu), ATT (Ile) and CTA (Leu) codons in the PCGs likely contributes to the AT-rich nature of this mitogenome. In the CR, ten microsatellites were detected but no tandem repeats were found. Stem-and-loop secondary structures were common along the entire length of the CR. Ka/Ks values estimated for all PCGs were < 1, indicating that all the PCGs experience purifying selection. A phylomitogenomic analysis based on translated PCGs confirms the sister relationship between C. longimanus and C. obscurus. The analysis did not support the monophyly of the genus Carcharhinus.
Conclusions
The assembled mitochondrial genome of this pelagic shark can provide insight into the phylogenetic relationships in the genus Carcharhinus and aid conservation and management efforts in the Central Pacific Ocean.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11033-024-09780-3.
Keywords: Mitogenome assembly, Genomic resources, Purifying selection, Phylomitogenomics
Introduction
The Carcharhiniformes is the largest order of sharks consisting of 200 extant species, the majority of which belongs to the genus Carcharhinus in the family Carcharhinidae [1]. In this genus, the oceanic whitetip shark Carcharhinus longimanus is considered as the only true oceanic shark [2]. With a robust build and large rounded dorsal and long paddle-like pectoral fins, these sharks are further distinguished by the presence of white mottled markings on the tips of their pectoral, dorsal and tail fins, and black tips on their anal and ventral surface of pectoral fins [3, 4].
Oceanic whitetip sharks are epipelagic, found in shallow waters to at least 152 m deep [5, 6] in all tropical and subtropical ocean basins (above 20 °C) between 30° N and 35° S [2, 5]. Maximum body mass in this species can exceed 150 kg [7] and specimens can reach up to nearly 395 cm in total length (TL) [8, 9]. This highly migratory shark is reported to have a lifespan of about 17 years [8]. No major differences in growth rate have been observed between male and female oceanic whitetip sharks and sexual maturity is reached at 6–7 years in the two sexes [2]. Mating typically occurs in June and July, while parturition takes place between February and July [10]. Oceanic whitetip shark litter size ranges between 1 and 14, sharks are 55–75 cm in total length (TL) at birth, and reach maturity at approximately 170–200 cm TL [2]. A weak positive correlation exists between female size and litter size in C. longimanus [10]. Carcharhinus longimanus is recognized as one of the most prevalent top-level predators in open waters [2] playing a crucial role in maintaining the structure and function of coastal and marine ecosystems [11]. Their diet mostly consists of oceanic teleost fishes and cephalopods [2].
Oceanic whitetip sharks share many life history traits with other elasmobranchs (sharks, rays, and skates), including late sexual maturity, low fecundity, slow growth rate, as well as long gestation periods and lifespan [6, 11–14]. Given the aforementioned life-history traits, Carcharhinus longimanus is highly vulnerable to fishing pressure and is expected to experience prolonged recovery periods following population decline [9]. Carcharhinus longimanus is one of the most common bycatch species in tuna fisheries in offshore tropical waters [2] and has experienced major population decline during the last several decades [4]. This population decline is also due to their demand in the global shark fin trade [15–17]. Given the increasing fishing pressure and high catchability, the species is likely to experience a decline of more than 80% in population size within three generations’ time [5] Today, over 30% of all shark species face imminent risk of extinction primarily due to overfishing [18]. Despite the global implementation of no-retention policies for C. longimanus in tuna longline fisheries, this species remains highly vulnerable to longline fishing practices [9]. Considering all these factors, this species, which was previously labeled as ‘vulnerable’ (VU) [5], is now categorized as ‘critically endangered’ (CR) by the IUCN [19], raise concerns of their conservation and management status [20].
Several studies on the biology and ecology of this imperiled shark have been conducted [2, 5, 9, 10, 16, 19], but very few genetic and genomic resources exist in this and other congeneric species of conservation concern. Studies based on short mitochondrial gene markers (CR) have found low levels of genetic diversity in the Indian and Atlantic Oceans, and restricted gene flow between the western and eastern Atlantic Ocean [6]. In a more recent study, using the entire mitochondrial DNA CR, a segment of the mitochondrial PCG nad4, and 12 nuclear microsatellite loci, weak but statistically significant differentiation was reported between the Western Atlantic and Indo-Pacific Oceans, with additional significant matrilineal structure between Indian and Pacific Oceans but no population structure within the Western Atlantic [21].
In this study, we have sequenced, assembled, and described in detail the complete mitochondrial genome of the Oceanic Whitetip Shark Carcharhinus longimanus. Following the protocols in Baeza [22], we analyzed nucleotide composition of the entire mitochondrial genome as well as codon usage profiles of and selective constraints in protein coding genes. We also explored the secondary structure of each identified tRNA gene and investigated the architecture of the control region (CR). We note that Li [23] did sequence the mitochondrial genome of C. longimanus from the South China Sea. However, this previous study did not characterize the mitochondrial genome of the species in detail as we have done here. By characterizing the complete mitochondrial genome of the Oceanic Whitetip shark, C. longimanus, we are aiming to support management and conservation strategies in this critically endangered shark.
Methods
To assemble the mitochondrial genome of the Oceanic Whitetip Shark Carcharhinus longimanus, we extracted genomic DNA (gDNA) from a specimen (SIO:1734e3c7-b223-40cc-b5eb-72d3b23579eb) deposited at the fish collection of the National Museum of Natural History, Smithsonian Institution, Washington DC, USA. The specimen was collected in the tropical Central Pacific Ocean (07° 45.0ʹ N, 141° 47.0ʹ W), southeast of Hawaii, on September 9, 1997, while onboard the RV Townsend Cromwell. gDNA was extracted from muscle tissue using an AutoGenPrep 965 automated DNA extraction robot (AutoGen, Holliston, MA, USA) according to the manufacturer’s guidelines. Next, an Illumina library was prepared following the standard NEB Ultra II DNA library prep kit (New England Biolabs, Ipswich, MA, USA) protocol. The library was sequenced on an Illumina NovaSeq (Illumina, San Diego, CA, USA) using a 2 × 150 cycle sequencing strategy. A total of 8,521,356 pairs of reads were employed to assemble ‘de novo’ the mitochondrial genome of Carcharhinus longimanus using the pipeline GetOrganelle v. 1.6.4 [24]. For the assembly, we used as a seed the mitochondrial genome of the congeneric C. falciformis, available in NCBI’s GenBank (accession number: OM885432). The run used k-mer sizes of 21, 55, 85, and 115. The sequence data are part of a project to sequence mitochondrial genomes of marine fishes occurring in the Exclusive Economic Zone of the United States based on voucher specimens (BioProject: PRJNA720393) and data are deposited on GenBank (BioSample: SAMN31811566).
The assembled genome of C. longimanus was first annotated using the webserver MITOS2 (https://mitos2.bioinf.uni-leipzig.de/—[25]) and the nucleotide composition of the whole mitochondrial genome was analyzed using the software MEGAX [26]. This first in silico annotation was manually curated using the web server Expasy (https://web.expasy.org/—[27]) in order to correct the start and stop codons of the protein coding genes. The entire mitochondrial genome was visualized using the web server Chloroplot (https://irscope.shinyapps.io/Chloroplot/—[28]). The transfer RNA genes (tRNA) were identified using the software MiTFi [29] as implemented in the web server MITOS2 and the secondary structure of each tRNA was visualized using the web server Forna (http://rna.tbi.univie.ac.at/forna/—[30]). The number and frequency of each codon in all protein codon genes was estimated using the vertebrate mitochondrial code in the web server Sequence Manipulation Suite (SMS) (https://www.bioinformatics.org/sms2/—[31]). Relative synonymous codon usage (RSCU), defined as the ratio of the observed frequency of codons to the expected frequency, of all concatenated protein coding genes was estimated and visualized using the EZcodon tool in the web server EZmito (https://ezmito.unisi.it/ezcodon—[32]). MEGAX was also used to analyze the nucleotide composition of ribosomal RNA (rRNA). Selective pressures acting on each mitochondrial PCG were examined while estimating rates of non-synonymous substitutions per non-synonymous site (Ka), synonymous substitutions per synonymous site (Ks) and the Ka/Ks ratio (ω) for each PCG using the program KaKs_calculator Toolbox 2.0 [33] with C. leucas (KF646785) as the outgroup. PCGs with Ka/Ks values below 1 experience negative (purifying) selection, whereas values above 1 indicate positive (diversifying) selection [33].
The long, non-coding control region (CR) was studied in detail. Repeats within the region were found using the BioPHP Microsatellite Repeats Finder web server (http://insilico.ehu.es/mini_tools/microsatellites/—[34]) and the Tandem Repeat Finder: 4.09 Version web server (https://tandem.bu.edu/trf/trf.basic.submit.html—[35]). Predictions of secondary structure of these regions were provided by RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi—[36]) to observe the presence of hairpin and loop structures. While RNAfold calculates minimum free energy (MFE) and relies on experimental data for scoring parameters, it fails to identify unconventional RNA structures arising from tandem repeats in mitochondrial genomes [37]. To overcome this limitation, we opted to explore the secondary structure of the same region using the hybrid method MXFold2 (http://ws.sato-lab.org/mxfold2/—[37]) which provides a more accurate prediction by incorporating folding scores obtained from deep-neural network trained on extensive data and avoids overfitting by using thermodynamic parameters to evaluate previously unobserved structures.
Phylomitogenomics of the genus Carcharhinus
To reveal the phylogenetic position of C. longimanus within the genus Carcharhinus, the newly assembled mitochondrial genome together with other 18 mitogenomes available in GenBank belonging to congeneric species were used for maximum likelihood (ML) phylogenetic inference. Given the evolutionary history of the genus Carcharhinus, which has been found to display incomplete lineage sorting and polytomies in previous phylogenetic inferences due to rapid radiation of its lineages [38–41], we carefully considered the phylogenetic pipeline used in this study. To address the previously reported phylogenetic complexities in the genus, dividing the alignment into gene partitions and selecting evolutionary models for each gene is necessary. Therefore, we decided to employ an ML analysis using a translated alignment of the protein coding genes.
A total of 19 other mitochondrial genomes were used in this study, including representatives from various genera of the family Carcharhinidae such as Galeocerdo (n = 2 species, G. cuvier), Glyphis (n = 5 species), Lamiopsis (n = 2 species), Loxodon (n = 2 species), Rhizoprionodon (n = 1 species), Scoliodon (n = 3 species), Triaenodon (n = 3 species, T. obesus), and the blue shark Prionace glauca and lemon shark Negaprion brevirostris. Additionally, mitochondrial genomes from closely related families Scyliorhinidae (belonging to the genera Cephaloscyllium (n = 2 species), Scyliorhinus (n = 2 species), and Poroderma pantherinum), Triakidae (n = 4 species, Hemitriakis japonica + Mustelus spp.), and Pentanchidae (Galeus melastomus, Halaelurus buergeri, and Parmaturus melanobranchus) were used as outgroups.
The analysis proceeded in a manner identical to that detailed in Baeza [22]. We first extracted all 13 PCG nucleotide sequences from all mitochondrial genomes and translated them to amino acids using the programs MEGA X and Clustal Omega [42], respectively. Poorly aligned regions in each PCG alignment were removed with trimAl [43] and best fitting models of sequence evolution for each PCG selected with ProtTest [44]. The best model selected was mtMAM + I + G4, applied to each of 13 partitions (one per PCG). Lastly, the concatenated and partitioned PCG amino acid dataset was used to perform a ML analysis in the program IQ-TREE version 1.6.10 using the default options [45]. The robustness of the ML tree topology was assessed by 1,000 bootstrap (method: UFboot) iterations of the observed dataset.
Results and discussion
Mitochondrial genome assembly and description
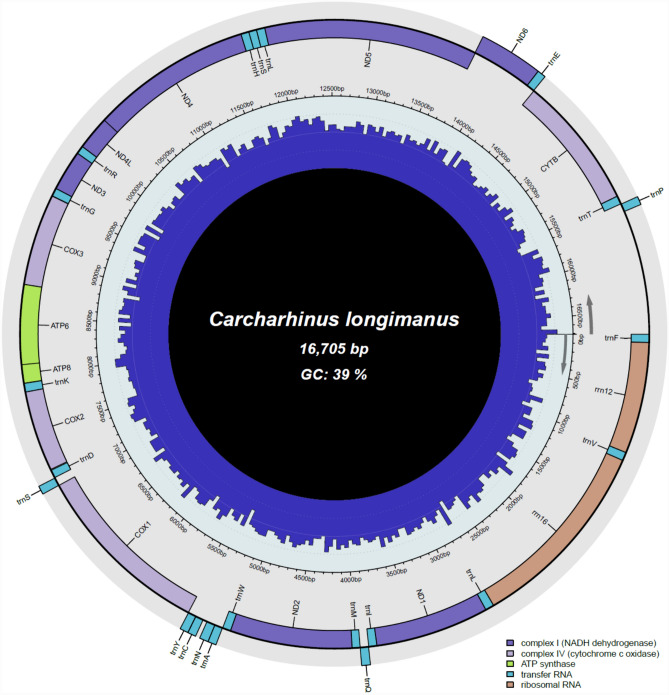
The pipeline GetOrganelle assembled a complete mitochondrial genome of Carcharhinus longimanus (OP057117) with an average coverage of 12.6 × and 51.6 × per k-mer and base, respectively. The mitochondrial genome of C. longimanus is 16,705 bp in length and contains 22 tRNA genes, 2 rRNA genes, 13 protein coding genes and a non-coding control region (CR) (Fig. 1; Table 1). Most genes reside on the heavy, positive strand of the genome, while nad6 and eight tRNAs (tRNA-Gln, tRNA-Asp, tRNA-Ala, tRNA-Cys, tRNA-Tyr, tRNA-Ser2, tRNA-Pro, and tRNA-Glu) are located on the light, negative strand. Other Carcharhinus spp. exhibit similar mitochondrial genome lengths ranging between 16,701 bp (C. falciformis—[46], C. macloti—[47] and 16,719 bp (C. acronotus—[47]). The mitochondrial gene order herein described for C. longimanus is identical to that documented before for other congeneric and cofamilial species [46–50].
Fig. 1.
Circular DNA mitochondrial genome map of Carcharhinus longimanus. The annotated map depicts 22 transfer RNA (tRNA) genes, 13 protein-coding genes (PCGs), 2 ribosomal RNA genes (rrnS: 12S ribosomal RNA and rrnL: 16S ribosomal RNA), and a putative control region
Table 1.
Mitochondrial genome of Carcharhinus longimanus
| Name | Type | Start | Stop | Strand | Length (bp) | Start | Stop | Anticodon | Continuity |
|---|---|---|---|---|---|---|---|---|---|
| trnF(gaa) | tRNA | 1 | 70 | + | 70 | GAA | 1 | ||
| rrnS | rRNA | 72 | 1029 | + | 958 | -3 | |||
| trnV(tac) | tRNA | 1027 | 1098 | + | 72 | TAC | 23 | ||
| rrnL | rRNA | 1122 | 2768 | + | 1647 | -1 | |||
| trnL2(taa) | tRNA | 2768 | 2842 | + | 75 | TAA | 0 | ||
| nad1 | PCG | 2843 | 3817 | + | 975 | TAA | ATG | 0 | |
| trnI(gat) | tRNA | 3818 | 3887 | + | 70 | GAT | 1 | ||
| trnQ(ttg) | tRNA | 3889 | 3960 | − | 72 | TTG | -1 | ||
| trnM(cat) | tRNA | 3960 | 4028 | + | 69 | CAT | 0 | ||
| nad2 | PCG | 4029 | 5075 | + | 1047 | ATG | TAG | -2 | |
| trnW(tca) | tRNA | 5074 | 5144 | + | 71 | TCA | 1 | ||
| trnA(tgc) | tRNA | 5146 | 5214 | − | 69 | TGC | 0 | ||
| trnN(gtt) | tRNA | 5215 | 5287 | − | 73 | GTT | 5 | ||
| OL | 5293 | 5322 | + | 30 | 0 | ||||
| trnC(gca) | tRNA | 5323 | 5390 | − | 68 | GCA | 1 | ||
| trnY(gta) | tRNA | 5392 | 5460 | − | 69 | GTA | 1 | ||
| cox1 | PCG | 5462 | 7018 | + | 1557 | GTG | TAA | 0 | |
| trnS2(tga) | tRNA | 7019 | 7089 | − | 71 | TGA | 3 | ||
| trnD(gtc) | tRNA | 7093 | 7162 | + | 70 | GTC | 7 | ||
| cox2 | PCG | 7170 | 7860 | + | 691 | ATG | T(AA) | 0 | |
| trnK(ttt) | tRNA | 7861 | 7934 | + | 74 | TTT | 1 | ||
| atp8 | PCG | 7936 | 8103 | + | 168 | ATG | TAA | -10 | |
| atp6 | PCG | 8094 | 8777 | + | 684 | ATG | TAA | -1 | |
| cox3 | PCG | 8777 | 9562 | + | 786 | ATG | TAA | 2 | |
| trnG(tcc) | tRNA | 9565 | 9634 | + | 70 | TCC | 0 | ||
| nad3 | PCG | 9635 | 9985 | + | 351 | ATG | TAG | -2 | |
| trnR(tcg) | tRNA | 9984 | 10,053 | + | 70 | TCG | 0 | ||
| nad4l | PCG | 10,054 | 10,350 | + | 297 | ATG | TAA | 7 | |
| nad4 | PCG | 10,344 | 11,724 | + | 1381 | ATG | T(AA) | 0 | |
| trnH(gtg) | tRNA | 11,725 | 11,793 | + | 69 | GTG | 0 | ||
| trnS1(gct) | tRNA | 11,794 | 11,860 | + | 67 | GCT | 0 | ||
| trnL1(tag) | tRNA | 11,861 | 11,932 | + | 72 | TAG | 0 | ||
| nad5 | PCG | 11,933 | 13,762 | + | 1830 | ATG | TAA | -5 | |
| nad6 | PCG | 13,758 | 14,279 | − | 522 | ATG | AGG | 0 | |
| trnE(ttc) | tRNA | 14,280 | 14,349 | − | 70 | TTC | 2 | ||
| cob | PCG | 14,352 | 15,497 | + | 1146 | ATG | TAG | -1 | |
| trnT(tgt) | tRNA | 15,497 | 15,568 | + | 72 | TGT | 2 | ||
| trnP(tgg) | tRNA | 15,571 | 15,639 | − | 69 | TGG | 33 | ||
| CR | 15,673 | 16,704 | + | 1032 | 1 |
Arrangement and annotation
The nucleotide composition of the studied mitochondrial genome is: A = 31.5%, T = 30.1%, G = 13.1%, C = 25.3%, with a high A + T content (61.5%) similar to that reported before for other congeneric and cofamilial species. In the genus Carcharhinus, A + T content has been reported to range between 59.9% in the Silky shark Carcharhinus falciformis [46] and 62.57% in the Bull shark Carcharhinus leucas [42]. In the order Carcharhiniformes, the lowest and highest reported A + T composition is 52.86% in the blotchy swellshark Cephaloscyllium umbratile [47] and 63.62% in the false catshark Pseudotriakis microdon [47], respectively. The high mutation rate from G to A in mitochondrial DNA may explain, in part, the A + T rich nature of this and other related mitochondrial genomes [51].
The PCGs of C. longimanus contain 3811 codons and range in length between 168 bp (atp8) and 1,830 bp (nad5) (Table 1). Start codons included ATG in 12 different PCGs and GTG in cox1 (Table 1). The stop codon TAA was used in nine different PCGs, TAG was used in cob, nad2 and nad3, and AGG was used in nad6. The incomplete termination codon T was used in cox2 and nad4. This occurrence of incomplete stop codons is common in mitochondrial PCGs of eumetazoans, including sharks [52].
In the mitochondrial PCGs of the species under study, there is bias in codon usage. Excluding start and stop codons, the most frequently used codons in the PCGs of the examined species were TTA (Leu), used 202 times (5.3%), followed by ATT (Ile), used 199 times (5.22%); and CTA (Leu), used 179 times (4.69%) (Online Resource 1). The least frequently used codons were CAG (Gln), used once (0.026%) followed by CGG (Arg), TCG (Ser) and ACG (Thr) each used twice at 0.052%. Relative synonymous codon usage (RSCU) analysis of PCGs in C. longimanus revealed that codons encoding Alanine, Serine, Leucine, Threonine, Glycine, Arginine and Proline are the most frequently used, whereas codons coding for Asparagine, Glutamine, Cysteine and Lysine were rare (Fig. 2). Codons starting with A or T are commonly used in comparison to other synonymous codons, for example, the codon for glutamine CAG was rare, which is consistent with previous observations of shark species in the order Carcharhiniformes, including the congeneric Carcharhinus acronotus [47].
Fig. 2.
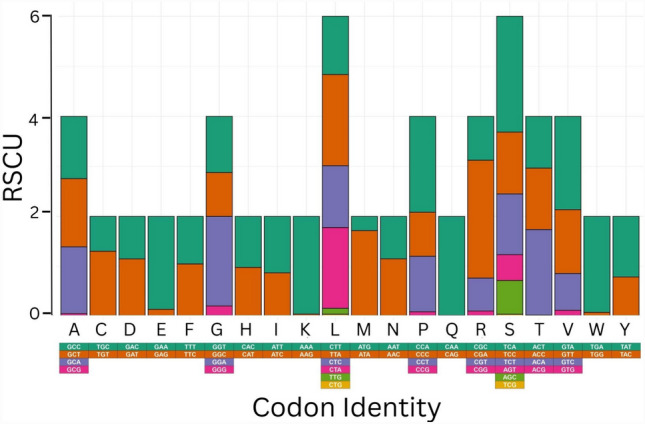
Codon usage analysis of PCGs in the mitochondrial genome of Carcharhinus longimanus. All 20 amino acids-alanine (A), cysteine (C), aspartic acid (D), glutamic acid (E), phenylalanine (F), glycine (G), histidine (H), isoleucine (I), lysine (K), leucine (L), methionine (M), asparagine (N), proline (P), glutamine (Q), arginine (R), serine (S), threonine (T), valine (V), tryptophan (W), tyrosine (Y) are listed by their one-letter abbreviations along the horizontal axis
In C. longimanus, all 13 mitochondrial PCGs exhibited Ka/Ks ratios < 1, indicating that these genes are exposed to ‘negative’ (= purifying) selection. The cox2 gene featured the highest Ka/Ks ratio (Ka/Ks = 0.508, P = 0.066) while nad1 exhibited the lowest Ka/Ks ratio (Ka/Ks = 0.007, P = 3.78E−45) (Table 2). Previous research has confirmed that mitochondrial PCGs exhibit higher mutation rates compared to nuclear genes [53]. Consistent with this notion, our results reveal a prevalence of PCGs undergoing negative selection, which is expected to eliminate deleterious mutations, rather than diversifying selection. This gradual accumulation of mutations over time suggests that negative or purifying selection is substantial in the evolution of mitogenomes [54]. Previous studies describing the mitochondrial genome of congeneric sharks have not explored selective pressures in PCGs. However, purifying selection affecting all 13 mitochondrial PCGs have been reported before in sharks belonging to the families Scyliorhinidae (i.e., Cephalloscyllium umbratile and Scyliorhinus canicula) and Proscylliidae (i.e., Proscyllium habereri—[43]), among others (e.g., in the Lemon shark Negaprion brevirostris—[22]).
Table 2.
Selective pressure analysis in the protein coding genes (PCGs) of Carcharhinus longimanus indicating purifying selection in all genes with Ka/Ks < 1
| Sequence | Ka | Ks | Ka/Ks | P-value |
|---|---|---|---|---|
| nad1 | 0.004086 | 0.548578 | 0.007448 | 3.78E−45 |
| nad2 | 0.00774 | 0.678907 | 0.011401 | 2.23E−52 |
| nad3 | 0.008117 | 0.4696 | 0.017285 | 6.55E−15 |
| nad4 | 0.0333873 | 0.232658 | 0.143504 | 4.27E−20 |
| nad5 | 0.0143924 | 0.686124 | 0.0209764 | 3.53E−77 |
| nad6 | 0.012766 | 0.476056 | 0.0268161 | 4.93E−18 |
| nad4l | 0.013756 | 0.593875 | 0.023162 | 5.38E−13 |
| cob | 0.013866 | 0.402372 | 0.03446 | 1.63E−36 |
| atp6 | 0.005916 | 0.422197 | 0.014012 | 1.26E−26 |
| atp8 | 0.016093 | 0.500994 | 0.032121 | 1.17E−06 |
| cox1 | 0.003239 | 0.335226 | 0.009663 | 6.16E−46 |
| cox2 | 0.003565 | 0.231419 | 0.015405 | 2.06E−16 |
| cox3 | 0.004864 | 0.361279 | 0.013463 | 2.81E−25 |
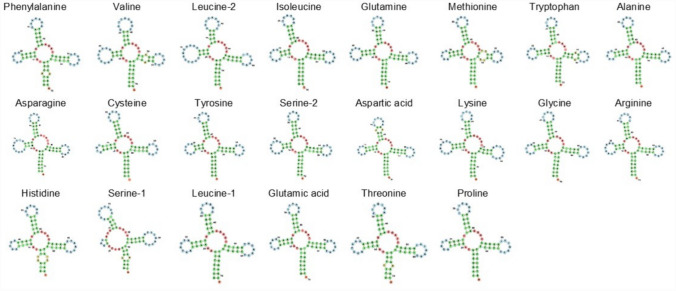
In the mitochondrial genome of C. longimanus, the length of the tRNA genes ranged between 67 bp (tRNA-Ser1) and 75 bp (tRNA-Leu2) and all of them but one exhibited a typical ‘cloverleaf’ secondary structure (Fig. 3). The software MITFI predicted that the tRNA-Ser1 gene was missing the dihydrouridine loop. Our observations coincide with that observed in most representatives of the genus Carcharhinus in which all tRNA genes exhibit a cloverleaf secondary structure except tRNA-Ser1 that is truncated (i.e., C. albimarginatus—[55], C. amblyrhynchoides—[48], C. perezi—[56], C. brachyurus—[49], C. limbatus—[50]). Interestingly, C. amblyrhynchoides [57] and C. melanopterus [58] exhibit a truncated tRNA-Ser2 (lacking the D-arm) instead of tRNA-Ser1. While the cloverleaf shape of tRNA molecules generally contributes to the overall stability of their tertiary structure, a deviation from the typical cloverleaf structure, particularly in the tRNA-Ser1 gene, is commonly observed in almost all mitochondrial genomes of eumetazoans [25, 59].
Fig. 3.
Secondary structure of tRNAs in the mitochondrial genome of Carcharhinus longimanus
In C. longimanus, the two ribosomal RNA (rRNA) genes are found in the positive strand. The 16S rRNA, 1,647 bp long, is located between tRNA-Va l and tRNA-Leu2, while 12S rRNA, 958 bp long, is located between tRNA-Val and tRNA-Phe. The nucleotide composition estimated for 16S rRNA is A = 36.06%, T = 26.59%, G = 16.69% and C = 20.64% and for 12S rRNA is A = 33.82%, T = 24%, G = 18.68% and C = 23.48%. The two rRNA genes are AT-rich in line to that reported for other congeneric species, including the Blacktip reef shark Carcharhinus melanopterus and the Bull shark Carcharhinus leucas [47].
The 1,065 bp long putative CR is located between the genes tRNA-Pro and tRNA-Phe. The nucleotide composition of the CR is A = 31.3%, T = 35.3%, C = 19.9%, and G = 13.5%, which is within range reported for the CR of other congeneric sharks [46, 47, 50]. In the genus Carcharhinus, the lowest and highest A + T content reported for the CR is 66.16% in the Blacktip reef shark C. leucas and 68.2% in the Hardnose shark C. macloti, respectively [47]. Stem-loop structures as well as microsatellite repeats were found within the CR. The web server microsatellite repeats-finder reported 10 microsatellites along this region, most of them AT-rich, repeated between 2 and a maximum of 5 times (Online Resource 2). Most microsatellites contain AA or TT dinucleotide repeats, with few CC and TA dinucleotides found towards the 3ʹ-end of the CR. The Tandem Repeats finder web server did not detect any repeats in the control region of C. longimanus. However, in the family Triakidae, instances of tandem repeats have been identified in the CR of species belonging to the genera Galeorhinus, Triakis and Mustelus (G. galeus, T. megalopterus M. palumbes, M. asterias and M. mosis) [41], but no tandem repeats have been reported in M. canis and M. norrisi [60]. Also, the RNAfold web server determined two possible secondary structures for the control region (Online Resource 3). Predicted Gibbs free energy (ΔG) values for the optimal and centroid RNA predicted secondary structures were ΔG = − 202.10 kcal/mol and ΔG = − 162.14 kcal/mol, respectively. According to this analysis, numerous stem-and-loop structures of different sizes were distributed along the entire CR. The MXFold2 web server predicted a more accurate analysis of the secondary structure of CR that also included multiple stem-and-loop structure (Online Resource 4). No previous study has analyzed in detail the CR of congeneric sharks. However, microsatellites are commonly observed in the CR of other sharks and the predicted secondary structure invariably exhibit stem-and-loops, as reported before in the closely related sharks Galeus melastomus, Negaprion brevirostris, Odontaspis ferox, and Prionace glauca [22, 52] and other distantly related species (i.e., the Grey bamboo shark Chiloscyllium griseum—[61]). The control region exhibits higher evolutionary rate compared to the other mitochondrial regions, making it an ideal tool for studying genetic diversity and population structure in representatives of the family Carcharhinidae.
Phylomitogenomics of the genus Carcharhinus
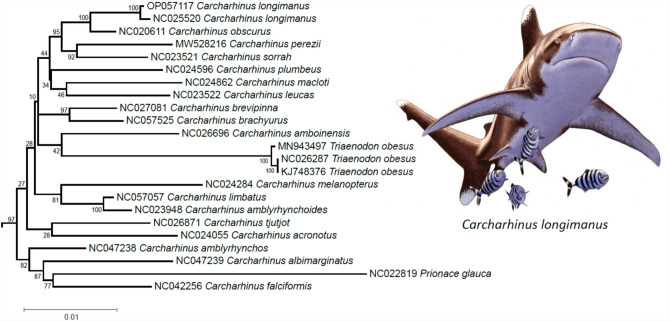
Our ML analysis (51 terminals, 3,799 amino acid characters, 737 informative sites) did not support the monophyly of the genus Carcharhinus (Fig. 4) due to the position of the blue shark Prionace glauca and the genus Trianodon, represented by 3 mitochondrial genomes belonging to the same species, T. obesus, in our study, which nested deep within a well-supported clade (bootstrap support value [bv] = 93) composed of all sharks belonging to the genus Carcharhinus used in the phylogenetic analysis. Specifically, T. obesus, formed a moderately supported (bv = 72) clade with C. amboinensis and C. melanopterus within a larger well supported clade (bv = 93) containing all other representatives of the genus Carcharhinus and the blue shark P. glauca. In turn, P. glauca formed a well-supported (bv = 93) clade with C. acronotus, C. albimarginatus, C. amblyrhynchos, C. falciformis, and C. tjutjot. Within the Carcharhinus + Trianodon + Prionace clade, the newly assembled mitochondrial genome of C. longimanus was sister (bv = 100) to a second mitochondrial genome of C. longimanus (NC025520). In turn, C. longimanus was sister to C. obscurus (bv = 100). Most of the internal relationships within the genus Carcharhinus were not resolved in our phylogenetic analysis based on translated mitochondrial PCGs. Nonetheless, fully or well supported sister relationships included Carcharhinus limbatus + Carcharhinus amblyrhynchoides (bv = 100), C brevipinna + C brachyurus (bv = 97), and C perezii + C sorrah (bv = 92). In line with Baeza [22] and Winn [41], our results suggest that the genus Carcharhinus, among others in the family Carcharhinidae, is in need of systematic re-arrangements. We argue in favor of additional studies assembling mitochondrial genomes in other representatives of this family to resolve internal relationships in the remarkable clade of sharks that is currently experiencing major environmental challenges.
Fig. 4.
Total evidence phylogenetic tree obtained from ML analysis based on a concatenated alignment of amino acids of the 13 protein-coding genes present in the mitochondrial genome of the Oceanic Whitetip shark Carcharhinus longimanus and other representatives of the genus Carcharhinus and family Carcharhinidae. The robustness of the ML tree topology was ascertained by 1000 bootstrap pseudoreplicates (numbers above or below the nodes) of the tree search. Depiction of C. longimanus by Kókay Szabolcs (used with permission)
Conclusion
This study assembled the complete mitochondrial genome of the oceanic whitetip shark, C. longimanus, which is considered by the IUCN (International Union for Conservation of Nature) Red List of Threatened Species as “vulnerable” throughout its range and “critically endangered” in the western north Atlantic. This genomic resource can serve as a baseline for biomonitoring and bioprospecting of this epipelagic shark using environmental DNA (eDNA) metabarcoding and/or metagenomic strategies. Also, the assembled mitochondrial genome plus others sequenced for other closely and distantly related species can be used as references to accurately detect the presence of imperilled species in the marketplace and flag mislabeling so to ensure compliance with trade regulations. By employing genomic tracking, the illegal trade of oceanic whitetip sharks and other imperilled species in the marketplace can be minimized. Furthermore, insights into the patterns of selective pressures and their effects on PCGs can enhance our understanding of mitogenome evolution and shed light on broader concepts of adaptation in shark species. Considering the significance of analyzing evolutionary patterns and addressing phylogenetic complexities, we emphasize the need for expanding genomic resources for this and other representatives of the family Carcharhinidae.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
JAB thanks Dr. Vincent P. Richards for bioinformatic support. This study was supported by Creative Inquiry, Clemson University. We thank Kókay Szabolcs for permission to use his remarkable illustration. Sequencing of the mitochondrial genome analyzed here was provided by Katherine Bemis (that was not able to participate in the preparation of this manuscript due to major time constraints) and a collaborative partnership between NOAA Fisheries, National Oceanic and Atmospheric Administration and the National Museum of Natural History, Smithsonian Institution to develop voucher-based reference libraries for mitochondrial genomes.
Abbreviations
- IUCN
International Union for Conservation of Nature
- NCBI
National Center for Biotechnology Information
- rRNA
Ribosomal RNA
- tRNA
Transfer RNA
- PCGs
Protein coding genes
- cox
Cytochrome c oxidase
- nad
Nicotinamide adenine dinucleotide
- cytb
Cytochrome b
- Ka
Number of non-synonymous substitutions per non-synonymous site
- Ks
Number of synonymous substitutions per synonymous site
- CR
Control region
- gDNA
Genomic DNA
- ML
Maximum likelihood
Author contributions
All authors contributed to all aspects of this manuscript.
Funding
Open access funding provided by the Carolinas Consortium. The author(s) reported there is no funding associated with the work featured in this article.
Data availability
The sequence data are part of a project to sequence mitochondrial genomes of marine fishes occurring in the Exclusive Economic Zone of the United States based on voucher specimens (BioProject: PRJNA720393) and data are deposited on GenBank (BioSample: SAMN31811566).
Declarations
Competing interests
The authors declare no conflict of interest.
Ethical approval
No approval from Ethical committees and Internal Review Boards were needed because specimens for sequencing were available at a museum (fixed specimens).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mcdiarmid M (1996) Shark attack. Parrogon Books Limited, Bristol, pp 70–80 [Google Scholar]
- 2.Bonfil R, Clarke S, Nakano H (2009) The biology and ecology of the oceanic whitetip shark, Carcharhinus Longimanus. In: Pitcher TJ, Camhi MD, Pikitch EK, Babcock EA (eds) Sharks of the open ocean. Wiley, Hoboken. 10.1002/9781444302516.ch11 [Google Scholar]
- 3.Ramsoomair KA (2016) The online guide to the animals of Trinidad and Tobago. University of the West Indies, Ecology [Google Scholar]
- 4.Passerotti MS, Andrews AH, Natanson LJ (2020) Inferring life history characteristics of the oceanic whitetip shark Carcharhinus longimanus from vertebral bomb radiocarbon. Front Mar Sci 7:581775. 10.3389/fmars.2020.581775 [Google Scholar]
- 5.Baum J, Medina E, Musick JA, Smale M (2015) Carcharhinus longimanus. The IUCN Red List of Threatened Species 2015: e.T39374A85699641. 10.2305/IUCN.UK.2015.RLTS.T39374A85699641.en
- 6.Camargo SM, Coelho R, Chapman D, Howey-Jordan L, Brooks EJ, Fernando D, Mendes NJ, Hazin FHV, Oliveira C, Santos MN, Foresti F, Mendonça FF (2016) Structure and genetic variability of the oceanic whitetip shark, Carcharhinus longimanus determined using mitochondrial DNA. PLoS ONE 11(5):e0155623. 10.1371/journal.pone.0155623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrzejaczek S, Gleiss AC, Jordan LKB, Pattiaratchi CB, Howey LA, Brooks JB (2018) Temperature and the vertical movements of oceanic whitetip sharks Carcharhinus longimanus. Sci Rep 8:8351. 10.1038/s41598-018-26485-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lessa R, Santana FM, Paglerani R (1999) Age, growth and stock structure of the oceanic whitetip shark, Carcharhinus longimanus, from the southwestern equatorial Atlantic. Fish Res 42:21–30. 10.1016/S0165-7836(99)00045-4 [Google Scholar]
- 9.D’Alberto BM, Chin A, Smart JJ, Baje L, White WT, Simpfendorfer CA (2017) Age, growth and maturity of oceanic whitetip shark (Carcharhinus longimanus) from Papua New Guinea. Mar Freshw Res 68:1118–1129 [Google Scholar]
- 10.Babcock EA, Camhi MD, Pikitch EK (2008) Sharks of the open ocean: biology fisheries and conservation. Blackwell Publishing Ltd, Hoboken [Google Scholar]
- 11.Kottillil S, Rao C, Bowen BW, Shanker K (2023) Phylogeography of sharks and rays: a global review based on life history traits and biogeographic partitions. PeerJ 11:e15396. 10.7717/peerj.15396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dulvy NK, Baum JK, Clarke S, Compagno LJV, Cortés E, Domingo A, Fordham S, Fowler S, Francis MP, Gibson C, Martínez J, Musick JA, Soldo A, Stevens JD, Valenti S (2008) You can swim but you can’t hide: the global status and conservation of oceanic pelagic sharks and rays. Aquat Conserv 18:459–482. 10.1002/aqc.975 [Google Scholar]
- 13.Dulvy NK, Fowler SL, Musick JA, Cavanagh RD, Kyne PM, Harrison LR, Carlson JK, Davidson LNK, Fordham SV, Francis MP, Pollock CM, Simpfendorfer CA, Burgess GH, Carpenter KE, Compagno LJ, Ebert DA, Gibson C, Heupel MR, Livingstone SR, Sanciangco JC, Stevens JD, Valenti S, White WT (2014) Extinction risk and conservation of the world’s sharks and rays. Elife 3:e00590. 10.7554/eLife.00590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canfield SJ, Galván-Magaña F, Bowen BW (2022) Little sharks in a big world: mitochondrial DNA reveals small-scale population structure in the California horn shark (Heterodontus francisci). J Hered 113(3):298–310. 10.1093/jhered/esac008 [DOI] [PubMed] [Google Scholar]
- 15.Howey-Jordan LA, Brooks EJ, Abercrombie DL, Jordan LKB, Brooks A, Williams S, Gospodarczyk E, Chapman DD (2013) Complex movements, philopatry and expanded depth range of a severely threatened pelagic shark, the oceanic whitetip (Carcharhinus longimanus) in the Western North Atlantic. PLoS ONE 8(2):e56588. 10.1371/journal.pone.0056588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young CN, Carlson JK (2020) The biology and conservation status of the oceanic whitetip shark (Carcharhinus longimanus) and future directions for recovery. Rev Fish Biol Fisheries 30:293–312. 10.1007/s11160-020-09601-3 [Google Scholar]
- 17.Scott M, Royer M, Hutchinson M (2023) Time of death: behavioral responses of an oceanic whitetip shark, Carcharhinus longimanus, to capture by a longline fishing vessel. Anim Biotelemetry 11:34. 10.1186/s40317-023-00346-x [Google Scholar]
- 18.Shea BD, Gallagher AJ, Bomgardner LK, Ferretti F (2023) Quantifying longline bycatch mortality for pelagic sharks in western Pacific shark sanctuaries. Sci Adv. 10.1126/sciadv.adg3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rigby CL, Barreto R, Carlson J, Fernando D, Fordham S, Francis MP, Herman K, Jabado RW, Liu KM, Marshall A, Pacoureau N, Romanov E, Sherley RB, Winker H (2019) Carcharhinus longimanus. The IUCN Red List of Threatened Species 2019: e.T39374A2911619. 10.2305/IUCN.UK.2019-3.RLTS.T39374A2911619.en
- 20.Sreelekshmi S, Sukumaran S, Kishor TG, Wilson S, Gopalakrishnan A (2020) Population genetic structure of the oceanic whitetip shark, Carcharhinus longimanus, along the Indian coast. Mar Biodivers. 10.1007/s12526-020-01104-5 [Google Scholar]
- 21.Ruck CL, Shivji MS, Jabado RW, Bernard AM (2024) Cross ocean-basin population genetic dynamics in a pelagic top predator of high conservation concern, the oceanic whitetip shark, Carcharhinus longimanus. Conserv Genetics 25:1–19. 10.1007/s10592-023-01596-1 [Google Scholar]
- 22.Baeza JA, Stephens NC, Baker A, Lyons A, Franks B, Pirro S, Feldheim KA (2024) Insights into the nuclear and mitochondrial genome of the Lemon shark Negaprion brevirostris using low-coverage sequencing: genome size, repetitive elements, mitochondrial genome, and phylogenetic placement. Gene 894:147939. 10.1016/j.gene.2023.147939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li W, Dai X, Xu Q, Wu F, Gao C, Zhang Y (2016) The complete mitochondrial genome sequence of oceanic whitetip shark, Carcharhinus longimanus (Carcharhiniformes: Carcharhinidae). Mitochondrial DNA Part A 27:1775–1776 [DOI] [PubMed] [Google Scholar]
- 24.Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ (2020) GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol 21:241. 10.1186/s13059-020-02154-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF (2013) MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol 69:313–319. 10.1016/j.ympev.2012.08.023 [DOI] [PubMed] [Google Scholar]
- 26.Kumar S, Stecher G, Li M, Knyaz C, Tamura K, Battistuzzi FU (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov D, Liechti R, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H (2012) ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res 40:W597–W603. 10.1093/nar/gks400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng S, Poczai P, Hyvönen J, Tang J, Amiryousefi A (2020) Chloroplot: an online program for the versatile plotting of organelle genomes. Front Genet 11:576124. 10.3389/fgene.2020.576124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jühling F, Pütz J, Bernt M, Donath A, Middendorf M, Florentz C, Stadler PF (2012) Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res 40(7):2833–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerpedjiev P, Hammer S, Hofacker IL (2015) Forna (force-directed RNA): simple and effective online RNA secondary structure diagrams. Bioinformatics 31:3377–3379. 10.1093/bioinformatics/btv372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stothard P (2018) The sequence manipulation suite: javascript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28:1102–1104. 10.2144/00286ir01 [DOI] [PubMed] [Google Scholar]
- 32.Cucini C, Leo C, Iannotti N, Boschi S, Brunetti C, Pons J, Fanciulli PP, Frati F, Carapelli A, Nardi F (2021) EZmito: a simple and fast tool for multiple mitogenome analyses. Mitochondrial DNA Part B 6:1101–1109. 10.1080/23802359.2021.1899865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang D, Zhang Y, Zhang Z, Zhu J, Yu J (2010) KaKs_Calculator 2.0: a toolkit incorporating gamma series methods and sliding window strategies. Genomics Proteomics Bioinf 8:77–80. 10.1016/S1672-0229(10)60008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bikandi J, San Millán R, Rementeria A, Garaiza J (2004) In silico analysis of complete bacterial genomes: PCR, AFLP–PCR and endonuclease restriction. Bioinformatics 20:798–799. 10.1093/bioinformatics/btg491 [DOI] [PubMed] [Google Scholar]
- 35.Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acid Res 27(2):573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL (2008) The Vienna RNA website. Nucleic Acids Res 36(2):70–74. 10.1093/nar/gkn188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato K, Akiyama M, Sakakibara Y (2021) RNA secondary structure prediction using deep learning with thermodynamic integration. Nat Commun 12:94. 10.1038/s41467-021-21194-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Díaz-Jaimes P, Bayona-Vásquez NJ, Adams DH, Uribe-Alcocer M (2016) Complete mitochondrial DNA genome of bonnethead shark, Sphyrna tiburo, and phylogenetic relationships among main superorders of modern elasmobranchs. Meta Gene 7:48–55. 10.1016/j.mgene.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Da Cunha DB, Da Silva Rodrigues-Filho LF, De Luna Sales JB (2017) A review of the mitogenomic phylogeny of the Chondrichthyes. InTech eBooks. 10.5772/intechopen.70028 [Google Scholar]
- 40.Amaral CRL, Pereira F, Silva DA, Amorim A, De Carvalho EF (2017) The mitogenomic phylogeny of the Elasmobranchii (Chondrichthyes). Mitochondrial DNA Part A 29(6):867–878. 10.1080/24701394.2017.1376052 [DOI] [PubMed] [Google Scholar]
- 41.Winn JC, Maduna SN, Merwe AEBD (2024) A comprehensive phylogenomic study unveils evolutionary patterns and challenges in the mitochondrial genomes of Carcharhiniformes: a focus on Triakidae. Genomics 116(1):110771. 10.1016/j.ygeno.2023.110771 [DOI] [PubMed] [Google Scholar]
- 42.Sievers F, Higgins DG (2014) Clustal Omega, accurate alignment of very large numbers of sequences. In: Russell DJ (ed) Multiple sequence alignment methods. Humana Press, Totowa, pp 105–116 [DOI] [PubMed] [Google Scholar]
- 43.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darriba D, Taboada GL, Doallo R, Posada D (2011) ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics (Oxford, England) 27(8):1164–1165. 10.1093/bioinformatics/btr088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johri S, Chapple TK, Dinsdale EA, Schallert R, Block BA (2020) Mitochondrial genome of the silky shark Carcharhinus falciformis from the British Indian Ocean Territory Marine Protected Area. Mitochondrial DNA Part B 5(3):2416–2417. 10.1080/23802359.2020.1775147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu KC, Liang YY, Wu N, Guo HY, Zhang N, Jiang SG, Zhang DC (2017) Sequencing and characterization of the complete mitochondrial genome of Japanese Swellshark (Cephalloscyllium umbratile). Sci Rep 7:15299. 10.1038/s41598-017-15702-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patel E, Bernard AM, Mehlrose M, Harned S, Finnegan KA, Fitzpatrick CK, Lea JS, Shivji MS (2020) The complete mitochondrial genome of a gray reef shark, Carcharhinus amblyrhynchos (Carcharhiniformes: Carcharhinidae), from the Western Indian Ocean. Mitochondrial DNA Part B 5(3):3498–3499. 10.1080/23802359.2020.1827064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim SW, Park SY, Kwon H, Giri SS, Kim SG, Kang JW, Kwon J, Lee SB, Jung WJ, Lee JM, Park SC, Kim JH (2021) Complete mitochondrial genome and phylogenetic analysis of the copper shark Carcharhinus brachyurus (Günther, 1870). Mitochondrial DNA Part B 6(6):1659–1661. 10.1080/23802359.2021.1920863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang X, Zhou Z, Lai T, He B, Zhang D (2022) Characterization of the complete mitochondrial genome of blacktip shark Carcharhinus limbatus (Carcharhiniformes: Carcharhinidae). Mitochondrial DNA Part B 7(2):385–386. 10.1080/23802359.2021.1914214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haag-Liautard C, Coffey N, Houle D, Lynch M, Charlesworth B, Keightley P (2008) Direct the estimation of the mitochondrial DNA mutation rate in Drosophila melanogaster. PLoS Biol 6(8):e204. 10.1371/journal.pbio.0060204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kousteni V, Mazzoleni S, Vasileiadou K, Rovatsos M (2021) Complete mitochondrial DNA genome of nine species of sharks and rays and their phylogenetic placement among modern elasmobranchs. Genes 12(3):324. 10.3390/genes12030324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ballard JWO, Whitlock MC (2004) The incomplete natural history of mitochondria. Mol Ecol 13:729–744. 10.1046/j.1365-294x.2003.02063.x [DOI] [PubMed] [Google Scholar]
- 54.Monsanto DM, Main DC, Janion-Scheepers C, Emami-Khoyi A, Deharveng L, Bedos A, Poapov M, Parbhu SP, Roux JJL, Teske PR (2022) Mitogenome selection in the evolution of key ecological strategies in the ancient hexapod class Collembola. Sci Rep 12:14810. 10.1038/s41598-022-18407-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johri S, Dunn N, Chapple TK, Curnick D, Savolainen V, Dinsdale EA, Block BA (2020) Mitochondrial genome of the silvertip shark Carcharhinus albimarginatus, from the British Indian Ocean Territory. Mitochondrial DNA Part B 5(3):2085–2086. 10.1080/23802359.2020.1765210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gallagher AJ, Shipley ON, Reese B, Singh V (2021) Complete mitochondrial genome of the Caribbean reef shark, Carcharhinus perezi (Carcharhinformes: Carcharhinidae). Mitochondrial DNA Part B 6(9):2662–2664. 10.1080/23802359.2021.1964394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feutry P, Pillans RD, Kyne PM, Chen X (2016) Complete mitogenome of the Graceful Shark Carcharhinus amblyrhynchoides (Carcharhiniformes: Carcharhinidae). Mitochondrial DNA Part A 27(1):314–315. 10.3109/19401736.2014.892094 [DOI] [PubMed] [Google Scholar]
- 58.Chen X, Shen X, Arunrugstichai S, Ai W, Xiang D (2016) Complete mitochondrial genome of the blacktip reef shark Carcharhinus melanopterus (Carcharhiniformes: Carcharhinidae). Mitochondrial DNA Part A 27(2):873–874. 10.3109/19401736.2014.919483 [DOI] [PubMed] [Google Scholar]
- 59.Watanabe Y, Suematsu T, Ohtsuki T (2014) Losing the stem-loop structure from metazoan mitochondrial tRNAs and co-evolution of interacting factors. Front Genet 5:109. 10.3389/fgene.2014.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kiser H, Skufca K, Bemis KE, Baeza JA (2024) Comparative analysis of the mitochondrial genomes of Smoothhound sharks provide insight into the phylogenetic relationships within the family Triakidae. Gene Rep. 10.1016/j.genrep.2024.101957 [Google Scholar]
- 61.Chen X, Ai W, Ye L, Wang X, Lin C, Yang S (2013) The complete mitochondrial genome of the grey bamboo shark (Chiloscyllium griseum) (Orectolobiformes: Hemiscylliidae): genomic characterization and phylogenetic application. Acta Oceanol Sin 32:59–65. 10.1007/s13131-013-0298-0 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data are part of a project to sequence mitochondrial genomes of marine fishes occurring in the Exclusive Economic Zone of the United States based on voucher specimens (BioProject: PRJNA720393) and data are deposited on GenBank (BioSample: SAMN31811566).