Abstract
BACKGROUND
Few studies have investigated the association between gestational age, birth weight, and esophageal cancer risk; however, causality remains debated. We aimed to establish causal links between genetic gestational age and birth weight traits and gastroesophageal reflux disease (GERD), Barrett’s esophagus (BE), and esophageal adenocarcinoma (EA). Additionally, we explored if known risk factors mediate these links.
AIM
To analyze of the relationship between gestational age, birth weight and GERD, BE, and EA.
METHODS
Genetic data on gestational age and birth weight (n = 84689 and 143677) from the Early Growth Genetics Consortium and outcomes for GERD (n = 467253), BE (n = 56429), and EA (n = 21271) from genome-wide association study served as instrumental variables. Mendelian randomization (MR) and mediation analyses were conducted using MR-Egger, weighted median, and inverse variance weighted methods. Robustness was ensured through heterogeneity, pleiotropy tests, and sensitivity analyses.
RESULTS
Birth weight was negatively correlated with GERD and BE risk [odds ratio (OR) = 0.78; 95% confidence interval (CI): 0.69-0.8] and (OR = 0.75; 95%CI: 0.60-0.9), respectively, with no significant association with EA. No causal link was found between gestational age and outcomes. Birth weight was positively correlated with five risk factors: Educational attainment (OR = 1.15; 95%CI: 1.01-1.31), body mass index (OR = 1.06; 95%CI: 1.02-1.1), height (OR = 1.12; 95%CI: 1.06-1.19), weight (OR = 1.13; 95%CI: 1.10-1.1), and alcoholic drinks per week (OR = 1.03; 95%CI: 1.00-1.06). Mediation analysis showed educational attainment and height mediated the birth weight-BE link by 13.99% and 5.46%.
CONCLUSION
Our study supports the protective role of genetically predicted birth weight against GERD, BE, and EA, independent of gestational age and partially mediated by educational attainment and height.
Keywords: Birth weight, Barrett’s esophagus, Esophageal adenocarcinoma, Gestational age, Gastroesophageal reflux disease, Preterm birth
Core Tip: Our study demonstrates that low birth weight, rather than prematurity, is associated with increased risk of gastroesophageal reflux disease (GERD) and Barrett’s esophagus (BE) in adulthood, with no association found for esophageal adenocarcinoma. Furthermore, this relationship is influenced by the mediating effects of educational attainment and height. Attention to educational attainment and height during the growth process of low-birth-weight individuals is necessary to reduce the incidence of GERD and BE.
INTRODUCTION
Esophageal cancer, ranking as the eighth most prevalent cancer globally, exhibits a 5-year survival rate of less than 20%. In 2020, over 604000 new cases were diagnosed worldwide[1,2]. Esophageal adenocarcinoma (EA), as one subtype of esophageal cancer, has witnessed a rapid increase in incidence in Western countries in recent years[3,4]. Barrett’s esophagus (BE), characterized by the metaplasia of esophageal squamous epithelium, serves as a precancerous lesion and a high-risk factor for EA. Approximately 3%-5% of BE cases eventually progress to EA[5]. The risk of BE development is associated with recurrent episodes of gastroesophageal reflux disease (GERD), affecting approximately 20% of adults in developed countries[6]. Furthermore, previous research indicates that educational level, socioeconomic status, mental health, obesity, and smoking contribute to varying degrees of EA risk[7-11].
In contemporary research, there is considerable attention on the association between perinatal factors and adult-onset diseases[12]. GERD, a prevalent condition in newborns, exhibits notably elevated risks in preterm infants and those with small-for-gestational-age (SGA)[13]. The debate continues regarding the correlation of preterm birth and SGA with adult-onset GERD, BE, or EA. In a large-scale follow-up study of preterm and SGA individuals, only 8 participants developed esophageal cancer during adulthood, while the standardized incidence of EA increased by more than 7-fold[14]. Conversely, an alternative perspective posits that preterm birth is the primary factor contributing to the elevated risk of EA in adulthood, with SGA having no impact on this risk[15]. In investigations into the risk of developing BE in adulthood, both preterm and SGA individuals demonstrate a significant association with adult BE risk[16]. Notably, in the Swedish BE population, it was observed that SGA individuals, rather than preterm ones, experienced a threefold increase in the risk of developing BE in adulthood[17].
Due to the significant time span from infancy to adulthood, participants’ lifestyle habits (smoking, alcohol consumption, obesity, etc.) may impact the ultimate development of esophageal diseases through alternative causal pathways. As a result, traditional cohort studies and retrospective research face substantial limitations in this process, potentially leading to misleading interpretations. The principle of Mendelian randomization (MR), involving the random assortment of genes from parents to offspring during gamete formation and fertilization, can significantly mitigate measurement errors and confounding pathways. MR provides an effective means of evaluating the causal nature of modifiable behaviors or environmental exposures[18,19].
We employed genetic information from a comprehensive alliance of European populations to elucidate the causal associations between fetal prenatal characteristics, birth weight, and the risk of GERD, BE, and EA in adulthood[20-29]. Our methodology incorporated a two-step MR approach, encompassing univariable MR (UVMR) analysis and multivariable MR (MVMR) analysis, mediation analyses, and sensitivity assessments[30]. The primary objective of this study extends beyond elucidating direct causal relationships to actively seeking intermediary factors influencing the development of esophageal diseases. This research endeavors to contribute novel insights for future investigations and clinical practices related to GERD, BE, and EA.
MATERIALS AND METHODS
Study design
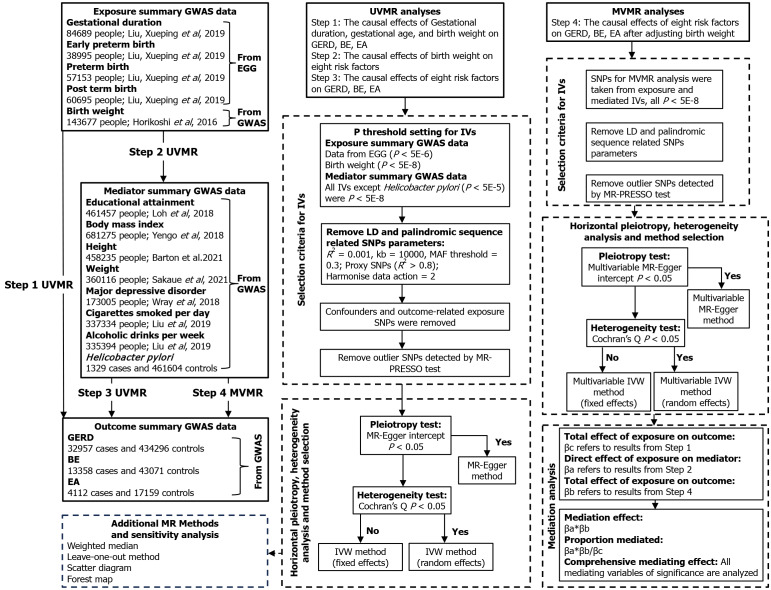
Multiple publicly available datasets from genome-wide association study (GWAS) were applied in this study[20-29]. All studies included in the analysis were ethically approved or patient consent, as described in Table 1. The univariate, multivariate, and two-step MR mediating analyses were performed to investigate whether genetically predicted gestational age characteristics, fetal birth weight, and adult susceptibility to GERD, BE, and EA were causally related. An attempt was made to find the mediating risk factors in the above associations and to calculate the corresponding mediating effect size. The route of this study is shown in Figure 1.
Table 1.
Genome-wide association study data sources and information included in the current study
|
Phenotype
|
Unit
|
Sample size
|
Ancestry
|
GWAS ID/data source
|
PMID
|
| Educational attainment (years of education) | SD increase in years of education | 461457 | European | ebi-a-GCST90029013 | 29892013 |
| Body mass index | SD increase in body mass index | 681275 | European | ieu-b-40 | 30124842 |
| Height | SD increase in cm of height | 458235 | European | ebi-a-GCST90025949 | 34226706 |
| Weight | SD increase in kg of birth adult weight | 360116 | European | ebi-a-GCST90018949 | 34594039 |
| Major depressive disorder | Binary categorical variable | 173005 | European | ieu-a-1188 | 29700475 |
| Cigarettes smoked per day | SD increase in cigarettes smoked per day | 337334 | European | ieu-b-25 | 30643251 |
| Alcoholic drinks per week | SD increase in alcoholic drinks per week | 335394 | European | ieu-b-73 | 30643251 |
| Birth weight | SD increase in kg of birth weight | 143677 | European | ieu-a-1083 | 27680694 |
| Gestational duration | SD increase in days of gestational duration | 84689 | European | EGG Consortium | 31477735 |
| Early preterm birth | Binary categorical variable | 38995 | European | EGG Consortium | 31477735 |
| Preterm birth | Binary categorical variable | 57153 | European | EGG Consortium | 31477735 |
| Post term birth | Binary categorical variable | 60695 | European | EGG Consortium | 31477735 |
| Helicobacter pylori | Binary categorical variable | 462933 | European | ukb-b-531 | / |
| Esophageal adenocarcinoma | Binary categorical variable | 21271 | European | ebi-a-GCST003739 | 27527254 |
| Barrett’s esophagus | Binary categorical variable | 56429 | European | ebi-a-GCST90000515 | 25826379 |
| Gastroesophageal reflux disease | Binary categorical variable | 467253 | European | ebi-a-GCST90018848 | 34594039 |
EGG: Early Growth Genetics; GWAS: Genome-wide association study.
Figure 1.
Flowchart of Mendelian randomization analyses in this study. Step 1: The causal effects of gestational duration, gestational age, and birth weight on gastroesophageal reflux disease (GERD), Barrett’s esophagus (BE), esophageal adenocarcinoma (EA); Step 2: The causal effects of birth weight on eight risk factors; Step 3: The causal effects of eight risk factors on GERD, BE, EA; Step 4: The causal effects of eight risk factors on GERD, BE, EA after adjusting birth weight. BE: Barrett’s esophagus; EA: Esophageal adenocarcinoma; EGG: Early Growth Genetics; GWAS: Genome-wide association study; IVs: Instrumental variables; IVW: Inverse variance weighted; MR-PRESSO: MR-Pleiotropy RESidual sum and outlier; UVMR: Univariable Mendelian randomization; SNPs: Single nucleotide polymorphisms; GERD: Gastroesophageal reflux disease; MVMR: Multivariable Mendelian randomization; LD: Linkage disequilibrium.
GWAS data for exposure
Genetic variation information pertaining to 84689 individuals of European descent, specifically related to the timing of delivery, was sourced from the collaborative efforts of the Early Growth Genetics (EGG) Consortium and the Integrative Psychiatric Research Consortium[25]. This dataset encompasses four fetal traits associated with the fetal genotype. The logistic regression analysis was applied to examine dichotomous outcomes, namely early preterm birth (< 34 wk), preterm birth (< 37 wk), postterm birth (≥ 42 wk). Additionally, gestational duration, subjected to regression on infant sex, had its resulting residuals quantile transformed to a standard normal distribution before undergoing linear regression to assess their association with fetal single nucleotide polymorphisms (SNPs) genotypes. Following gender-specific transformation and adjustments for gestational week and study-specific ancestry covariates, the genetic information pertaining to birth weight was included in the study only for a European population of 143677 individuals from the EGG Consortium[27].
GWAS data for mediator
In previous observational studies, risk factors associated with GERD, BE, and EA have been considered as potential mediators. Utilizing the most recent and largest-scale European GWAS meta-analysis data, we incorporated eight variables, namely, educational attainment (years of education), body mass index (BMI), height, weight, major depressive disorder (MDD), cigarettes smoked per day, alcoholic drinks per week, and Helicobacter pylori (H. pylori) infection. Genetic information for educational attainment (years of education) was sourced from Loh et al[24], employing a mixed-model association approach with a cohort of 461457 European individuals. The genetic characteristics of BMI were derived from the Genetic Investigation of ANthropometric Traits Consortium, encompassing a large-scale meta-analysis of 681275 individuals of European descent[20]. Height’s genetic data originated from an exome array association study based on 458235 Europeans from the United Kingdom Biobank[29]. Weight’s genetic summary data involved 360116 individuals of European descent[23]. Genetic information for MDD was obtained from the Psychiatric Genomics Consortium through a comprehensive genome-wide association meta-analysis with 59851 cases and 113154 controls[21]. Daily smoking quantity and weekly alcohol consumption data were sourced from the GWAS and Sequencing Consortium of Alcohol and Nicotine use, comprising 337334 and 335394 European individuals, respectively[26]. The genetic characteristics of H. pylori infection were derived from GWAS involving 1329 cases and 461604 controls of European ancestry[31,32].
GWAS data for outcome
The summary data for GERD (32957 cases and 434296 controls) was extracted from a large-scale human genetic phenotypic study conducted by Sakaue et al[23]. The genetic characteristics of BE (13358 cases and 43071 controls) were amalgamated from a comprehensive data synthesis derived from a large prospective etiological study of complex diseases in middle and older-aged individuals by Sudlow et al[22]. As for EA (4112 cases and 17159 controls), the genetic features originated from a large-scale meta-analysis of the entire genome-wide association data. All participants involved in the GERD, BE, and EA studies were of European descent[28].
Statistical analysis
Selection of genetic instrumental variables: Initially, due to stringent criteria (P < 5E-8), the genomic significant SNPs acquired for four fetal gestational features (preterm birth, preterm birth, postterm birth, gestational duration) did not yield sufficient power for subsequent MR analyses. Consequently, a more lenient screening threshold (P < 5E-6) was incorporated, which has been demonstrated as feasible in previous studies[33]. Similarly, a P < 5E-5 threshold was employed in constructing instrumental variables (IVs) for H. pylori infection.
Subsequently, SNPs failing to meet the independence criterion were removed through linkage disequilibrium (LD) to obtain independently associated SNPs (r2 < 0.001, kb = 10000, MAF < 0.3). Proxy SNPs (r2 > 0.8) were utilized to address the inability to extract SNPs associated with exposure or mediating IVs in the outcomes of esophageal diseases, achieved through the LD link tool[34]. In this process, SNPs with ambiguous palindromic sequences were excluded.
Based on previous evidence from observational studies, educational attainment, BMI, smoking, alcohol consumption, depression, or anxiety are considered confounding variables that may violate the assumption of causality[8-10,35]. To mitigate potential horizontal pleiotropy and violations of the correlation assumption in the study of exposure on outcomes, the PhenoScanner network tool was employed to exclude confounding SNPs that might represent alternative pathways, aligning with the exclusivity assumption in the core principles of MR[36]. The MR-PRESSO test was utilized to detect and remove outliers[37], resulting in the final set of IVs for MR analysis. To ensure the adherence to the core assumptions of MR, specifically the exclusion restriction assumption, identified IVs underwent scrutiny with criteria including F-statistic values > 10 and statistical power > 0.8, significantly minimizing potential bias introduced by weak IVs[38-40].
UVMR analysis: We conducted multiple rounds of UVMR analyses. Initially, causal estimates were performed for gestational duration-related genetic characteristics, birth weight, and their associations with GERD, BE, and EA. Subsequently, significant fetal traits identified in the initial analysis were included in subsequent UVMR analyses involving eight risk factors. The causal effects between these eight risk factors and esophageal diseases were also estimated using UVMR. Each UVMR analysis underwent tests for heterogeneity and pleiotropy. If the MR-Egger intercept test indicated horizontal pleiotropy (P < 0.05)[41], the MR-Egger regression method was employed to mitigate bias introduced by pleiotropy[42]; alternatively, in the absence of such indication, the initial consideration was given to the inverse-variance weighted (IVW) method[43]. If Cochran’s Q statistic suggested the presence of heterogeneity (P < 0.05)[30], the IVW fixed-effects model, rather than the IVW random-effects model, was utilized to minimize bias.
MVMR analysis: To accurately assess the effects of eight risk factors on three esophageal diseases, MVMR analysis method was employed[19]. Adjustments were made for significant neonatal characteristics identified in UVMR analysis. All relevant SNPs were included under a threshold of P < 5E-8, and a reevaluation of LD, alignment of effect alleles, removal of palindromic sequences, and multivariate tests for heterogeneity and pleiotropy were conducted.
Mediation analysis: As depicted in the intermediate analysis steps illustrated in Figure 1, UVMR initially computed the total effect of exposure on the outcome (βc), the direct effect of exposure on the mediator (βa), and the causal effect of the mediator on the outcome. The subsequent MVMR revealed the results of analyzing the direct effect of exposure on the mediator (βb) after adjusting for the exposure[19]. The mediation effect was considered established when both the total effect and the mediator effect (βa*βb) acted in the same direction and remained statistically significant[44]. Subsequently, we calculated the proportion of mediation by dividing the mediator effect by the total effect. The delta method was employed to estimate confidence intervals for independent samples[44,45]. Through the integration of mediation analysis with MVMR, concurrently adjusting for multiple intermediary risk factors, we derived an estimation of the comprehensive mediation effect.
Sensitivity analysis: Sensitivity analysis was employed to assess the robustness of MR results. In addition to IVW method (both fixed-effects and random-effects models) and the MR-Egger method, the weighted median method was also utilized as a reference[41]. Ensuring consistency in the direction of effects among different MR methods, with statistical significance met, enhances the credibility of the results. Supplementary scatter plots, forest plots, and Leave-One-Out (LOO) analysis were incorporated to ensure the validity of causal inferences in this study[46]. In the LOO analysis results, SNPs that were statistically significant and led to a reversal in the direction of effects were excluded. Statistical analyses were carried out using R version 4.2.1, and the R packages TwoSampleMR (version 0.5.7), MendelianRandomization (version 0.3.0), and forestploter (version 1.1.1) were employed for these analyses.
RESULTS
Causal links between four gestational traits and birth weight on GERD, BE, and EA
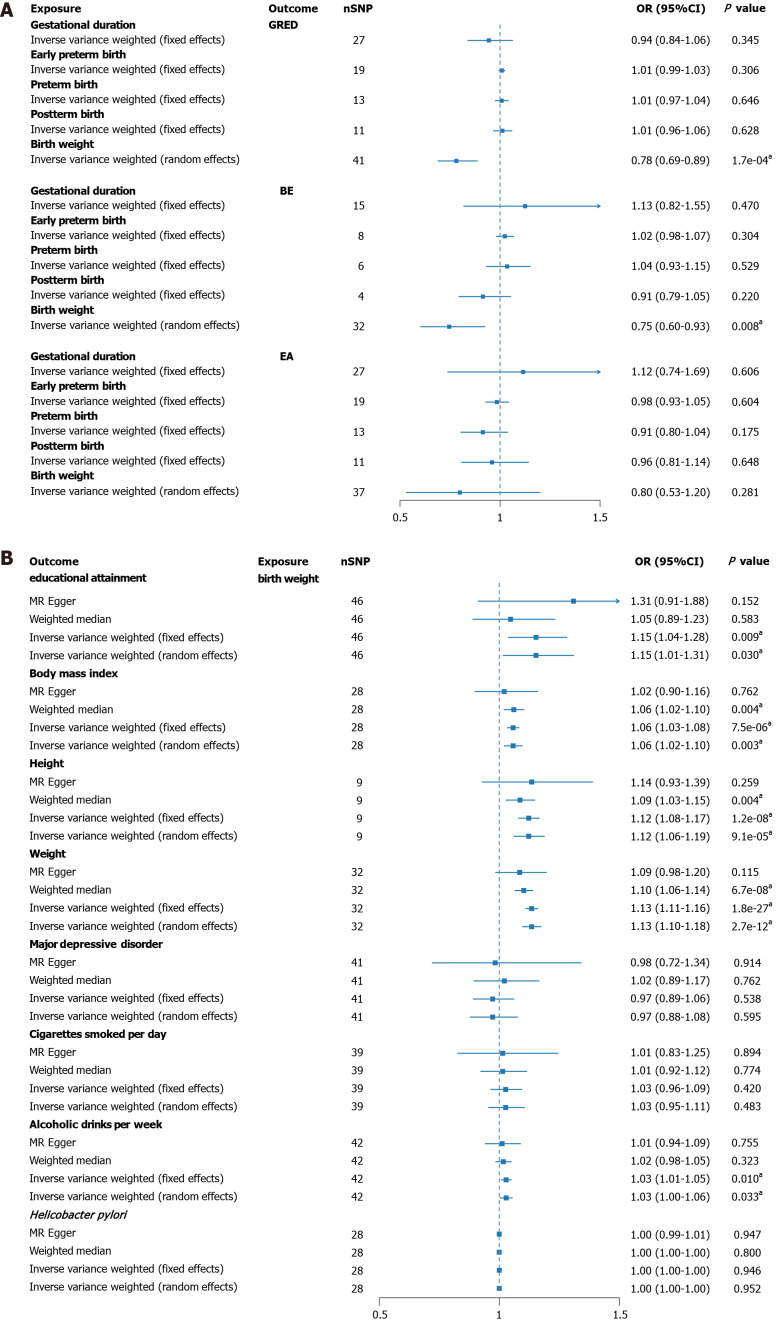
After rigorously excluding outlier SNPs, the final IVs associated with four gestational phenotypes (gestational duration, early preterm birth, preterm birth, postterm birth) was 2.38%, with F-statistics ranging from 26.99 to 199.63. For the IVs related to birth weight, the explained variance was 2.23%, with F-statistics between 607.81 and 740.38 (Supplementary Tables 1 and 2). Figure 2A visually presents the results of UVMR analysis. An increase in birth weight demonstrated a significant effect in reducing the risk of GERD and BE, while showing no association with EA risk. GERD (OR = 0.78, 95%CI: 0.69-0.8) and BE (OR = 0.75, 95%CI: 0.60-0.9). However, no significant associations were detected for gestational duration, early preterm birth, preterm birth, or postterm birth in the outcomes of the three esophageal disorders. Additional results from other MR methods are reported in Supplementary Table 3. No horizontal pleiotropy was observed in all analyses (PMR-Egger Intercept > 0.05), and random-effects models were applied to correct for heterogeneity. Sensitivity analysis indicated robust results, with no significant outliers identified in the LOO analysis (Supplementary Figures 1-15).
Figure 2.
Univariable Mendelian randomization analysis. A: Univariable Mendelian randomization analysis of gestational age-related genetic characteristics, birth weight and causality of esophageal disease; B: Univariable Mendelian randomization analysis of causal effects of genetic birth weight on risk factors. aP < 0.05. BE: Barrett’s esophagus; CI: Confidence interval; EA: Esophageal adenocarcinoma; GERD: Gastroesophageal reflux disease; nSNPs: Number of single nucleotide polymorphisms; OR: Odds ratio.
Causal links between birth weight on multiple risk factors
Due to statistical significance observed only between birth weight and the outcome in UVMR, subsequent analyses were focused solely on birth weight. The explanatory power of birth weight among various risk factors was 3.64%, with F-statistics ranging from 160.61 to 833.99 (Supplementary Tables 4 and 5). Figure 2B illustrates a significant positive correlation between birth weight and five risk factors, while showing no association with others. Educational attainment (OR = 1.15, 95%CI: 1.01-1.31), BMI (OR = 1.06, 95%CI: 1.02-1.1), height (OR = 1.12, 95%CI: 1.06-1.19), weight (OR = 1.13, 95%CI: 1.10-1.1), and alcoholic drinks per week (OR = 1.03, 95%CI: 1.00-1.06) all exhibited significant associations (Supplementary Table 6). No evidence of pleiotropy was detected, and all MR methods showed consistent effect directions. Sensitivity analysis also revealed no significant outliers, ensuring the reliability of the results (Supplementary Figures 16-23).
Causal links of risk factors on GERD, BE, and EA considering adjusted and unadjusted birth weight
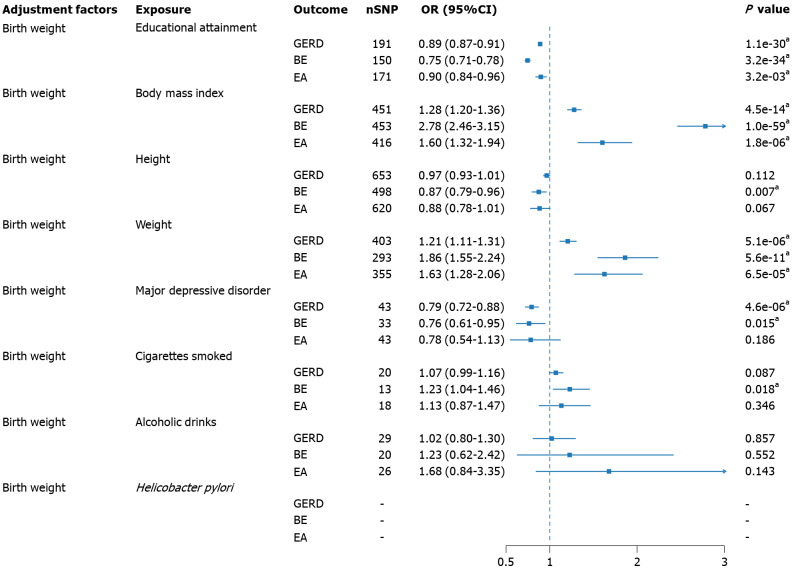
Through UVMR, the causal effects of risk factors on GERD, BE, and EA were estimated without adjusting for birth weight (Supplementary Tables 7-9), with corresponding sensitivity analyses presented in Supplementary Figures 24-47. The application of MVMR yielded the direct effects of various risk factors on GERD, BE, and EA after adjusting for birth weight (Figure 3, Supplementary Table 10). Only weight and partially adjusted BMI showed evidence of horizontal pleiotropy, and MR-Egger was employed to report the results. Educational attainment significantly reduced the risk of GERD, BE, both before adjustment (OR = 0.89, 95%CI: 0.87-0.91; OR = 0.76, 95%CI: 0.73-0.81), and after adjustment (OR = 0.89, 95%CI: 0.87-0.91; OR = 0.75, 95%CI: 0.71-0.78), and EA (OR = 0.90, 95%CI: 0.84-0.96). BMI increased the susceptibility to all three esophageal diseases, both before adjustment (OR = 1.29, 95%CI: 1.21-1.39; OR = 2.64, 95%CI: 2.32-3.00; OR = 1.50; 95%CI: 1.22-1.86), and after adjustment (OR = 1.28, 95%CI: 1.20-1.36; OR = 2.78, 95%CI: 2.46-3.15; OR = 1.60, 95%CI: 1.32-1.94). Height demonstrated a protective effect in BE (OR = 0.95, 95%CI: 0.92-0.99) and EA (OR = 0.86, 95%CI: 0.79-0.93), with a significant protective effect observed only in BE after adjustment (OR = 0.87, 95%CI: 0.79-0.96). Conversely, weight significantly reduced the likelihood of GERD (OR = 0.73, 95%CI: 0.55-0.97) and BE before adjustment (OR = 0.27, 95%CI: 0.14-0.54). However, after adjustment, weight exhibited an opposite effect, with significant differences observed for GERD (OR = 1.21, 95%CI: 1.11-1.31), BE (OR = 1.86, 95%CI: 1.55-2.24), and EA (OR = 1.63, 95%CI: 1.28-2.06). A similar contrast phenomenon was observed in MDD, where MDD, before adjustment, acted as a factor increasing the risk of GERD (OR = 1.37, 95%CI: 0.94-1.98) and BE (OR = 2.70, 95%CI: 1.91-3.83). However, after adjustment, MDD exhibited a protective effect (OR = 0.79, 95%CI: 0.72-0.88; OR = 0.76, 95%CI: 0.61-0.95). Daily smoking quantity, before adjustment, did not show significance in any outcome, but after adjustment, smoking increased the risk of BE (OR = 1.23, 95%CI: 1.04-1.46). Before adjustment, a substantial increase in weekly alcohol consumption raised the risk of EA (OR = 3.88, 95%CI: 1.53-9.89), but in the adjusted results, no significant effect was observed. H. pylori infection was not reported to be significant before adjustment, and in the analysis adjusting for birth weight, due to sample size limitations, independent SNPs were not obtained for MVMR analysis at the threshold of P < 5E-5.
Figure 3.
Multivariable Mendelian randomization analysis of causal effects of risk factors on esophageal disease after adjusting for birth weight. aP < 0.05. Helicobacter pylori phenotypes do not acquire sufficient single nucleotide polymorphisms in multivariable Mendelian randomization. GERD: Gastroesophageal reflux disease; BE: Barrett’s esophagus; EA: Esophageal adenocarcinoma; OR: Odds ratio; CI: Confidence interval; nSNPs: Number of single nucleotide polymorphisms.
Mediation effect analysis
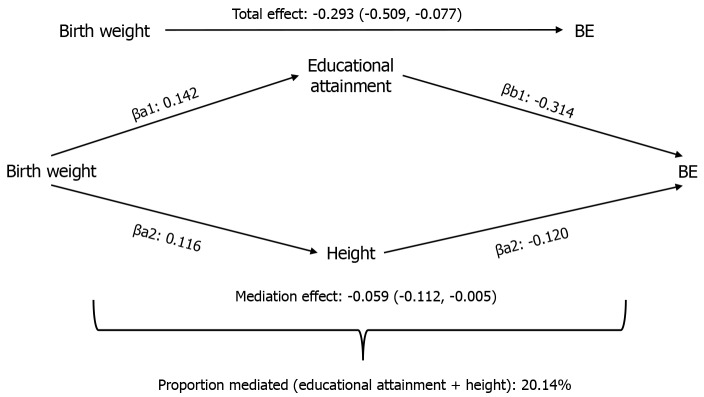
For factors demonstrating consistent and statistically significant total and mediation effects, we conducted confidence interval calculations and estimated mediation proportions (Supplementary Table 11). Despite the consistent negative association of educational attainment with GERD observed in both UVMR and MVMR, the mediation effect did not reach statistical significance (β = -0.017, 95%CI: -0.052 to 0.019). Conversely, education level (proportion mediated = 13.99%, β = -0.041, 95%CI: -0.080, to -0.003) and height (proportion mediated = 5.46%, β = -0.016; 95%CI: -0.029 to -0.002) mediated the impact of birth weight on BE, both exhibiting negative correlations. Subsequently, Figure 4 and Supplementary Tables 12 and 13 present the comprehensive mediation effects of education level and height on BE. For educational attainment (proportion mediated = 15.36%, β = -0.045; 95%CI: -0.086 to -0.003) and height (proportion mediated = 4.78%, β = -0.014, 95%CI: -0.026 to -0.002), the results indicate significant and consistent mediation effects. No evidence of horizontal pleiotropy was found, with both multivariable IVW and MVMR-Egger results demonstrating significance and consistent direction, supporting the robustness of our findings.
Figure 4.
Combined effects of multiple mediators on esophageal diseases. The combined proportions mediated by educational attainment and height for the effect of birth weight on Barrett’s esophagus. βa: The effect of birth weight on the risk of modifiable risk factors; βb: The effect of risk factors on the risk of Barrett’s esophagus after combination of mediating factors; Total effect: The effect of birth weight on the risk of Barrett’s esophagus; BE: Barrett’s esophagus.
DISCUSSION
In this study, we employed a large-scale genetic dataset and conducted UVMR, MVMR, and mediation analyses to comprehensively assess the causal relationships between fetal genetic characteristics during pregnancy, birth weight, and the onset risk of GERD, BE, and EA in adulthood. Additionally, we analyzed potential mediation effects of known risk factors. Our research confirms that gestational age does not impact the risk of GERD, BE, and EA in adulthood. This conclusion remains consistent whether analyzed as a continuous variable based on gestational weeks or as a binary variable distinguishing early preterm, preterm, and post-term births. Lower birth weight is causally associated with an increased risk of GERD and BE in adults. Conversely, higher birth weight shows causal relationships with higher education levels, obesity (high BMI), increased height and weight, and a higher weekly alcohol intake. Furthermore, within these factors, education level and height mediate 15.36% and 4.78%, respectively, of the total effect of birth weight on BE. All analysis results withstand sensitivity and multicollinearity tests, ensuring the reliability of our findings.
Considering the strong concealment of EA, it remains a challenging subtype in the early diagnosis and prevention of esophageal cancer worldwide, with an increasing incidence in many Western countries[4,47]. Most EA patients face a poor prognosis, with the overall 5-year survival rate for esophageal cancer still below 20%[2]. Meanwhile, BE, as the only precancerous lesion of EA, has an annual incidence rate of approximately 0.2%-0.5%[5]. In developed countries, adult GERD accounts for about 20%[6], showing an association with increased risks of BE and EA. Therefore, understanding the high-risk populations and risk factors for the onset of GERD, BE, and EA is crucial for early screening and prevention decisions regarding esophageal cancer.
In recent years, limited research has focused on the potential connections between neonatal characteristics and the risk of GERD, BE, and EA in adulthood. However, these studies have yielded inconsistent viewpoints. Although GERD is common in preterm or SGA infants[13], there is no reported association between GERD risk in adults and a history of preterm birth in cohort studies. Shiota et al[16] proposed that preterm birth increases the risk of adult BE, with this risk decreasing in infants larger than the gestational age, and this phenomenon was not mediated by known high-risk factors. In contrast, Forssell et al[17] suggested a different perspective, indicating that the increased risk of BE in adulthood is due to the impact of SGA and is unrelated to preterm birth. This aligns with our study’s findings, where individuals with higher birth weight have a 25% lower risk of developing BE, with no evidence supporting the influence of preterm birth on BE. In a long-term follow-up involving 3364 preterm/SGA infants, eight individuals developed esophageal cancer in adulthood compared to only one in the control group. This resulted in an 11.5-fold increase in EA risk in SGA, but concerns were raised about potential false associations due to the small sample size[14]. Some argue that the increased risk of adult EA is unrelated to birth weight but associated with gestational weeks[15], while prospective studies, including ours, find no correlation between gestational weeks, birth weight, and EA risk[48]. These phenomena may be due to the adaptive response of SGA in the early stage of the disease, which leads to the lagging growth and development of tissue and organ function through metabolic or hormonal changes, resulting in an increased risk of disease. It is also possible that perinatal promoter methylation affects gene expression in pathways associated with a range of physiological processes and continues into adulthood[49].
These long-term cohort studies lack sufficient exploration of other risk factors’ potential effects on GERD, BE, and EA, and are burdened by significant limitations. It remains unclear if preterm/SGA individuals exhibit growth tendencies or engage in high-risk behaviors like smoking, drinking, or obesity[17,50]. Additionally, the increased risk of adult depression linked to low birth weight complicates inferences[51], as depression or anxiety further heightens gastrointestinal disease risk[15,52]. Scarce research on preterm/SGA individuals regarding esophageal diseases and a lack of reports on intermediate factors prompted our study. Leveraging large-scale genetic data from the European population (84689 for gestational age, 143677 for birth weight)[25,27] and utilizing MR analysis, we overcame confounding factors, facilitating precise causal inferences[18].
Past studies on risk factors for esophageal diseases did not consider the potential impact of neonatal characteristics. This research offers new insights into the role of adjusted risk factors after birth weight modification in esophageal diseases. Firstly, the increase in education level, as found by Jansson et al[9] and Zhang et al[11], is associated with a decreased risk of GERD, BE, and esophageal cancer. Our results align with this, showing that education level remains a protective factor after adjusting for birth weight. Obesity (high BMI/overweight) has been widely recognized for its detrimental effects on gastrointestinal diseases. In our study, adjusting for birth weight increased the risks of GERD, BE, and EA by 28%, more than double, and 60%, respectively. The association between these factors may be attributed to visceral obesity, resulting in increased abdominal fat thickness and heightened intra-abdominal pressure. Mechanical damage to the esophagus is more common in obese individuals, increasing the chances of acid exposure and thereby elevating the risk of GERD. Additionally, elevated levels of serum adipocyte-derived cytokines, intensified by inflammatory responses mediated through adiponectin or leptin, contribute to an increased risk of BE[53]. Thrift et al[54] found that increased height contributes to a lower probability of BE and EA in males, with no significant effect in females. In our study, adjusted height was only significantly associated with BE, possibly due to the genetic information related to height explaining the overall effects in both male and female populations, minimizing the impact of gender subgroups. Regarding the impact of MDD on esophageal diseases, some argue that MDD is unrelated to BE and EA but is associated with an increased risk of GERD[35]. In our study, after adjusting for birth weight, MDD had statistical significance for both GERD and BE, but caution is needed in interpreting this due to inconsistent directions in effects before and after adjustment and the limited number of SNPs obtained. Smoking is associated with a 23% increased risk of BE in this study, aligning with findings by Yuan and Larsson[55]. Smoking has been shown to induce damage to esophageal epithelial cells, exacerbating metaplasia. Infection with H. pylori (adjusted before) and alcohol consumption (adjusted after) is unrelated to the occurrence of GERD, BE, and EA, aligning with some research conclusions[56,57] but differing from others[1,6,58]. Perhaps this is due to the impact of low socioeconomic status and insufficient education levels in SGA individuals during adulthood, leading to increased risks of smoking, drinking, H. pylori infection, etc[59]. Neglecting birth weight in observational studies masks the true effects of relevant factors, potentially resulting in incorrect inferences.
This study provides support for education level being an intermediate factor in the increased risk of BE associated with high birth weight. In several large birth cohort studies, early improvements in birth weight have been shown to effectively increase education levels, cognitive abilities, and social status, subsequently reducing the risk of chronic diseases in adulthood[60,61]. Higher education levels can further reduce the harms brought by adverse habits[24,62]. The protective intermediate effect of height may be due to growth restriction in the womb for SGA individuals, resulting in a relatively small stature during growth, increasing the risk of gastrointestinal diseases such as GERD due to increased intra-abdominal pressure[54,63]. This process may eventually evolve into BE through repeated stimulation.
The findings of this study address previous research limitations by proposing the impact of birth weight-adjusted relevant risk factors on esophageal diseases and highlighting the intermediate roles of education level and height. New perspectives on the effects of pathogenic factors like smoking and drinking have been introduced. This study is expected to enhance our understanding of the complex mechanisms of genetic factors in esophageal diseases. Early intervention measures and personalized treatment should be developed for SGA individuals, and early screening and prevention of esophageal diseases should be conducted in adulthood for these high-risk populations. However, the mediating mechanism of height between SGA children and precancerous lesions of esophageal cancer remains unclear, requiring further research to explain this effect.
Limitations
This study focused solely on the European population, and the generalization of findings to other ethnic groups remains inconclusive. In the MVMR analysis after adjusting for birth weight, H. pylori infection lacked a sufficient number of SNPs for subsequent analysis. Despite incorporating multiple known risk factors into our analysis, we cannot eliminate the potential effects of unknown factors.
CONCLUSION
Our research supports the association between low birth weight and an increased risk of GERD and BE, but not EA. Gestational weeks are unrelated to all three esophageal diseases. The causal effect of birth weight on BE is jointly mediated by education level and height, accounting for approximately 20%. Furthermore, our study provides a comprehensive effect of known risk factors on esophageal diseases after adjusting for birth weight.
ACKNOWLEDGEMENTS
The authors thank the staff and participants of the United Kingdom Biobank, Genetic Investigation of ANthropometric Traits Consortium, Psychiatric Genomics Consortium, genome-wide association study and Sequencing Consortium of Alcohol and Nicotine, and Integrative Psychiatric Research Consortium for making the data publicly available (Supplementary Table 14).
Footnotes
Conflict-of-interest statement: All the authors report no relevant conflicts of interest for this article.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country of origin: China
Peer-review report’s classification
Scientific Quality: Grade A
Novelty: Grade A
Creativity or Innovation: Grade A
Scientific Significance: Grade A
P-Reviewer: Skrypnyk I, Ukraine S-Editor: Wang JJ L-Editor: A P-Editor: Zheng XM
Contributor Information
Lian-Cheng Ruan, Department of Thoracic Surgery, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Yang Zhang, Department of Thoracic Surgery, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Lang Su, Department of Thoracic Surgery, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Ling-Xiao Zhu, Department of Thoracic Surgery, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Si-Lin Wang, Department of Thoracic Surgery, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Qiang Guo, Department of Thoracic Surgery, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Bin-Gen Wan, Department of Thoracic Surgery, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Sheng-Yu Qiu, Department of Thoracic Surgery, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Sheng Hu, Department of Thoracic Surgery, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Yi-Ping Wei, Department of Thoracic Surgery, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China. weiyip2000@hotmail.com.
Qiao-Ling Zheng, Nanchang Medical College, Nanchang 330004, Jiangxi Province, China.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Thrift AP. Global burden and epidemiology of Barrett oesophagus and oesophageal cancer. Nat Rev Gastroenterol Hepatol. 2021;18:432–443. doi: 10.1038/s41575-021-00419-3. [DOI] [PubMed] [Google Scholar]
- 3.Arshad HMS, Farooq U, Cheema A, Arshad A, Masood M, Vega KJ. Disparities in esophageal cancer incidence and esophageal adenocarcinoma mortality in the United States over the last 25-40 years. World J Gastrointest Endosc. 2023;15:715–724. doi: 10.4253/wjge.v15.i12.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McColl KEL. What is causing the rising incidence of esophageal adenocarcinoma in the West and will it also happen in the East? J Gastroenterol. 2019;54:669–673. doi: 10.1007/s00535-019-01593-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma P. Barrett Esophagus: A Review. JAMA. 2022;328:663–671. doi: 10.1001/jama.2022.13298. [DOI] [PubMed] [Google Scholar]
- 6.Maret-Ouda J, Markar SR, Lagergren J. Gastroesophageal Reflux Disease: A Review. JAMA. 2020;324:2536–2547. doi: 10.1001/jama.2020.21360. [DOI] [PubMed] [Google Scholar]
- 7.Uhlenhopp DJ, Then EO, Sunkara T, Gaduputi V. Epidemiology of esophageal cancer: update in global trends, etiology and risk factors. Clin J Gastroenterol. 2020;13:1010–1021. doi: 10.1007/s12328-020-01237-x. [DOI] [PubMed] [Google Scholar]
- 8.Thrift AP, Shaheen NJ, Gammon MD, Bernstein L, Reid BJ, Onstad L, Risch HA, Liu G, Bird NC, Wu AH, Corley DA, Romero Y, Chanock SJ, Chow WH, Casson AG, Levine DM, Zhang R, Ek WE, MacGregor S, Ye W, Hardie LJ, Vaughan TL, Whiteman DC. Obesity and risk of esophageal adenocarcinoma and Barrett's esophagus: a Mendelian randomization study. J Natl Cancer Inst. 2014;106 doi: 10.1093/jnci/dju252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansson C, Nordenstedt H, Johansson S, Wallander MA, Johnsen R, Hveem K, Lagergren J. Relation between gastroesophageal reflux symptoms and socioeconomic factors: a population-based study (the HUNT Study) Clin Gastroenterol Hepatol. 2007;5:1029–1034. doi: 10.1016/j.cgh.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Jansson C, Johansson AL, Nyrén O, Lagergren J. Socioeconomic factors and risk of esophageal adenocarcinoma: a nationwide Swedish case-control study. Cancer Epidemiol Biomarkers Prev. 2005;14:1754–1761. doi: 10.1158/1055-9965.EPI-05-0140. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Yang X, Zhang T, Yin X, Man J, Lu M. Association of educational attainment with esophageal cancer, Barrett's esophagus, and gastroesophageal reflux disease, and the mediating role of modifiable risk factors: A Mendelian randomization study. Front Public Health. 2023;11:1022367. doi: 10.3389/fpubh.2023.1022367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miranda JO, Ramalho C, Henriques-Coelho T, Areias JC. Fetal programming as a predictor of adult health or disease: the need to reevaluate fetal heart function. Heart Fail Rev. 2017;22:861–877. doi: 10.1007/s10741-017-9638-z. [DOI] [PubMed] [Google Scholar]
- 13.Omari TI, Barnett CP, Benninga MA, Lontis R, Goodchild L, Haslam RR, Dent J, Davidson GP. Mechanisms of gastro-oesophageal reflux in preterm and term infants with reflux disease. Gut. 2002;51:475–479. doi: 10.1136/gut.51.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaijser M, Akre O, Cnattingius S, Ekbom A. Preterm birth, low birth weight, and risk for esophageal adenocarcinoma. Gastroenterology. 2005;128:607–609. doi: 10.1053/j.gastro.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 15.Forssell L, Cnattingius S, Bottai M, Edstedt Bonamy AK, Lagergren J, Agréus L, Akre O. Risk of oesophageal adenocarcinoma among individuals born preterm or small for gestational age. Eur J Cancer. 2013;49:2207–2213. doi: 10.1016/j.ejca.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 16.Shiota S, El-Serag HB, Thrift AP. Premature Birth and Large for Gestational Age Are Associated with Risk of Barrett's Esophagus in Adults. Dig Dis Sci. 2016;61:1139–1147. doi: 10.1007/s10620-015-3967-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forssell L, Cnattingius S, Bottai M, Edstedt Bonamy AK, Lagergren J, Agréus L, Akre O. Increased risk of Barrett's esophagus among individuals born preterm or small for gestational age. Clin Gastroenterol Hepatol. 2013;11:790–794. doi: 10.1016/j.cgh.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 18.Davey Smith G, Ebrahim S. What can mendelian randomisation tell us about modifiable behavioural and environmental exposures? BMJ. 2005;330:1076–1079. doi: 10.1136/bmj.330.7499.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanderson E. Multivariable Mendelian Randomization and Mediation. Cold Spring Harb Perspect Med. 2021;11 doi: 10.1101/cshperspect.a038984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, Frayling TM, Hirschhorn J, Yang J, Visscher PM GIANT Consortium. Meta-analysis of genome-wide association studies for height and body mass index in ~700000 individuals of European ancestry. Hum Mol Genet. 2018;27:3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, Adams MJ, Agerbo E, Air TM, Andlauer TMF, Bacanu SA, Bækvad-Hansen M, Beekman AFT, Bigdeli TB, Binder EB, Blackwood DRH, Bryois J, Buttenschøn HN, Bybjerg-Grauholm J, Cai N, Castelao E, Christensen JH, Clarke TK, Coleman JIR, Colodro-Conde L, Couvy-Duchesne B, Craddock N, Crawford GE, Crowley CA, Dashti HS, Davies G, Deary IJ, Degenhardt F, Derks EM, Direk N, Dolan CV, Dunn EC, Eley TC, Eriksson N, Escott-Price V, Kiadeh FHF, Finucane HK, Forstner AJ, Frank J, Gaspar HA, Gill M, Giusti-Rodríguez P, Goes FS, Gordon SD, Grove J, Hall LS, Hannon E, Hansen CS, Hansen TF, Herms S, Hickie IB, Hoffmann P, Homuth G, Horn C, Hottenga JJ, Hougaard DM, Hu M, Hyde CL, Ising M, Jansen R, Jin F, Jorgenson E, Knowles JA, Kohane IS, Kraft J, Kretzschmar WW, Krogh J, Kutalik Z, Lane JM, Li Y, Lind PA, Liu X, Lu L, MacIntyre DJ, MacKinnon DF, Maier RM, Maier W, Marchini J, Mbarek H, McGrath P, McGuffin P, Medland SE, Mehta D, Middeldorp CM, Mihailov E, Milaneschi Y, Milani L, Mill J, Mondimore FM, Montgomery GW, Mostafavi S, Mullins N, Nauck M, Ng B, Nivard MG, Nyholt DR, O'Reilly PF, Oskarsson H, Owen MJ, Painter JN, Pedersen CB, Pedersen MG, Peterson RE, Pettersson E, Peyrot WJ, Pistis G, Posthuma D, Purcell SM, Quiroz JA, Qvist P, Rice JP, Riley BP, Rivera M, Saeed Mirza S, Saxena R, Schoevers R, Schulte EC, Shen L, Shi J, Shyn SI, Sigurdsson E, Sinnamon GBC, Smit JH, Smith DJ, Stefansson H, Steinberg S, Stockmeier CA, Streit F, Strohmaier J, Tansey KE, Teismann H, Teumer A, Thompson W, Thomson PA, Thorgeirsson TE, Tian C, Traylor M, Treutlein J, Trubetskoy V, Uitterlinden AG, Umbricht D, Van der Auwera S, van Hemert AM, Viktorin A, Visscher PM, Wang Y, Webb BT, Weinsheimer SM, Wellmann J, Willemsen G, Witt SH, Wu Y, Xi HS, Yang J, Zhang F eQTLGen; 23andMe, Arolt V, Baune BT, Berger K, Boomsma DI, Cichon S, Dannlowski U, de Geus ECJ, DePaulo JR, Domenici E, Domschke K, Esko T, Grabe HJ, Hamilton SP, Hayward C, Heath AC, Hinds DA, Kendler KS, Kloiber S, Lewis G, Li QS, Lucae S, Madden PFA, Magnusson PK, Martin NG, McIntosh AM, Metspalu A, Mors O, Mortensen PB, Müller-Myhsok B, Nordentoft M, Nöthen MM, O'Donovan MC, Paciga SA, Pedersen NL, Penninx BWJH, Perlis RH, Porteous DJ, Potash JB, Preisig M, Rietschel M, Schaefer C, Schulze TG, Smoller JW, Stefansson K, Tiemeier H, Uher R, Völzke H, Weissman MM, Werge T, Winslow AR, Lewis CM, Levinson DF, Breen G, Børglum AD, Sullivan PF; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, Pell J, Silman A, Young A, Sprosen T, Peakman T, Collins R. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakaue S, Kanai M, Tanigawa Y, Karjalainen J, Kurki M, Koshiba S, Narita A, Konuma T, Yamamoto K, Akiyama M, Ishigaki K, Suzuki A, Suzuki K, Obara W, Yamaji K, Takahashi K, Asai S, Takahashi Y, Suzuki T, Shinozaki N, Yamaguchi H, Minami S, Murayama S, Yoshimori K, Nagayama S, Obata D, Higashiyama M, Masumoto A, Koretsune Y FinnGen, Ito K, Terao C, Yamauchi T, Komuro I, Kadowaki T, Tamiya G, Yamamoto M, Nakamura Y, Kubo M, Murakami Y, Yamamoto K, Kamatani Y, Palotie A, Rivas MA, Daly MJ, Matsuda K, Okada Y. A cross-population atlas of genetic associations for 220 human phenotypes. Nat Genet. 2021;53:1415–1424. doi: 10.1038/s41588-021-00931-x. [DOI] [PubMed] [Google Scholar]
- 24.Loh PR, Kichaev G, Gazal S, Schoech AP, Price AL. Mixed-model association for biobank-scale datasets. Nat Genet. 2018;50:906–908. doi: 10.1038/s41588-018-0144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Helenius D, Skotte L, Beaumont RN, Wielscher M, Geller F, Juodakis J, Mahajan A, Bradfield JP, Lin FTJ, Vogelezang S, Bustamante M, Ahluwalia TS, Pitkänen N, Wang CA, Bacelis J, Borges MC, Zhang G, Bedell BA, Rossi RM, Skogstrand K, Peng S, Thompson WK, Appadurai V, Lawlor DA, Kalliala I, Power C, McCarthy MI, Boyd HA, Marazita ML, Hakonarson H, Hayes MG, Scholtens DM, Rivadeneira F, Jaddoe VWV, Vinding RK, Bisgaard H, Knight BA, Pahkala K, Raitakari O, Helgeland Ø, Johansson S, Njølstad PR, Fadista J, Schork AJ, Nudel R, Miller DE, Chen X, Weirauch MT, Mortensen PB, Børglum AD, Nordentoft M, Mors O, Hao K, Ryckman KK, Hougaard DM, Kottyan LC, Pennell CE, Lyytikainen LP, Bønnelykke K, Vrijheid M, Felix JF, Lowe WL Jr, Grant SFA, Hyppönen E, Jacobsson B, Jarvelin MR, Muglia LJ, Murray JC, Freathy RM, Werge TM, Melbye M, Buil A, Feenstra B. Variants in the fetal genome near pro-inflammatory cytokine genes on 2q13 associate with gestational duration. Nat Commun. 2019;10:3927. doi: 10.1038/s41467-019-11881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, Datta G, Davila-Velderrain J, McGuire D, Tian C, Zhan X 23andMe Research Team; HUNT All-In Psychiatry, Choquet H, Docherty AR, Faul JD, Foerster JR, Fritsche LG, Gabrielsen ME, Gordon SD, Haessler J, Hottenga JJ, Huang H, Jang SK, Jansen PR, Ling Y, Mägi R, Matoba N, McMahon G, Mulas A, Orrù V, Palviainen T, Pandit A, Reginsson GW, Skogholt AH, Smith JA, Taylor AE, Turman C, Willemsen G, Young H, Young KA, Zajac GJM, Zhao W, Zhou W, Bjornsdottir G, Boardman JD, Boehnke M, Boomsma DI, Chen C, Cucca F, Davies GE, Eaton CB, Ehringer MA, Esko T, Fiorillo E, Gillespie NA, Gudbjartsson DF, Haller T, Harris KM, Heath AC, Hewitt JK, Hickie IB, Hokanson JE, Hopfer CJ, Hunter DJ, Iacono WG, Johnson EO, Kamatani Y, Kardia SLR, Keller MC, Kellis M, Kooperberg C, Kraft P, Krauter KS, Laakso M, Lind PA, Loukola A, Lutz SM, Madden PAF, Martin NG, McGue M, McQueen MB, Medland SE, Metspalu A, Mohlke KL, Nielsen JB, Okada Y, Peters U, Polderman TJC, Posthuma D, Reiner AP, Rice JP, Rimm E, Rose RJ, Runarsdottir V, Stallings MC, Stančáková A, Stefansson H, Thai KK, Tindle HA, Tyrfingsson T, Wall TL, Weir DR, Weisner C, Whitfield JB, Winsvold BS, Yin J, Zuccolo L, Bierut LJ, Hveem K, Lee JJ, Munafò MR, Saccone NL, Willer CJ, Cornelis MC, David SP, Hinds DA, Jorgenson E, Kaprio J, Stitzel JA, Stefansson K, Thorgeirsson TE, Abecasis G, Liu DJ, Vrieze S. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51:237–244. doi: 10.1038/s41588-018-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horikoshi M, Beaumont RN, Day FR, Warrington NM, Kooijman MN, Fernandez-Tajes J, Feenstra B, van Zuydam NR, Gaulton KJ, Grarup N, Bradfield JP, Strachan DP, Li-Gao R, Ahluwalia TS, Kreiner E, Rueedi R, Lyytikäinen LP, Cousminer DL, Wu Y, Thiering E, Wang CA, Have CT, Hottenga JJ, Vilor-Tejedor N, Joshi PK, Boh ETH, Ntalla I, Pitkänen N, Mahajan A, van Leeuwen EM, Joro R, Lagou V, Nodzenski M, Diver LA, Zondervan KT, Bustamante M, Marques-Vidal P, Mercader JM, Bennett AJ, Rahmioglu N, Nyholt DR, Ma RCW, Tam CHT, Tam WH CHARGE Consortium Hematology Working Group, Ganesh SK, van Rooij FJ, Jones SE, Loh PR, Ruth KS, Tuke MA, Tyrrell J, Wood AR, Yaghootkar H, Scholtens DM, Paternoster L, Prokopenko I, Kovacs P, Atalay M, Willems SM, Panoutsopoulou K, Wang X, Carstensen L, Geller F, Schraut KE, Murcia M, van Beijsterveldt CE, Willemsen G, Appel EVR, Fonvig CE, Trier C, Tiesler CM, Standl M, Kutalik Z, Bonas-Guarch S, Hougaard DM, Sánchez F, Torrents D, Waage J, Hollegaard MV, de Haan HG, Rosendaal FR, Medina-Gomez C, Ring SM, Hemani G, McMahon G, Robertson NR, Groves CJ, Langenberg C, Luan J, Scott RA, Zhao JH, Mentch FD, MacKenzie SM, Reynolds RM; Early Growth Genetics (EGG) Consortium, Lowe WL Jr, Tönjes A, Stumvoll M, Lindi V, Lakka TA, van Duijn CM, Kiess W, Körner A, Sørensen TI, Niinikoski H, Pahkala K, Raitakari OT, Zeggini E, Dedoussis GV, Teo YY, Saw SM, Melbye M, Campbell H, Wilson JF, Vrijheid M, de Geus EJ, Boomsma DI, Kadarmideen HN, Holm JC, Hansen T, Sebert S, Hattersley AT, Beilin LJ, Newnham JP, Pennell CE, Heinrich J, Adair LS, Borja JB, Mohlke KL, Eriksson JG, Widén EE, Kähönen M, Viikari JS, Lehtimäki T, Vollenweider P, Bønnelykke K, Bisgaard H, Mook-Kanamori DO, Hofman A, Rivadeneira F, Uitterlinden AG, Pisinger C, Pedersen O, Power C, Hyppönen E, Wareham NJ, Hakonarson H, Davies E, Walker BR, Jaddoe VW, Jarvelin MR, Grant SF, Vaag AA, Lawlor DA, Frayling TM, Davey Smith G, Morris AP, Ong KK, Felix JF, Timpson NJ, Perry JR, Evans DM, McCarthy MI, Freathy RM. Genome-wide associations for birth weight and correlations with adult disease. Nature. 2016;538:248–252. doi: 10.1038/nature19806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gharahkhani P, Fitzgerald RC, Vaughan TL, Palles C, Gockel I, Tomlinson I, Buas MF, May A, Gerges C, Anders M, Becker J, Kreuser N, Noder T, Venerito M, Veits L, Schmidt T, Manner H, Schmidt C, Hess T, Böhmer AC, Izbicki JR, Hölscher AH, Lang H, Lorenz D, Schumacher B, Hackelsberger A, Mayershofer R, Pech O, Vashist Y, Ott K, Vieth M, Weismüller J, Nöthen MM Barrett's and Esophageal Adenocarcinoma Consortium (BEACON); Esophageal Adenocarcinoma GenEtics Consortium (EAGLE); Wellcome Trust Case Control Consortium 2 (WTCCC2), Attwood S, Barr H, Chegwidden L, de Caestecker J, Harrison R, Love SB, MacDonald D, Moayyedi P, Prenen H, Watson RGP, Iyer PG, Anderson LA, Bernstein L, Chow WH, Hardie LJ, Lagergren J, Liu G, Risch HA, Wu AH, Ye W, Bird NC, Shaheen NJ, Gammon MD, Corley DA, Caldas C, Moebus S, Knapp M, Peters WHM, Neuhaus H, Rösch T, Ell C, MacGregor S, Pharoah P, Whiteman DC, Jankowski J, Schumacher J. Genome-wide association studies in oesophageal adenocarcinoma and Barrett's oesophagus: a large-scale meta-analysis. Lancet Oncol. 2016;17:1363–1373. doi: 10.1016/S1470-2045(16)30240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barton AR, Sherman MA, Mukamel RE, Loh PR. Whole-exome imputation within UK Biobank powers rare coding variant association and fine-mapping analyses. Nat Genet. 2021;53:1260–1269. doi: 10.1038/s41588-021-00892-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greco M FD, Minelli C, Sheehan NA, Thompson JR. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat Med. 2015;34:2926–2940. doi: 10.1002/sim.6522. [DOI] [PubMed] [Google Scholar]
- 31.Lyon MS, Andrews SJ, Elsworth B, Gaunt TR, Hemani G, Marcora E. The variant call format provides efficient and robust storage of GWAS summary statistics. Genome Biol. 2021;22:32. doi: 10.1186/s13059-020-02248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elsworth B, Lyon MS, Alexander T, Liu Y, Matthews P, Hallett J, Bates P, Palmer T, Haberland V, Smith GD, Zheng J, Haycock P, Gaunt T, Hemani G. The MRC IEU OpenGWAS data infrastructure. 2020 Preprint. Available from: bioRxiv:2020.08.10.244293v1.
- 33.Jones DP, Wootton RE, Gill D, Carter AR, Gunnell D, Munafò MR, Sallis HM. Mental Health as a Mediator of the Association Between Educational Inequality and Cardiovascular Disease: A Mendelian Randomization Study. J Am Heart Assoc. 2021;10:e019340. doi: 10.1161/JAHA.120.019340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ong JS, An J, Han X, Law MH, Nandakumar P 23andMe Research team; Esophageal cancer consortium, Schumacher J, Gockel I, Bohmer A, Jankowski J, Palles C, Olsen CM, Neale RE, Fitzgerald R, Thrift AP, Vaughan TL, Buas MF, Hinds DA, Gharahkhani P, Kendall BJ, MacGregor S. Multitrait genetic association analysis identifies 50 new risk loci for gastro-oesophageal reflux, seven new loci for Barrett's oesophagus and provides insights into clinical heterogeneity in reflux diagnosis. Gut. 2022;71:1053–1061. doi: 10.1136/gutjnl-2020-323906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, Paul DS, Freitag D, Burgess S, Danesh J, Young R, Butterworth AS. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32:3207–3209. doi: 10.1093/bioinformatics/btw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burgess S, Thompson SG CRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40:755–764. doi: 10.1093/ije/dyr036. [DOI] [PubMed] [Google Scholar]
- 39.Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013;42:1497–1501. doi: 10.1093/ije/dyt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papadimitriou N, Dimou N, Tsilidis KK, Banbury B, Martin RM, Lewis SJ, Kazmi N, Robinson TM, Albanes D, Aleksandrova K, Berndt SI, Timothy Bishop D, Brenner H, Buchanan DD, Bueno-de-Mesquita B, Campbell PT, Castellví-Bel S, Chan AT, Chang-Claude J, Ellingjord-Dale M, Figueiredo JC, Gallinger SJ, Giles GG, Giovannucci E, Gruber SB, Gsur A, Hampe J, Hampel H, Harlid S, Harrison TA, Hoffmeister M, Hopper JL, Hsu L, María Huerta J, Huyghe JR, Jenkins MA, Keku TO, Kühn T, La Vecchia C, Le Marchand L, Li CI, Li L, Lindblom A, Lindor NM, Lynch B, Markowitz SD, Masala G, May AM, Milne R, Monninkhof E, Moreno L, Moreno V, Newcomb PA, Offit K, Perduca V, Pharoah PDP, Platz EA, Potter JD, Rennert G, Riboli E, Sánchez MJ, Schmit SL, Schoen RE, Severi G, Sieri S, Slattery ML, Song M, Tangen CM, Thibodeau SN, Travis RC, Trichopoulou A, Ulrich CM, van Duijnhoven FJB, Van Guelpen B, Vodicka P, White E, Wolk A, Woods MO, Wu AH, Peters U, Gunter MJ, Murphy N. Physical activity and risks of breast and colorectal cancer: a Mendelian randomisation analysis. Nat Commun. 2020;11:597. doi: 10.1038/s41467-020-14389-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slob EAW, Burgess S. A comparison of robust Mendelian randomization methods using summary data. Genet Epidemiol. 2020;44:313–329. doi: 10.1002/gepi.22295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burgess S, Daniel RM, Butterworth AS, Thompson SG EPIC-InterAct Consortium. Network Mendelian randomization: using genetic variants as instrumental variables to investigate mediation in causal pathways. Int J Epidemiol. 2015;44:484–495. doi: 10.1093/ije/dyu176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carter AR, Gill D, Davies NM, Taylor AE, Tillmann T, Vaucher J, Wootton RE, Munafò MR, Hemani G, Malik R, Seshadri S, Woo D, Burgess S, Davey Smith G, Holmes MV, Tzoulaki I, Howe LD, Dehghan A. Understanding the consequences of education inequality on cardiovascular disease: mendelian randomisation study. BMJ. 2019;365:l1855. doi: 10.1136/bmj.l1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burgess S, Bowden J, Fall T, Ingelsson E, Thompson SG. Sensitivity Analyses for Robust Causal Inference from Mendelian Randomization Analyses with Multiple Genetic Variants. Epidemiology. 2017;28:30–42. doi: 10.1097/EDE.0000000000000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mansour NM, Groth SS, Anandasabapathy S. Esophageal Adenocarcinoma: Screening, Surveillance, and Management. Annu Rev Med. 2017;68:213–227. doi: 10.1146/annurev-med-050715-104218. [DOI] [PubMed] [Google Scholar]
- 48.Akre O, Forssell L, Kaijser M, Norén-Nilsson I, Lagergren J, Nyrén O, Ekbom A. Perinatal risk factors for cancer of the esophagus and gastric cardia: a nested case-control study. Cancer Epidemiol Biomarkers Prev. 2006;15:867–871. doi: 10.1158/1055-9965.EPI-05-0590. [DOI] [PubMed] [Google Scholar]
- 49.Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med. 2008;359:61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larsson SC, Carter P, Kar S, Vithayathil M, Mason AM, Michaëlsson K, Burgess S. Smoking, alcohol consumption, and cancer: A mendelian randomisation study in UK Biobank and international genetic consortia participants. PLoS Med. 2020;17:e1003178. doi: 10.1371/journal.pmed.1003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loret de Mola C, de França GV, Quevedo Lde A, Horta BL. Low birth weight, preterm birth and small for gestational age association with adult depression: systematic review and meta-analysis. Br J Psychiatry. 2014;205:340–347. doi: 10.1192/bjp.bp.113.139014. [DOI] [PubMed] [Google Scholar]
- 52.Ruan X, Chen J, Sun Y, Zhang Y, Zhao J, Wang X, Li X, Yuan S, Larsson SC. Depression and 24 gastrointestinal diseases: a Mendelian randomization study. Transl Psychiatry. 2023;13:146. doi: 10.1038/s41398-023-02459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lagergren J. Influence of obesity on the risk of esophageal disorders. Nat Rev Gastroenterol Hepatol. 2011;8:340–347. doi: 10.1038/nrgastro.2011.73. [DOI] [PubMed] [Google Scholar]
- 54.Thrift AP, Risch HA, Onstad L, Shaheen NJ, Casson AG, Bernstein L, Corley DA, Levine DM, Chow WH, Reid BJ, Romero Y, Hardie LJ, Liu G, Wu AH, Bird NC, Gammon MD, Ye W, Whiteman DC, Vaughan TL. Risk of esophageal adenocarcinoma decreases with height, based on consortium analysis and confirmed by Mendelian randomization. Clin Gastroenterol Hepatol. 2014;12:1667–76.e1. doi: 10.1016/j.cgh.2014.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan S, Larsson SC. Adiposity, diabetes, lifestyle factors and risk of gastroesophageal reflux disease: a Mendelian randomization study. Eur J Epidemiol. 2022;37:747–754. doi: 10.1007/s10654-022-00842-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tramacere I, Pelucchi C, Bagnardi V, Rota M, Scotti L, Islami F, Corrao G, Boffetta P, La Vecchia C, Negri E. A meta-analysis on alcohol drinking and esophageal and gastric cardia adenocarcinoma risk. Ann Oncol. 2012;23:287–297. doi: 10.1093/annonc/mdr136. [DOI] [PubMed] [Google Scholar]
- 57.Niknam R, Lankarani KB, Moghadami M, Taghavi SA, Zahiri L, Fallahi MJ. The association between helicobacter pylori infection and erosive gastroesophageal reflux disease; a cross-sectional study. BMC Infect Dis. 2022;22:267. doi: 10.1186/s12879-022-07278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kountouras J, Zavos C, Chatzopoulos D, Katsinelos P. Helicobacter pylori and gastro-oesophageal reflux disease. Lancet. 2006;368:986; author reply 986–986; author reply 987. doi: 10.1016/S0140-6736(06)69405-1. [DOI] [PubMed] [Google Scholar]
- 59.Orri M, Pingault JB, Turecki G, Nuyt AM, Tremblay RE, Côté SM, Geoffroy MC. Contribution of birth weight to mental health, cognitive and socioeconomic outcomes: two-sample Mendelian randomisation. Br J Psychiatry. 2021;219:507–514. doi: 10.1192/bjp.2021.15. [DOI] [PubMed] [Google Scholar]
- 60.Adair LS, Fall CH, Osmond C, Stein AD, Martorell R, Ramirez-Zea M, Sachdev HS, Dahly DL, Bas I, Norris SA, Micklesfield L, Hallal P, Victora CG COHORTS group. Associations of linear growth and relative weight gain during early life with adult health and human capital in countries of low and middle income: findings from five birth cohort studies. Lancet. 2013;382:525–534. doi: 10.1016/S0140-6736(13)60103-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barker DJ, Forsén T, Uutela A, Osmond C, Eriksson JG. Size at birth and resilience to effects of poor living conditions in adult life: longitudinal study. BMJ. 2001;323:1273–1276. doi: 10.1136/bmj.323.7324.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nejatinamini S, Godley J, Minaker LM, Sajobi TT, McCormack GR, Cooke MJ, Nykiforuk CIJ, Koning L, Olstad DL. Quantifying the contribution of modifiable risk factors to socio-economic inequities in cancer morbidity and mortality: a nationally representative population-based cohort study. Int J Epidemiol. 2021;50:1498–1511. doi: 10.1093/ije/dyab067. [DOI] [PubMed] [Google Scholar]
- 63.Wakelin DE, Al-Mutawa T, Wendel C, Green C, Garewal HS, Fass R. A predictive model for length of Barrett's esophagus with hiatal hernia length and duration of esophageal acid exposure. Gastrointest Endosc. 2003;58:350–355. doi: 10.1067/s0016-5107(03)00007-5. [DOI] [PubMed] [Google Scholar]