Abstract
Human foamy virus (HFV) is the prototype member of the spumaviruses. While similar in genomic organization to other complex retroviruses, foamy viruses share several features with their more distant relatives, the hepadnaviruses such as human hepatitis B virus (HBV). Both HFV and HBV express their Pol proteins independently from the structural proteins. However unlike HBV, Pol is not required for assembly of HFV core particles or for packaging of viral RNA. These results suggest that the assembly of Pol into HFV particles must occur by a mechanism different from those used by retroviruses and hepadnaviruses. We have examined possible mechanisms for HFV Pol incorporation, including the role of proteolysis in assembly of Pol and the role of initiation of reverse transcription. We have found that proteolytic activity is not required for Pol incorporation. p4 Gag and the residues immediately upstream of the cleavage site in Gag are also not important. Deletion of the primer binding site had no effect on assembly, ruling out early steps of reverse transcription in the process of Pol incorporation.
Human foamy virus (HFV) is the best-characterized member of the Spumavirus genus of the family Retroviridae (33). Foamy viruses are complex retroviruses, encoding the canonical retroviral genes gag, pol, and env, as well as several accessory genes (13). Despite clear sequence and genomic structural homology with other retroviruses, several features of HFV replication are similar to more distant relatives, the hepadnaviruses, which are the only other mammalian reverse transcriptase (RT)-encoding viruses (1, 40).
Like hepatitis B virus (HBV), HFV expresses its Pol protein independently of structural proteins (11, 27, 40). Foamy viruses express Pol from a spliced mRNA, whereas hepadnaviruses use either ribosomal scanning or internal initiation (20). The resulting HFV Pol polyprotein contains no Gag domains and must therefore be assembled into particles by a mechanism different from those used by other known retroviruses, where Gag-Pol fusion proteins are incorporated into particles via Gag-Gag interactions (14, 21, 35). We have previously demonstrated that the requirement for HFV Pol during assembly is similar to what has been found for other retroviruses and different from that found for hepadnaviruses. As with other retroviruses, abrogation of HFV Pol expression has no effect on assembly of particles, packaging of viral genomic RNA, or release of virus from the cell (1). For hepadnaviruses however, the P protein is required to initiate proper assembly and is essential for genome encapsidation (3, 5, 31, 32). While these data suggest an HFV assembly pathway which is initiated as in other retroviruses, they give no hint of how the Pol protein might be incorporated into the particles.
The pathway of HFV reverse transcription also follows the retroviral paradigm in which the initiation at the primer binding site (PBS) requires complex formation with a specific tRNA. The HFV PBS contains 18 nucleotides of perfect homology to the 3′ end of both rat and human tRNA1,2Lys, and there is evidence for synthesis of strong-stop DNA (23, 24). In contrast, the HBV P protein binds in cis to a secondary structure (ɛ) in the genomic RNA (3, 5, 31, 32), an event which initiates both reverse transcription and assembly, processes which are intimately coupled (32). Priming of HBV reverse transcription uses a tyrosine residue on the N-terminal domain of the P protein (37). A HFV Pol deletion mutant still packages RNA (1), demonstrating a clear difference from the assembly pathway of HBV.
The activities of the HFV Pol domains have been studied in vitro. The RT activity from a foamy virus was first demonstrated in 1971 (29), and later template-primer sets for endogenous RT activity were optimized for simian foamy virus 1 (7), and the foamy virus “strain H4188” (26). The HFV RT domain has been expressed in Escherichia coli and shown to have DNA polymerase activity in in situ RT gel assays (23, 24). The RNase H (RH) domain has also been shown to be active (6, 23). An unusual feature of HFV is that Pol is activated before or during viral assembly and release. About 25% of HFV particles contain full-length DNA (40, 42), and experiments with RT inhibitors such as zidovudine are consistent with the fact that DNA is the infectious genome (28, 40). It is unclear exactly when reverse transcription begins. The finding that the RNA genome is packaged by Gag (1) does not rule out the possibility that reverse transcription is initiated early in assembly. Therefore, it is also possible that complex formation between Pol, tRNA, and the PBS is required for packaging Pol protein into virions.
Although HFV Pol contains a protease (PR) domain, the proteolytic processing of Gag and Pol by the HFV PR is different from that in other retroviruses. Only two cleavage events are known to occur (25). The 78-kDa HFV Gag protein is processed once at its C terminus, to release a 4-kDa peptide, an event which occurs in approximately 50% of the Gag precursor molecules. Recent work suggests that this cleavage is required for efficient replication, since mutants which lack cleavage site replicate less well and revert to wild type in culture. Mutants lacking the C-terminal p4 protein can also replicate, but at very low levels (10). The exact Gag cleavage site has been identified biochemically by using recombinant PR (30). PR also cleaves the 127-kDa Pol polyprotein once to release a 45-kDa integrase protein (IN), but the exact site of cleavage between the reverse transcriptase (RT/RH) and IN remains unknown. This cleavage is probably essential, as a PR active site mutant (HFV-D/A) is not replication competent (25). There are no data bearing on the role of proteolytic processing for packaging of Pol proteins into virions.
In this report, we investigate the role of proteolytic cleavage, complex formation of RT at the PBS, and initiation of reverse transcription with respect to their importance for the incorporation of the Pol protein into particles. We demonstrate that neither PR activity nor the PBS is required for Pol assembly.
MATERIALS AND METHODS
Recombinant plasmid DNAs.
A shuttle vector was generated to facilitate cloning and manipulation of five unique restriction-derived fragments from the original molecular clone of HFV, human spumaretrovirus clone 13 (HSRV13) (33). An annealed set of kinase-treated linker oligonucleotides was added to the NEB193 (New England Biolabs) polylinker at the PacI site to generate plasmid HFVLink2 (L2). The insertion destroyed the existing PacI site. These oligonucleotides were Linktop (5′-CGGCCGATTTAAATTAATTAATCCGGAGCTGAGCTTAAGCCTAGGGATATCATGCATAT-3′) and Linkbot (5′-ATGCAT GATATCCCTAGGCTTAAGCTCAGCTCCGGATTAATTAATTTAAATCG GCCGAT-3′). This insert contains all of the unique sites from the viral sequence of HSRV13. The insert was screened for orientation such that the enzyme sites were in the following order with respect to the NEB193 polylinker: BamHI (NEB193), EagI, SwaI, PacI, BspEI, BlpI, AflII, AvrII, EcoRV, NsiI, XbaI (NEB193), SalI (NEB193).
Five unique fragments of the HFV genome from HSRV13 were then cloned into the L2 vector. Each subclone was named according to the position of the unique fragment in the genome. Sub1 contains EagI-SwaI, Sub2 contains SwaI-PacI, Sub3 contains PacI-BspEI, Sub4 contains BspEI-BlpI, and Sub5 contains BlpI-SalI. These subclones were subsequently used for PCR mutagenesis.
PCR mutagenesis.
The general strategy for mutagenesis was as follows. Two oligonucleotides corresponding to sequences outside the NEB193 polylinker were used in all mutagenesis reactions. The external primers were 193(+), corresponding to positions 21 to 40 in NEB193 (5′-GGTGAAACCTCTGACACAT-3′), and 193(−), corresponding to positions 577 to 558 (5′-CCCAGGCTTTACACTTTATG-3′). Internal primers were designed to contain mutations and a unique NheI site for ligation of PCR products. Ligated PCR products were digested with enzymes unique to the L2 polylinker and subcloned into L2.
A protein kinase A (PKA) phosphorylation site was introduced in the C terminus of the IN domain of Pol. The consensus PKA sequence is Arg-Arg-X-Ser-X (RRxSx), where x is preferably a small hydrophobic amino acid (4, 5, 38). Nucleotide positions 6262 to 6265 were changed from ATT to CGT, converting isoleucine to arginine. Positions 6269 to 6275 were changed from ACTTCT to GCTAGC, converting a threonine to alanine such that the amino acid sequence IRTSL was changed to RRASL. The oligonucleotides for PCR were Nhe1top (5′-TTACAGGAACGTCGTGCTAGCTTATACCATCCATCCACCCCTCCAGCC-3′) with 193(−) and Nhe1bot (5′-ATGGTATAAGCTAGCACGACGTTCCTGTAAAAGAGAAAGTTCTTCTTC-3′) with 193(+); 5 ng of Sub3 was used as a template for PCR. PCR products were digested with NheI and ligated to each other. The ligation product was digested with BamHI and SalI and subsequently cloned into BamHI/SalI-digested L2 vector. The PacI/BspEI fragment containing the PKA (Sub3-PKA) site was then cloned back into HSRV13 and named HFV-PKA. The same PacI/BspEI fragment was then cloned into different mutant backgrounds.
The PR active site mutant HFV-D/A contains an aspartic acid-to-alanine mutation at the active site of the viral PR (25). Sub3-PKA was cloned into the D/A background as described above and named HFV-D/A-PKA. The negative control for assembly, ΔATG was generated by digesting the Sub1 vector with MfeI and NcoI, filling in with Klenow, and religating. This was then transferred to the PKA and the D/A-PKA backgrounds, which were called ΔATG-PKA and ΔATG-D/A-PKA, respectively. Three Gag mutants were constructed in a similar fashion, using the Sub2 vector; these are called 78T/A, 74Stop, and 68Stop. For each mutant, two separate PCRs were performed with mutagenic oligonucleotides and the vector-based oligonucleotides 193(+) and 193(−). Both PCR products contained an NheI site in the region downstream of the desired mutation in Gag. PCR products were digested with NheI, ligated together, and redigested with enzymes unique to the polylinker flanking the HSRV13 unique sites. The Sub2 mutants were then cloned back into the viral background with the enzymes SwaI and PacI. The oligonucleotides used for these reactions were 78T/A-1 (5′-AGCCTTGCTAGCCAGAGTGCCACGTCCTCCACAGATC-3′), 78T/A-2 (5′-CAGTTCGCTAGCTGCGGCGACAGCGCGTGAGTCACCAGC-3′), 74STOP-2 (5′-GTACGCTAGCTTACTAATTGACAGCGCGTGAGTCACCAGC-3′), 68STOP-1 (5′-AGCCTTGCTAGCCGCGGAGGAAGAGGTAACCACAACCG-3′), and 68STOP-2 (5′-GTACGCTAGCTTACTAAGCTGGTCTGGGAGTTTGTGACTG-3′). Primer pairs were as follows for the initial reactions: 78T/A, 193(+) plus 78T/A-2 and 193(−) plus 78T/A-1; 74STOP, 193(+) plus 78T/A-2 and 193(−) plus 74STOP-1); 68STOP, 193(+) plus and 68STOP-2 and 193(−) and 68STOP-1. For the STOP mutants, the coding sequence was unaltered prior to the TAA insertion. The cleavage site mutant 78T/A contains four mutations at and near the cleavage site, changing the amino acid sequence from NTVT to AAAS. The amino acids alanine and serine (AS) correspond to the coding sequence of the NheI site (GCTAGC) used to ligate the PCR products. These mutants were generated twice, once in the wild-type HFV-PKA [HFV (wt)] background and once in the D/A-PKA background.
The PBS was deleted from Sub1 and subsequently recloned into various viral backgrounds. The PBS sequence (5′-TGGCGCCCAACGTGGGG-3′; positions 1124 to 1142 of the proviral DNA) was replaced with an NheI site (AGCGCT) by the PCR strategy described above. The oligonucleotides used for the PBS deletion were PBS-NHE-1 (5′-AGTGATGCTAGCCTCGAATATAAGTCGGGTTTATTTG-3′) and PBS-NHE-2 (5′-TCAGATGCTAGCATTGTCATGGAATTTTGTATATTG-3′). Primer sets were 193(+) plus PBS-NHE-2 and 193(−) plus PBS-NHE-1.
Cell culture and transient transfection.
FAB indicator cells expressing β-galactosidase from the HFV long terminal repeat were maintained in Dulbecco’s modified Eagle’s medium supplemented with 5% fetal bovine serum. Transfection and infection efficiencies were measured directly with FAB cells by fixing and staining with a colorometric substrate for β-galactosidase (41). FAB cells stain blue only when the HFV transactivator Tas (present in all proviral constructs) is expressed. Human embryonic lung fibroblasts, COS cells, and 293T cells were maintained in Dulbecco’s modified Eagle’s medium–10% fetal bovine serum. Transfections were performed with Lipofectamine reagent (Gibco-BRL) according to the manufacturers instructions and optimized for our cells and DNA as previously described (1).
Gradient purification of HFV particles.
Transfected cell supernatants were clarified of cell debris by low-speed centrifugation (2,000 rpm; IEC clinical HN-SII tabletop centrifuge) and filtered through a Nalgene 0.45 μm-pore-size syringe filter. Virus was pelleted through standard buffer (50 mM Tris [pH 7.5], 1 mM EDTA, 140 mM NaCl) containing 20% sucrose by ultracentrifugation in an SW28 rotor (Beckman Instruments) at 24,000 rpm for 2 h. Virus was resuspended in standard buffer and placed on a 10 to 40% step gradient of iodixanol (Optiprep; Nycomed Pharma). The 5-ml gradients were formed by sequentially underlaying 1-ml aliquots of increasing concentrations of iodixanol at 10, 20, 30, and 40%. Gradients were centrifuged from 4 to 12 h at 36,000 rpm in a Ti55 rotor (Beckman Instruments). Gradients were made in 5-ml Beckman Ultraclear tubes (13 by 51 mm); 0.7-ml fractions were collected from the top of the gradient, and proteins were precipitated from each by adding trichloroacetic acid (TCA) to a final concentration of 10%. The precipitates were pelleted at 14,000 rpm in a microcentrifuge (Eppendorf), washed once with 10% TCA to remove the iodixanol, and finally washed with acetone to remove the TCA. Pellets were resuspended in Tris-EDTA (TE) containing 1% sodium dodecyl sulfate (SDS) and boiled to solubilize the precipitate; 10% of each fraction was then analyzed by Western blotting for viral Gag protein, using polyclonal anti-Gag antibody derived from the central (capsid) domain of Gag (1), and the remaining 90% was diluted 1:10 in TE (pH 8.0) (0.1%, final concentration) for immunoprecipitation (IP)-PKA analysis of the Pol proteins (see below).
Protein kinase assay for detection of Pol.
The catalytic subunit of PKA (Sigma) was used to phosphorylate viral and cellular Pol proteins containing the recognition sequence RRxSx (4, 5, 38). For all figures in this report, Pol proteins were immunoprecipitated from cell lysates or viral pellets by using polyclonal anti-RH rabbit serum (1). Pol expression was also verified by two independent criteria. The same Pol proteins were detected by IP-kinase reactions using either anti-IN serum (Martin Löchelt, Heidelberg, Germany) or anti-RH serum. In addition, radioimmunoprecipitation with the anti-RH serum as previously described (1) (data not shown) verified that the 127-kDa band seen in the IP-kinase assays was in fact the Pol protein. Cellular Pol protein was immunoprecipitated from one transfected plate of FAB cells. Lysates for IP were prepared by resuspending cells or virus in antibody buffer (20 mM Tris [pH 7.5], 50 mM NaCl, 0.5% Nonidet P-40 [NP-40], 0.5% SDS, 0.5% sodium deoxycholate [DOC], 0.5% aprotinin), cellular nucleic acids were sheared with a 23-gauge needle, and insoluble materials were pelleted and discarded; 2 to 4 μl of antiserum was added to the lysate and vortexed. The immune complexes were precipitated with protein A-Sepharose for 3 h at 10°C, washed twice with high-stringency radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris [pH 7.4], 150 mM NaCl, 1% NP-40, 1% DOC, 0.1% SDS, 0.5% aprotinin), washed once with high-salt buffer (10 mM Tris [pH 7.4], 2 M NaCl, 1% NP-40, 1% DOC), and finally washed with 1× PKA buffer (20 mM Tris [pH 7.5], 100 mM NaCl, 12 mM MgCl2, 4 mM dithiothreitol). PKA was added in 1× PKA buffer (20 U/reaction) in the presence of 10 to 25 μCi of [γ-32P or 33P]ATP. Reactions were carried out at 37°C for 30 to 60 min. The complex was then washed twice with RIPA buffer to remove most of the unincorporated label. A second IP was performed by boiling the complex in TE–1% SDS, removing the supernatant, and diluting it 1:10 in TE (pH 8.0). Fresh anti-Pol antiserum and protein A-Sepharose were added for 3 h. This complex was again washed three times with RIPA buffer and once with TE. Then 1× SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer was added directly, and the samples were boiled for 5 to 10 min. Proteins were separated by SDS-PAGE, dried onto Whatman 3MM, and exposed to film or a PhosphorImager screen.
RESULTS
Proteolytic processing of Gag or Pol is not required for assembly of Pol into HFV particles.
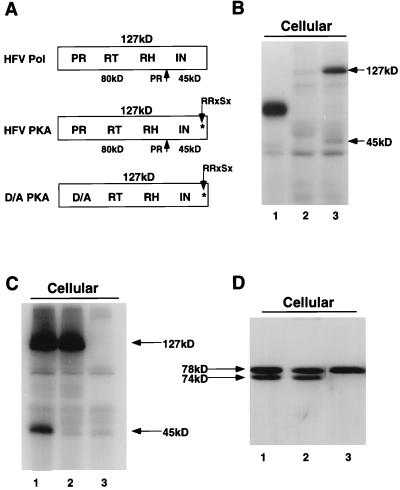
To better understand the role of the viral PR during assembly, we wanted to test whether the HFV PR active site mutant (HFV-D/A) was capable of assembling Pol into virions. However there are inherent difficulties in detecting the small amount of Pol protein present in virions from transiently transfected cells. Since traditional methods such as Western blot analysis and radioimmunoprecipitation analysis failed to convincingly detect Pol, we introduced a consensus recognition sequence for the catalytic subunit of PKA (HFV-PKA) into the pol gene. This method had previously been used to detect the HBV P protein (4, 5). The PKA site was introduced near the C terminus of IN via site-directed PCR mutagenesis, by changing the amino acid sequence IRTSL to RRASL (Fig. 1A). Using antibodies raised against the RH domain of Pol, we were able to immunoprecipitate and specifically phosphorylate the 127-kDa Pol precursor from transfected cells (Fig. 1B, lane 3; Fig. 1C, lane 1). Although we used anti-RH serum, we also detected the cleaved form of IN (45 kDa) by SDS-PAGE. This suggests that either cleavage can occur in vitro after IP or that the complex between the cleaved Pol proteins is stable under the conditions used for the IP. Interactions between RT and IN have been demonstrated for other retroviral Pol proteins (19, 39). In the wild-type HFV-transfected cells (Fig. 1B, lane 2), no specific bands were seen, indicating that phosphorylation of Pol requires the engineered PKA site. Analysis of the Gag proteins demonstrated that cleavage of Gag by the HFV-PKA Pol is similar to that seen for HFV (wt) (Fig. 1D, lanes 1 and 2). While the expression and processing of Gag and Pol appear normal for HFV-PKA, infectivity was reduced 1,000-fold with respect to HFV (wt) (data not shown). We presume but have not established that this difference is due to a defect in integration. Since all of our analyses are done during the first round of replication, before reinfection can occur, the integration status is not important.
FIG. 1.
IP-PKA and Western blot analysis of HFV-PKA and HFV-D/A-PKA. Assay conditions are as described in Materials and Methods except that the second IP step was omitted for PKA analysis of cellular Pol proteins. Anti-RH antiserum was used to immunoprecipitate Pol, and anticapsid antiserum was used for Gag Western blots. (A) Schematic diagram of HFV (wt) and HFV-PKA Pol proteins. (B) IP-PKA analysis of HFV-PKA. Lanes: 1, positive control for PKA phosphorylation; purified interleukin-1 receptor (38); 2, HFV (wt)-transfected cells; 3, HFV-PKA-transfected cells. (C) PKA analysis of cell lysates mock transfected (lane 3) or transfected with HFV-PKA and HFV-D/A-PKA (lanes 1 and 2, respectively). (D) Western blot analysis of HFV-PKA Gag proteins. Lanes: 1, HFV (wt); 2, HFV-PKA; 3, HFV-D/A-PKA.
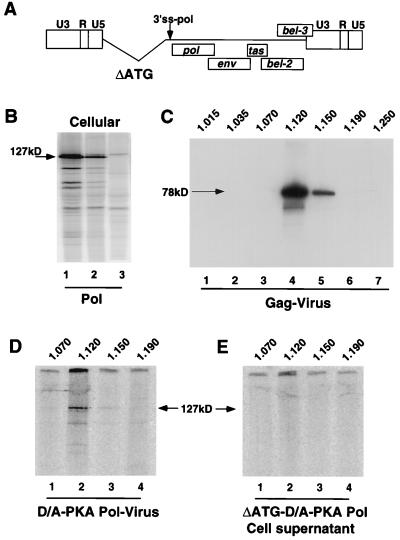
We next subcloned the PKA mutation into the PR active site mutant background, HFV-D/A, in order to study the role of proteolysis during assembly. As expected for HFV-D/A-PKA, we no longer detected cleavage of either Pol (by the IP-kinase assay [Fig. 1C, lane 2]) or Gag (by Western blot analysis [Fig. 1D, lane 3]). As a negative control for virus assembly, we constructed a mutant with a deletion spanning the initiation codon for Gag. This mutant, ΔATG-D/A-PKA (Fig. 2A), contains a deletion of 1,240 bases beginning at position 1120 of the proviral sequence and ending at position 2368, upstream of the 3′ splice site for pol. ΔATG-D/A-PKA makes no detectable Gag by Western blot analysis (Fig. 3C, lane 1) but regularly expresses Pol at higher levels than Gag-expressing proviral constructs such as HFV-D/A-PKA (Fig. 2B, lanes 1 and 2) when transfection efficiencies are comparable as measured by β-galactosidase activity in FAB cells.
FIG. 2.
Optiprep gradient purification and analysis of Gag and Pol proteins from HFV-D/A-PKA virus particles. Gradients and analyses were performed as described in Materials and Methods. (A) Schematic of negative control for virus assembly and Pol incorporation, ΔATG-PKA. (B) IP-PKA analysis of cell-associated Pol proteins from ΔATG-PKA and HFV-D/A-PKA, using anti-RH serum. Cells were transfected with ΔATG-D/A-PKA (lane 1) or HFV-D/A-PKA (lane 2) or mock transfected (lane 3). (C) Western blot analysis of purified HFV-D/A-PKA particles, using anti-Gag antiserum. Fraction densities (in grams per cubic centimeter) are listed above the lanes. Lanes 1 to 7 correspond to fractions 1 to 7 from the gradient; 78 kDa is the expected size for unprocessed Gag from HFV-D/A-expressing constructs (Fig. 3A). (D) IP-PKA analysis of gradient-purified HFV-D/A-PKA virus particles. Lanes 1 to 4 correspond to fractions 3 to 6 from the gradient. Fraction densities (in grams per cubic centimeter) are listed above the lanes. (E) IP-PKA analysis of gradient fractions from cell supernatants of the negative control for virus assembly, ΔATG-D/A-PKA. Lanes 1 to 4 correspond to fractions 3 to 6.
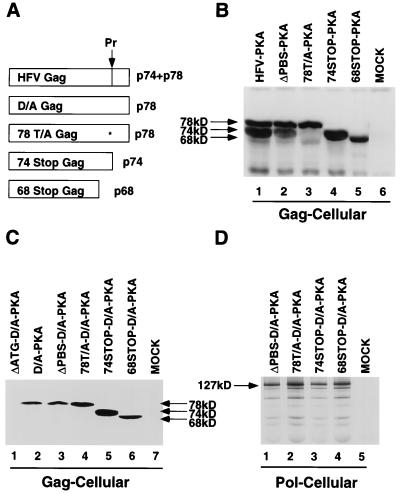
FIG. 3.
Analysis of cellular Gag and Pol expression from HFV-PKA and HFV-D/A-PKA mutants. (A) Schematic of expected Gag protein products from mutant proviral constructs. (B) Western blot analysis of Gag proteins from cells mock transfected (lane 6) or transfected with the constructs indicated at the top. (C) Western blot analysis of Gag proteins from cells mock transfected (lane 7) or transfected with the HFV-D/A-PKA-expressing constructs indicated at the top. (D) IP-PKA analysis of cellular Pol proteins. Shown are the mutants in the D/A-PKA background which were tested for Pol incorporation into virus particles. These experiments included a second IP step after the phosphorylation reaction.
We then purified HFV-D/A-PKA virus particles of the correct density from cell supernatants to determine whether Pol is encapsidated. To this end, viral pellets were resuspended and centrifuged to density equilibrium on gradients of iodixanol (Optiprep). Iodixanol gradients have previously been used to study the incorporation of Vif into human immunodeficiency virus type 1 virions (9). We found that HFV-D/A-PKA particles sedimented at the appropriate density of 1.12 to 1.15 g/cm3 (Fig. 2C, lanes 4 and 5) as detected by Gag Western blot analysis of the gradient fractions. When we analyzed the same fractions for the presence of Pol, we were able to detect HFV-D/A-PKA Pol cosedimenting with Gag (Fig. 2D, lane 2). Importantly, extracellular Pol was not detected in gradient fractions of comparable density from cells expressing Pol but no Gag (Fig. 2E). Thus, we conclude that detection of Pol in fractions at 1.12 to 1.15 g/cm3 requires particle formation. Taken together, these data indicate that proteolytic activity is not necessary for the specific incorporation of HFV Pol into virions.
The primary structure at the cleavage site in Gag is not important for Pol incorporation into particles.
We were interested in determining whether the binding event which initiates the single cleavage of Gag by PR might be responsible for recruiting Pol into particles. We tested several mutations at or near the cleavage site (Fig. 3A). The Gag cleavage site was mutated to block cleavage (78T/A); two Gag truncation mutants, 74Stop (truncated exactly at the cleavage site) and 68Stop (truncated 25 amino acid residues upstream of the cleavage site) were also generated. We introduced these mutations into the HFV-PKA (wtPR) background. We found that Gag proteins of the expected sizes were expressed by all of these clones after transfection (Fig. 3B, lanes 3 to 5).
Detection and quantitation of HFV-PKA Pol in virions proved difficult due to HFV PR activity either before or during the IP-kinase assay. Although the signal-to-noise ratio was greatly improved by the use of a second IP after the IP-kinase reaction, cleaved forms of IN might be lost during the second IP, preventing quantitative comparison of the Gag mutants to the PR active site mutant. As PR activity was not required for Pol incorporation, the same Gag mutations were generated in the D/A-PKA background. When the resultant mutant viruses were analyzed by Western blotting, Gag proteins of the correct size were detected (Fig. 3C, lanes 4 to 6). No Gag was synthesized by the Gag deletion mutant, ΔATG-D/A-PKA (Fig. 3C, lane 1). In the IP-kinase assay, Pol proteins were expressed at similar levels in cell extracts after transfection with the Gag mutant constructs (Fig. 3D, lanes 2 to 4).
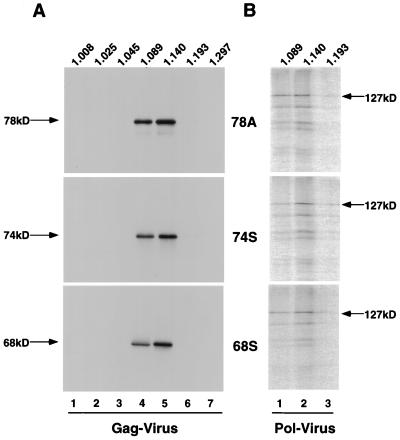
Virus particles produced after transfection with the Gag mutants were purified on iodixanol gradients and probed for Gag by Western blotting (Fig. 4A, lanes 4 and 5). All of the mutants analyzed sedimented at a density similar to that of the D/A-PKA parental virus (Fig. 2B). When the corresponding fractions were analyzed for Pol, we were able to detect Pol in each of the Gag mutants (Fig. 4B, lanes 1 and 2). No Pol could be detected in other fractions of the gradient (data not shown). These results demonstrate that while the cleavage of Gag seems to be an important step in the replication cycle of HFV (10), neither the Gag cleavage site itself nor the primary sequence immediately surrounding it is required for incorporation of Pol into virus particles.
FIG. 4.
Gradient purification and analysis of Gag mutant viruses. (A) Western blot analysis of gradient fractions for viral Gag. Lanes 1 to 7 correspond to gradient fractions 1 to 7. (B) IP-PKA analysis of fractions 4 to 6 from the gradients shown in panel A. Lanes 1 to 3 correspond to fractions 4 to 6. 78A, 78T/A; 74S, 74Stop; 68S, 68Stop (see Fig. 3B for schematic). Fraction densities (in grams per cubic centimeter) are listed above the lanes.
The PBS is not essential for Pol incorporation.
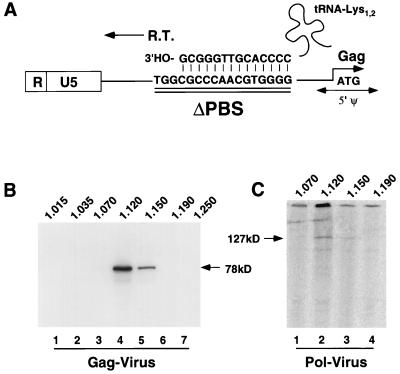
Binding of Pol to the PBS occurs at an early stage of HFV assembly, and it is possible that this step is responsible for its incorporation into particles. Such a scenario would be similar to what is found for HBV, where P protein binds to ɛ and initiates both reverse transcription and assembly (3, 5, 31, 32). Since HFV Pol is active during assembly, rather than at an early stage after infection, the mechanism of HFV Pol assembly could involve a specific stage of reverse transcription. We deleted the PBS by PCR mutagenesis (Fig. 5A). In the ΔPBS constructs, Gag was expressed as expected. Gag was processed in the HFV-PKA background (Fig. 3B, lane 2) and not processed in the D/A-PKA background (Fig. 3C, lane 3). ΔPBS-D/A-PKA Pol was expressed in transfected cells at levels similar to those of other D/A-PKA-expressing mutants (Fig. 3D, lane 1). Gradient-purified particles from the ΔPBS-D/A-PKA-transfected cells sedimented at 1.12 to 1.15 g/cm3, as seen for other mutant viruses (Fig. 5B, lanes 4 and 5), and Pol was detected in the same fractions (Fig. 5C, lanes 2 and 3), demonstrating that Pol was indeed present in these particles. Thus, the PBS is not required for Pol assembly.
FIG. 5.
Gradient purification and analysis of ΔPBS-D/A-PKA. (A) Detailed schematic of the PBS deletion on the RNA genome. Shown is the homology with the 3′ end of tRNA1,2Lys from which reverse transcription is initiated. Deleted nucleotides are underlined; a putative 5′ encapsidation signal is labeled Ψ. (B) Western blot analysis of Gag. Lanes 1 to 7 correspond to fractions 1 to 7. Fraction densities (in grams per cubic centimeter) are listed above the lanes. (C) IP-PKA analysis of Pol proteins. Lanes 1 to 4 correspond to fractions 3 to 6 shown in panel B.
DISCUSSION
During the course of these studies on the mechanisms of Pol expression and assembly, we noticed that HFV Pol expression is quite low, both at the mRNA level (40) and at the protein level (1, 2). We were unable to detect Pol in virus particles without radiolabeling in vitro. The regulation of Gag and Pol stoichiometry is an important aspect of retroviral replication. Conventional retroviruses have evolved sophisticated mechanisms to regulate both Pol expression and activation (reviewed in references 8 and 21). Synthesis of Pol as a Gag-Pol fusion protein keeps the level of Pol protein low, allows incorporation of Pol into particles via Gag domains, and prevents activation of Pol until a subsequent round of infection. It could be detrimental to the host cell to have active RT in the cytoplasm where mRNA substrates are abundant. In the case of conventional retroviruses, overexpression of Gag-Pol and the resulting activation of PR in the cytoplasm has been shown to negatively affect virus assembly (22). While on average, retroviruses contain two copies of their genomic RNA per virion, they contain one Gag-Pol fusion for every 20 Gag molecules (21), or about 100 Pol proteins per virion (36). Hepadnaviruses have a much lower ratio of P protein to core protein per virion. Since P binds to the ɛ signal in the RNA, there are only one or two P proteins per virion (5). It is not known how many Pol molecules there are per HFV particle. If Pol-RNA interactions are required, then the ratio of Pol to Gag in virions could be as low as one to two Pol proteins per virion, which is consistent with difficulty in detecting Pol in particles. However, even if Gag-Pol interactions are responsible for assembly, low levels of Pol synthesis could be the limiting factor. Quantitative analyses of the ratio of Gag and Pol in cells and particles have not yet been done.
The mechanisms for regulation of expression and activation of HFV Pol are not known. Splicing to generate pol mRNA is one step where regulation could occur (11, 27, 40). Translation of the spliced mRNA may also be regulated, as the pol gene contains a very long 5′ untranslated region. We were unable to get Pol expression from cytomegalovirus promoter constructs lacking the bona fide splice junction (data not shown), consistent with a role for the untranslated region in protein expression. It has recently been shown for HBV that the dicistronic message which results in P protein expression is regulated at the translational level (20). The ΔATG mutant, however, leads to higher levels of Pol expression than Gag-expressing proviruses (Fig. 2C; compare lanes 1 and 2). Perhaps for the ΔATG mutant, Pol can be translated from both spliced and unspliced mRNAs, whereas for wild-type HFV, gag translation downmodulates pol expression during viral replication. Another outstanding question is how Pol is activated. Our data suggest (Fig. 1 and 2) that PR activity is not important for Pol incorporation, but how are the activities of PR and RT temporally controlled? It is likely that cleavage of Pol plays a role in activation, a step which appears to be initiated after Pol incorporation. This is the case for other retroviruses where RT activity is dependent on cleavage by PR during virion maturation. For avian leukosis virus, Gag-Pol precursors have very little RT activity until processed by PR in trans after assembly and during maturation (34). While the mechanisms regulating retroviral maturation remain a mystery, the regulation of RT activation is clearly an important issue to both the virus and its host. Perhaps a molecular chaperone, such as one of the Hsp family members, sequesters the HFV Pol protein until it can find its binding partners. In the case of HBV RT, the chaperone Hsp90 binds to and is essential for the activity of the enzyme during assembly (16–18).
In this study, we examined the mechanism of Pol assembly in HFV. We have considered both Pol-protein and Pol-RNA interactions, as well as the role of proteolysis. We have found that neither the activity of the viral PR nor the PBS is required for assembly of Pol into particles. Our studies to date, therefore, do not answer the question of whether Pol incorporation occurs through protein-protein or protein-RNA interactions. If protein-protein interactions are important, the PR domain remains a likely candidate. Delineation of a region in Gag critical for Pol incorporation is an important next step.
If Pol-RNA interactions are important, the region of the aHFV genome located at the 3′ end of the pol gene, which has been reported to be critical for HFV vector transfer (12, 15), is a good candidate. While the role that this genome region plays in vector transfer is not known, it might contain sequence information or secondary structure which directs Pol binding. If the pol region of the genome is important for Pol assembly, the spliced pol mRNA might interfere, but this mRNA should not be packaged since it does not contain the putative Ψ region required for vector transfer. Alternatively, the presence of ribosomes on the pol message might block the ability of the RNA to form the appropriate secondary structure for Pol recognition. Additionally, posttranslational translocation of Pol protein would also be required. Pol proteins would then have a much higher probability of encountering the abundant viral genome than the spliced pol message in the cytoplasm. If Pol-RNA interactions are important for assembly, then genome dimerization could also play a role in secondary structure formation and hence Pol assembly. Dimerization of Pol proteins and viral genomes might even act in concert to assemble Pol and RNA. It is also possible that Env plays a role in the assembly of Pol since Gag-Env interactions are required for very late stages of assembly (1).
While these studies do not reveal the actual mechanism of Pol assembly, they have ruled out two essential processes in the replication pathway. In the future, it will be important to study domains of Gag and Pol and to look at the effects of uncoupling RNA packaging and Pol incorporation in Gag mutants. Delineation of a mechanism could come from Gag mutants lacking only one of these functions.
ACKNOWLEDGMENTS
D.N.B. was supported by Public Health Service National Research Service Award T32 GM07270 from the National Institute of General Medical Sciences. This investigation was also supported by grant CA18282 from the National Cancer Institute to M.L.L.
We thank Christopher Meiering for thoughtful discussions and help with the design and implementation of subcloning and mutagenesis strategies. We thank Michael Emerman (FHCRC) for valuable discussion and critical reading of the manuscript, Markus Karl Dettenhofer (Johns Hopkins University, Baltimore, Md.) for input regarding the Optiprep reagents, and Martin Löchelt and Rolf Flügel (Heidelberg, Germany) for kindly providing IN antiserum.
REFERENCES
- 1.Baldwin D N, Linial M L. The roles of Pol and Env in the assembly pathway of human foamy virus. J Virol. 1998;72:3658–3665. doi: 10.1128/jvi.72.5.3658-3665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin, D. N., and M. L. Linial. 1998. Unpublished results.
- 3.Bartenschlager R, Junker-Niepmann M, Schaller H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol. 1990;64:5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager R, Kuhn C, Schaller H. Expression of the P-protein of the human hepatitis B virus in a vaccinia virus system and detection of the nucleocapsid-associated P-gene product by radiolabelling at newly introduced phosphorylation sites. Nucleic Acids Res. 1992;20:195–202. doi: 10.1093/nar/20.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartenschlager R, Schaller H. Hepadnavirus assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992;11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benzair A B, Rhodes-Feuillette A, Emanoil-Ravicovitch R, Peries J. Characterization of RNase H activity associated with reverse transcriptase in simian foamy virus type 1. J Virol. 1983;47:249–252. doi: 10.1128/jvi.47.1.249-252.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benzair A B, Rhodes-Feuillette A, Emanoil-Ravicovitch R, Peries J. Reverse transcriptase from simian foamy virus serotype 1: purification and characterization. J Virol. 1982;44:720–724. doi: 10.1128/jvi.44.2.720-724.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craven R C, Bennett R P, Wills J W. Role of the avian retroviral protease in the activation of reverse transcriptase during virion assembly. J Virol. 1991;65:6205–6217. doi: 10.1128/jvi.65.11.6205-6217.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dettenhofer M, Yu X-F. Highly purified human immunodeficiency virus type I reveals a virtual absence of Vif in virions. J Virol. 1999;73:1460–1467. doi: 10.1128/jvi.73.2.1460-1467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enssle J, Fischer N, Moebes A, Mauer B, Smola U, Rethwilm A. Carboxy-terminal cleavage of the human foamy virus Gag precursor molecule is an essential step in the viral life cycle. J Virol. 1997;71:7312–7317. doi: 10.1128/jvi.71.10.7312-7317.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enssle J, Jordan I, Mauer B, Rethwilm A. Foamy virus reverse transcriptase is expressed independently from the Gag protein. Proc Natl Acad Sci USA. 1996;93:4137–4141. doi: 10.1073/pnas.93.9.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erlwein O, Bieniasz P D, McClure M O. Sequences in pol are required for transfer of human foamy virus-based vectors. J Virol. 1998;72:5510–5516. doi: 10.1128/jvi.72.7.5510-5516.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flügel R M. Spumaviruses: a group of complex retroviruses. J Acquired Immune Defic Syndr Hum Retrovirol. 1991;4:739–750. [PubMed] [Google Scholar]
- 14.Hatfield D L, Levin J G, Rein A, Oroszlan S. Translation suppression in retroviral gene expression. Adv Virus Res. 1992;41:193–239. doi: 10.1016/S0065-3527(08)60037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinkelein M, Schmidt M, Fischer N, Moebes A, Lindemann D, Enssle J, Rethwilm A. Characterization of a cis-acting sequence in the Pol region required to transfer human foamy virus vectors. J Virol. 1998;72:6307–6314. doi: 10.1128/jvi.72.8.6307-6314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu J, Seeger C. Expression and characterization of hepadnavirus reverse transcriptases. Methods Enzymol. 1996;275:195–208. doi: 10.1016/s0076-6879(96)75013-9. [DOI] [PubMed] [Google Scholar]
- 17.Hu J, Seeger C. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc Natl Acad Sci USA. 1996;93:1060–1064. doi: 10.1073/pnas.93.3.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu J, Toft D O, Seeger C. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 1997;16:59–68. doi: 10.1093/emboj/16.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu S C, Court D L, Zweig M, Levin J G. Murine leukemia virus pol gene products: analysis with antisera generated against reverse transcriptase and endonuclease fusion proteins expressed in Escherichia coli. J Virol. 1986;60:267–274. doi: 10.1128/jvi.60.1.267-274.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang W L, Su T S. Translational regulation of hepatitis B virus polymerase gene by termination-reinitiation of an upstream minicistron in a length-dependent manner. J Gen Virol. 1998;79:2181–2189. doi: 10.1099/0022-1317-79-9-2181. [DOI] [PubMed] [Google Scholar]
- 21.Jacks T. Translational suppression in gene expression in retroviruses and retrotransposons. In: Swanstrom R, Vogt P K, editors. Retroviruses; strategies of replication. 1st ed. Berlin, Germany: Springer-Verlag; 1990. pp. 93–124. [DOI] [PubMed] [Google Scholar]
- 22.Karacostas V, Wolffe E J, Nagashima K, Gonda M A, Moss B. Overexpression of the HIV-1 gag-pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology. 1993;193:661–671. doi: 10.1006/viro.1993.1174. [DOI] [PubMed] [Google Scholar]
- 23.Kögel D, Aboud M, Flügel R M. Molecular biological characterization of the human foamy virus reverse transcriptase and ribonuclease H domains. Virology. 1995;213:97–108. doi: 10.1006/viro.1995.1550. [DOI] [PubMed] [Google Scholar]
- 24.Kögel D, Aboud M, Flügel R M. Mutational analysis of the reverse transcriptase and ribonuclease H domains of the human foamy virus. Nucleic Acids Res. 1995;23:2621–2625. doi: 10.1093/nar/23.14.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konvalinka J, Löchelt M, Zentgraf H, Flügel R M, Kräusslich H G. Active foamy virus proteinase is essential for virus infectivity but not for formation of a Pol polyprotein. J Virol. 1995;69:7264–7268. doi: 10.1128/jvi.69.11.7264-7268.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W T, Natori T, Chang K S, Wu A M. Reverse transcriptase of foamy virus. Purification of the enzymes and immunological identification. Arch Virol. 1977;55:187–200. doi: 10.1007/BF01319905. [DOI] [PubMed] [Google Scholar]
- 27.Löchelt M, Flügel R M. The human foamy virus pol gene is expressed as a Pro-Pol polyprotein and not as a Gag-Pol fusion protein. J Virol. 1996;70:1033–1040. doi: 10.1128/jvi.70.2.1033-1040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moebes A, Enssle J, Bieniasz P D, Heinkelein M, Lindemann D, Bock M, McClure M O, Rethwilm A. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J Virol. 1997;71:7305–7311. doi: 10.1128/jvi.71.10.7305-7311.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parks W P, Todaro G J, Scolnick E M, Aaronson S A. RNA dependent DNA polymerase in primate syncytium-forming (foamy) viruses. Nature. 1971;229:258–260. doi: 10.1038/229258a0. [DOI] [PubMed] [Google Scholar]
- 30.Pfrepper K I, Rackwitz H R, Schnölzer M, Heid H, Löchelt M, Flügel R M. Molecular characterization of proteolytic processing of the Pol proteins of human foamy virus reveals novel features of the viral protease. J Virol. 1998;72:7648–7652. doi: 10.1128/jvi.72.9.7648-7652.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollack J R, Ganem D. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J Virol. 1993;67:3254–3263. doi: 10.1128/jvi.67.6.3254-3263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollack J R, Ganem D. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J Virol. 1994;68:5579–5587. doi: 10.1128/jvi.68.9.5579-5587.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rethwilm A, Darai G, Rosen A, Maurer B, Flügel R M. Molecular cloning of the genome of human spumaretrovirus. Gene. 1987;59:19–28. doi: 10.1016/0378-1119(87)90262-9. [DOI] [PubMed] [Google Scholar]
- 34.Stewart L, Vogt V M. trans-acting viral protease is necessary and sufficient for activation of avian leukosis virus reverse transcriptase. J Virol. 1991;65:6218–6231. doi: 10.1128/jvi.65.11.6218-6231.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swanstrom R, Wills J W. Synthesis, assembly, and processing of viral proteins. In: Coffin J, Hughes S, Varmus H, editors. Retroviruses. 1st ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 36.Vogt V M, Simon M N. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy. J Virol. 1999;73:7050–7055. doi: 10.1128/jvi.73.8.7050-7055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang G H, Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. J Virol. 1993;67:6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitehorn E A, Tate E, Yanofsky S D, Kochersperger L, Davis A, Mortensen R B, Yonkovich S, Bell K, Dower B, Barrett R W. A generic method for expression and use of “tagged” soluble versions of cell surface receptors. Bio/Technology. 1995;13:1215–1219. doi: 10.1038/nbt1195-1215. [DOI] [PubMed] [Google Scholar]
- 39.Wu X, Liu H, Xiao H, Conway J A, Hehl E, Kalpana G V, Prasad V, Kappes J C. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J Virol. 1999;73:2126–2135. doi: 10.1128/jvi.73.3.2126-2135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu S F, Baldwin D N, Gwynn S R, Yendapalli S, Linial M L. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science. 1996;271:1579–1582. doi: 10.1126/science.271.5255.1579. [DOI] [PubMed] [Google Scholar]
- 41.Yu S F, Linial M L. Analysis of the role of the bel and bet open reading frames of human foamy virus by using a new quantitative assay. J Virol. 1993;67:6618–6624. doi: 10.1128/jvi.67.11.6618-6624.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu S F, Sullivan M D, Linial M L. Evidence that the foamy virus genome is DNA. J Virol. 1999;73:1565–1672. doi: 10.1128/jvi.73.2.1565-1572.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]