Abstract
The Serp2 protein encoded by the leporipoxvirus myxoma virus is essential for full virulence (F. Messud-Petit, J. Gelfi, M. Delverdier, M. F. Amardeilh, R. Py, G. Sutter, and S. Bertagnoli, J. Virol. 72:7830–7839, 1998) and, like crmA of cowpox virus (CPV), is reported to inhibit the interleukin-1β-converting enzyme (ICE, caspase-1) (F. Petit, S. Bertagnoli, J. Gelfi, F. Fassy, C. Boucraut-Baralon, and A. Milon, J. Virol. 70:5860–5866, 1996). Serp2 and CrmA both contain Asp at the P1 position within the serpin reactive site loop and yet are only 35% identical overall. Serp2 protein was cleaved by ICE but, unlike CrmA, did not form a stable complex with ICE that was detectable by native gel electrophoresis. Attempts to covalently cross-link ICE-serpin inhibitory complexes were successful with CrmA, but no complex between ICE and Serp2 was visible after cross-linking. Purified His10-tagged Serp2 protein was a relatively poor inhibitor of ICE, with a Ki of 80 nM compared to 4 pM for CrmA. Serp2 protein resembled CrmA in that a stable complex with the serine proteinase granzyme B was detectable after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. However, Serp2 was less effective at inhibiting granzyme B activity (Ki = 420 nM) than CrmA (Ki = 100 nM). Finally, Serp2 was tested for the ability to replace CrmA and inhibit apoptosis in LLC-PK1 cells infected with a CPV recombinant deleted for CrmA but expressing Serp2. Unlike wild-type-CPV-infected cells, apoptosis was readily observed in cells infected with the recombinant virus, as indicated by the induction of both nuclear fragmentation and caspase-mediated cleavage of DEVD-AMC [acetyl-Asp-Glu-Val-Asp-(amino-4-methyl coumarin)]. These results indicate that Serp2 is unable to functionally substitute for CrmA within the context of CPV and that the inhibition spectra for Serp2 and CrmA are distinct.
Myxoma virus (MYX) is the causative agent of myxomatosis in the European rabbit Oryctolagus cuniculus. This disease is almost invariably lethal and involves fulminating lesions and immunosuppression leading to severe gram-negative bacterial secondary infections of the respiratory tract (26). However, MYX exists in a nonpathogenic symbiotic relationship with its natural host, the South American rabbit (Sylvilagus sp.), and other leporipoxviruses, such as the Shope fibroma virus, induce only a mild disease. Many of the genes involved in the virulence, pathogenesis, and host range of poxviruses are nonessential for growth in cell culture and typically reside at either end of the linear viral genome, outside the central conserved core of genes devoted to housekeeping functions (29). Examples of such genes include those which subvert the host immune response by interfering with cytokine action (27, 36, 42). Leporipoxviruses, such as MYX, encode secreted receptors for tumor necrosis factor alpha (TNF-α) (gene T2), gamma interferon (IFN-γ) (gene T7) (27), and for chemokines (gene T1 [14] and gene T7 [18]). Orthopoxviruses such as cowpox virus (CPV) and vaccinia virus (VV) express a secreted receptor for interleukin-1β (IL-1β) (VV open reading frame [ORF] B15R) and a complement control protein homolog (ORF C21L), in addition to other immune modulators, including a variety of soluble cytokine receptors (viroceptors) and a cytokine mimic (virokine) which belongs to the epidermal growth factor superfamily.
Many genera of poxviruses also encode serpins (serine proteinase inhibitors), some of which inhibit inflammation by interfering both with the processing of cellular cytokines from precursors and with apoptosis (53). Typical of serpins, the P1 residue of a given serpin is located within the reactive site loop (RSL) close to the C terminus. It is this residue which largely determines proteinase specificity (38). The prototypic poxvirus serpin is the cowpox virus CrmA protein (37), which has aspartic acid at the P1 position in the RSL, consistent with the ability of CrmA to inhibit caspases (cysteine proteinases which cleave after aspartic acid) and granzyme B. ICE is the prototypic member of the family of caspases that in mammals has at least thirteen members (3, 51). CrmA serves to regulate inflammation by blocking the proteolytic activation of the precursor proIL-1β by inhibition of IL-1β converting enzyme (ICE; caspase-1) (40). CrmA additionally inhibits caspase-8 (FLICE) (43, 45, 57), a pro-apoptotic proteinase thought to be at the apex of the proteolytic cascade which is activated after signaling via engagement of the Fas or TNF receptors. The CrmA–caspase-8 interaction probably accounts for the antiapoptotic activity of CrmA that has been demonstrated in several heterologous systems (for a review, see reference 10) and in cowpox virus infection of certain cells (20, 41). The CrmA protein also inhibits the serine proteinase granzyme B (39), a major component of the granules of cytotoxic T lymphocytes and natural killer (NK) cells that is an aspase associated with programmed cell death.
MYX encodes a serpin named Serp1 that has arginine at the P1 position of the RSL (55). The Serp1 protein is a secreted glycoprotein that has anti-inflammatory activity both within the context of MYX infection of rabbits (22) and as an exogenous protein in animal models of restenosis (19) and arthritis (23). MYX strain T1 contains a second serpin gene at the right end of the genome known as Serp2 (35). The Serp2 protein is synthesized throughout the virus infection and, unlike Serp1, is intracellular. Although the Serp2 protein is not significantly more related to CrmA in terms of overall sequence identity than it is to other members of the serpin superfamily, the P1 residue in the Serp2 RSL is aspartic acid, suggesting that Serp2, like CrmA, may inhibit caspases. When the Serp2 protein was overexpressed by means of a baculovirus system, extracts containing Serp2 were reported to be able to inhibit ICE-mediated cleavage of a fluorogenic “ICE-like” peptide substrate and activation of in vitro translated proIL-1β when compared with control extracts lacking Serp2 (35). A complex between Serp2 derived from recombinant baculovirus-infected cell extracts and ICE was visualized by native polyacrylamide gel electrophoresis (PAGE) and immunoblotting (35). Serp2 is required for the occurrence of the full symptoms of myxomatosis in infected rabbits, as a MYX serp2 mutant was strongly attenuated compared with wild-type MYX, giving 30% mortality compared with 100% lethality for wild-type MYX (wtMYX) (28).
We have examined the Serp2 protein in comparison with CrmA. Serp2 from the Lausanne strain of MYX was expressed by coupled transcription-translation in vitro in order to study the interactions of Serp2 with ICE in terms of cleavage and complex formation. Also, a purified His10-tagged derivative of Serp2 was examined for its ability to inhibit human ICE, human caspase-2 through caspase-9, and granzyme B. Our results indicate that, like CrmA, Serp2 inhibits both ICE and granzyme B. However, in contrast to crmA, the inhibition of ICE and granzyme B by Serp2 was relatively weak, raising the question as to whether these proteinases are likely to be natural targets for Serp2 in vivo.
MATERIALS AND METHODS
Viruses and cells.
The Lausanne strain of myxoma virus was obtained from Grant McFadden (Robarts Research Institute, London, Ontario, Canada) and was propagated on RK-13 rabbit kidney cells (ATCC CCL-37).
Plasmids and nucleotide sequencing.
A genomic clone of the serp2 gene from MYX strain T1 was provided by Frederique Petit (INRA-ENVT, Toulouse, France). pGEM-5Zf(+)/serp2 was constructed as follows. The MYX serp2 gene was PCR amplified from genomic DNA of strain Lausanne by using Vent polymerase (New England Biolabs) with primers RM541 (5′-GCGACCATGGAGCTTTTCAA GCATTTC-3′) at the 5′ end of the ORF containing an added NcoI site (underlined) and RM542 (5′-GCCCTCGAGT TAGTAATTGG GAGAAGTGAC TC-3′) at the 3′ end engineered to contain a XhoI site. The PCR product was digested with NcoI and XhoI and inserted into pGEM5Zf(+) that had been digested with NcoI and SalI, so that the serp2 gene was oriented correctly with respect to the T7 promoter. Plasmid DNA purified by using the Qiagen maxiprep kit and PCR products were sequenced in an MJ Research, Inc., PTC-100 thermal cycler by using the ABI Prism dye terminator kit (Perkin-Elmer).
Coupled transcription-translation in vitro.
35S-labeled Serp2 and CrmA proteins were synthesized by transcribing plasmid DNAs with T7 RNA polymerase and translating them in the same buffer with added Tran35S-Label (ICN) as the source of [35S]methionine. Both reactions were carried out with the Promega TNT T7 Quick Coupled Transcription/Translation System as suggested by the manufacturer except that an additional 0.5 mM Mg2+ (magnesium acetate) was added for the synthesis of Serp2 (52). Radiolabeled CrmA protein was made in the TNT system from pALTER-Ex1/crmA constructed by P. Y. Musy (32a), and did not require additional magnesium.
Native and sodium dodecyl sulfate (SDS)-PAGE analysis of serpin-proteinase interactions.
Radiolabeled serpins synthesized in the TNT system were tested for cleavage and complex formation after treatment with the proteinases ICE and granzyme B. In each case, control reactions without enzyme were incubated in the same buffer as that used for the enzyme at the same temperature for an equivalent time. Unpurified transcription-translation reaction products containing 35S-labeled serpins were incubated with ICE in caspase buffer (100 mM HEPES, pH 7.5; 10% sucrose; 0.1% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}; 10 mM dithiothreitol [DTT]) (50) for 15 min at 37°C. Radiolabeled Serp2 and CrmA were incubated with purified granzyme B for 30 min at 37°C in 0.1 M HEPES (pH 7.5)–10 mM CaCl2 (33). In both cases the products were separated by electrophoresis on SDS and native 10% acrylamide gels. Native PAGE was done exactly as for standard SDS-PAGE, but with SDS and DTT omitted (11). Cross-linking of the reaction products after treatment of CrmA or Serp2 with ICE in caspase buffer was achieved by adding 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) in 0.1 M potassium phosphate (pH 7) buffer to a final concentration of 40 mM and incubating the mixture for 30 min at room temperature prior to analysis by SDS-PAGE. Gels were treated with Amplify (Amersham) to enhance the autoradiographic detection of 35S-labeled proteins.
Expression of His-tagged Serp2 protein.
A His10 tag was added to the N terminus of Serp2 by recloning the Serp2 gene into pTM1-His (46), a derivative of the vaccinia-T7 expression vector pTM1 (30) which contains the His tag inserted from pET-16b. The Serp2 ORF was excised from pGEM-5Zf(+)/serp2 by digestion with NcoI (5′ end) and NsiI (3′ end) and recloned into NcoI- and PstI-digested pTM1-His (NsiI and PstI have compatible sticky ends). VV-His-serp2, a VV recombinant containing the His-tagged serp2 gene inserted into the thymidine kinase (TK) gene, was constructed by transfecting pTM1-His-serp2 plasmid DNA and wtVV genomic DNA into a VV mutant dependent for growth on IBT (isatin-β-thiosemicarbazone) and then selecting for IBT-independent 5-bromo-2′-deoxyuridine-resistant plaques (12, 54).
His-tagged Serp2 protein was prepared from suspension HeLa cells after coinfection with vTF7-3 (13), a VV derivative expressing the T7 RNA polymerase, and VV-His-Serp2. Cytoplasmic extracts were prepared; His-Serp2 protein was purified by immobilized metal affinity chromatography by using His-Bind Resin (Novagen) (4, 46) and then quantified by the Bradford assay.
ICE assays.
Purified His-tagged Serp2 or CrmA was preincubated with 50 U of recombinant human ICE (∼1 pmol) (kindly provided by Nancy Thornberry) for 5 min at room temperature in ICE buffer, and the fluorogenic substrate Ac-YVAD-AMC [acetyl-Tyr-Val-Ala-Asp-(amino-4-methyl coumarin)] was added to 14 μM. Cleavage of the peptide substrate was monitored by fluorometry to detect free amino methyl coumarin, with an excitation at 380 nm and an emission at 460 nm. A Hoefer/Pharmacia DyNA Quant 200 fluorometer, a Turner Designs TD-700 fluorometer, and a Tecan SpectraFluor microplate reader were used.
Granzyme B enzymatic assay.
Native mouse granzyme B was purified from cytoplasmic granules of the cytolytic cell line MTL2.8.2. Cell pellets were washed in phosphate-buffered saline containing 5 mM EGTA–1 mM MgCl2 and resuspended in 20 ml of the same buffer. A crude lysate was obtained by subjection to three cycles of freeze-thawing at −70 and 37°C, followed by centrifugation for 5 min at 4°C and 12,000 × g. The granules in this supernatant were lysed by the addition of NaCl to 2 M with freezing as described above. The granule lysate was cleared by centrifugation for 60 min at 4°C and 90,000 × g. Chromatography on Heparin HiTrap columns (Pharmacia) was performed after 10-fold dilution with 50 mM MES (morpholineethanesulfonic acid; pH 6.1). Granule proteins were applied to 1-ml columns, which were developed with a linear gradient of 0.5 to 1.0 M NaCl in 50 mM MES (pH 6.1). Granzyme B was eluted at approximately 0.7 M.
Purified Serp2 or CrmA protein was preincubated with purified granzyme B from the mouse cell line MTL2.8.2 or from the human YT cell line in 0.1 M HEPES (pH 7.5)–10 mM CaCl2 containing 1 mg of bovine serum albumin per ml for 15 min at 37°C in a total volume of 50 μl. Loss of granzyme B activity during incubation at 37°C was controlled for by incubating granzyme B without serpin but with an equivalent volume of serpin solvent for 15 min at 37°C. Then, 450 μl of 0.1 M HEPES (pH 7.5)–10 mM CaCl2 containing 0.1 mM tert-butyloxycarbonyl-Ala-Ala-Asp-thiobenzyl ester substrate (Enzyme Systems Products, Dublin, Calif.) and 0.11 mM dithiobis(2-nitrobenzoic acid) was added. Substrate cleavage was monitored by measuring the absorbance at 405 nm for 20 min at room temperature.
Construction and analysis of a cowpox virus recombinant deleted for CrmA that expresses Serp2.
An isogenic series of recombinant cowpox viruses derived from CPVΔcrmA (2) was constructed by cloning into the plasmid vector pSC65 (9), which contains a synthetic early-late poxvirus promoter for expression, flanking sequences from the TK gene to facilitate insertion into and inactivation of the viral TK gene, and a β-galactosidase cassette for selection of recombinant poxviruses. The serp2 and crmA genes were recloned separately into pSC65 by standard techniques, and recombinant viruses were generated after the transfection of plasmid DNA into CV-1 cells infected with CPVΔcrmA. TK− viruses were selected by their resistance to 5-bromo-2′-deoxyuridine on Rat2 (TK−) cells, and plaques were screened for blue color on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (4). The “empty” plasmid vector was used to transfect cells, generating a control CrmA− TK− virus, CPVΔcrmA TK::lacZ. Transfection with pSC65-serp2 gave CPVΔcrmA TK::serp2, in which serp2 and lacZ have been inserted into the TK gene. The control virus CPVΔcrmA TK::crmA was constructed by recombination between CPVΔcrmA virus and plasmid pSC65-crmA, and has crmA reinserted into the TK gene with the lacZ marker.
Rabbit antiserum produced under contract by HTI Bio-Products, Inc., against purified His-Serp2 protein, and a monoclonal antibody against CrmA were used to evaluate the expression of Serp2 and CrmA, respectively, by wtCPV, CPVΔcrmA TK::lacZ, CPVΔcrmA TK::serp2, and CPVΔcrmA TK::crmA. Immunoblotting and DAPI (4′,6-diamidino-2-phenylindole) staining of cells infected with CPVΔcrmA derivatives were as described previously (20). Extracts of infected LLC-PK1 cells were made by resuspending cell pellets from 35-mm wells in 100 μl of extract buffer (10 mM HEPES, pH 7.5; 2 mM EDTA; 0.1% CHAPS; 1 mM DTT), freeze-thawing four times, and removing the insoluble material by centrifugation. The protein concentration of the supernatants was measured by the Bradford assay in a microplate reader. The amount of DEVD-AMC cleaving activity in 2.5 μg of total protein was measured by the increase in fluorescence with time by using the substrate Ac-DEVD-AMC [acetyl-Asp-Glu-Val-Asp-(amino-4-methyl coumarin)] at 10 μM in 200 μl of caspase buffer.
Nucleotide sequences.
The DNA sequence for the Serp2 ORF of MYX strain Lausanne has been deposited in the GenBank database under GenBank accession no. AF141941.
RESULTS
Sequence of the MYX strain Lausanne serp2 gene.
The serp2 ORF was introduced into the vector pGEM-5Zf(+) containing the T7 promoter in order to facilitate expression of the Serp2 protein in vitro. Primers against the 5′ and 3′ ends of the published serp2 sequence for the T1 strain of MYX (35) were used to amplify the serp2 gene from genomic DNA of the MYX strain Lausanne, as described in Materials and Methods. Three clones were sequenced in their entirety and were found to be identical to one another but different from the published sequence for the T1 serp2 gene at four positions. To rule out strain differences, we sequenced a genomic clone of the T1 serp2 gene in pBluescript and found that the serp2 gene from T1 was identical to the serp2 gene from Lausanne. The published T1 serp2 sequence (35) therefore contains four single-base errors, each leading to a different amino acid substitution. The correct nucleotides, with amino acid differences indicated in parentheses, are as follows: at nucleotide position 215, A(Lys) instead of C(Thr); at position 227, C(Ala) rather than T(Val); at position 380, C(Ala) not A(Asp); and at position 424, T(Phe) not A(Ile). The fact that the serp2 DNA sequences were identical for the T1 and Lausanne strains is consistent with conservation of the protein and suggests that Serp2 is likely to function as a proteinase inhibitor and to contribute to virus growth or virulence.
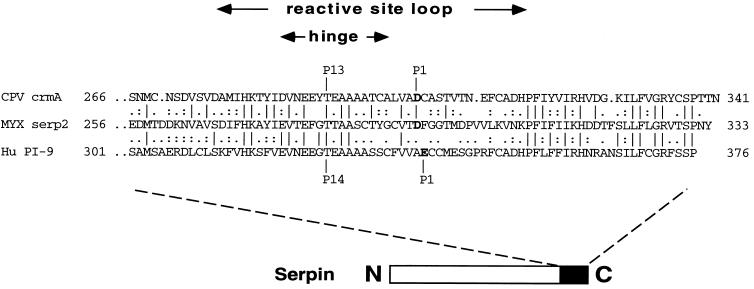
The Serp2 protein is 35.1% identical overall to the CPV serpin CrmA (cytokine response modifier A) (37), and identity to other poxvirus serpins ranges from 28.9% for VV SPI-3 (ORF K2L) to 33.4% for the swinepox virus SPI-7 (24). The human serpin PI-9 (44), which is a granzyme B inhibitor, shares 32.9% identity with Serp2 overall. A comparison of the reactive site loops of Serp2 with the granzyme B inhibitors CrmA and PI-9 is shown in Fig. 1. Within the 40-amino-acid region centered on the P1/P1′ residues of serp2, the Serp2 protein is 35.9% identical and 61.5% similar to CrmA and 30% identical and 42.5% similar to PI-9. Over this 40-amino-acid region, the CrmA and PI-9 proteins are 53.8% identical and 71.8% similar, indicating that within this region the cellular serpin PI-9 is more closely related to CPV CrmA than is the viral protein Serp2.
FIG. 1.
Alignment of the Serp2 reactive site loop with the corresponding regions of cowpox virus CrmA and human PI-9 serpins. The RSL regions of Serp2, CrmA, and PI-9 were compared by using the GAP program (Wisconsin Package version 8.1; Genetics Computer Group). Identity is indicated by vertical lines, highly similar amino acids are indicated by colons, somewhat similar residues are indicated by dots, and dissimilar amino acids are indicated by gaps. The regions of each protein shown are numbered at each end; the P1 residue (Asp for Serp2 and CrmA, Glu for PI-9) is labeled. The position of the RSL (solid rectangle) toward the C terminus of the serpin is indicated in the diagram below the alignment.
The fact that the natural ICE inhibitors CrmA and the baculovirus p35 protein inhibit other aspases, in addition to ICE (8, 56, 57), led us to expect that the inhibition spectrum of Serp2 might extend beyond the reported activity against ICE (35). We set out to characterize the interactions between Serp2 and caspases or granzyme B by first assessing the ability of unpurified 35S-labeled serpin protein synthesized in vitro to form an inhibitory complex with a given proteinase (16) and later by expressing and purifying a deca-histidine-tagged derivative of Serp2 to test directly for inhibitory activity.
Cleavage of Serp2 by ICE (caspase-1) and lack of complex detection by PAGE.
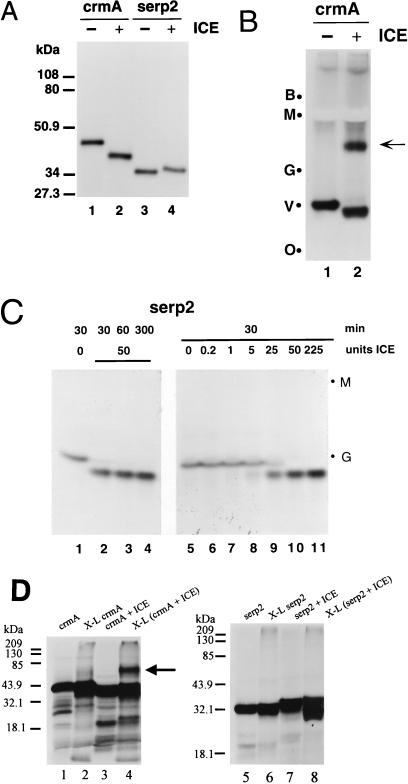
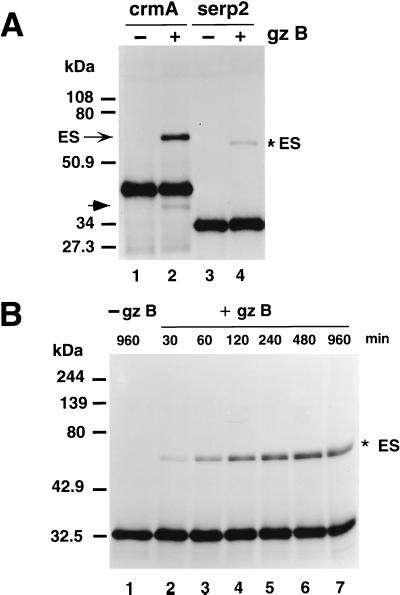
Coupled transcription-translation of pGEM-5Zf(+)/serp2 by means of the Promega TNT T7 Quick system gave a single radiolabeled protein with an apparent mobility of about 34 kDa (Fig. 2A, lane 3). The migration of Serp2 on SDS gels was somewhat anomalous, being substantially faster than expected for a protein with a calculated size of 38 kDa. Maximum expression of Serp2 in vitro was found to be dependent on the addition to the standard transcription-translation reaction mix of 0.5 mM magnesium acetate (52). The behavior of Serp2 after incubation with ICE (caspase-1) was monitored by using both SDS and native PAGE and then compared with radiolabeled CrmA protein synthesized in the TNT system. Analysis of the reaction products by PAGE allows detection of stable complexes between the serpin and proteinase, a hallmark of inhibitory serpins. Treatment of CrmA with ICE resulted in the generation of a faster-migrating cleaved form of the serpin on a denaturing SDS gel (Fig. 2A, lanes 1 and 2). Cleavage of CrmA by ICE was also readily apparent on a native gel (Fig. 2B). In contrast, incubation of Serp2 with ICE gave a band that unexpectedly migrated slightly slower than untreated Serp2 by SDS-PAGE (Fig. 2A, lanes 3 and 4). However, ICE-treated Serp2 protein migrated faster than the untreated Serp2 in a native gel, (Fig. 2C, lane 2 versus lane 1), a finding in agreement with a previous study (35), indicating that cleavage had indeed occurred. Cleavage adjacent to the Asp residue within the RSL would generate a product that should run faster in this native gel system than the intact Serp2 as the result of a smaller molecular mass (33.6 versus 38 kDa) coupled with a lower isoelectric point (calculated pI of 5.95 for residues 1 to 294 of Serp2 versus a pI of 6.33 for the entire protein). We have observed that the predicted pI is an excellent indicator of mobility in native gels for different poxvirus serpins, all of which are approximately 40 kDa in size. Uncleaved CrmA migrated much faster on native gels than did Serp2 in our native gel system (made with Tris buffer at pH 8.8), as the pI for CrmA is 4.4, considerably lower than the calculated value of 6.3 for Serp2.
FIG. 2.
Electrophoresis of radiolabeled CrmA and Serp2 proteins after incubation with ICE (caspase-1). 35S-labeled CrmA and Serp2 proteins from transcription and translation in vitro were treated with ICE, and the products were resolved by electrophoresis on SDS or native (nondenaturing, nonreducing) 10% acrylamide gels and visualized by autoradiography. (A) SDS-polyacrylamide gel of CrmA incubated without ICE (lane 1), crmA with 50 U of ICE incubated for 15 min at 37°C (lane 2), Serp2 without ICE (lane 3), and Serp2 with ICE (lane 4). The positions of Kaleidoscope-prestained standards (Bio-Rad) are indicated to the left. (B) Native polyacrylamide minigel of CrmA without ICE (lane 1) or incubated with 50 U of ICE for 15 min (lane 2). The CrmA-ICE complex is indicated by the arrow. The positions of the Kaleidoscope markers are shown on the left: B, blue (myosin); M, magenta (β-galactosidase); G, green (bovine serum albumin); V, violet (carbonic anhydrase); and O, orange (soybean trypsin inhibitor). (C) Section of native polyacrylamide gel showing results of incubation of Serp2 with 50 U of ICE for various times at 37°C (lanes 1 to 4) or for 30 min at 37°C with various amounts of ICE (lanes 5 to 11) as shown above the lanes. Serp2 was left without ICE (lane 1) or was treated with 50 U of ICE for 30, 60, or 300 min (lanes 2 to 4, respectively). Serp2 was also incubated for 30 min with 0, 0.2, 1, 5, 25, 50, and 225 U of ICE (lanes 5 to 11, respectively). The position of the magenta (M) and green (G) Kaleidoscope markers are shown. (D) SDS gel of radiolabeled CrmA (lanes 1 to 4) and Serp2 (lanes 5 to 8) after treatment with ICE and/or the cross-linking agent EDC. Lanes: 1, untreated crmA; 2, EDC-treated (cross-linked) CrmA; 3, ICE-treated CrmA; 4, ICE- and EDC-treated CrmA; 5, untreated Serp2; 6, cross-linked Serp2; 7, ICE-treated Serp2; 8, ICE- and EDC-treated Serp2. The cross-linked ICE-CrmA complex in lane 4 is indicated by the arrow.
A stable complex between CrmA and ICE was readily seen in native gels (Fig. 2B, arrow), a finding in agreement with previous reports (17). The complex of the serine proteinase inhibitor CrmA with the cysteine proteinase ICE is not sufficiently stable to withstand boiling in the presence of SDS and reducing agent, like similar complexes involving other cysteine proteinases (38), and is therefore not visualized by SDS-PAGE (Fig. 2A, lane 2). In view of the detection of an ICE-Serp2 complex by immunoblotting and the reported inhibition of ICE by Serp2 (35), we were surprised that no complex between ICE and radiolabeled Serp2 from the TNT system was seen in either denaturing (Fig. 2A) or native (Fig. 2C) gels. Incubation of Serp2 with 50 U of ICE for 30, 60, or 300 min (Fig. 2C, lanes 2 to 4) resulted in cleavage but gave no evidence of complex formation. To preclude the possibility that a complex was formed but then degraded by the presence of excess proteinase, we incubated a fixed quantity of Serp2 with amounts of ICE that varied from 0.2 to 225 U for 30 min and then analyzed the products in native gels (Fig. 2C, lanes 6 to 11). Although the extent of cleavage increased with greater amounts of ICE, we detected no evidence for complex formation between 35S-labeled Serp2 protein and ICE under any of the conditions tested (Fig. 2C).
However, it remains formally possible that a Serp2-ICE complex could comigrate with uncomplexed Serp2 on native gels and thereby be undetected. We looked for an ICE-Serp2 complex by using a novel method for detecting serpin-caspase complexes that involves treatment with the cross-linking agent EDC prior to separation of the reaction products under denaturing and reducing PAGE conditions. Incubation of the reaction products of ICE and radiolabeled CrmA with EDC prior to SDS-PAGE resulted in the appearance of a band at approximately 60 kDa (Fig. 2D, lane 4, arrow), which presumably represents CrmA covalently linked to the p20 subunit of ICE. No complex between Serp2 and ICE could be visualized by SDS-PAGE after cross-linking, (Fig. 2D, lane 8), indicating that any association of ICE and Serp2 was not sufficiently long-lived or stable to allow linkage of Serp2 to ICE. Collectively, these results suggest that Serp2 is a much weaker inhibitor of ICE than CrmA. We assessed this hypothesis directly by testing the ability of purified Serp2 protein to inhibit ICE in an enzymatic assay in vitro.
Inhibition of ICE by purified Serp2 protein.
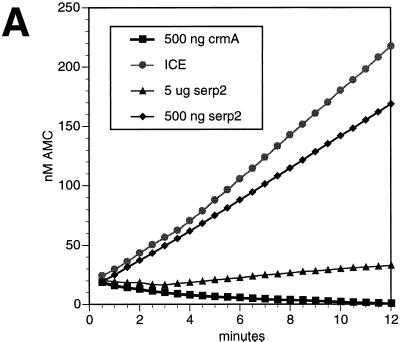
N-terminal fusions of CrmA and Serp2 to a decahistidine tag were expressed by the vaccinia-T7 system and purified by immobilized metal affinity chromatography. After SDS-PAGE analysis, preparations of His-CrmA and His-Serp2 each gave a single band by silver staining that migrated slightly more slowly than the corresponding untagged serpin, suggesting that both proteins were pure and intact. In a standard assay for the ability of serpins to inhibit ICE activity (50), ICE and serpin were preincubated in 1 ml of ICE buffer for 5 min at room temperature before the addition of the fluorogenic peptide substrate Ac-YVAD-AMC to measure residual enzyme activity (Fig. 3A). A total of 40 U of ICE (33 ng) was used, which is equivalent to ca. 1.1 pmol of active sites (49). Under these conditions, 500 ng of CrmA protein (12 pmol) completely inhibited ICE, but 500 ng (12 pmol) of Serp2 protein only reduced the activity to 68% of the uninhibited rate. A 500-ng amount of His-crmA protein (molecular weight of 40,830) should represent a quantity of functional inhibitor equivalent to 500 ng of His-Serp2 protein (molecular weight of 40,799), if we assume that a similar proportion of each protein preparation is active as an inhibitor. Increasing the amount of Serp2 protein to 5 μg in this assay failed to show complete inhibition, with a residual ICE activity of 8.4% remaining (Fig. 3A).
FIG. 3.
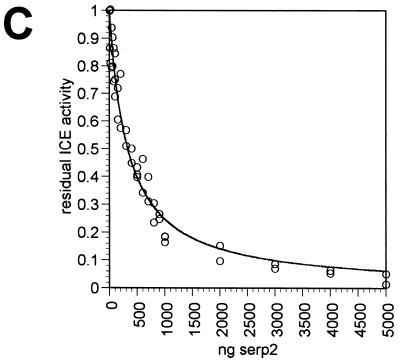
Inhibition of ICE by CrmA and Serp2 proteins. (A) Purified human recombinant ICE was incubated with His-tagged CrmA or Serp2 protein, and the residual ICE activity was determined by cleavage of the fluorogenic substrate Ac-YVAD-AMC. First, 40 U of ICE (1.1 pmol) were incubated for 5 min at room temperature in 1 ml of ICE buffer without any added protein (circles) or with 500 ng of CrmA (squares), 500 ng of Serp2 (diamonds) or 5 μg of Serp2 (triangles). (B) Titration of CrmA against 20 U of ICE. The amount of active ICE remaining after incubation with CrmA in 1 ml of ICE buffer is expressed as the proportion of initial activity. (C) Titration of Serp2 against 20 U of ICE. Enzyme and serpin were preincubated in 100 μl of ICE buffer before the residual activity was determined. The curve was fitted by using DeltaGraph 4.0 as described in the text.
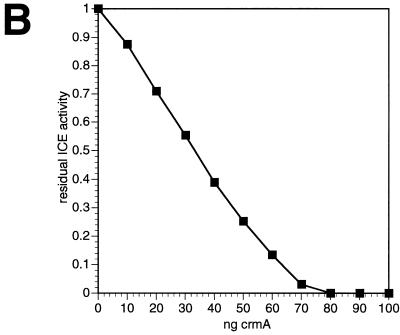
The inhibition of a fixed quantity of ICE as a function of different amounts of CrmA and of Serp2 is shown in Fig. 3B and C, respectively. In the case of CrmA there was an almost linear relationship between the extent of inhibition and the quantity of serpin added, indicating a tight association between ICE and CrmA as reported previously (17). By plotting [crmA]/i versus 1/(1 − i), where i is the proportion of ICE inhibited (6), the Ki of CrmA for ICE was estimated from the slope to be 4 pM (not shown). The intercept gave the amount of CrmA required for complete inhibition of 0.6 pmol of ICE active sites as ca. 68 ng, or 1.7 pmol of CrmA. These data can be reconciled with the 1:1 stoichiometry between enzyme and inhibitor that has been reported for serpins (38), including CrmA (17), if we assume that the proportion of the purified serpin that is active as an inhibitor is significantly less than 100%, with the remaining material acting as a substrate rather than as an inhibitor (38).
When Serp2 was titrated against 20 U of ICE and the residual enzymatic activity was plotted (Fig. 3C), a very different result was obtained. Instead of a linear relationship between the residual ICE activity and the amount of Serp2 added, the points fell on a curve. By using nonlinear regression analysis with DeltaGraph 4.0 and the formula activity = Ki/(Ki + S) for loose binding, where Ki is the inhibition constant for Serp2 binding to ICE, and S is the concentration of Serp2, the curve shown in Fig. 3C was fitted to the data points. The data were consistent with a Ki of 80 nM and indicate that Serp2 inhibits ICE weakly compared with CrmA.
Complex formation between Serp2 and granzyme B in vitro.
Based on the observed inhibition of ICE by Serp2 (Fig. 3) and the fact that the P1 residue is aspartic acid (Fig. 1), it seemed likely that the proteinase target for Serp2 was an aspase. We therefore studied the interaction between Serp2 and the serine proteinase granzyme B, which is also inhibited by CrmA but less efficiently than ICE (39). The activity of Serp2 against murine granzyme B was first assessed by looking for the formation of a stable complex between the serpin and the proteinase. CrmA was used as a positive control that gives a complex with granzyme B (a serine proteinase) that can be detected after resolving the reaction products in denaturing gels under reducing conditions (16). Radiolabeled CrmA protein was synthesized by coupled transcription-translation in vitro and incubated with granzyme B in a total volume of 50 μl for 30 min at 37°C; the products were then resolved by denaturing, reducing gels, and autoradiography. Under these conditions CrmA formed a complex with granzyme B (Fig. 4A, lane 2, arrow) that migrated with an apparent size of 64 kDa, in approximate agreement with the sum of RSL-cleaved CrmA (33.8 kDa) and granzyme B (31 kDa). The complex observed by SDS-PAGE represents a covalent linkage between cleaved CrmA and granzyme B which is formed after the denaturation and collapse of the tetrahedral complex between uncleaved serpin and enzyme that is thought to occur naturally and to be responsible for inhibition (38). A small proportion of the CrmA treated with granzyme B migrated faster than intact CrmA (Fig. 4A, lane 2, arrowhead), consistent with cleavage within the RSL. After incubation of 35S-labeled Serp2 protein with granzyme B for 30 min, a band of apparent mobility of 60 kDa was seen (Fig. 4A, lane 4, asterisk) that appeared to represent a 1:1 complex between Serp2 and granzyme B. The observed size of 60 kDa for the complex of Serp2 and granzyme B compared with an expected size of 64.4 kDa (31 kDa for granzyme B plus 33.6 kDa for cleaved Serp2) and presumably reflects the anomalous fast migration of Serp2 alone in SDS-PAGE. Although cleavage of Serp2 by granzyme B was not evident by SDS-PAGE (Fig. 4), some cleavage was seen by electrophoresis in native gels (data not shown).
FIG. 4.
Complex formation between granzyme B and either CrmA or Serp2. Radiolabeled CrmA or Serp2 protein was synthesized by coupled transcription-translation in vitro, incubated with granzyme B, and analyzed by electrophoresis and autoradiography. (A) Denaturing (SDS) gel showing CrmA protein without granzyme B (lane 1), CrmA treated with granzyme B (lane 2), and Serp2 without (lane 3) or with (lane 4) granzyme B. The CrmA-granzyme B complex is indicated by the arrow at left, and cleaved CrmA is indicated by the arrowhead. The Serp2-granzyme B complex is marked by the asterisk on the right. (B) Time course of complex formation between granzyme B and Serp2. Serp2 protein was incubated without granzyme B (lane 1) or with granzyme B (lanes 2 to 7) for the time indicated above each lane in minutes.
The intensity of the Serp2-granzyme B complex after 30 min of incubation was weaker than the corresponding CrmA-granzyme B complex formed under the same conditions (compare lanes 4 and 2 in Fig. 4A), suggesting that the Serp2-granzyme B complex was less stable than the CrmA-granzyme B complex and/or that Serp2 associated with granzyme B more slowly than CrmA. Increasing the time of incubation of Serp2 with granzyme B was found to give a complex of greater intensity (Fig. 4B, lanes 2 through 7), indicating that a relatively slow association rate constant accounts, at least in part, for the low amount of Serp2-granzyme B complex seen after incubation for 30 min (Fig. 4A, lane 4, and 4B, lane 2). Nevertheless, the appearance of a complex of Serp2 and granzyme B that could be visualized by electrophoresis after the boiling in SDS sample buffer with the reducing agent DTT indicated that Serp2 was likely to be a functional inhibitor of granzyme B.
Purified Serp2 protein inhibits cleavage of a peptide substrate by granzyme B.
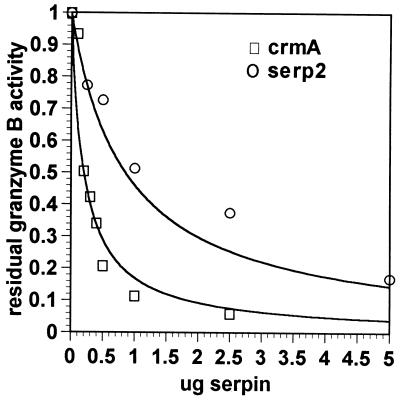
His-tagged Serp2 protein was tested directly for its ability to inhibit granzyme B in vitro by using the chromogenic substrate Boc-Ala-Ala-Asp-thiobenzyl ester, again using similarly tagged and purified CrmA protein as a known control inhibitor. CrmA protein was able to inhibit granzyme B activity (Fig. 5), although binding of CrmA to granzyme B appeared to be weak, in contrast to the association seen with ICE (Fig. 3B). By nonlinear regression, the Ki of granzyme B for CrmA was estimated to be 100 nM. Serp2 protein appears to be a weaker inhibitor of granzyme B than CrmA, requiring a greater amount of serpin for comparable inhibition of the same quantity of granzyme B under identical assay conditions (Fig. 5). Analysis of the data in Fig. 5 gave a Ki value of granzyme B for serp2 of approximately 420 nM.
FIG. 5.
Inhibition of granzyme B by CrmA and by Serp2. Inhibition of a fixed quantity of granzyme B by CrmA or by Serp2. Residual enzyme activity is expressed as a proportion of initial activity. The curves were fitted by using nonlinear regression analysis as described in the text.
Serp2 cannot replace CrmA to inhibit apoptosis in infected LLC-PK1 cells.
We next evaluated Serp2 for ability to inhibit purified human caspase-2 through caspase-9, reasoning that perhaps Serp2 was likely to be a stronger inhibitor of members of the ICE/ced-3 family other than ICE (caspase-1). Purified His-tagged Serp2 (at concentrations ranging from 1.4 nM to 1 μM) was incubated with 1 nM concentrations of each enzyme separately, and then an appropriate fluorogenic tetrapeptide substrate was added to measure the residual enzyme activity. Complete inhibition was not observed in any instance (data not shown). Serp2 was unable to inhibit FLICE (caspase-8), unlike CrmA, which is an efficient inhibitor of caspase-8 (43, 45, 57). ICErel-III (caspase-5) was partially inhibited by Serp2, with the residual activity decreasing progressively with increasing concentrations of Serp2 to a plateau of ca. 50% activity (data not shown), although this inhibition of 1 nM caspase-5 by Serp2 required a large (at least 100-fold) molar excess of Serp2.
Our biochemical data suggest that CrmA and Serp2 differ functionally in vitro. However, a more meaningful question relates to how the two proteins perform relative to each other within the context of a viral infection in the presence of other cellular and viral gene products. Serp2 is reported to function to inhibit apoptosis within the context of a MYX infection of rabbits (28). Under certain conditions, CrmA has a similar function during CPV infections (20, 41). Therefore, we sought to determine whether Serp2 could substitute for CrmA within cowpox virus to provide the antiapoptosis activity normally specified by CrmA. The experiment makes use of the system involving a CrmA mutant of cowpox virus and LLC-PK1 cells (41). LLC-PK1 cells infected with CPVΔcrmA but not wtCPV give many of the hallmarks of apoptosis (41), including caspase activation (20). A derivative of CPVΔcrmA was constructed with the serp2 gene inserted into the CPV TK gene under the control of a synthetic poxvirus early-late promoter (9). The presence of lacZ within the shuttle plasmid within the TK flanks but outside of the serpin cloning site facilitated recombinant selection and allowed a control for the effects of disrupting the CPV TK gene by using the same plasmid shuttle vector devoid of either serpin. The recombinant viruses CPVΔcrmA TK::serp2 and CPVΔcrmA TK::lacZ (control) were isolated after infection and transfection. In addition, CPVΔcrmA TK::crmA was constructed where the replaced crmA, like serp2, was within the TK gene rather than at the original crmA locus.
Expression of the Serp2 protein from the PE/L promoter in CPVΔcrmA TK::serp2 was confirmed by immunoblot analysis by using extracts from infected CV-1 cells (Fig. 6). A strong band with a mobility of approximately 34 kDa was observed on infection with CPVΔcrmA TK::serp2 (Fig. 6, lane 3, arrowhead) and was absent from uninfected CV-1 cells (lane 1) and from cells infected with the control virus CPVΔcrmA TK::lacZ (lane 2). Similar results were obtained when Serp2 expression was assessed in extracts made from infected LLC-PK1 cells (data not shown). In parallel experiments, CrmA expression was restored in the recombinant CPVΔcrmA TK::crmA (data not shown).
FIG. 6.
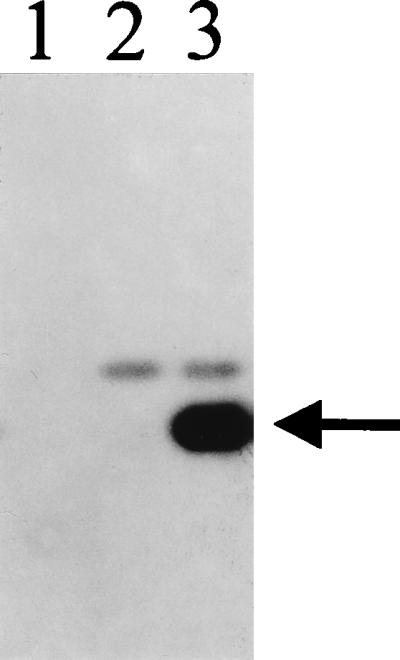
Expression of Serp2 protein by CPVΔcrmA TK::serp2. Extracts of infected CV-1 cells harvested at 16 h postinfection were electrophoresed on SDS-acrylamide gels and immunoblotted with rabbit antisera against Serp2. Lanes: 1, uninfected CV-1; 2, cells infected with CPVΔcrmA TK::lacZ; 3, cells infected with CPVΔcrmA TK::serp2. The intense band in lane 3 (arrow) migrated with an apparent size of ca. 34 kDa and indicated that Serp2 protein was being expressed in the derivative of CPVΔcrmA with Serp2 inserted into the TK gene under the control of a synthetic early-late promoter.
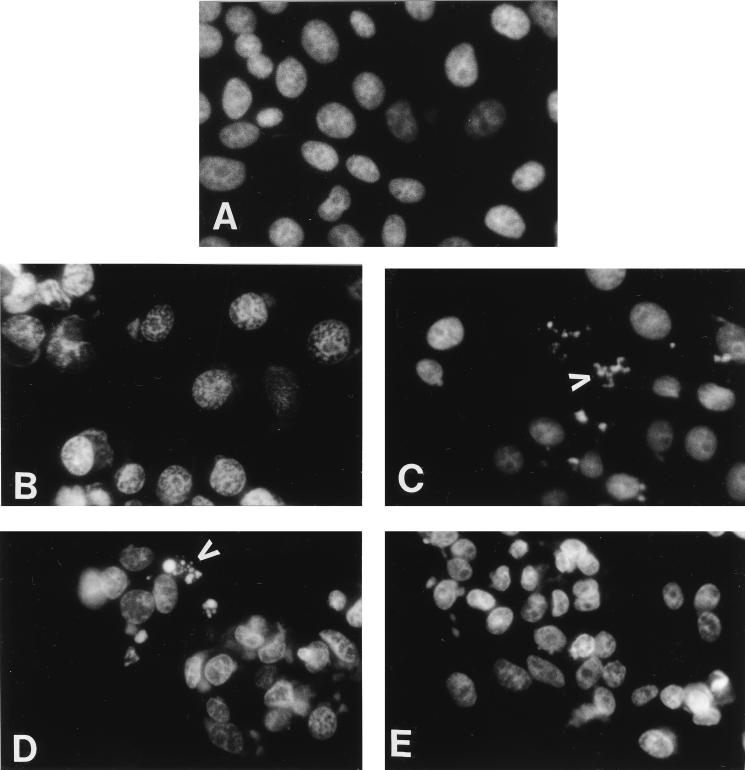
The ability of Serp2 to substitute for CrmA in blocking apoptosis of CPVΔcrmA-infected LLC-PK1 cells was studied both by morphological criteria and by measuring the amount of caspase activation by cleavage of a synthetic peptide substrate. DAPI staining of wtCPV-infected cells at 16 h postinfection (Fig. 7B) indicated a nuclear morphology similar to that observed for uninfected cells (Fig. 7A). However, LLC-PK1 cells infected with CPVΔcrmA TK::lacZ (Fig. 7C) gave extensive nuclear condensation and blebbing indicative of apoptosis, as described earlier for CPVΔcrmA (20, 41). The induction of apoptosis was solely dependent on deletion of the crmA gene and was independent of the presence of the lacZ gene within the TK locus. LLC-PK1 cells infected with CPVΔcrmA TK::serp2 also resulted in nuclear fragmentation indicative of apoptosis (Fig. 7D) at levels similar to that seen in cells infected with CPVΔcrmA TK::lacZ, indicating that the expression of Serp2 protein was unable to substitute for CrmA and prevent induction of the apoptotic morphology. By contrast the control virus CPVΔcrmA TK::crmA (Fig. 7E) gave DAPI-stained nuclei that were indistinguishable from those infected with wtCPV (Fig. 7B). Reintroduction of the crmA gene but not serp2 into TK under the PE/L promoter therefore resulted in full suppression of apoptosis by this criterion. These results also indicate that crmA was fully functional when expressed from the PE/L promoter after insertion into the TK gene.
FIG. 7.
DAPI staining of LLC-PK1 cells infected with derivatives of CPVΔcrmA expressing Serp2 or CrmA. Cells were left uninfected (A) or were infected with CPVΔcrmA derivatives (B to E) and then stained with DAPI at 16 h postinfection to visualize nuclei. (A) Uninfected LLC-PK1 cells. (B) infected with wtCPV. (C) CPVΔcrmA TK::lacZ. (D) CPVΔcrmA TK::serp2. (E) CPVΔcrmA TK::crmA. Apoptotic bodies are indicated by arrowheads in panels C and D.
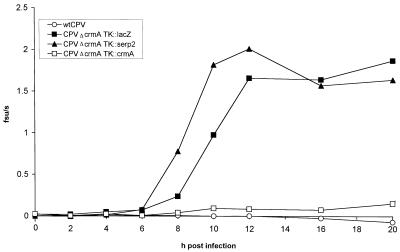
Unlike nuclear morphology, which allows a qualitative evaluation of apoptosis, a more quantitative measure of apoptosis is the activation of caspases. The extent of caspase activation was measured in a time course experiment by monitoring the activity against DEVD-AMC, a fluorogenic substrate for caspase-3. Pilot experiments indicated that cell extracts made from LLC-PK1 cells infected with CPVΔcrmA at 16 h postinfection contained high levels of DEVD-AMC cleaving activity, but cells infected with wtCPV contained none, nor did uninfected cells (data not shown). Extracts were made from LLC-PK1 cells infected with wtCPV, CPVΔcrmA TK::lacZ, CPVΔcrmA TK::serp2, and CPVΔcrmA TK::crmA at various times up to 20 h postinfection, and the DEVD-AMC cleaving activity was measured. At time points later than 24 h significant lysis of infected cells occurred and cytoplasmic extracts could not be reliably prepared. The results (Fig. 8) indicate that effector apoptotic caspases able to cleave DEVD-AMC were not present in extracts from wtCPV-infected cells at any time point examined. However, in cells infected with CPVΔcrmA TK::lacZ, high levels of DEVD-AMC cleaving activity were present by 12 h postinfection, with induction beginning during the late phase of infection between 6 and 8 hours postinfection. Similar results were observed for cells infected with CPVΔcrmA TK::serp2, showing that the expression of Serp2 did not significantly affect the extent or timing of caspase activation compared with CPVΔcrmA TK::lacZ. Reinsertion of CrmA into CPVΔcrmA TK::crmA completely suppressed the induction of DEVD-AMC cleaving activity. Based on this assay Serp2 is therefore unable to substitute for CrmA in preventing apoptosis of LLC-PK1 cells infected with CPV derivatives. If Serp2 and CrmA were cytoplasmic proteinase inhibitors with similar or identical target proteinase specificities, the two serpins would be expected to be functionally interchangeable. However, consistent with our in vitro data, the results with infected LLC-PK1 cells clearly indicate that this is not the case.
FIG. 8.
DEVD-AMC cleaving activity in extracts of infected LLC-PK1 cells. LLC-PK1 cells were infected with the indicated viruses at a multiplicity of infection of 20 and were harvested at various times postinfection. The protein concentration of each extract was determined, and the rate of DEVD-AMC cleavage was measured for samples containing 2.5 μg of total protein. The results are averages of two independent experiments and are expressed as rates of fluorescence increase per second plotted against the time postinfection.
DISCUSSION
Despite originating in two distinct viruses, the leporipoxvirus (myxoma)-encoded Serp2 and the orthopoxvirus (cowpox)-encoded CrmA are similar in the sense that both are intracellular serpins and contain aspartic acid as the P1 residue within the reactive site loop. Both are reported to inhibit ICE (35, 40) and inflammation (28, 34). Despite the apparent parallels between the two serpins, our studies suggest that the proteinases targeted by CrmA and by Serp2 may well be different.
Comparisons of the behavior of purified proteins in vitro indicate that Serp2 is a relatively poor inhibitor of ICE compared to CrmA (Fig. 3). The low affinity of purified His-Serp2 for ICE is presumably related to our failure to observe an inhibitory complex between radiolabeled Serp2 and ICE, either via the standard assay for caspase-serpin complexes with native gels or by SDS-PAGE after cross-linking (Fig. 2). The absence of a Serp2-ICE complex in our assays would seem to conflict with earlier results indicating interaction of Serp2 with ICE (35). However, Serp2-ICE complexes were detected in those experiments under conditions with far higher levels of ICE (1 μg) and most likely Serp2 as well (unlabeled protein from 20 μl of a baculovirus extract in which Serp2 was overexpressed) (35). When smaller amounts of ICE (200 or 100 ng) were used (35), these workers also failed to detect any inhibitory complexes. Our test for complex formation uses low concentrations of both ICE (ca. 30 ng or 1 pmol in a 20-μl volume) and radiolabeled Serp2 (estimated at less than 1 ng), which would work against detection of a low-affinity serpin-ICE complex. In addition, the unlabeled rabbit reticulocyte proteins of the TNT system are present during the incubation of ICE and 35S-labeled serpin. We did, however, detect the high-affinity CrmA-ICE complex under these conditions. Indeed, the weak interactions of Serp2 compared to CrmA with ICE detailed in Fig. 3 could explain why, in the earlier study, baculovirus extracts containing unusually high levels of Serp2 still failed to completely inhibit ICE-mediated tetrapeptide substrate cleavage (35).
We also analyzed Serp2 for the ability to inhibit granzyme B, a further activity ascribed to CrmA. Although Serp2 apparently associates with granzyme B at a slower rate than that seen for CrmA, a Serp2-granzyme B complex was readily detected (Fig. 4). Purified Serp2 does inhibit enzymatic activity of murine and human granzyme B (Fig. 5) and may be even more effective against rabbit granzyme B. The interactions of Serp2 with granzyme B suggest that Serp2 should be ectopically expressed in tissue culture cells and tested for inhibition of killing by cytotoxic T lymphocytes as has been demonstrated for CrmA (21, 47).
Our studies examining interaction of Serp2 with ICE and with human caspase-2 through caspase-9 clearly indicate that Serp2 from MYX does not have the broad inhibition spectrum against human caspases that is seen for cowpox virus CrmA (57) and for the baculovirus p35 protein (8, 56). The most intriguing experimental finding, one consistent with the lack of activity of Serp2 against caspase-2 through caspase-9 in vitro (data not shown), is that Serp2 cannot substitute for CrmA within the context of the entire cowpox virus genome to prevent apoptosis in CPV-infected LLC-PK1 cells (Fig. 7 and 8). The inability of Serp2 to mimic the protection against apoptosis provided by CrmA in LLC-PK1 cells highlights the intrinsic differences between the two serpins.
The effects of Serp2 on apoptosis within the context of MYX are mixed. A MYX Serp2 mutant did not lead to apoptosis in infected rabbit RL5 CD4+ lymphocytes, although in contrast MYX mutants deleted for the M-T2 (TNF receptor), the host range genes M-T5 or M11L (25), or the M-T4 gene (5) each cause apoptosis after infection of RL5 cells. However, in infected rabbits Serp2 has been reported to block apoptosis, i.e., more apoptosis was observed in the lymphocytes derived from the parotid lymph nodes of animals infected with the MYX Serp2 mutant than with wtMYX (28).
One possible explanation for the inability of Serp2 to inhibit human caspases efficiently and to substitute for CrmA could be related to species specificity. Serp2 may have evolved within the context of leporipoxvirus infections of rabbits so that only rabbit proteinases are inhibited effectively by Serp2. Strict species specificity for rabbit IFN-γ has been noted in the case of the MYX-secreted M-T7 protein, which is unable to bind human or murine IFN-γ (32). In contrast, the orthopoxvirus-secreted IFN-γ receptor homologs are much less fastidious, showing interactions with IFN-γ from a variety of species (1, 31). A second, more interesting possibility is that the intracellular targets recognized by Serp2 are neither caspase-1 through caspase-9 nor granzyme B but some as-yet-unknown proteinase.
There are striking differences between CrmA and Serp2 evidenced by the behavior of virus knockout mutants in animal systems. In mice, CrmA appears to have relatively little effect on the virulence of orthopoxviruses (15, 48). Serp2, on the other hand, is necessary for full virulence of MYX, as rabbits infected with a MYX mutant in which the serp2 gene has been deleted are clearly attenuated (28). The histopathology of lesions from infected rabbits suggested that the inflammatory response of the rabbit to the MYXΔserp2 mutant was more rapid than that observed for wtMYX (28), indicating that Serp2 has anti-inflammatory activity. In contrast, mice infected intranasally with a CPV CrmA mutant showed less inflammation compared with wtCPV-infected mice (48).
Other than the presence of aspartic acid at the P1 position in the reactive site loops of CrmA and Serp2, the sequences of the two proteins are quite divergent, arguing against a common ancestor. The human serpin PI-9 is the closest cellular homolog of CrmA yet identified and is a good inhibitor of granzyme B but a poor inhibitor of caspases, including ICE, and does not protect against Fas-mediated apoptosis (7). The cellular serpin from which Serp2 arose might also be expected to be primarily a granzyme B inhibitor which inhibits cell killing by misdirected granzyme B rather than a caspase inhibitor. To date, cellular serpins that inhibit caspases have not been found and may not exist in nature.
Finally, the possibility remains that the intracellular viral serpins such as CrmA and Serp2 do not act independently but rather act as part of a complex with other viral and/or cellular proteins. If so, then the properties of these serpins determined in vitro as isolated purified proteins may differ significantly from their behavior in vivo. We are currently searching for novel proteins that interact with poxvirus serpins.
ACKNOWLEDGMENTS
We gratefully acknowledge Nancy Thornberry (Merck) for human recombinant ICE and for testing Serp2 against caspase-2 through caspase-9. We thank Pierre Musy and Liping Zhang for construction of pSC65-crmA and pSC65-serp2, respectively, and David Silverman and Chingkuang Tu for assistance with kinetic analysis. The DNA Sequencing and DNA Synthesis Cores of the University of Florida provided excellent technical services.
R.C.B. is supported by the Medical Research Council of Canada. R.W.M. was supported by grant AI-15722 from the NIH, and P.C.T. was supported by grant 9701732 from the American Heart Association Florida affiliate.
REFERENCES
- 1.Alcami A, Smith G L. Vaccinia, cowpox, and camelpox viruses encode soluble gamma interferon receptors with novel broad species specificity. J Virol. 1995;69:4633–4639. doi: 10.1128/jvi.69.8.4633-4639.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali A N, Turner P C, Brooks M A, Moyer R W. The SPI-1 gene of rabbitpox virus determines host range and is required for hemorrhagic pock formation. Virology. 1994;202:305–314. doi: 10.1006/viro.1994.1347. [DOI] [PubMed] [Google Scholar]
- 3.Alnemri E S, Livingston D J, Nicholson D W, Salvesen G, Thornberry N A, Wong W W, Yuan J. Human ICE/ced-3 protease nomenclature. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. Expression of proteins in mammalian cells using vaccinia viral vectors. [Google Scholar]
- 5.Barry M, Hnatiuk S, Mossman K, Lee S F, Boshkov L, McFadden G. The myxoma virus M-T4 gene encodes a novel RDEL-containing protein that is retained within the endoplasmic reticulum and is important for the productive infection of lymphocytes. Virology. 1997;239:360–377. doi: 10.1006/viro.1997.8894. [DOI] [PubMed] [Google Scholar]
- 6.Bieth J G. Theoretical and practical aspects of proteinase inhibition kinetics. Methods Enzymol. 1995;248:59–84. doi: 10.1016/0076-6879(95)48007-2. [DOI] [PubMed] [Google Scholar]
- 7.Bird C H, Sutton V R, Sun J, Hirst C E, Novak A, Kumar S, Trapani J A, Bird P I. Selective regulation of apoptosis: the cytotoxic lymphocyte serpin proteinase inhibitor 9 protects against granzyme B-mediated apoptosis without perturbing the Fas cell death pathway. Mol Cell Biol. 1998;18:6387–6398. doi: 10.1128/mcb.18.11.6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bump N J, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, Licari P, Mankovitch J, Shi L, Greenberg A H, Miller L K, Wong W W. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- 9.Chakrabarti S, Sisler J R, Moss B. Compact, synthetic, vaccinia virus early/late promoter for protein expression. BioTechniques. 1997;23:1094–1097. doi: 10.2144/97236st07. [DOI] [PubMed] [Google Scholar]
- 10.Cohen G M. Caspases: the executioners of apoptosis. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coligan J, Dunn B, Ploegh H, Speicher D, Wingfield P. Current protocols in protein science. New York, N.Y: John Wiley & Sons; 1998. [Google Scholar]
- 12.Fathi Z, Sridhar P, Pacha R F, Condit R C. Efficient targeted insertion of an unselected marker into the vaccinia virus genome. Virology. 1986;155:97–105. doi: 10.1016/0042-6822(86)90171-6. [DOI] [PubMed] [Google Scholar]
- 13.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham K A, Lalani A S, Macen J L, Ness T L, Barry M, Liu L-Y, Lucas A, Clark-Lewis I, Moyer R W, McFadden G. The T1/35kDa family of poxvirus secreted proteins bind chemokines and modulate leukocyte influx into virus infected tissues. Virology. 1997;229:12–24. doi: 10.1006/viro.1996.8423. [DOI] [PubMed] [Google Scholar]
- 15.Kettle S, Blake N W, Law K M, Smith G L. Vaccinia virus serpins B13R (SPI-2) and B22R (SPI-1) encode Mr 38.5 and 40K, intracellular polypeptides that do not affect virus virulence in a murine intranasal model. Virology. 1995;206:136–147. doi: 10.1016/s0042-6822(95)80028-x. [DOI] [PubMed] [Google Scholar]
- 16.Komiyama T, Quan L, Snipas S, Ray C A, Pickup D J, Salvesen G. In vitro expression of serpins. In: Crabb J W, editor. Techniques in protein chemistry V. San Diego, Calif: Academic Press, Inc.; 1994. pp. 305–312. [Google Scholar]
- 17.Komiyama T, Ray C A, Pickup D J, Howard A D, Thornberry N A, Peterson E P, Salvesen G. Inhibition of interleukin-1 beta converting enzyme by the cowpox virus serpin CrmA. An example of cross-class inhibition. J Biol Chem. 1994;269:19331–19337. [PubMed] [Google Scholar]
- 18.Lalani A S, Graham K, Mossman K, Rajarathnam K, Clark-Lewis I, Kelvin D, McFadden G. The purified myxoma virus gamma interferon receptor homolog M-T7 interacts with the heparin-binding domains of chemokines. J Virol. 1997;71:4356–4363. doi: 10.1128/jvi.71.6.4356-4363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lucas A, Liu L-Y, Macen J, Nash P, Dai E, Stewart M, Graham K, Etches W, Boshkov L, Nation P N, Humen D, Hobman M L, McFadden G. Virus-encoded serine proteinase-inhibitor SERP-1 inhibits atherosclerotic plaque development after balloon angioplasty. Circulation. 1996;94:2890–2900. doi: 10.1161/01.cir.94.11.2890. [DOI] [PubMed] [Google Scholar]
- 20.Macen J, Takahashi A, Moon K B, Nathaniel R, Turner P C, Moyer R W. Activation of caspases in pig kidney cells infected with wild-type and crmA/SPI-2 mutants of cowpox and rabbitpox viruses. J Virol. 1998;72:3524–3533. doi: 10.1128/jvi.72.5.3524-3533.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macen J L, Garner R S, Musy P Y, Brooks M A, Turner P C, Moyer R W, McFadden G, Bleackley R C. Differential inhibition of the Fas and granule-mediated cytolysis pathways by the orthopoxvirus cytokine response modifier A/SPI-2 and SPI-1 protein. Proc Natl Acad Sci USA. 1996;93:9108–9113. doi: 10.1073/pnas.93.17.9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macen J L, Upton C, Nation N, McFadden G. SERP1, a serine proteinase inhibitor encoded by myxoma virus, is a secreted glycoprotein that interferes with inflammation. Virology. 1993;195:348–363. doi: 10.1006/viro.1993.1385. [DOI] [PubMed] [Google Scholar]
- 23.Maksymowych W P, Nation N, Nash P, Macen J, Lucas A, McFadden G, Russell A S. Amelioration of antigen induced arthritis in rabbits treated with a secreted viral serine proteinase inhibitor. J Rheumatol. 1996;23:878–882. [PubMed] [Google Scholar]
- 24.Massung R F, Jayarama V, Moyer R W. DNA sequence analysis of conserved and unique regions of swinepox virus: identification of genetic elements supporting phenotypic observations including a novel G protein-coupled receptor homologue. Virology. 1993;197:511–528. doi: 10.1006/viro.1993.1625. [DOI] [PubMed] [Google Scholar]
- 25.McFadden G, Barry M. How poxviruses oppose apoptosis. Semin Virol. 1998;8:429–442. [Google Scholar]
- 26.McFadden G. Poxviruses: rabbit, hare, squirrel, and swinepox. In: Webster R G, Granoff A, editors. Encyclopedia of virology. London, England: Academic Press, Inc.; 1994. p. 1153. [Google Scholar]
- 27.McFadden G, Graham K, Ellison K, Barry M, Macen J, Schreiber M, Mossman K, Nash P, Lalani A, Everett H. Interruption of cytokine networks by poxviruses: lessons from myxoma virus. J Leukoc Biol. 1995;57:731–738. doi: 10.1002/jlb.57.5.731. [DOI] [PubMed] [Google Scholar]
- 28.Messud-Petit F, Gelfi J, Delverdier M, Amardeilh M F, Py R, Sutter G, Bertagnoli S. Serp2, an inhibitor of the interleukin-1β-converting enzyme, is critical in the pathobiology of myxoma virus. J Virol. 1998;72:7830–7839. doi: 10.1128/jvi.72.10.7830-7839.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. New York, N.Y: Raven Press; 1996. pp. 2637–2672. [Google Scholar]
- 30.Moss B, Elroy Stein O, Mizukami T, Alexander W A, Fuerst T R. Product review. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 31.Mossman K, Upton C, Buller R M, McFadden G. Species specificity of ectromelia virus and vaccinia virus interferon-gamma binding proteins. Virology. 1995;208:762–769. doi: 10.1006/viro.1995.1208. [DOI] [PubMed] [Google Scholar]
- 32.Mossman K, Upton C, McFadden G. The myxoma virus-soluble interferon-gamma receptor homolog, M-T7, inhibits interferon-gamma in a species-specific manner. J Biol Chem. 1995;270:3031–3038. doi: 10.1074/jbc.270.7.3031. [DOI] [PubMed] [Google Scholar]
- 32a.Musy, P. Y. Unpublished data.
- 33.Odake S, Kam C M, Narasimhan L, Poe M, Blake J T, Krahenbuhl O, Tschopp J, Powers J C. Human and murine cytotoxic T lymphocyte serine proteases: subsite mapping with peptide thioester substrates and inhibition of enzyme activity and cytolysis by isocoumarins. Biochemistry. 1991;30:2217–2227. doi: 10.1021/bi00222a027. [DOI] [PubMed] [Google Scholar]
- 34.Palumbo G J, Pickup D J, Fredrickson T N, McIntyre L J, Buller R M. Inhibition of an inflammatory response is mediated by a 38-kDa protein of cowpox virus. Virology. 1989;172:262–273. doi: 10.1016/0042-6822(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 35.Petit F, Bertagnoli S, Gelfi J, Fassy F, Boucraut-Baralon C, Milon A. Characterization of a myxoma virus-encoded serpin-like protein with activity against interleukin-1β-converting enzyme. J Virol. 1996;70:5860–5866. doi: 10.1128/jvi.70.9.5860-5866.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickup D J. Poxviral modifiers of cytokine responses to infection. Infect Agents Dis. 1994;3:116–127. [PubMed] [Google Scholar]
- 37.Pickup D J, Ink B S, Hu W, Ray C A, Joklik W K. Hemorrhage in lesions caused by cowpox virus is induced by a viral protein that is related to plasma protein inhibitors of serine proteases. Proc Natl Acad Sci USA. 1986;83:7698–7702. doi: 10.1073/pnas.83.20.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Potempa J, Korzus E, Travis J. The serpin superfamily of proteinase inhibitors: structure, function, and regulation. J Biol Chem. 1994;269:15957–15960. [PubMed] [Google Scholar]
- 39.Quan L T, Caputo A, Bleackley R C, Pickup D J, Salvesen G S. Granzyme B is inhibited by the cowpox virus serpin cytokine response modifier A. J Biol Chem. 1995;270:10377–10379. doi: 10.1074/jbc.270.18.10377. [DOI] [PubMed] [Google Scholar]
- 40.Ray C A, Black R A, Kronheim S R, Greenstreet T A, Sleath P R, Salvesen G S, Pickup D J. Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1 beta converting enzyme. Cell. 1992;69:597–604. doi: 10.1016/0092-8674(92)90223-y. [DOI] [PubMed] [Google Scholar]
- 41.Ray C A, Pickup D J. The mode of death of pig kidney cells infected with cowpox virus is governed by the expression of the crmA gene. Virology. 1996;217:384–391. doi: 10.1006/viro.1996.0128. [DOI] [PubMed] [Google Scholar]
- 42.Smith G L. Virus proteins that bind cytokines, chemokines or interferons. Curr Opin Immunol. 1996;8:467–471. doi: 10.1016/s0952-7915(96)80032-x. [DOI] [PubMed] [Google Scholar]
- 43.Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Litwack G, Alnemri E S. Molecular ordering of the Fas-apoptotic pathway: the Fas/APO-1 protease Mch5 is a CrmA-inhibitable protease that activates multiple Ced-3/ICE-like cysteine proteases. Proc Natl Acad Sci USA. 1996;93:14486–14491. doi: 10.1073/pnas.93.25.14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun J, Bird C H, Sutton V, McDonald L, Coughlin P B, De J T, Trapani J A, Bird P I. A cytosolic granzyme B inhibitor related to the viral apoptotic regulator cytokine response modifier A is present in cytotoxic lymphocytes. J Biol Chem. 1996;271:27802–27809. doi: 10.1074/jbc.271.44.27802. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi A, Hirata H, Yonehara S, Imai Y, Lee K-K, Moyer R W, Turner P C, Mesner P W, Okazaki T, Sawai H, Kishi S, Yamamoto K, Okuma M, Sasada M. Affinity labeling displays the stepwise activation of ICE-related proteases by Fas, staurosporine, and CrmA-sensitive caspase-8. Oncogene. 1997;14:2741–2752. doi: 10.1038/sj.onc.1201131. [DOI] [PubMed] [Google Scholar]
- 46.Takahashi A, Musy P Y, Martins L M, Poirier G G, Moyer R W, Earnshaw W C. CrmA/SPI-2 inhibition of an endogenous ICE-related protease responsible for lamin A cleavage and apoptotic nuclear fragmentation. J Biol Chem. 1996;271:32487–32490. doi: 10.1074/jbc.271.51.32487. [DOI] [PubMed] [Google Scholar]
- 47.Tewari M, Telford W G, Miller R A, Dixit V M. CrmA, a poxvirus-encoded serpin, inhibits cytotoxic T-lymphocyte-mediated apoptosis. J Biol Chem. 1995;270:22705–22708. doi: 10.1074/jbc.270.39.22705. [DOI] [PubMed] [Google Scholar]
- 48.Thompson J P, Turner P C, Ali A N, Crenshaw B C, Moyer R W. The effects of serpin gene mutations on the distinctive pathobiology of cowpox and rabbitpox virus following intranasal inoculation of BALB/c mice. Virology. 1993;197:328–338. doi: 10.1006/viro.1993.1594. [DOI] [PubMed] [Google Scholar]
- 49.Thornberry N A. Interleukin-1 beta converting enzyme. Methods Enzymol. 1994;244:615–631. doi: 10.1016/0076-6879(94)44045-x. [DOI] [PubMed] [Google Scholar]
- 50.Thornberry N A, Bull H G, Calaycay J R, Chapman K T, Howard A D, Kostura M J, Miller D K, Molineaux S M, Weidner J R, Aunins J, et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 51.Thornberry N A, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 52.Turner P C. Analysis of interactions between proteinases and serpins expressed in vitro using the TNT T7 Quick system. Promega Notes. 1997;60:11–13. [Google Scholar]
- 53.Turner P C, Musy P Y, Moyer R W. Poxvirus serpins. In: McFadden G, editor. Viroceptors, virokines and related immune modulators encoded by DNA viruses. R. G. Austin, Tex: Landes Co.; 1995. pp. 67–88. [Google Scholar]
- 54.Turner P C, Young D C, Flanegan J B, Moyer R W. Interference with vaccinia virus growth caused by insertion of the coding sequence for poliovirus protease 2A. Virology. 1989;173:509–521. doi: 10.1016/0042-6822(89)90563-1. [DOI] [PubMed] [Google Scholar]
- 55.Upton C, Macen J L, Wishart D S, McFadden G. Myxoma virus and malignant rabbit fibroma virus encode a serpin-like protein important for virus virulence. Virology. 1990;179:618–631. doi: 10.1016/0042-6822(90)90129-f. [DOI] [PubMed] [Google Scholar]
- 56.Xue D, Horvitz R. Inhibition of the Caenorhabditis elegans cell-death protease CED-3 by a CED-3 cleavage site in baculovirus p35 protein. Nature. 1995;377:248–251. doi: 10.1038/377248a0. [DOI] [PubMed] [Google Scholar]
- 57.Zhou Q, Snipas S, Orth K, Muzio M, Dixit V M, Salvesen G S. Target protease specificity of the viral serpin CrmA. Analysis of five caspases. J Biol Chem. 1997;272:7797–7800. doi: 10.1074/jbc.272.12.7797. [DOI] [PubMed] [Google Scholar]