Abstract
A full-length clone of the 142-kb pseudorabies virus (PRV) genome was constructed as a stable F plasmid in Escherichia coli. The clone, pBecker1, was colinear with PRV-Becker genomic DNA, lacking detectable rearrangements, deletions, or inversions. The transfection of pBecker1 into susceptible eukaryotic cells resulted in productive viral infection. Virus isolated following transfection was indistinguishable from wild-type virus in a rodent model of infection and spread to retinorecipient regions of the brain following inoculation in the vitreous body of the eye. Mutagenesis of pBecker1 in E. coli with a mini-Tn5-derived transposon enabled the rapid isolation of insertion mutants, identification of essential viral genes, and simplified construction of viral revertants. The serial passage of a viral insertion mutant demonstrated the transposon insertion to be stable. However, the F-plasmid insertion present in the viral gG locus was found to undergo a spontaneous deletion following transfection into eukaryotic cells. The implications of F-plasmid insertion into the viral genome with regard to phenotype and genomic stability are discussed.
The herpesviruses are a large group of viruses characterized, in part, by a double-stranded linear DNA genome ranging in size from approximately 80 to 250 kb. Representatives of this family cause recurrent infections in both humans and livestock animals, which are sometimes debilitating or lethal (34). The economic impact of these infections has encouraged research into the mechanisms by which these viruses disseminate and cause disease. Molecular techniques are often used to examine the roles of virally encoded gene products in viral growth and pathogenesis.
The primary method for investigating the function of individual herpesvirus genes is mutagenesis. Mutated viruses are usually constructed by homologous recombination following the cotransfection of viral genomic DNA and a mutated allele on a separate DNA fragment (9, 34). Recombinant viruses are either screened or selected during several sequential rounds of plaque purification. However, if the mutation results in a growth defect relative to wild-type parental virus, the mutant virus may be difficult or impossible to purify (for example, see reference 42). Lethal mutations often are only detected indirectly by the unsuccessful isolation of the desired mutant virus. Such mutants can only be isolated and confirmed as lethal by the construction of a complementing cell line expressing the wild-type allele (for example, see reference 31).
To overcome some of these limitations, several laboratories have cloned entire herpesvirus genomes in Escherichia coli. Because of the large size of the viral genomes, the clones comprise either overlapping cosmid sets or single full-length clones in F plasmids. Cosmid sets have been assembled for several viruses, including pseudorabies virus (PRV), herpes simplex virus type 1 (HSV-1), varicella-zoster virus, Epstein-Barr virus (EBV), and human cytomegalovirus (10, 11, 20, 39, 40). However, concerns about cosmid instability in E. coli and the requirement for the cosmid sets to recombine precisely and in some cases repair a truncation in a terminal repeat have slowed the adoption of this technology.
More recently, full-length F-plasmid clones (also referred to as bacterial artificial chromosomes) of mouse cytomegalovirus (MCMV), EBV, and HSV-1 have been constructed (12, 27, 35, 37). Because the entire viral genome is maintained in a single E. coli plasmid, these clones do not require repair or homologous recombination following transfection into susceptible eukaryotic cells. Furthermore, F-plasmid cloning technology has gained widespread acceptance for the construction of mammalian genomic libraries due to their stable maintenance of large foreign DNA inserts in E. coli (29, 36). Therefore, F-plasmid-based clones of herpesvirus genomes have three important benefits: (i) they are stable in E. coli, (ii) they are amendable to E. coli genetic methods, and (iii) they result in productive viral infection without the need for repair or homologous recombination following transfection of eukaryotic cells.
We report here the first construction of an F-plasmid-based infectious clone of PRV. PRV is a member of the alphaherpesvirus subfamily that includes the human pathogens HSV-1 and varicella-zoster virus (23). These viruses are neurotropic and share the ability to spread from local sites of infection to the central nervous system in a neuronal circuit-specific manner (17). PRV provides an attractive model for detailed analysis of the pathogenesis of this virus group because of its ease of laboratory manipulation, its ability to infect and cause similar disease in a wide variety of animals, and its inability to infect humans (14). Because we are primarily interested in investigating the pathogenesis of PRV in animals, the emphasis of our design was to maintain the wild-type virulence of the parental virus. We therefore inserted the F-plasmid sequences into the viral gG locus, which is thought to be dispensable for viral spread and virulence in both rodent and porcine models of infection (references 2 and 18 and references therein). Virus harvested from transfection with the infectious clone was characterized for genotype and phenotype, both in tissue cultures and in the rodent model, in an effort to determine if the F-plasmid clone had merit for studying viral pathogenesis.
The clone was also subjected to transposon mutagenesis as a means to quickly and efficiently produce random viral insertion mutants, which were easily classified by transfection of supercoiled plasmid DNA from E. coli into eukaryotic cells. The stabilities of both the F-plasmid insertion and a transposon insertion were examined.
MATERIALS AND METHODS
Virus and cells.
PRV-Becker is a virulent isolate of PRV and the parental strain of all recombinant viruses used in this study (7). PRV-Becker has been in continuous passage in cell culture for over 10 years. PRV-BeBlue is a PRV-Becker derivative in which the lacZ gene from E. coli is fused in frame after the seventh amino acid of the viral gG gene. PRV-BeBlue expresses beta-galactosidase during infection and has been previously described (3). All PRV strains were propagated in the PK15 (porcine kidney 15) epithelial cell line. Virus titers were determined in duplicate by plaque assay on PK15 cells. The cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), while viral infections were performed in DMEM supplemented with 2% FBS. All work involving the manipulation of virus or E. coli harboring the infectious plasmid was conducted in a biosafety level 2 facility.
Plasmids.
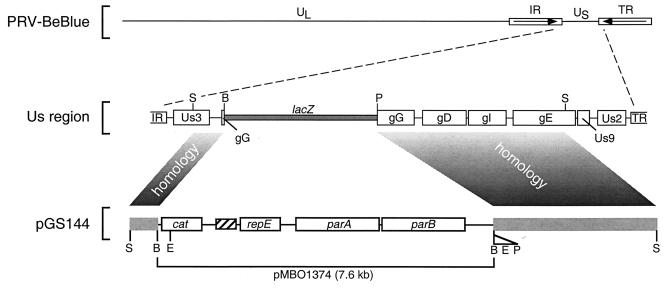
The mini-F plasmid pMBO1374 is a pMBO131 derivative in which a 645-bp HaeII fragment containing the multiple cloning site-embedded lacZ gene of pBluescript II KS(+) was subcloned into the unique SalI site of pMBO131 (29). This results in several unique cloning sites, including BamHI, which can be used in combination with a beta-galactosidase screen. pMBO1374 was a gift from Michael O’Connor. The gG gene of PRV-Becker was subcloned from pAK44, which contains a BamHI:EcoRI:XbaI:PstI linker subcloned into the endogenous BamHI and PstI sites in the gG open reading frame (ORF) and has previously been described (22). This was accomplished by releasing the region encoding gG from pAK44 as a 6.3-kb SphI fragment spanning the region from the Us3 gene to the gE gene. The SphI fragment was additionally digested with BamHI, resulting in two fragments of 0.8 and 5.3 kb. These fragments were religated at their SphI ends, thereby reversing their orientations relative to one another. The resulting 6.3-kb fragment was then subcloned into the unique BamHI site of pMBO1374, resulting in pGS144. This design allowed the linearization of pGS144 with SphI, such that the PRV-Becker sequences would have the appropriate orientation relative to the pMBO1374 vector sequence for subsequent recombination into viral genomic DNA (Fig. 1).
FIG. 1.
Construction of the recombinant virus PRV-251 is illustrated. The mini-F plasmid pMBO1374 was recombined into the gG locus of PRV-BeBlue from pGS144 following cotransfection into PK15 cells. The recombinant virus was plaque purified by screening for the loss of beta-galactosidase activity in a plaque overlay assay. UL, PRV unique long region; US, PRV unique short region; IR, PRV internal repeat; TR, PRV terminal repeat; lacZ, beta-galactosidase gene; cat, chloramphenicol resistance gene; repE, parA, and parB, replication and partitioning genes; hatched box, F-plasmid origin of replication; B, BamHI site; E, EcoRI site; P, PstI site; S, SphI site.
The transposon delivery plasmid pCGB12 is a derivative of pBSL202 (1). These plasmids encode RP4oriT and the R6K origin of replication. Both plasmids also contain the gene for ampicillin resistance (beta-lactamase) and Tn5-derived sequences. The Tn5 transposase gene is positioned outside of the transposable element, resulting in decreased transposon size and more stable integration (reviewed in reference 13). In pCGB12, the mini-Tn5 element of pBSL202 has been modified to carry a gene for kanamycin resistance (aphA-3) and a promoterless derivative of the gfp gene from Aequorea victoria. pCGB12 was a gift from Carlos Guzman.
A second delivery vector, pGS284, was used for allelic exchange. pGS284 is a pCVD442 derivative in which a synthetic oligonucleotide linker was inserted into the unique SphI and XbaI cloning sites of pCVD442 containing recognition sequences for NheI, NotI, NsiI, SalI, and BglII. The linker was a dimer comprised of two oligonucleotides: 5′CTAGCGGCCGCATGCATGTCGACAGAT3′ and 5′CTAGATCTGTCGACATGCATGCGGCCGCTAGCATG3′. The pCVD442 transfer vector is based on the RP4oriT and the R6K origin of replication of pGP704 (28). In addition, pCVD442 encodes beta-lactamase and carries the sacB gene from Bacillus amyloliquefaciens. The sacB gene provides a means of enrichment for recombinants which have lost the pCVD442 plasmid from an integrated merodiploid state, as has been previously described (33). pCVD442 was a gift from Michael Donnenberg. Repair of the pBecker1-1 mutation was accomplished with pGS294, which was constructed by cloning the ∼12-kb BglII-F fragment from PRV-Becker nucleocapsid DNA into the BamHI site of pGS284 (5).
Virus construction.
The PRV-251 virus was isolated from PK15 cells cotransfected with pGS144 linearized with SphI and PRV-BeBlue nucleocapsid DNA (Fig. 1). The desired recombinant virus was plaque purified by using the loss of beta-galactosidase activity as a screen, as previously described (22).
Isolation of viral DNA.
Linear viral DNA was isolated from nucleocapsids harvested from infected cells. Five 150-mm-diameter dishes of confluent PK15 cells were infected at a multiplicity of infection (MOI) of 5 PFU/cell each. After 1 h of absorption at 37°C, the inoculum was replaced with 20 ml of DMEM plus 10% FBS per dish. The dishes were incubated at 37°C for 10 to 15 h, and the cells were then harvested by scraping them into 2 ml of phosphate-buffered saline (PBS) per 150-mm-diameter dish. The combined sample was washed twice in PBS by pelleting in a Clay Adams brand Dynac centrifuge (Becton Dickinson) at 2,000 rpm (relative centrifugal force, 730), and the final pellet was resuspended in 10 ml of LCM buffer (130 mM KCl, 30 mM Tris [pH 7.4], 5 mM MgCl2, 0.5 mM EDTA, 0.5% Nonidet P-40 [NP-40], 0.043% 2-mercaptoethanol). The sample was extracted twice with 1.5 ml of Freon (1,1,2-trichloro-1,2,2-trifluoroethane) and then centrifuged through two LCM buffer-based glycerol step gradients (3.0 ml of 5% glycerol and 2.5 ml of 45% glycerol) with one-half of the sample loaded on top of each preestablished gradient. Centrifugation was done in a model L5-75 ultracentrifuge with a SW41 swinging-bucket rotor (Beckman) at 25,000 rpm for 1 h at 4°C. The nucleocapsid pellets were resuspended and combined in 9.5 ml of TNE (50 mM Tris [pH 7.5], 100 mM NaCl, 10 mM EDTA). The DNA was released with the addition of 0.5 ml of 10% sodium dodecyl sulfate (SDS) and 1 mg of proteinase K (Boehringer Mannheim). The sample was extracted twice with a 1:1 mixture of Tris-saturated phenol and chloroform, and the DNA was precipitated by the addition of 20 ml of ethanol prechilled to −20°C. The viral DNA was isolated in suspension with a glass hook, blotted dry, and resuspended in 0.5 ml of TE (10 mM Tris [pH 7.6], 1 mM EDTA).
For the transformation of E. coli, replicative intermediate (covalently closed circular) viral DNA was isolated from one 100-mm-diameter dish of confluent PK15 cells. The cells were infected at an MOI of 3 PFU/cell and incubated at 37°C for 5 h. The cells were harvested by scraping them into 1 ml of PBS prechilled to 4°C and washed once with an additional 10 ml of PBS and once with 10 ml of a solution containing 10 mM Tris (pH 8.0) and 10 mM EDTA (pH 8.0). The final cell pellet was resuspended in 2 ml of a solution containing 10 mM Tris (pH 8.0), 10 mM EDTA (pH 8.0), and 0.25 mg of proteinase K (Boehringer Mannheim) per ml. SDS was added to a final concentration of 0.6%, and the sample was immediately used to electroporate E. coli.
DNA transfections.
Transfections of viral DNA were done by the calcium phosphate precipitation method as previously described (38). In the case of bacterial infectious clones, plasmid DNA was isolated from 1 ml of a stationary-phase culture of E. coli by standard alkaline lysis procedures. The plasmid was suspended in 50 μl of water, and 45 μl of the preparation was used in the standard transfection protocol. The remaining 5 μl was examined following digestion with PstI, and the yield was compared to that of a pBecker1 plasmid preparation performed in parallel with the mutated plasmids. In all cases, yields of mutated plasmids were identical to yields of pBecker1, which averaged ∼0.1 μg. The pBecker1 sample was also included as a transfection control by which the cytopathic effect (CPE) for all mutant viruses was scored. By these methods, the transfection of pBecker1 and viable mutant derivatives typically yielded 50 to 100 infectious foci in a 100-mm-diameter dish of PK15 cells. This is equivalent to transfections of PRV nucleocapsid DNA, which average ∼1,000 foci/μg, indicating that supercoiled plasmid DNA and viral DNA are equally infectious.
Cloning PRV into bacteria.
E. coli DH10B (Research Genetics, Inc.) was transformed with 1 μl of fresh circular viral DNA isolated from infected PK15 cells (see above). The transformation was performed with a Gene Pulser II electroporation system with 0.1-cm Gene Pulser cuvettes (Bio-Rad). Settings were as follows: 1.8 kV, 200 Ω, and 25 μF. Bacteria were recovered in 0.45 ml of SOC (35a) and grown on Luria-Bertani (LB) plates containing 20 mg of chloramphenicol per ml.
Pulsed-field gel electrophoresis.
All pulsed-field gels were 1.0% agarose in a buffer of 40 mM Tris-acetate and 2 mM EDTA (TAE). Electrophoresis was conducted in a 15.5- by 15.5- by 3.5-in. chamber housing electrodes in an orthogonal configuration. Voltage was provided by a Hoefer PS 500XT power supply (Pharmacia) and was directed to the electrodes by a solenoid controlled by a ChronTrol electronic timer (Lindburg Enterprises, Inc.). Gels were typically run at 150 V with a switch time of 5 or 10 s.
Single-step growth curves.
Viral growth rates were determined by single-step growth analysis as previously described (38). Cells and supernatants were harvested at 2, 5, 8, 12, and 24 h following the removal of viral inoculum. Titers were determined in duplicate by plaque assay on PK15 cells.
Animal experiments, tissue processing, and immunohistochemistry.
Adult male Sprague-Dawley rats weighing 200 to 250 g at the time of the experiment were used in this study. Food and water were freely available during the course of the experiment, and the photoperiod was standardized to 14 h of light and 10 h of darkness. Experimental protocols were approved by the Princeton University Animal Welfare Committee and were consistent with the regulations stipulated by the American Association for Accreditation of Laboratory Animal Care and those in the Animal Welfare Act (Public Law 99-198). The animals were confined to a biosafety level 2 facility, and the experiments were conducted with specific safeguards as described previously (16).
For intraocular injections, 2.5 μl of virus suspension (approximately 109 PFU/ml) was injected into the vitreous humor of the left eye of an anesthetized animal. When symptoms of infection were overt, the animals were sacrificed and exsanguinated, and the brains were removed as described previously (15). Immunohistochemical analysis of coronal brain slices by using rabbit polyclonal antiserum to whole PRV (Rb133) has been described previously (15). Tissues were taken for analysis just prior to the estimated time of death.
For the recovery of PFU from the brain of an infected animal, no fixative was perfused through the animal following exsanguination. Instead, the unfixed brain was removed and immediately frozen in liquid nitrogen. A mortar and pestle were used to grind the frozen tissue in 4 ml of DMEM supplemented with 10% FBS. The homogenized sample was freeze-thawed for three consecutive rounds between −80 and 37°C and stored at −80°C. A stock of recovered virus was made by infecting one 10-cm-diameter dish of PK15 cells with a 0.1-ml sample.
Transposon mutagenesis.
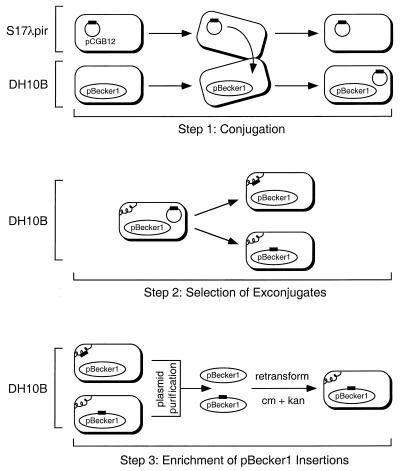
Transposon delivery and selection of exconjugates were performed essentially as previously described (13). The primary modification was the inclusion of 20 μg of chloramphenicol per ml and 50 μg of kanamycin per ml during selection of the exconjugates. E. coli S17-1λpir was used as the donor for the transposon delivery plasmid pCGB12 (see Table 1 in reference 13). The recipient was DH10B harboring pBecker1. Plasmids from the exconjugate population were purified by using Qiagen Plasmid Midi columns (Qiagen, Inc.) and electroporated into E. coli DH10B. Isolates harboring transposon insertions in pBecker1 were selected by a second round of growth in the presence of 20 μg of chloramphenicol per ml and 50 μg of kanamycin per ml (see Fig. 5).
FIG. 5.
Strategy for isolating transposon insertions in the pBecker1 plasmid. Transposon mutagenesis was carried out by conjugating a mini-Tn5-derived transposon into E. coli harboring pBecker1 (step 1). The delivery plasmid pCGB12 cannot replicate in DH10B. Therefore, kanamycin resistance (encoded by the transposon) and chloramphenicol resistance (encoded by pBecker1) together select for exconjugates. The resulting exconjugates harbor the transposon either in the E. coli chromosome or in pBecker1 (step 2). The isolation of transposon insertions in pBecker1 was accomplished by purifying F plasmids from the exconjugate pool, transforming the plasmid library into E. coli, and reselecting for pBecker1 and the transposon (step 3). The transposon is represented in all steps as a black rectangle. The E. coli chromosome is represented as a curled line.
Sequencing transposon and pBecker1 junctions was accomplished by subcloning the transposon-borne kanamycin resistance gene (aphA-3) along with the transposon I-end and pBecker1 flanking sequence into pSP72 (Promega). This was done by digesting full-length transposon insertion mutants of pBecker1 with EcoRI and SphI. EcoRI cuts the transposon once on the O-end side of aphA-3, and SphI cuts the viral sequences at a high frequency but does not cut the transposon. The desired clone was selected with 100 μg of ampicillin per ml and 50 μg of kanamycin per ml and used as a template for sequencing reactions with Sequenase 2.0 (Amersham) and a primer (5′-GACCCAAGTACCGCCACC3′) homologous to the transposon I end.
Allelic exchange.
Delivery of the wild-type UL36 allele and selection of exconjugates were performed essentially as for transposon mutagenesis. However, there were several important differences. The delivery vector used, pGS294 (see above), was carried in the donor strain S17-1λpir. The recipient was pBecker1 harbored in a derivative of DH10B in which the chromosomal recA1 allele was repaired to recA+ (E. coli GS243) (unpublished data). Conjugation was done by cross-streaking the donor and recipient on an LB plate. Exconjugates were isolated from the intersection of the two streaks and selected by growth in the presence of 20 μg of chloramphenicol per ml and 100 μg of ampicillin per ml. Unlike transposon delivery, exconjugates by this protocol resulted from integration of the entire delivery plasmid into pBecker1 by homologous recombination. Bacteria that spontaneously lost the integrated plasmid were enriched by growth in the presence of 5% sucrose in LB plates lacking NaCl at 30°C and confirmed by replica plating on three LB plates containing either 20 μg of chloramphenicol per ml, 100 μg of ampicillin per ml, or 50 μg of kanamycin per ml. Isolates with the repaired allele were chloramphenicol resistant, ampicillin sensitive, and kanamycin sensitive.
RESULTS
Construction of a full-length clone of PRV in E. coli.
The method for constructing an infectious clone of PRV was similar to the method previously used for MCMV (27). PRV sequences derived from a unique short SphI restriction fragment were introduced into the mini-F plasmid pMBO1374, such that the vector sequences were flanked by the gene encoding the gG glycoprotein (Fig. 1). The gG locus was chosen as the site of F-plasmid insertion because gG null mutants exhibit near wild-type virulence and spread in the vertebrate nervous system (2, 18, and references therein). The resulting construct was linearized by digestion with SphI and cotransfected into PK15 cells with purified viral DNA from PRV-BeBlue, a PRV-Becker-derived viral strain containing a lacZ insertion in the gG ORF (3). Recombinant virus was plaque purified by screening for the loss of beta-galactosidase activity and was designated PRV-251.
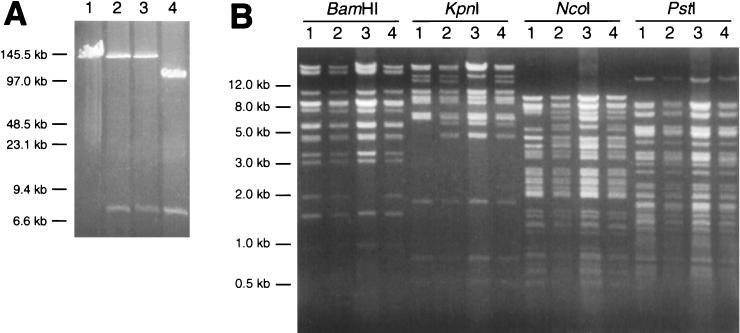
Viral DNA was isolated by treating PRV-251-infected cells with proteinase K and SDS. The resulting lysate was used to transform electrocompetent E. coli DH10B (Research Genetics, Inc.). Clones were selected by growth on LB plates containing chloramphenicol. Individual colonies were screened by digestion of plasmid DNA with NcoI, which cuts PRV DNA at a high frequency (data not shown). Isolates that appeared to have F plasmids with large inserts were examined further by pulsed-field gel electrophoresis, following digestion with EcoRI. Because sites for EcoRI are present within the pMBO1374 sequences derived from pGS144, but not in the PRV-Becker sequence (Fig. 1), full-length clones were initially identified by comigration with purified PRV-Becker nucleocapsid DNA. Of ten isolates screened in this way, three were chosen for examination by pulsed-field gel electrophoresis, and two appeared to be full-length (Fig. 2A).
FIG. 2.
(A) Pulse-field gel electrophoresis of EcoRI-digested E. coli F plasmids. The F plasmids were isolated from three chloramphenicol-resistant isolates of E. coli that were transformed with DNA recovered from PK15 cells infected with PRV-251. PRV-Becker DNA was isolated from viral nucleocapsids and provides a size standard for full-length linear viral DNA. Two of the three isolates appear to contain full-length viral DNA. The third isolate carries a deletion in the viral sequences. The 7.6-kb restriction fragment present in all three isolates is derived from the pMBO1374 sequences present in the gG ORF. The first isolate (lane 2) was designated pBecker1. Size standards are indicated. Lane 1, PRV-Becker nucleocapsid DNA; lanes 2 to 4, isolated E. coli F plasmids. (B) Restriction patterns of linear pBecker1 were compared to those from viral nucleocapsid DNAs. Each sample was digested with BamHI, KpnI, NcoI, and PstI, all of which cut PRV-Becker DNA at a high frequency. Lane 1, PRV-Becker viral nucleocapsid DNA; lane 2, PRV-251 viral nucleocapsid DNA; lane 3, pBecker1 E. coli plasmid DNA; lane 4, vBecker1 (virus harvested from pBecker1 transfection) viral nucleocapsid DNA.
One of these clones was designated pBecker1 and was further examined by restriction digestion with a panel of restriction enzymes that cut the plasmid at a high frequency. The comparison of these digests to those resulting from digestion of viral nucleocapsid DNA from PRV-Becker indicated that pBecker1 contained virus-derived sequence; however, several restriction fragment length polymorphisms were observed (Fig. 2B). The polymorphisms were determined to arise from two sources. First, pBecker1 contains the pMBO1374 sequence in gG that is absent in PRV-Becker genomic DNA (compare PRV-251 to PRV-Becker and pBecker1; Fig. 2B). Second, pBecker1 isolated from E. coli is a covalently closed circle, while viral DNA isolated from nucleocapsids is linear. When pBecker1 was transfected into eukaryotic cells and viral nucleocapsid DNA was isolated (see below), the polymorphisms that were not accounted for by pMBO1374 were absent (compare PRV-251, pBecker1, and vBecker1; Fig. 2B). Together, these data indicated that pBecker1 was a full-length isogenic clone of the PRV-Becker genome.
Characterization of vBecker1 in tissue culture.
The transfection of pBecker1 into PK15 cells resulted in productive viral infection. Virus harvested from cells transfected with pBecker1 was designated vBecker1. Viral titers of vBecker1 harvested from cells transfected with pBecker1 were typically on the order of 108 to 109 PFU per ml, which is comparable to standard titers of PRV-Becker.
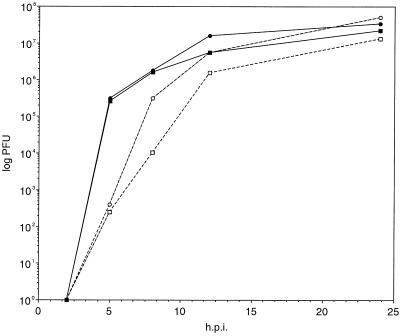
The growth properties of vBecker1 in tissue cultures were more closely examined by single-step growth curves. PK15 cells were infected with vBecker1 and PRV-Becker, and virus was recovered from both cells and supernatants at 2, 5, 8, 12, and 24 h postinfection. The titers of each sample were determined by plaque assay on PK15 cells (Fig. 3). Although both stocks had similar numbers of cell-associated PFU at all time points, a transient drop in vBecker1 relative to PRV-Becker was observed in the supernatants. This lag in PFU release was most dramatic at 8 h. A similar drop was seen with PRV-BeBlue, which contains a lacZ insertion in the gG locus (data not shown). Thus, insertions into the PRV-Becker gG locus appear to cause a small defect in viral release from cells.
FIG. 3.
Single-step growth curves of wild-type PRV-Becker and vBecker1. Virus was harvested from both the media and cells at 2, 5, 8, 12, and 24 h postwash, and titers were determined as described in Materials and Methods. Shown are data for cells (solid symbols), supernatants (open symbols), PRV-Becker (circles), and vBecker1 (squares). h.p.i., hours postinfection.
Characterization of vBecker1 in animals.
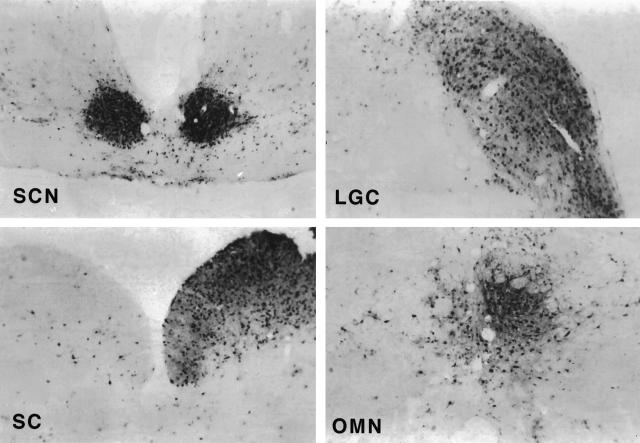
For pBecker1 to be used as a general tool for studying PRV biology, virus derived from it must be virulent and spread like the parental virus in the vertebrate nervous system. As we have previously reported, PRV-Becker spreads to the visual centers of the central nervous system in a circuit-specific manner following the inoculation of virus into the eye of an adult male Sprague-Dawley rat. We used this model to address the neuroinvasiveness (ability to spread to the central nervous system) of vBecker1. Three animals were inoculated into the vitreous body of one eye with 2.5 × 106 PFU of vBecker1. The animals were monitored for symptoms of infection and sacrificed when death was imminent. The mean time to terminal symptoms was 72.25 h (standard deviation, 1.75; n = 3), which is equivalent to the results in a previous report for PRV-Becker (6). Each animal was immediately perfused with fixative, and the brains were collected for examination of viral spread. Sections of each brain were examined for viral antigen by immunohistochemistry. In this model, PRV-Becker spreads to the retinorecipient regions of the brain, including the superior colliculus (SC), the dorsal and ventral geniculate nuclei (DGN and VGN), the intergeniculate leaflet (IGL), the suprachiasmatic nucleus (SCN), and the oculomotor nucleus (OMN) (8). Each of these regions was examined in serial coronal sections of the three brains, and vBecker1 was found to be capable of circuit-specific spread to each retinorecipient region (Fig. 4).
FIG. 4.
Immunohistochemistry of representative brain slices from a Sprague-Dawley rat infected with vBecker1. The presence of viral antigen is indicated by the dark stain and is shown in the SCN, lateral geniculate complex (DGN, VGN and IGL), SC, and OMN.
Transposon mutagenesis of pBecker1.
The production of herpesvirus mutants traditionally is a time-consuming process, taking several weeks from original transfection to final purified stock. However, the F-plasmid technology can speed this process up significantly. For example, we applied transposon mutagenesis of pBecker1 in E. coli to rapidly isolate random insertions of a mini-Tn5-derived cassette in the viral genome. The time from transposition in E. coli to final stocks of isolated viral insertion mutants was 8 days, and 10 to 15 viral stocks of novel insertion mutants could be processed simultaneously. The transposon was delivered to E. coli DH10B harboring pBecker1 by conjugation from E. coli S17-1λpir harboring pCGB12. The pCGB12 plasmid encodes three important elements necessary for delivery of the transposon: (i) the RP4oriT initiation of transfer locus that allows for conjugation of the plasmid in the presence of the RP4tra genes, which are integrated in the chromosome of S17-1λpir; (ii) an R6K origin of replication that is functional in the presence of the π protein expressed in the S17-1λpir strain but is inoperative in DH10B; (iii) the mini-Tn5 transposon that encodes kanamycin resistance in E. coli (reviewed in reference 13). Exconjugates were selected by growth in the presence of chloramphenicol, which selects for the presence of pBecker1, and kanamycin, which selects for the presence of the transposon (Fig. 5).
The resulting exconjugate pool should contain a mixture of isolates with transposon insertions in either pBecker1 or the DH10B chromosome. We enriched for the former by purifying the pBecker1 plasmids from the pool by alkaline lysis and by using the plasmid preparation to transform fresh DH10B. Transformants were selected for growth in the presence of both chloramphenicol and kanamycin. Only those exconjugates with the transposon inserted into pBecker1 can provide resistance to both antibiotics in the enrichment procedure (Fig. 5).
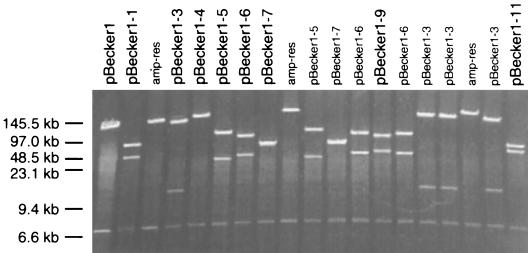
The site of the transposon insertion was mapped for a series of individual isolates by pulsed-field gel electrophoresis following digestion with EcoRI, which cuts the pMBO1374 sequence in the gG ORF and the transposon but not the viral sequences. An example of such a mapping experiment is shown in Fig. 6. Isolates that appeared similar were examined more closely by digestion with additional restriction enzymes, and siblings were discarded (data not shown). The exact site of insertion was determined by sequencing. In all cases, sequencing was performed with a primer specific to the transposon I end. The primer was recessed from the end of the transposon to easily resolve the fusion junction of the transposon and pBecker1 sequences and to avoid annealing to the indirect repeats of the transposon.
FIG. 6.
Pulsed-field gel electrophoresis of pBecker1::mini-Tn5 isolates digested with EcoRI. Two EcoRI sites are present in pBecker1, both of which are in the pMBO1374 sequences in the gG ORF. A third EcoRI site is carried by the mini-Tn5 transposon. Therefore, pBecker1::mini-Tn5 isolates release three DNA fragments following restriction with EcoRI: a ∼7-kb fragment derived from pMBO1374 and two PRV-Becker-derived fragments. The sizes of the latter two fragments provide preliminary information regarding the transposon location. Isolates labeled in black were saved for future study, and isolates labeled in gray were discarded, because they either were siblings of a previous isolate or were illegitimate recombinants between pCGB12 and pBecker1. The latter were identified as ampicillin resistant, indicating that the pCGB12 vector sequences were integrated into pBecker1. Size standards are indicated.
Each of the sequenced isolates was transfected into PK15 cells to produce recombinant virus and determine if the insertion was in an essential viral gene. Fresh plasmid DNA was used for all transfections, and a preparation of pBecker1 plasmid DNA was always included to control for plasmid yields and subsequent transfection efficiencies. If plasmid yields of mutated genomes were not equal to those of the pBecker1 control, they were not used for transfection. A summary of the mapping and transfection data is shown in Table 1. Because the PRV genome has not yet been fully sequenced, several isolates had transposon insertions in regions that have no homology to known PRV genes. One of these isolates had significant homology to the HSV-1 UL36 gene (24).
TABLE 1.
Summary of pBecker1::mini-Tn5 isolates
| E. coli plasmid | Distance (kb) from Fa | Locusb | % Intactc | CPEd |
|---|---|---|---|---|
| pBecker1-1 | ∼50 | UL36 homology | ? | − |
| pBecker1-2 | ∼60 | UL39 (RR1) | 97 | ++ |
| pBecker1-3 | ∼10 | IR (IE180) | 100e | ++l |
| pBecker1-4 | ∼3 | US6 (gD) | 5 | +m |
| pBecker1-5 | ∼35 | UL28 (ICP 18.5) | 46 | − |
| pBecker1-6 | ∼35 | UL13 (kinase) | 49 | + |
| pBecker1-7 | ∼70 | UL41 (VHS) | 100f | ++ |
| pBecker1-8 | ∼30 | UL5 (helicase) | 54 | − |
| pBecker1-9 | ∼40 | no homology | ? | − |
| pBecker1-10 | <2 | US4 (gG) | [89]g | ++ |
| pBecker1-11 | ∼60 | UL22 (gH) | 100h | + |
| pBecker1-12 | ∼15 | IR (IE180) | 100i | ++l |
| pBecker1-13 | ∼30 | UL5 (helicase) | 25 | − |
| pBecker1-14 | ∼45 | UL38 (VP19c) | 7 | − |
| pBecker1-15 | ∼60 | no homology | ? | ++ |
| pBecker1-16 | ∼50 | UL21 | 70 | + |
| pBecker1-17 | ∼20 | UL19 (VP5) | 100j | + |
| pBecker1-18 | ∼70 | UL44 (gC) | 100k | ++ |
| pBecker1-19 | ∼55 | no homology | ? | − |
| pBecker1-20 | ∼35 | UL28 (ICP 18.5) | 44 | − |
| pBecker1-21 | ∼40 | UL9 (origin bp) | 25 | − |
| pBecker1-22 | ∼40 | UL29 (ICP 8) | 36 | − |
| pBecker1-23 | ∼40 | UL13 (kinase) | 90 | + |
Approximate location of the transposon insertions is listed as the distance (in kilobases) from the F plasmid in the gG ORF, based on restriction digestion analysis. The distance listed is always no more than half of the total genome size, as the genome is circular in E. coli.
Determined by sequencing the I end of the transposon.
Percentage of ORF intact 5′ of the transposon insertion site, determined by sequencing the I end of the transposon. Insertions in previously unidentified PRV loci are indicated by question marks.
Each isolate was scored for CPE following transfection into PK15 cells. Designations are as follows: no CPE resulted (−), CPE manifested with notably slower kinetics than pBecker1 (+), or CPE was similar or identical to that of pBecker1 (++).
Transposon is 41 bp upstream from the transcript start site.
Transposon is 54 bp upstream from the ORF.
The gG ORF is interrupted at the 5′ end by the F plasmid; therefore, the number given is only an indication of the transposon insertion site.
Transposon is 19 bp downstream from the ORF.
Transposon is between the E4 enhancer element and the P1 promoter of IE180.
Transposon is 2 bp downstream from the ORF.
Transposon is 118 bp downstream from ORF.
Wild-type CPE probably results from the equalization of the inverted repeats during viral replication.
CPE was restricted to small infectious foci.
Reversion of a transposon mutant by allelic exchange.
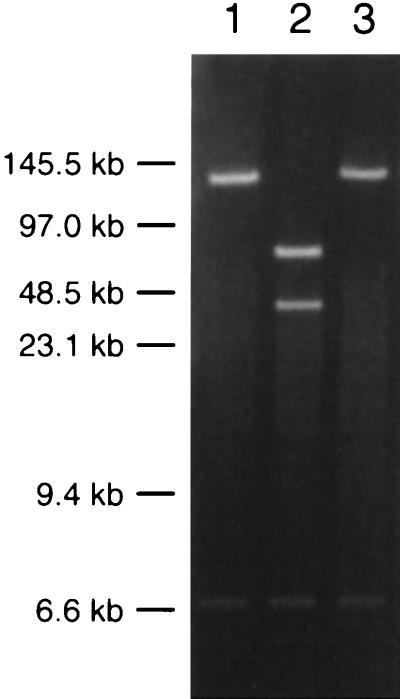
The mutagenesis of viral genes with a transposon simplifies and accelerates further modifications of the mutated gene, due to the selectable marker associated with the transposon. For example, homologous recombination in E. coli can be achieved by the method of allelic exchange. This provides a means for targeted mutagenesis of the herpesvirus infectious clone in E. coli, as previously demonstrated with the MCMV F-plasmid-based clone (27). We used allelic exchange to revert a transposon insertion mutation in the UL36 gene of pBecker1-1 to the wild type by using the loss of kanamycin resistance, which is encoded by the transposon, as a marker for successful recombination. We chose the UL36 mutant because it was a lethal insertion and reversion could be unambiguously confirmed by phenotype. The UL36 insertion allele was replaced by homologous recombination between the pBecker1-1 plasmid and a delivery plasmid containing the wild-type UL36 locus in a ∼12-kb PRV-Becker BglII-F fragment (5). Introduction of the wild-type UL36 allele was accomplished by conjugation to a recA+ strain of E. coli carrying pBecker1-1. Because the infectious clone, transposon, and delivery plasmid all have selectable markers, the desired recombinant was isolated by screening for growth or absence of growth in the presence of appropriate antibiotics and metabolites (see Materials and Methods). One such isolate was further examined by EcoRI digestion and was found to lack the EcoRI site in UL36 that was present in the transposon insertion of the parental pBecker1-1 plasmid (Fig. 7). This revertant was given the designation pBecker1-1R. The transfection of pBecker1-1R into PK15 cells resulted in productive viral infection, indicating that the UL36 gene was successfully repaired (Table 2).
FIG. 7.
Demonstration of reversion by allelic exchange. EcoRI digests of pBecker1-derived plasmids were examined by pulsed-field gel electrophoresis. Size standards are indicated. Lane 1, pBecker1; lane 2, pBecker1-1; lane 3, pBecker1-1R.
TABLE 2.
Titers of virus recovered from transfected F plasmids
Titers were determined for virus harvested after transfection of each E. coli plasmid. Virus was harvested when CPE was considered complete (5 days posttransfection).
The infection resulting from transfection of pBecker1-1 was harvested at the same time as pBecker1 and pBecker1-1R, although no CPE had occurred. No PFU were found in 3 ml of the harvest.
Stability of transposon and F-plasmid insertions in vBecker1.
For transposon mutagenesis to be useful in viral genetics, the insertions must be stable in the recombinant virus resulting from transfection. We examined the vBecker1-6 isolate for stability by serial passage in PK15 cells. The transposon insertion in the UL13 gene of mutant pBecker1-6 produced virus following transfection, but CPE developed at a reduced rate compared to the pBecker1 parent (Table 1). We reasoned that a spontaneous reversion to a wild-type phenotype would have a growth advantage and might enrich for spontaneous deletions of the transposon sequences if the insertion was not stable.
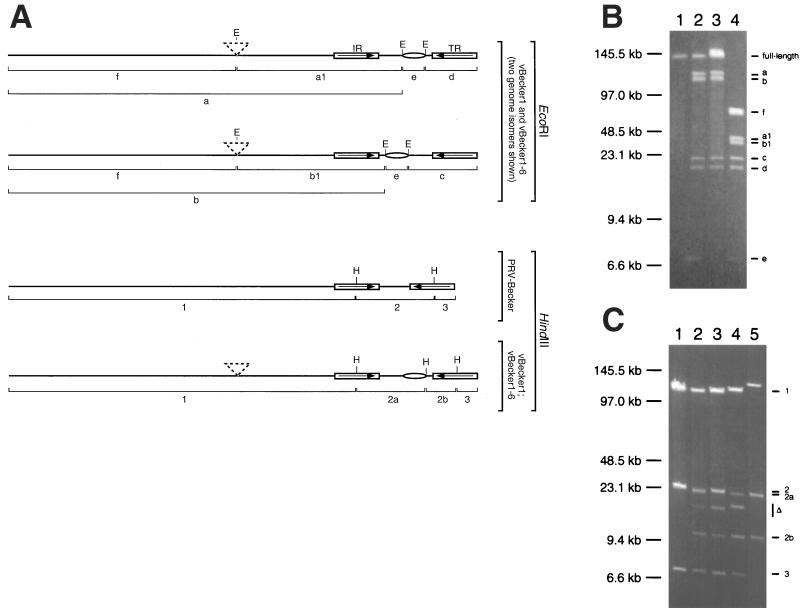
vBecker1-6 harvested from a transfection of pBecker1-6 was serial passaged in PK15 cells for five rounds at a low MOI (∼0.01 PFU/cell). Viral DNA recovered from the final infection was examined by EcoRI digestion and compared to digestions of vBecker1 and PRV-Becker DNAs, which are not cut by EcoRI. As in Fig. 7, EcoRI is only predicted to cut these DNAs in the pMBO1374 sequence in the gG ORF and in the transposon. However, the viral genomes are a more complex substrate than the plasmids isolated from E. coli, because they are linear molecules existing as two distinct isoforms. The genome isomerization occurs in eukaryotic cells as a result of the inversion of the unique short region of the genome, which is flanked by two large inverted repeats (referred to individually as the internal and terminal repeats) (5). The EcoRI sites present in the pMBO1374 sequence participate in this isomerization, as the gG ORF resides in the unique short region of the viral genome. The transposon in vBecker1-6, which is in the UL13 gene, is not affected by this isomerization. Therefore, the digestion of vBecker1 DNA is predicted to release five restriction fragments: the unique long region, internal repeat, and a varying piece of the unique short region depending upon isomerization (a and b); the terminal repeat and a varying piece of the unique short region depending upon isomerization (c and d); and the majority of pMBO1374 (e). The presence of the transposon in the UL13 gene of vBecker1-6 is predicted to truncate the a and b fragments (a1 and b1) and produce a sixth fragment (f) which consists of the majority of the unique long region (Fig. 8A). These fragments were all observed as expected, and there was no evidence of the untruncated a and b fragments in the vBecker1-6 sample, indicating that the transposon was stable (Fig. 8B).
FIG. 8.
Determination of genomic stability of recombinant viruses. (A) Schematic representation of vBecker1, vBecker1-6, and PRV-Becker genomes. EcoRI sites (E) with predicted restriction fragments are shown for vBecker1 and vBecker1-6 (upper half). The genomes are shown with the unique short region in two orientations, because the EcoRI fragment lengths are dependent upon genome isomerization. HindIII sites (H) with predicted fragments are shown for PRV-Becker, vBecker1, and vBecker1-6 (lower half). Isomerization of the genome does not affect HindIII restriction patterns. Also shown are the transposon insertion in vBecker1-6 (dashed triangles), inverted repeats (internal repeat [IR] and terminal repeat [TR]), and the pMBO1374 sequence (ellipse). (B) Pulsed-field gel electrophoresis of viral nucleocapsid DNAs digested with EcoRI. Lane 1, PRV-Becker; lane 2, vBecker1; lane 3, vBecker1 isolated from rat brain; lane 4, serially passaged vBecker1-6. Restriction fragments are labeled as in panel A. Size standards are indicated. (C) Pulsed-field gel electrophoresis of viral nucleocapsid DNAs digested with HindIII. Lane 1, PRV-Becker; lane 2, vBecker1; lane 3, vBecker1 isolated from rat brain; lane 4, serially passaged vBecker1-6; lane 5, pBecker1 F-plasmid DNA. Restriction fragments are labeled as in panel A. PRV-Becker provides size standards for fragments 1 and 3, and pBecker1 provides size standards for fragments 2a and 2b. In vBecker1-6, the size of fragment 1 is increased due to the presence of the transposon. In pBecker1, fragments 1 and 3 migrate as a single ligated band. Fragments harboring deletions are indicated (▵). Size standards are indicated.
Unexpectedly, the vBecker1 sample contained DNAs that were not cut by EcoRI (therefore comigrating with the PRV-Becker full-length viral genome sample). This indicated that vBecker1 was composed of a mixed population in which some of the virus had lost the pMBO1374 sequence in the gG ORF. Because the two EcoRI sites of the pMBO1374 insertion are ∼7 kb apart, deletions of at least this size must have occurred in the population. We also examined the restriction pattern of vBecker1 harvested from the brain of an infected Sprague-Dawley rat and found that the proportion of the deleted viruses had increased, but undeleted vBecker1 still appeared in the population (Fig. 8B). The deletion of the pMBO1374 sequence also occurred in vBecker1-6 but was not evident by EcoRI digestion, as the f fragment comigrated with the fragment lacking the vector sequence (see below).
To estimate the size of the deletion in vBecker1 and vBecker1-6, the DNAs were digested with HindIII, which cuts in each of the inverted repeats flanking the unique short region and once in the pMBO1374 sequence (Fig. 8A). In addition to the expected HindIII restriction fragments, new fragments appeared as a result of deletion of pMBO1374 in the viral populations (Fig. 8C). We estimated the deletion to be approximately 12.5 kb in the vBecker1 population and 11.5 kb in the vBecker1-6 population. Taking the 7.6 kb of pMBO1374 sequence into account, this indicated that approximately 5 to 6 kb of viral sequence was deleted along with the pMBO1374 sequence.
DISCUSSION
We describe here a full-length PRV infectious clone, pBecker1. By cloning the entire genome of PRV into E. coli and introducing further genomic modifications in the absence of viral growth and selective pressures, we have bypassed many of the difficulties with mutagenesis inherent to traditional cotransfection methods. Furthermore, we gain access to bacterial genetic techniques that were previously impossible or cumbersome to implement.
The insertion of F-plasmid sequences inevitably affects viral function, making the choice of the site of F-plasmid insertion critical. In the case of MCMV, the F-plasmid sequences replaced a ∼8-kb region of viral DNA that was nonessential for viral replication in culture (27). The EBV clone was derived from a mutant virus with a large deletion, and the deletion site was chosen as the insertion site for the F-plasmid sequence (21). Two HSV-1 clones have been made, both of which were designed as helper viruses for defective herpesvirus vectors. In one case, the F plasmid was inserted into the gene encoding the virus host shutoff function (UL41), and the other replaced portions of two genes encoding tegument phosphoproteins (UL46 and UL47) with the F-plasmid sequences (35, 37). The site of F-plasmid insertion was a minor concern with the published HSV-1 clones, as both are intended as helper viruses used for amplicon packaging, and many viral functions involved in virulence were dispensable (35, 37). However, for the genetic analysis of viral pathogenesis, a herpesvirus clone cannot be attenuated. Our initial goal was to design the PRV clone pBecker1 such that the F-plasmid sequences would not result in attenuation of the resulting recombinant virus. Previously, viruses containing lacZ insertions in the gG ORF were observed to have no detectable defect in viral spread or virulence in both rodent and porcine models of infection (references 2 and 18 and references therein). As such, we targeted the F-plasmid sequences to the gG locus.
The growth of vBecker1 was similar to that of PRV-Becker in cell culture. In the rat eye model of infection, vBecker1 killed host animals with a mean time to terminal symptoms indistinguishable from that of wild-type PRV-Becker (6). Additionally, vBecker1 spread to the SC, DGN, VGN, IGL, and SCN, all of which require anterograde transport of the virus from the somata of infected retinal ganglion cells, and to the OMN, which requires infection of axon terminals in the eye and retrograde transport to neuron cell bodies in the brain (32). The invasion of vBecker1 into the rat brain by the ocular route was indistinguishable from that of PRV-Becker, which has been previously described, and demonstrates that vBecker1 was capable of both anterograde and retrograde spread in the vertebrate nervous system (8). Also like PRV-Becker, circuit-specific spread was not limited to first- and second-order neurons, as instances of spread through at least three sequential synaptically linked neurons were indicated by the presence of infected cortical neurons (data not shown) (30). Thus, vBecker1 was indistinguishable from wild-type virus in our animal model of infection. Our first application of pBecker1 was to produce a collection of viral transposon mutants. Transposon mutagenesis offers a fast procedure for the isolation of random insertion mutations in E. coli. The use of Tn5 for insertion mutagenesis of a herpesvirus was first reported in 1987 (41). At that time, there were no cosmid- or F-plasmid-based clones of herpesviruses available. Instead, the authors limited their study to mutagenesis of pBR322/5 vectors carrying pieces of the HSV-1 unique short region in E. coli. The method required recombining the mutated unique short fragment into full-length viral DNA following cotransfection, which limited the study to three recombinant viruses with insertions in nonessential genes. Importantly, all three transposon insertions were reported to be stable in the recombinant viruses and did not interfere with other viral functions (41). This is in agreement with our finding that a transposon insertion that conveys an apparent growth defect is stable during serial passage of the virus.
A previous study using a mini-Mu phage to create random insertions in a cloned 10.4-kb fragment of the HSV-1 genome resulted in some instances of recombinant virus that possessed large unexpected deletions. In this case the mini-Mu phage contained an HSV-1 thymidine kinase (TK) gene, and recombination between the Mu TK gene and the endogenous HSV-1 TK gene was suggested to be the cause of this instability (19). In any case, the Tn5 transposons here do not contain any viral sequences. Because the transposons are stable in the recombinant viruses, the transfection of mutated pBecker1 derivatives into susceptible eukaryotic cells results in a purified mutant virus population without the need for plaque purification. Therefore, by using transposon mutagenesis in combination with the infectious clone we were able to simplify and speed up the isolation of mutated viruses. We sequenced 23 mutations in E. coli to determine the site of transposon insertion. From these, we isolated 13 individual mutant viruses. The remaining 10 genomes had lethal transposon insertion mutations. Transposon insertions occurred randomly in all regions of the viral genome, including the unique long region, unique short region, and inverted repeats. Several insertions were in noncoding regions, but the majority were in viral ORFs. The latter included genes encoding known or potential capsid (VP19c), tegument (UL13, UL21, and UL36), envelope (gD and gG), and nonstructural (RR1, ICP8, ICP18.5, UL5, and UL9) proteins. Several mutants had insertions in previously undocumented PRV loci, and one of these was a homologue of the HSV-1 gene UL36 (24). The HSV-1 UL36 gene encodes VP1/2, a 270-kDa tegument protein (25, 26). A PRV homologue of UL36 has not been previously reported. The transfection of pBecker1-1 did not result in productive infection, demonstrating that the gene is essential in PRV, in agreement with the occurrence of a temperature-sensitive mutation in the UL36 gene of HSV-1 (4).
We isolated multiple transposon insertions in the UL5, UL13, and UL28 genes. Although no two transposons were inserted in the same location of any gene, transfection yielded consistent results between viruses with different insertions in the same gene. Multiple insertions of transposons in a single viral gene have the potential to produce a series of truncation mutants which could be useful to the study of protein function; however, the presence of the transposon sequence may destabilize viral transcripts. For example, the transfection of pBecker1-17, which has a transposon insertion 2 bp downstream from the stop codon of the major capsid protein VP5, results in disseminated CPE at a notably reduced rate compared to that of pBecker1. Therefore, the study of truncated gene products is probably best approached by site-directed mutagenesis and allelic exchange. However, further allelic exchange will be simplified in E. coli by the transposon insertion mutation.
The stability of F plasmids in viral genomes has not previously been addressed. We were surprised to find that the F-plasmid sequence could be lost spontaneously from vBecker1 in eukaryotic cells. Although virus lacking the F-plasmid sequence was amplified during infection of the rat, the deletion event appears to occur during the transfection of the DNA into eukaryotic cells. PRV-251, the source of the viral genome used to establish the pBecker1 clone in E. coli, shows no signs of instability based on EcoRI digestion of isolated nucleocapsid DNA (data not shown). We have recently isolated a second infectious clone of PRV-Becker that has the F plasmid inserted at another locus, and this virus is stable (work in progress). This implies that the size of the F-plasmid insertion alone is not the problem but rather the location of the insertion is an important factor. Instability may be sequence specific as well, as the lacZ insertion in the gG gene of PRV-BeBlue, which is in the same location as the F plasmid in vBecker1, shows no signs of instability based on beta-galactosidase activity in passaged virus (our unpublished observations). At this time, we cannot fully explain why the F-plasmid DNA is deleted in vBecker1. The deletions do not appear to be the result of site-specific or homologous recombination, as the deletion varied in size between viruses harvested from independent transfections. Clearly, as new herpesvirus infectious clones are constructed, their stabilities will have to be determined empirically.
In conclusion, we have described a full-length herpesvirus clone amenable to the study of viral pathogenesis. While we found this clone to produce virus closely approximating PRV-Becker in phenotype, the insertion of F-plasmid sequences is not without effect. Although pBecker1 was very stable in E. coli, we found spontaneous deletions of the F-plasmid sequences upon transfection into eukaryotic cells. Nevertheless, these mutations did not result in appreciable phenotypic defects and therefore do not preclude the use of the clone for studies of neurotropism and neurovirulence. By applying the clone to the method of transposon mutagenesis, mutant viruses could be rapidly produced. We intend to examine the neurovirulence and neurotropism of viruses harboring transposon insertions in future studies.
ACKNOWLEDGMENTS
We thank Michael O’Connor, Carlos Guzman, and Michael Donnenberg for generously sharing their plasmids, Jean Schwarzbauer for extensive use of her electroporator, and Tom Silhavy for informative discussions during the course of the work.
This work was supported by NINDS grant 1RO133506 to L.W.E. G.A.S. is a Lilly Fellow of the Life Sciences Research Foundation.
REFERENCES
- 1.Alexeyev M F, Shokolenko I N, Croughan T P. New mini-Tn5 derivatives for insertion mutagenesis and genetic engineering in gram-negative bacteria. Can J Microbiol. 1995;41:1053–1055. doi: 10.1139/m95-147. [DOI] [PubMed] [Google Scholar]
- 2.Babic N, Mettenleiter T C, Ugolini G, Flamand A, Coulon P. Propagation of pseudorabies virus in the nervous system of the mouse after intranasal inoculation. Virology. 1994;204:616–625. doi: 10.1006/viro.1994.1576. [DOI] [PubMed] [Google Scholar]
- 3.Banfield B W, Yap G S, Knapp A C, Enquist L W. A chicken embryo eye model for the analysis of alphaherpesvirus neuronal spread and virulence. J Virol. 1998;72:4580–4588. doi: 10.1128/jvi.72.6.4580-4588.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batterson W, Furlong D, Roizman B. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Porat T, Kaplan A S. Molecular biology of pseudorabies virus. In: Roizman B, editor. The herpesviruses. New York, N.Y: Plenum Publishing Corp.; 1985. pp. 105–173. [Google Scholar]
- 6.Card J P, Dubin J R, Whealy M E, Enquist L W. Influence of infectious dose upon productive replication and transynaptic passage of pseudorabies virus in rat central nervous system. J Neurovirol. 1995;1:349–358. doi: 10.3109/13550289509111024. [DOI] [PubMed] [Google Scholar]
- 7.Card J P, Rinaman L, Schwaber J S, Miselis R R, Whealy M E, Robbins A K, Enquist L W. Neurotropic properties of pseudorabies virus: uptake and transneuronal passage in the rat central nervous system. J Neurosci. 1990;10:1974–1994. doi: 10.1523/JNEUROSCI.10-06-01974.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Card J P, Whealy M E, Robbins A K, Moore R Y, Enquist L W. Two α-herpesvirus strains are transported differentially in the rodent visual system. Neuron. 1991;6:957–969. doi: 10.1016/0896-6273(91)90236-s. [DOI] [PubMed] [Google Scholar]
- 9.Coen D M, Ramig R F. Viral genetics. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 101–139. [Google Scholar]
- 10.Cohen J I, Seidel K E. Generation of varicella-zoster virus (VZV) and viral mutants from cosmid DNAs: VZV thymidylate synthetase is not essential for replication in vitro. Proc Natl Acad Sci USA. 1993;90:7376–7380. doi: 10.1073/pnas.90.15.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham C, Davison A J. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology. 1993;197:116–124. doi: 10.1006/viro.1993.1572. [DOI] [PubMed] [Google Scholar]
- 12.Delecluse H J, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci USA. 1998;95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 14.Enquist L W. Circuit-specific infection of the mammalian nervous system. ASM News. 1995;61:633–638. [Google Scholar]
- 15.Enquist L W, Card J P. Pseudorabies virus: a tool for tracing neuronal connections. In: Enquist L W, Lowenstein P R, editors. Protocols for gene transfer in neuroscience: towards gene therapy of neurological disorders. New York, N.Y: John Wiley & Sons, Inc.; 1996. pp. 333–348. [Google Scholar]
- 16.Enquist L W, Dubin J, Whealy M E, Card J P. Complementation analysis of pseudorabies virus gE and gI mutants in retinal ganglion cell neurotropism. J Virol. 1994;68:5275–5279. doi: 10.1128/jvi.68.8.5275-5279.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enquist L W, Husak P J, Banfield B W, Smith G A. Infection and spread of alphaherpesviruses in the nervous system. Adv Virus Res. 1998;51:237–347. doi: 10.1016/s0065-3527(08)60787-3. [DOI] [PubMed] [Google Scholar]
- 18.Heffner S, Kovacs F, Klupp B G, Mettenleiter T C. Glycoprotein gp50-negative pseudorabies virus: a novel approach toward a nonspreading live herpesvirus vaccine. J Virol. 1993;67:1529–1537. doi: 10.1128/jvi.67.3.1529-1537.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkins F J, Casadaban M J, Roizman B. Application of the mini-Mu-phage for target-sequence-specific insertional mutagenesis of the herpes simplex virus genome. Proc Natl Acad Sci USA. 1985;82:4773–4777. doi: 10.1073/pnas.82.14.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kemble G, Duke G, Winter R, Spaete R. Defined large-scale alterations of the human cytomegalovirus genome constructed by cotransfection of overlapping cosmids. J Virol. 1996;70:2044–2048. doi: 10.1128/jvi.70.3.2044-2048.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kempkes B, Pich D, Zeidler R, Sugden B, Hammerschmidt W. Immortalization of human B lymphocytes by a plasmid containing 71 kilobase pairs of Epstein-Barr virus DNA. J Virol. 1995;69:231–238. doi: 10.1128/jvi.69.1.231-238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knapp A C, Enquist L W. Pseudorabies virus recombinants expressing functional virulence determinants gE and gI from bovine herpesvirus 1.1. J Virol. 1997;71:2731–2739. doi: 10.1128/jvi.71.4.2731-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthews R E. Classification and nomenclature of viruses. Intervirology. 1982;17:1–199. doi: 10.1159/000149278. [DOI] [PubMed] [Google Scholar]
- 24.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 25.McNabb D S, Courtney R J. Analysis of the UL36 open reading frame encoding the large tegument protein (ICP1/2) of herpes simplex virus type 1. J Virol. 1992;66:7581–7584. doi: 10.1128/jvi.66.12.7581-7584.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNabb D S, Courtney R J. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology. 1992;190:221–232. doi: 10.1016/0042-6822(92)91208-c. [DOI] [PubMed] [Google Scholar]
- 27.Messerle M, Crnkovic I, Hammerschmidt W, Ziegler H, Koszinowski U H. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Connor M, Peifer M, Bender W. Construction of large DNA segments in Escherichia coli. Science. 1989;244:1307–1312. doi: 10.1126/science.2660262. [DOI] [PubMed] [Google Scholar]
- 30.Paxinos G. The rat nervous system. 2nd ed. San Diego, Calif: Academic Press; 1995. [Google Scholar]
- 31.Peeters B, de Wind N, Hooisma M, Wagenaar F, Gielkens A, Moormann R. Pseudorabies virus envelope glycoproteins gp50 and gII are essential for virus penetration, but only gII is involved in membrane fusion. J Virol. 1992;66:894–905. doi: 10.1128/jvi.66.2.894-905.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rassnick S, Enquist L W, Sved A F, Card J P. Pseudorabies virus-induced leukocyte trafficking into the rat central nervous system. J Virol. 1998;72:9181–9191. doi: 10.1128/jvi.72.11.9181-9191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ried J L, Collmer A. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene. 1987;57:239–246. doi: 10.1016/0378-1119(87)90127-2. [DOI] [PubMed] [Google Scholar]
- 34.Roizman B, Sears E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1043–1107. [Google Scholar]
- 35.Saeki Y, Ichikawa T, Saeki A, Chiocca E A, Tobler K, Ackermann M, Breakefield X O, Fraefel C. Herpes simplex virus type 1 DNA amplified as bacterial artificial chromosome in Escherichia coli: rescue of replication-competent virus progeny and packaging of amplicon vectors. Hum Gene Ther. 1998;9:2787–2794. doi: 10.1089/hum.1998.9.18-2787. [DOI] [PubMed] [Google Scholar]
- 35a.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Shizuya H, Birren B, Kim U-J, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stavropoulos T A, Strathdee C A. An enhanced packaging system for helper-dependent herpes simplex virus vectors. J Virol. 1998;72:7137–7143. doi: 10.1128/jvi.72.9.7137-7143.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tirabassi R S, Enquist L W. Role of envelope protein gE endocytosis in the pseudorabies virus life cycle. J Virol. 1998;72:4571–4579. doi: 10.1128/jvi.72.6.4571-4579.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomkinson B, Robertson E, Yalamanchili R, Longnecker R, Kieff E. Epstein-Barr virus recombinants from overlapping cosmid fragments. J Virol. 1993;67:7298–7306. doi: 10.1128/jvi.67.12.7298-7306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Zijl M, Quint W, Briaire J, de Rover T, Gielkens A, Berns A. Regeneration of herpesviruses from molecularly cloned subgenomic fragments. J Virol. 1988;62:2191–2195. doi: 10.1128/jvi.62.6.2191-2195.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber P C, Levine M, Glorioso J C. Rapid identification of nonessential genes of herpes simplex virus type 1 by Tn5 mutagenesis. Science. 1987;236:576–579. doi: 10.1126/science.3033824. [DOI] [PubMed] [Google Scholar]
- 42.Whealy M E, Robbins A K, Enquist L W. Pseudorabies virus glycoprotein gIII is required for efficient virus growth in tissue culture. J Virol. 1988;62:2512–2515. doi: 10.1128/jvi.62.7.2512-2515.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]