Abstract
Myosins are important motor proteins that associate with the actin cytoskeleton. Structurally, myosins function as heteromeric complexes where smaller light chains, such as calmodulin (CaM), bind to isoleucine–glutamine (IQ) domains in the neck region to facilitate mechano-enzymatic activity. We recently identified Arabidopsis CaM-like (CML) proteins CML13 and CML14 as interactors of proteins containing multiple IQ domains, including a myosin VIII. Here, we demonstrate that CaM, CML13, and CML14 bind the neck region of all four Arabidopsis myosin VIII isoforms. Among CMLs tested for binding to myosins VIIIs, CaM, CML13, and CML14 gave the strongest signals using in planta split-luciferase protein interaction assays. In vitro, recombinant CaM, CML13, and CML14 showed specific, high-affinity, calcium-independent binding to the IQ domains of myosin VIIIs. CaM, CML13, and CML14 co-localized to plasma membrane-bound puncta when co-expressed with red fluorescent protein–myosin fusion proteins containing IQ and tail domains of myosin VIIIs. In vitro actin motility assays using recombinant myosin VIIIs demonstrated that CaM, CML13, and CML14 function as light chains. Suppression of CML13 or CML14 expression using RNA silencing resulted in a shortened-hypocotyl phenotype, similar to that observed in a quadruple myosin mutant, myosin viii4KO. Collectively, our data indicate that Arabidopsis CML13 and CML14 are novel myosin VIII light chains.
Keywords: Calmodulin-like proteins, cytoskeleton, motor protein, myosin light chains, plant myosins
Myosins are key proteins in the plant cytoskeleton, but the identity of their light chain components is unknown. Here, we show that calmodulin-like proteins function as novel myosin light chains.
Introduction
The cytoskeleton is a dynamic and complex system that functions in the organization of the intracellular environment. A ubiquitous component of the cytoskeleton is the acto-myosin network that contains myosin motor proteins bound to actin polymers. The myosin superfamily is broadly organized into two groups: conventional and unconventional. Conventional myosins form filaments and mainly act in animal muscle contraction and cell motility (Altman, 2013). In contrast, unconventional myosins do not form filaments and are involved in various processes such as organelle trafficking, cell and organism growth, and nuclear rearrangements (Hartman et al., 2011; Altman, 2013; Nebenführ and Dixit, 2018). These unconventional myosins possess four distinct domains: the motor, neck, and tail domains, and may also contain coiled-coil domains.
Although there are 79 phylogenetic classes of myosins (Kollmar and Mühlhausen, 2017), only two of these, VIII and XI, are present in plants (Nebenführ and Dixit, 2018). The Arabidopsis genome encodes four class VIII and 13 class XI myosins (Avisar et al., 2009; Peremyslov et al., 2010; Haraguchi et al., 2018). Myosin XIs are involved in a range of processes including endo- and exocytosis, cytoplasmic streaming, nuclear shape and positioning, gravitropism, and biotic stress responses (Peremyslov et al., 2010; Tominaga and Nakano, 2012; Buchnik et al., 2015; Abu-Abied et al., 2018; Nebenführ and Dixit, 2018). In contrast, the roles of myosin VIIIs are not as clear. A study using a quadruple knockout line for all of the myosin VIIIs did not observe any strong phenotype (Talts et al., 2016). Nevertheless, class VIII myosins have been implicated in processes such as plasmodesmatal function and endocytosis (Baluška et al., 2001; Golomb et al., 2008; Sattarzadeh et al., 2008). Their enzymatic profile suggests tension sensor and/or tension generator activity rather than fast movement (Haraguchi et al., 2014; Henn and Sadot, 2014; Rula et al., 2018; Olatungi et al., 2023). In addition, recent reports show the involvement of myosin VIIIs in the movement of viral particles of tobacco mosaic, rice stripe, and rice grassy stunt viruses, and the VirE2 protein from Agrobacterium tumefaciens (Pitzalis and Heinlein, 2017; Liu et al., 2023, Preprint). In the moss Physcomitrella patens, myosin VIIIs function during cytokinesis (Wu et al., 2014). Recently, the myosin VIII isoform, ATM1, was found to function in hypocotyl elongation as well as root apical meristem development in a sugar-dependent manner (Olatunji and Kelley 2020; Olatunji et al., 2023).
In a generalized model of myosin architecture, the catalytic motor (head) domain of myosins interacts with actin filaments and is responsible for ATP hydrolysis, the neck domain binds one or more myosin light chains (MLCs) via IQ (isoleucine–glutamine) motifs, the coiled-coil domain (when present) functions in homodimerization, and the globular tail domain binds to various cargo (Peremyslov et al., 2013; Kurth et al., 2017; Duan and Tominaga, 2018). Very little is known about the neck domains of plant myosins, but they serve as the lever arm during mobility and are thus essential for proper myosin function. Myosin necks possess one or more IQ domains, defined by the consensus motif IQXXXRGXXXR, that are arranged in proximity in the primary structure and act as the binding sites for MLCs that provide the rigidity essential for myosin movement (Heissler and Sellers, 2014). Plant myosin VIIIs are predicted to possess 3–4 IQ domains, whereas myosin XIs have 5–6 (Nebenführ and Dixit, 2018). MLCs are thus critical myosin partners and components of the cytoskeleton network. Historically, MLCs have been referred to by various names; however, upon their discovery, the MLC occupying the first IQ motif of muscle myosin II was called the ‘essential’ light chain (ELC). The term ‘essential’ was chosen as the ELC could only be removed under harsh conditions and dissociation inactivated the holoenzyme (Heissler and Sellers, 2014). Similarly, the MLC bound to the second IQ motif was termed the ‘regulatory’ light chain (RLC), as this MLC was involved in regulating myosin II’s motor activity (Heissler and Sellers, 2014). However, the categorization of MLCs as ELC versus RLC was based on the properties of conventional myosins, and thus is not necessarily applicable to unconventional myosins such as the myosin VIIIs and XIs found in plants. Regardless, MLCs described to date are either the evolutionarily conserved, Ca2+-binding protein calmodulin (CaM) or members of the EF-hand-containing superfamily (Heissler and Sellers, 2014).
It is thought that Ca2+ modulates acto-myosin dynamics in plants by decreasing myosin XI motility in response to an increased Ca2+ concentration (Yokota et al., 1999), but the model is based primarily on studies of mammalian class V myosins (Tominaga and Nakano, 2012). MLCs such as CaM regulate class V myosins by binding to the IQ domains of the neck region in a Ca2+-independent manner, and Ca2+-induced dissociation of MLCs from the neck domain in vivo is thought to be the main mode of regulation (Manceva et al., 2007; Tominaga and Nakano, 2012). In this respect, IQ domains are distinct from most other (non-IQ) CaM-binding domains where CaM binds in the Ca2+-bound (holo-CaM) form (Clapham, 2007). CaM is well established as an MLC in animals, and myosins from both lily pollen tubes and a green alga (Chara corallina) co-purified with CaM and other, unidentified, Ca2+-binding, non-CaM proteins that may be alternative MLCs (Kakei et al., 2012). This suggests that CaM acts as a light chain for some plant myosins in vivo. However, some recombinant Arabidopsis myosin XIs co-expressed solely with CaM in insect cells do not exhibit smooth motility in vitro (Haraguchi et al., 2018), suggesting that, in addition to CaM, other plant-specific MLCs may be needed for mechano-enzyme function. By comparison, various non-CaM MLCs have been identified and characterized in mammalian and fungal species, where they often perform vital roles (Heissler and Sellers, 2014).
Despite their potential importance in myosin function, very little is known about plant MLCs and whether any of the CaM-related proteins found in plants may function as MLCs. In addition to the evolutionarily conserved CaM, plants also possess a large family of unique CaM-like (CML) proteins, with 50 isoforms in Arabidopsis that range from ~20% to 80% sequence identity with CaM (McCormack and Braam, 2003; Zhu et al., 2015). The only functional domains in CMLs are Ca2+-binding EF-hands, and thus they are predicted to function like CaM as regulatory proteins via interaction with downstream targets. While the functions of most CMLs remain unclear, several have been shown to participate in abiotic and biotic stress responses as well as during various stages of development (DeFalco et al., 2009; La Verde et al., 2018; Zeng et al., 2023). However, unlike CaM, where many targets have been characterized, most CML targets remain unknown (La Verde et al., 2018).
Several previous studies speculated that CaM and other EF-hand-containing proteins may function as MLCs in plants. For example, CaM was proposed to be an MLC of myosin ATM1 (also known as myosin VIII-1) as it bound to IQ1 and increased the actin sliding velocity in vitro (Haraguchi et al., 2014). However, when additional IQs in the neck domain of ATM1 were included, the motility experiments were not successful in the presence of CaM alone, suggesting that additional MLCs are probably needed for the function of ATM1 and possibly other myosins (Haraguchi et al., 2014, 2018). Similarly, a screen for cytoskeletal modulators using fibroblast cells isolated CML24 (aka TCH2) as a putative interactor of the neck domain of ATM1 in vitro (Abu-Abied et al., 2006). Recently, proteome analysis of complexes associated with Arabidopsis SnRK1-α1 and -γ1 identified myosin XI isoforms as well as CML13 and CML14 (Van Leene et al., 2022). These studies provide circumstantial evidence that some CMLs may function as MLCs in plants. We recently reported that CML13 and CML14 interact with proteins that possess multiple IQ domains, including members of the IQ67-domain family (IQDs), CaM-binding transcriptional activators (CAMTAs), and class VIII myosins (Teresinski et al., 2023). Moreover, we found that CML13 and CML14 are important for normal plant growth and development (Symonds et al., 2023) and participate in the salinity stress response (Hau et al., 2023). Here, we expand on these previous findings to investigate whether CML13 and CML14 are functional MLCs. We focused on the four members of the Arabidopsis myosin VIII family and used a combination of in vitro and in vivo approaches to assess the interaction of these myosins with CaM and various CMLs. Our data strongly suggest that conserved CaM, as well as CML13 and CML14, are novel MLCs in Arabidopsis.
Materials and methods
Plant material and growth conditions
Nicotiana benthamiana and Arabidopsis thaliana (Col-0) seeds were sown in Sunshine mix #1 (Sun Gro Horticulture Canada Ltd) and transferred to a growth chamber under short-day (SD) or long-day (LD) conditions, respectively (Conviron MTR30; SD, 12 h photoperiod; LD, 18 h photoperiod, 22 °C, ~150 μmol m–2 s–1). For N. benthamiana, 14-day-old seedlings were transplanted to independent 10 cm pots and returned to the growth chamber. All pots were sub-irrigated as needed, with N-P-K fertilizer (20-20-20, 1 g l–1) applied every other week. For fluorescence microscopy assays, N. benthmaniana growth conditions were as previously described (Belausov et al., 2023). For hypocotyl assays, Arabidopsis seeds were sown onto 0.5× Murashige and Skoog (MS) agar plates with or without 5 µM dexamethasone (Dex), as previously described, before cold stratifying in the dark at 4 °C for 48 h (Symonds et al., 2023). Plates were then moved to the growth chamber in the dark and hypocotyls were allowed to elongate for 5 d before measuring the hypocotyl lengths with ImageJ (Rueden et al., 2017). The Dex-inducible hpRNAi lines were produced previously (Symonds et al., 2023). The Myosin VIII 4KO line, hereafter referred to as myosin viii4KO, was previously described (Talts et al., 2016).
Plasmid constructs and recombinant protein expression
For PCR and cloning, oligonucleotide primers used are listed in Supplementary Table S1. See Supplementary Table S2 for a description of plasmid constructs used in this study and the corresponding regions of proteins encoded by the respective cDNAs, and respective locus identifiers. The cDNAs encoding a representative group of AtCMLs (CML6, 8, 15, 19, 24, 35, 38, and 42), as well as CML13, CML14, and petunia CaM81 were cloned as full-length ORFs into the pCambia1300-C-Luciferase (CLuc) vector downstream of the firefly luciferase enzyme as described previously (Chen et al., 2008). CaM81 (hereafter, referred to as CaM) is a conserved isoform of CaM from petunia (GenBank accession no. M80836) that is identical at the protein level to Arabidopsis CaM7. cDNAs encoding the full region or truncations of the Arabidopsis myosin class VIII neck domains were cloned upstream of the N-terminus of firefly luciferase in the pCambia1300-N-Luciferase (NLuc) binary vector. For fluorescent protein fusions, we used the Golden Gate system (Engler et al., 2014). The cDNAs of CML13, CML14, and CaM with flanking adapters having a BsaI site (5' TGGTCTCAAATG and 3' GGTTCGTGAGACCA) and lacking the stop codon were synthesized by Twist Bioscience. The cDNA of green fluorescent protein (GFP) containing a linker sequence at the 5' end (GGATCAACGGGTTCT encoding 5AA-GSTGS) was synthesized by Twist Bioscience. The BsaI adapters for GFP marker were TGGTCTCATTCG at the 5' end and GCTTTGAGACCA at the 3' end. The plasmids coding for 35S×2pro and 35S terminator were pich41295 and pich41276, respectively. All the level-0 plasmids were mixed to create a level-1 backbone binary plasmid (pICH47742) by a cut and ligate reaction using BsaI restriction enzyme and T4 DNA ligase from New England BioLabs (NEB). All plasmid constructs were confirmed by DNA sequencing. The plasmid encoding red fluorescent protein (RFP)–ATM1, RFP–ATM2, RFP–VIIIA, and RFP–VIIIB were previously described (Avisar et al., 2009). Agrobacterium infiltration was performed as described (Belausov et al., 2023).
For recombinant protein expression, the cDNA sequences of interest were subcloned into expression vectors (Supplementary Table S2). cDNAs encoding the neck regions of myosin ATM1 (At3g19960) and ATM2 (At5g54280) were cloned into pET28b, pET28b-GB1, and pGEX-4T-3 vectors (Novagen) for expression in Escherichia coli strain BL21 (DE3) CPRIL (Novagen). pET28b-GB1 encodes GB1, the IgG-binding B1 domain of the G protein found in Streptococcus. It is a highly stable and soluble protein used to improve the expression and solubility of recombinant fusion proteins (Cheng and Patel, 2004). We tested binding of CaM, CML13, and CML14 to GB1 as a negative control in our binding assays. Recombinant proteins corresponding to the full neck regions of myosin VIIIs were solubilized using 8 M urea and purified using an Ni-NTA His60 column (Takara), or for glutathione S-transferse (GST)–multiple IQ fusions, using a glutathione–Sepharose (Sigma) column, as per the manufacturer’s instructions. Myosin full neck proteins were maintained in 8 M urea as they were unstable in aqueous buffers, whereas proteins comprised of multiple IQ domains were dialyzed overnight against Tris-buffered saline (TBS; 50 mM Tris–HCl, 150 mM NaCl, pH 7.5). Recombinant CaM, CML13, and CML14 were expressed and purified as described (Teresinski et al., 2023).
For the myosin in vitro sliding assays, the ATM1 construct encodes the motor domain and native neck regions (four IQ motifs) and the ATM2 construct encodes a motor domain and native neck regions (three IQ motifs) of ATM2. Full-length cDNAs of ATM1 (At3g19960) and ATM2 (At5g54280) were provided by the RIKEN BioResource Center and Dr Motiko Tominaga, respectively. A baculovirus transfer vector for ATM1 and ATM2 was generated using PCR (Supplementary Table S1). PCR products were ligated into the NcoI–AgeI restriction sites of pFastBac MD (Ito et al., 2009). The resulting constructs, pFastBac ATM1 and ATM2, encode N-terminal amino acids (MDYKDDDDKRS) containing the FLAG tag (DYKDDDDK), amino acid residues 1–935 and 1–968 of ATM1 4IQ and ATM2 3IQ, respectively, and C-terminal amino acids (GGGEQKLISEEDLHHHHHHHHSRMDEKTTGWRGGHVVEGLAGELEQLRARLEHHPQGQREPSR) containing a flexible linker (GGG), a Myc-epitope sequence (EQKLISEEDL), a His tag (HHHHHHHH), and a streptavidin-binding peptide (SBP) tag (MDEKTTGWRGGHVVEGLAGELEQLRARLEHHPQGQREP). ATM1 and ATM2 were expressed using a baculovirus expression system in HighFive™ insect cells. They were purified using nickel-affinity and FLAG-affinity resins as previously described (Haraguchi et al., 2022).
Split-luciferase assay
The split-luciferase (SL) protein–protein interaction assays were performed as described (Chen et al., 2008; Teresinski et al., 2023). In brief, full cDNAs encoding ‘bait’ proteins (CaM and CMLs) were cloned into the CLuc binary vector for expression as fusion proteins in-frame with the C-terminal domain of firefly luciferase. The cDNAs encoding ‘prey’ proteins (regions of myosin VIIIs as indicated in the figures) were cloned into the NLuc binary vector for expression as fusion proteins in-frame with the N-terminal domain of firefly luciferase. Transformation and growth of A. tumefaciens strain GV3101 were performed as described (Teresinski et al., 2023). Six-week-old N. benthamiana leaves were infiltrated by A. tumefaciens strain GV3101 carrying bait and prey constructs as indicated in the figures. Whole leaves were removed after 4 d incubation and leaves were sprayed with a 1 mM d-luciferin (GoldBio) and 0.01% Silwett L-77 (Lehle Seeds Inc.) solution, and incubated in the dark for 10 min. Leaves were imaged on the ChemiDoc™ Touch Imaging System (Biorad Inc.). Alternatively, leaf discs were taken after 4 d, incubated with 100 μl of water containing 1 mM luciferin in a 96-well plate for 10–15 min, and luminescence was captured with the SpectraMax Paradigm multimode detection platform (Molecular Devices Inc.). The expression of CLuc fusions was verified by immunoblot (Santa-Cruz Biotech).
Microscopy and image analysis
For Förster resonance energy transfer (FRET)-fluorescence lifetime imaging microscopy (FLIM) analysis, images were acquired by the Leica Stellaris 8 Falcon confocal microscope containing the white laser WLL/supercon. The fluorescence lifetime of the donor GFP (CML13–GFP, CML14–GFP, or CaM–GFP) was measured in leaves expressing the donor only. Fluorescence lifetime of the GFP donor was then measured in the presence of the co-expressed acceptors (mCherry–ATM1, mCherry–ATM2, mCherry–VIIIA, or mCherry–VIIIB). Image acquisition parameters were as follows; resolution 512 × 512, excitation 489 nm, emission 497–553 nm, 20% laser power, line accumulation ×8, laser pulse frequency 80 MHz (12.5 ns), objective HC PL APO CS2 × 63/1.2 water immersion. Fluorescence lifetime was extracted from pixel clusters on a Phasor plot, with GFP of the donor only behaving as a monoexponential component. FRET was calculated by the equation E=1–(tDA/tD) where t is GFP fluorescence lifetime, D is donor, and DA is donor in the presence of the acceptor. For each treatment, five measurements were performed from five different cells. For donor only, images were acquired from the cytoplasm; for donor+acceptor, the focus was adjusted on a region of interest (ROI) at the membrane according to the acceptor signal and then the image was acquired from the GFP channel only.
For co-localization live-cell imaging, a Leica SP8 confocal microscope was used. Imaging was performed using HyD detectors, HC PL APO CS ×63/1.2 water immersion objective (Leica, Wetzlar, Germany), and an OPSL 488 laser for GFP excitation with 500–530 nm emission range, and an OPSL 552 laser for RFP with 565–640 nm emission light detection. Co-localization analysis was done using the coloc function of Imaris (Oxford Instruments) where a minimum of 10 cells were analyzed from each treatment. Rate of co-localization was calculated by the function of Pearson’s coefficient analysis in Imaris. Graphs and statistics analysis were performed by GraphPad Prism 9.5.1 using one-way ANOVA.
Fluorescent protein–protein interaction overlay assays
CaM/CML binding overlay assays were performed as previously described (DeFalco et al., 2010) with minor changes. Briefly, pure recombinant myosin VIII fusion proteins were blotted onto nitrocellulose membranes and blocked overnight at 4 °C with TBST [TBS and 0.01% Tween-20 (v/v)] supplemented with 5% casein (w/v). The blots were washed and then incubated with 200 nM of pure, recombinant CaM or CMLs that had been covalently tagged with 680RD-NHS Ester (Li-Cor Biosciences) as per the manufacturer’s instructions. Protein–protein overlay assays were conducted in the presence of 2 mM CaCl2 or 5 mM EGTA for 1 h at room temperature. The blots were washed several times in excess buffer without probe and imaged on the Odyssey XF Imager (Li-Cor Biosciences) on the 700 nm channel with a 120 s acquisition time. Images were exported to ImageJ (Rueden et al., 2017) and the mean gray-scale per unit area was quantified for each of the C aM/CML–myosin pairs.
Steady-state dansyl fluorescence spectroscopy
Recombinant CaM and CMLs were covalently labeled using dansyl fluoride as described (Alaimo et al., 2013). Samples of 600 nM dansylated CaM (D)-CaM or 3 µM CMLs were incubated in TBS with 1 mM CaCl2 or 1 mM EGTA with or without a 10 M excess of IQ motif peptide, for 30 min at room temperature. Fluorescence spectra were collected using an excitation of 360 nm and emission wavelengths of 400–600 nm for CaM (Alaimo et al., 2013), and 400–650 nm for CML13 and CML14 at 25 °C with a SpectaMax Paradigm instrument. Peptides were synthesized commercially by GenScript with the following sequences; ATM1-IQ1, LHGILRVQSSFRGYQARCLLKEL; and ATM2-IQ1, LQGIVGLQKHFRGHLSRAYFQNM.
Actin gliding assays
Actin sliding velocity was measured using an anti-Myc antibody-based version of the in vitro actin gliding assay as described (Ito et al., 2007). The velocity of actin filaments was measured in 25 mM KCl, 4 mM MgCl2, 25 mM HEPES-KOH (pH 7.4), 1 mM EGTA, 3 mM ATP, 10 mM DTT, and an oxygen scavenger system (120 μg ml–1 glucose oxidase, 12.8 mM glucose, and 20 μg ml–1 catalase) at 25 °C. CaM, CML13, and CML14 alone or in different combinations of two putative MLCs were added in the assay buffer. Average sliding velocities were determined by measuring the displacements of actin filaments that were smoothly moving for distances >10 μm using ImageJ (Rueden et al., 2017) with the MTrackJ plugin (Miejering et al., 2012). Ionic strength was calculated using CALCON based on Goldstein’s algorithm (Goldstein, 1979).
Results
Sequence comparisons of Arabidopsis CaM, CML13, and CML14 and the myosin VIII neck domains
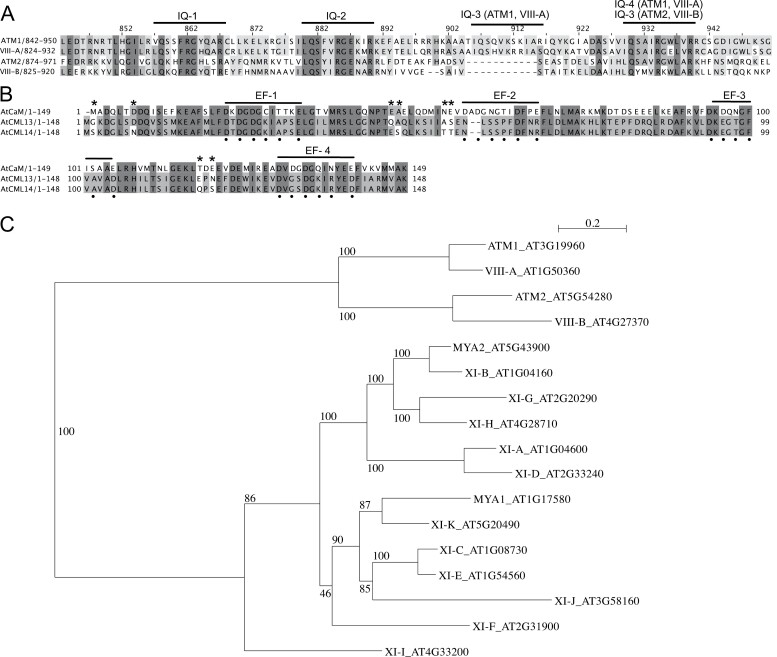
Comparisons of the primary structure of the neck regions of Arabidopsis myosin VIII isoforms are presented in Fig. 1A and of Arabidopsis CML13, CML14, and CaM in Fig. 1B. A phylogenetic tree (Fig. 1C) based on the 17 Arabidopsis myosins shows the clear subgrouping of class VIII members. A sequence comparison and phylogenetic tree of CML13/14 orthologs from several plant taxa are presented (Supplementary Fig. S1) and are also compared with MLCs from a range of plant and non-plant species (Supplementary Fig. S2). Arabidopsis paralogs CML13 and CML14 are 95% identical and differ by only eight residues while sharing ~50% identity with CaM. Where CaM possesses four EF-hands, only the first EF-hand of CML13 and CML14 is predicted to be functional according to InterPro analysis (Paysan-Lafosse et al., 2023) and a recent study on CML14 (Vallone et al., 2016). Typically, positions 1, 3, 5, 7, 9, and 12 within the 12 residue loop of the helix–loop–helix of an EF-hand are responsible for Ca2+ coordination (Gifford et al., 2007). In both CML13 and CML14 there are several unusual residues in positions that correspond to Ca2+ coordination residues in EF-hands 3 and 4 of CaM (Fig. 1B). In addition, the gaps within the EF-hand 2 region, as aligned with CaM, make this highly degenerate in CML13 and CML14 and unlikely to function in Ca2+ binding.
Fig. 1.
Protein sequence alignments of (A) the neck regions (residues ~800–1000) for Arabidopsis class VIII myosins, and (B) Arabidopsis CaM (AtCaM7, AT3G43810), CML13, and CML14. Amino acid residues were shaded based on their percentage identity, dark gray if identical, and progressively lighter gray through to white as unconserved. In (A), predicted IQ motif consensus sequences are overlined. The EF-hands of CaM in (B) are overlined, and Ca2+-coordinating residues in CaM are marked with a dot beneath the alignment. Residues that differ between CML13 and CML14 are indicated with an asterisk. ClustalΩ was used for alignment (Sievers and Higgins, 2014; Gouy et al., 2021), and images were generated using Jalview Version 2.11.2.6. (C) Phylogenetic tree of Arabidopsis class VIII and XI myosins. The tree was constructed using Seaview version 5.0 (Sievers and Higgins, 2014; Gouy et al., 2021) from a complete alignment of proteins using PhyML with bootstrapping analysis (1000 replicates).
In the myosin VIII neck alignment, the core IQ consensus motifs are underlined and indicate that the neck regions of ATM1 and VIII-A are each predicted to possess four IQ domains, whereas ATM2 and VIII-B possess three, having lost the IQ domain that corresponds to the third IQ domain of ATM1 and VIII-A (Fig. 1A). IQ3 is quite degenerate in ATM1 and VIII-A, and is missing several of the five core residues within the IQXXXRGXXXR canonical motif. Conversely, the most conserved IQ motif among myosin VIII isoforms, IQ2, deviates from the consensus myosin IQ sequence in that it possesses a negatively charged residue (Glu) at position 8 in place of hydrophobic residues which typically occupy this position (Houdusse et al., 2006). The most C-terminal IQ domain is relatively conserved among the isoforms and matches the typical CaM-binding IQ consensus, the exception being VIII-A which has a Glu in place of a large hydrophobic residue (Fig. 1A). Given these sequence differences between CML13/14 and CaM, and among the IQ domains of myosin VIIIs, we sought to assess the interaction of these proteins and the potential of CaM, CML13, and CML14 to function as MLCs.
CMLs bind the neck domain of Arabidopsis class VIII myosins in planta
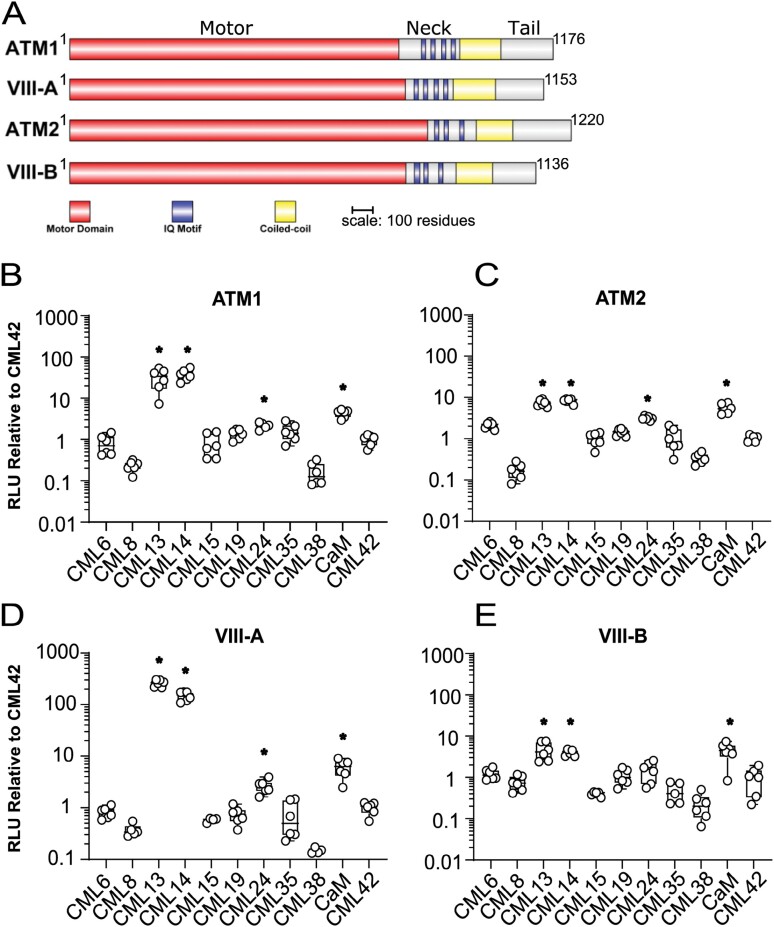
To assess the association of Arabidopsis CaM and CMLs with the neck domains (i.e. the IQ domain regions) of the myosin VIIIs, we used leaves of N. benthamiana and the SL protein interaction assay (Fig. 2). A comparative schematic showing the domains of the myosin VIIIs is presented (Fig. 2A). Immunoblotting, using antisera against the C-terminus of firefly luciferase confirmed the expression of the CML–luciferase fusion constructs in planta (Supplementary Fig. S3). A representative full-leaf image of an SL assay testing interaction of ATM1 with CaM, CML13, CML14, and CML42 is presented in Supplementary Fig. S4 and emphasizes the variability of the SL system as the level of bacterial infiltration, Agrobacterium infection efficiency, and transient protein expression are all aspects that cannot be controlled (Bashandy et al., 2015). Although common in the literature, empty vectors do not make suitable negative controls in the SL system. As such, we assigned CML42 a value of 1.0 as our base-level (negative) control, indicative of non-specific, background luciferase activity in the SL assay based on our previous study which indicated that CML42 does not associate with IQ domains (Teresinski et al., 2023). In pairwise tests, we observed the interaction of CaM with all four myosin VIII isoforms at a level ~3- to 4-fold (note log scale) above CML42 controls (Fig. 2B–E), consistent with reports that CaM is an MLC in plants (Ma and Yen, 1989; Yokota and Shimmen, 1994; Haraguchi et al., 2014, 2018). CML24 gave weaker signals, ~2-fold above background, with ATM1, VIII-A, and ATM2, but did not interact with VIII-B (Fig. 2B–E). By comparison, CML13 and CML14 showed interaction signals with ATM2 and VIII-B of ~3- to 5-fold above controls, similar to those for CaM, but displayed markedly stronger signals with ATM1 and VIII-A, at levels ~35- and 200-fold, respectively, above CML42 controls (Fig. 2B–E). These observations corroborate a previous report of CML24 as a putative interactor of ATM1 (Abu-Abied et al., 2006) and demonstrate that CaM, CML13, and CML14 are able to bind to myosin VIIIs in planta.
Fig. 2.
Split-luciferase protein interaction of Arabidopsis CMLs with the neck region of class VIII myosins in planta. N. benthamiana leaves were infiltrated with Agrobacterium harboring respective NLuc–prey (myosin VIII neck regions) and CLuc–bait (CaM, CMLs) vectors and tested for luciferase activity 4 d after inoculation, as described in the Materials and methods. (A) Schematic representation of myosin VIIIs showing the relative positions of myosin domains. In planta analysis of various CMLs with the neck regions of myosin (B) ATM1 (residues 848–943), (C) ATM2 (residues 878–967), (D) VIII-A (residues 828–930), and (E) VIII-B (residues 834–913), respectively. Split-luciferase data are expressed as a fold difference relative to the RLU signal (log10 scale) observed using the negative control bait CML42 which was set to an RLU of 1.0. Boxes contain each data point for six technical replicates, means are shown by a horizontal bar, the gray region is the 95% confidence interval, and whiskers extend to maximum and minimum data points. Asterisks indicate a significantly higher signal versus CLuc–CML42 as a negative control bait (one-way ANOVA against CML42 with Sidak’s test for multiple comparisons, P-value <0.05). Data are representative of at least three independent experiments. RLU, relative light units.
We further explored the specificity of the myosin VIII/MLC interaction by testing the neck region of all four myosin VIIIs with representative isoforms from each of the nine CML subfamilies in Arabidopsis (CML6, 8, 13, 14, 15, 19, 24, 35, and 38, with CML42 as a negative control) (Fig. 2B–E). Only CaM, CML13, CML14, and CML24 showed significant signals relative to CML42 controls. CML13 and CML14 typically gave the strongest signals in these protein interaction tests. Taken together, these data indicate a clear specificity among the Arabidopsis CML family for interaction with myosin VIIIs, with CaM, CML13, CML14, and CML24 representing candidate MLCs.
CML13 and CML14 co-localize to the plasma membrane with class VIII myosins
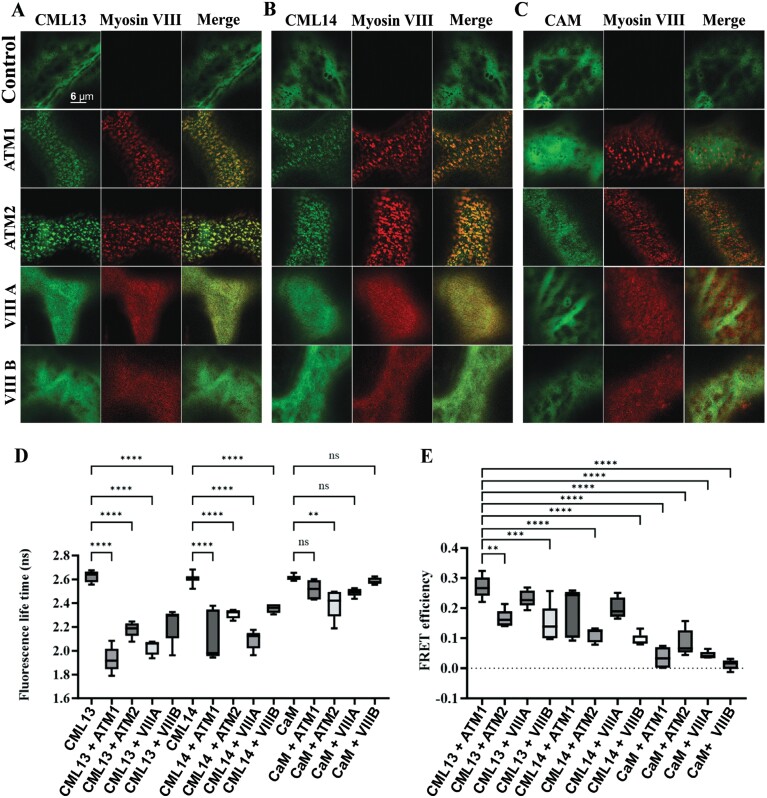
To further test whether CML13, CML14, and CaM can interact with the IQ domain of myosin VIIIs within cells, FRET-FLIM analyses were performed (Fig. 3). We used RFP fused to truncated myosin VIIIs that included the neck and tail region because the subcellular localization of these RFP–myosins has been previously reported (Golomb et al., 2008; Bar-Sinai et al., 2022). GFP–CaM or GFP–CML constructs were transiently overexpressed in the presence or absence of RFP–myosin fusion constructs in N. benthamiana (Fig. 3). When expressed alone, we observed the localization of CML13 and CML14 to the cytoplasm, whereas myosin VIIIs localized to discrete puncta at the plasma membrane, consistent with previous reports (Golomb et al., 2008; Teresinski et al., 2023). Interestingly, when GFP–CMLs and RFP–myosins were co-expressed, CML13 and CML14 distinctly altered their localization and were recruited to the punctate structures at the plasma membrane populated by the RFP–myosin fusion proteins (Fig. 3A, B) (Kumari et al., 2021). CaM, however, showed variable co-localization with myosin VIIIs, mainly co-localizing with ATM2 and to a lesser extent the other myosin VIII isoforms (Fig. 3C). To consolidate the interaction, we analyzed the fluorescence lifetime of GFP fused to CML13, CML14, or CaM and performed FRET-FLIM analysis (Fig. 3D, E; Supplementary Fig. S5). The fluorescence lifetime of GFP experienced a significant reduction upon fusion with CML13 or CML14 in the presence of RFP fused to all four myosins VIII isoforms. Conversely, GFP fused to CaM showed only a slight decrease in its lifetime, and this occurred only in the presence of RFP–ATM2. The highest FRET efficiency was observed between CML13 and ATM1, suggesting a relatively strong association as detected by this method.
Fig. 3.
FRET-FLIM analysis of myosin VIII IQ-tail fragments with CML13, CML14, and CAM. RFP–ATM1IQ-tail, RFP–ATM2 IQ-tail, RFP–VIII-A IQ-tail, or RFP–VIIIB IQ-tail were transiently expressed in N. benthamiana leaves with (A) GFP–CML13, (B) GFP–CML14, or (C) GFP–CaM. (D) Fluorescence lifetime. (E) FRET efficiency. Microscopy was performed 48 h after agro-infiltration using a Leica SP8 confocal microscope or Leica Stellaris 8 with a white laser and Falcon application. Statistical analysis was by one-way ANOVA, **P<0.01, *** P<0.001, ****P<0.0001. All microscopy images are at the same magnification as indicated in panel A.
Class VIII myosin IQ motifs have different specificities for CML13, CML14, and CaM
As CaM, CML13, and CML14 gave the strongest interaction signals in the SL system, we explored whether they exhibit specificity for distinct IQ domains within a given myosin. ATM1 and ATM2 were chosen as representatives among the two class VIII subgroups (Fig. 1C), and their neck domains were sequentially truncated and tested in SL assays for interaction with CaM, CML13, and CML14. Data were again expressed relative to CML42 as the negative control. Myosin IQ motif pairs, which have been speculated to facilitate cooperative MLC interactions in other myosins (Terrak et al., 2005; Powell et al., 2017), as well as single IQ motifs were tested for interaction with the putative MLCs. For ATM1, the IQ pairs ATM1-IQ1 + 2 and ATM1-IQ3 + 4, and the single IQ domains ATM1-IQ3 and ATM1-IQ4, bound to CML13, CML14, and CaM (Fig. 4A). ATM1-IQ1 gave the strongest signal with CaM in the SL assays, suggesting a possible preference for CaM over CML13 and CML14. In contrast to ATM1-IQ1, ATM1-IQ2 when tested alone showed a statistically significant interaction exclusively with CML13 (Fig. 4A).
Fig. 4.
Split-luciferase protein interaction of Arabidopsis CaM, CML13, and CML14 with single and paired IQ domains of myosin ATM1 and ATM2 in planta. N. benthamiana leaves were infiltrated with Agrobacterium harboring respective NLuc–prey (myosin VIII IQ domains) and CLuc–bait (CaM, CMLs) vectors, and tested for luciferase activity 4 d later, as described in the Materials and methods. (A) Split-luciferase data are expressed as a fold increase or decrease relative to the RLU signal (log10 scale) observed using the negative control bait CML42 which was set to an RLU of 1. Boxes contain each data point for six technical replicates, means are shown by a horizontal bar, the colored region is the 95% confidence interval, and whiskers extend to maximum and minimum data points. Asterisks indicate a significantly higher signal versus CLuc–CML42 as a negative control bait (one-way ANOVA against CML42 with Sidak’s test for multiple comparisons, *P<0.05, **P<0.01, ***P<0.001). Data are representative of at least three independent experiments. RLU, relative light units. Range of residues from each myosin tested were: ATM1 (894–943), ATM1-IQ1 + 2 (848–905), ATM1-IQ1 (848–879), ATM1-IQ3 + 4 (890–943), ATM1-IQ3 (890–928), ATM1-IQ4 (916–943), ATM2 (878–967), ATM2-IQ1 + 2 (878–949), ATM2-IQ1 (878–911), ATM2-IQ2 (899–949), and ATM2-IQ3 (922–967).
Interestingly, for ATM2 binding assays, ATM2-IQ1 + 2 interacted with CML13 and CML14 but not with CaM (Fig. 4B). We did not test the combination of ATM2-IQ2 + 3 as these IQ domains are not arranged in close proximity and are thus not classified as a pair (Fig. 1A). Delineation of the individual IQ domains of ATM2 revealed that CML13 and CML14, but not CaM, interacted significantly with ATM2-IQ2, whereas the ATM2-IQ1 domain alone did not associate with any of the putative MLCs tested (Fig. 4B). ATM2-IQ3 bound to each of CaM, CML13, or CML14. In general, the delineation analysis of IQ domains within the neck regions of ATM1 and ATM2 strongly suggests specificity for different MLCs among some of the myosin VIII IQ motifs.
Most myosin VIII multi-IQ motifs have calcium-insensitive interactions with MLCs
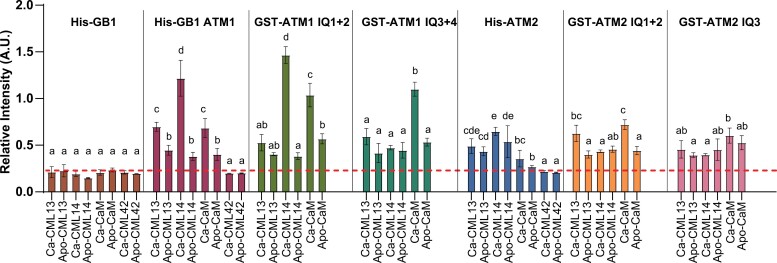
We tested the effects of Ca2+ on the binding of putative MLCs to ATM1 and ATM2 using in vitro protein interaction overlay assays (Supplementary Fig. S6; Fig. 5). As previously reported for other myosins, we observed poor solubility of recombinant fusion proteins that contained multiple IQ domains, and thus used myosin fusion proteins solubilized in urea to test binding to the full neck regions of ATM1 and ATM2 (Teresinski et al., 2023). Shorter regions of the neck domains were soluble as fusion proteins or synthetic peptides. We spotted 200 ng of pure myosin fusion proteins onto nitrocellulose and probed these for interaction with fluorescently labeled CaM, CML13, or CML14 at a final concentration of 200 nM in the presence of excess Ca2+ or EGTA (apo-CaM/CML). Coomassie-stained blots in the uppermost panels of Supplementary Fig. S6 indicate equivalent protein loading in all overlay assays. We did not observe the binding of any of these labeled proteins to a negative control, GB1, which was used as a fusion protein (pET28b-GB1; Fig. 5; Supplementary Fig. S6). Each of these putative MLCs interacted with the paired IQ domains of ATM1 and ATM2 independently of Ca2+, although the signals in some cases were notably stronger when Ca2+ was present in the binding assays, such as His-GB1-ATM1 and GST–ATM1-IQ1 + 2 with CML14, and CaM with GST–ATM1-IQ3 + 4 (Fig. 5). However, we did observe some exceptions to this pattern. For example, the fluorescent signal of CML14 interaction with the full neck region of ATM2 was not significantly different in either the presence or the absence of Ca2+. Interestingly, in contrast to our observations with the in planta SL assay (Fig. 4), CaM interacted with the ATM2-IQ1 + 2 peptide in overlay assays. Whether this reflects a difference in cellular versus in vitro conditions is unclear at this point. As an additional control, we fluorescently labeled CML42 and tested its interaction with the full neck region of ATM1 and ATM2. We observed a faint but discernible signal only for CML42 with ATM2 under Ca2+ conditions, and these signals were notably weaker than those observed using CaM, CML13, or CML14 with ATM2, validating our use of CML42 as a negative control (Supplementary Fig. S6).
Fig. 5.
In vitro protein interaction overlay assays of CaM, CML13, CML14, and CML42 with IQ domains of ATM1 and ATM2. A representative bar graph showing the mean ±SD of three technical replicates of each CaM/CML–myosin interaction is shown. The IQ regions of ATM1 and ATM2 were tested as His-tagged, His-GB1-tagged, or GST-tagged fusion proteins as indicated above each set of graphs. Triplicate samples (200 ng) of pure, recombinant fusion proteins corresponding to the ATM1 or ATM2 full neck region (ATM1, ATM2), paired IQ domains (ATM1-IQ1 + 2, ATM1-IQ3 + 4, ATM2-IQ1 + 2), or the isolated IQ domain of ATM2 (ATM2-IQ3) were spotted onto nitrocellulose, blocked with 5% casein in TBST, and incubated with 200 nM CaM, CML13, CML14, or CML42, as indicated, each of which was covalently labeled with the infra-red dye, 680RD-NHS as described in the Materials and methods. Recombinant GB1 protein (pET28-GB1) was tested as a negative control. Protein–protein interaction was assayed in the presence of 2 mM CaCl2 (Ca) or 5 mM EGTA (Apo) and detected using the LI-COR Odyssey-XF infra-red imager. Data are representative of a minimum of three independent experiments with three technical replicates each. Data were analyzed using one-way ANOVA with Tukey’s test for multiple comparisons of different CMLs for a given myosin fusion protein, where different letters indicate statistical differences (P-values <0.05) between treatments within each graph set. See Supplementary Table S2 and Supplementary Fig. S6 for a description of the primary sequence from the neck regions of ATM1 and ATM2 that were tested for binding and the dot blot binding assay image, respectively.
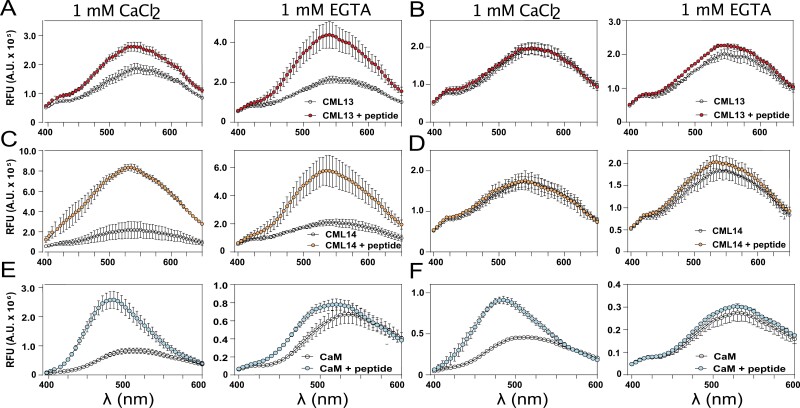
As an additional test of in vitro interaction, we labeled CaM, CML13, and CML14 with dansyl chloride and examined their binding in solution to synthetic peptides corresponding to the single IQ domains, ATM1-IQ1 or ATM2-IQ1. The fluorescence spectra of D-CaM, D-CML13, and D-CML14 in the presence or absence of ATM1-IQ1 and ATM2-IQ1 peptides under Ca2+ versus EGTA conditions are shown in Fig. 6. In agreement with our in planta SL and in vitro overlay assays, ATM1-IQ1 associated with CML13, CML14, and CaM in both the presence and absence of Ca2+, as seen by the blue peak shift and increase in fluorescence intensity in the emission spectra (Fig. 6A). Interestingly, for ATM2-IQ1, a change in the spectrum of D-CaM was observed in the presence of 1 mM CaCl2 but not in EGTA, and the presence of ATM2-IQ1 did not alter the spectra of D-CML13 and D-CML14 (Fig. 6B). The apparent lack of interaction of CML13 and CML14 with ATM2-IQ1 here is consistent with our observations using the SL system (Fig. 4B). The effect of Ca2+ versus EGTA was most apparent in the case of CaM in the presence of either ATM1-IQ1 (Fig. 6E) or ATM2-IQ1 peptide (Fig. 6F), where Ca2+ elicited a marked increase in dansyl fluorescence and a blue shift of the emission spectrum.
Fig. 6.
In vitro interaction of dansyl-CaM, -CML13, or -CML14 with IQ domain synthetic peptides of ATM1-IQ1 and ATM2-IQ1. Dansyl fluorescence was measured over an emission wavelength (λ) window from 400 nm to 600 nm or 650 nm and an excitation wavelength of 360 nm. Samples of 3 µM dansyl-CML13, dansyl-CML14, or using 600 nM dansyl-CaM, were separately tested for fluorescence alone, or in the presence (A, C, E, respectively) of ATM1-IQ1 or (B, D, E, respectively) ATM2-IQ1 peptide under conditions of Ca2+ (left panels) or EGTA (right panels). Peptide concentrations were used at a 10-fold molar excess. Spectra were collected for dansyl-CaM and dansyl-CMLs in the presence or absence of IQ peptides as indicated. The intensity was measured in arbitrary relative fluorescence units (RFU). The fluorescence traces are the mean ±SD of three independent experiments performed in triplicate.
CML13, CML14, and CaM act as MLCs for the motility of myosin VIIIs
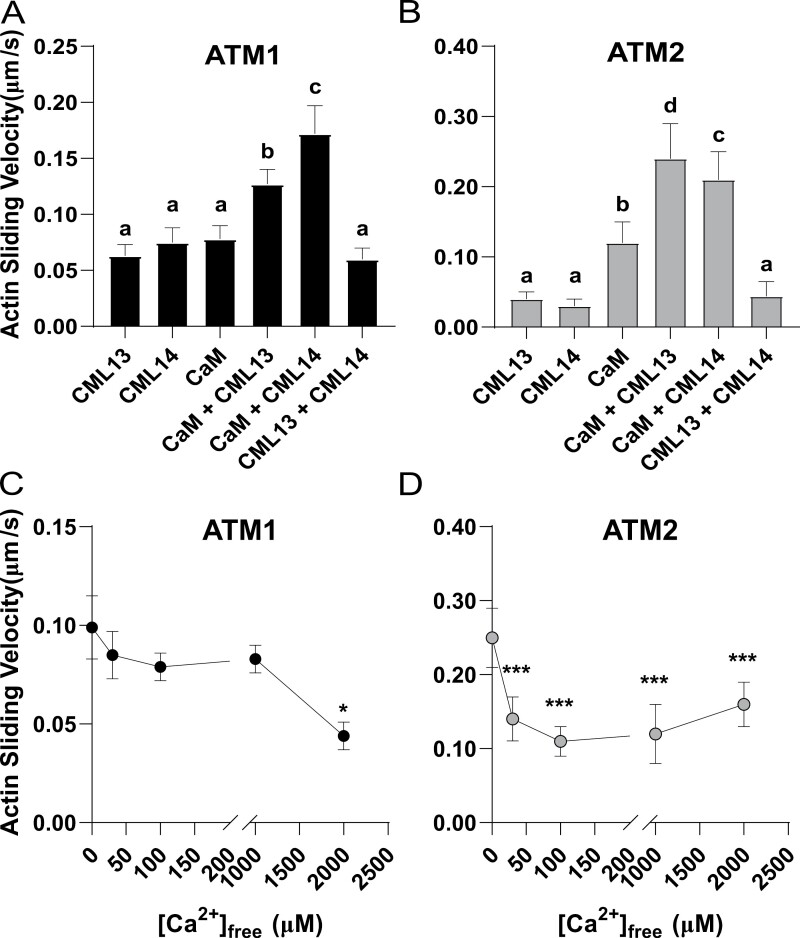
We generated two constructs to measure the velocities of ATM1 and ATM2. The ATM1 construct encodes a protein with the motor domain and native neck regions (four IQ motifs) of ATM1. Similarly, the ATM2 construct encodes the motor domain and native neck regions (three IQ motifs) of ATM2. These constructs were expressed using a baculovirus system in High Five insect cells and purified by affinity chromatography. The velocities of ATM1 and ATM2 were measured using an anti-Myc antibody-based version of the in vitro actin gliding assay (Ito et al., 2007). The velocities of ATM1 and ATM2 were tested in the presence of CaM, CML13, and CML14 alone or in different combinations of two putative MLCs (Fig. 7A, B). The in vitro velocities of ATM1 with any of the single MLCs were not statistically different and were comparable with the velocity when tested with both CML13 and CML14 together (Fig. 7A). However, the combination of CaM and CML13 or CML14 showed increased motility and the fastest motility was observed using the combination of CaM with CML14 (Fig. 7A). In contrast, the velocity of ATM2 with either CML13 or CML14 alone was significantly slower compared with assays using CaM as the solo MLC. This is consistent with our interaction data that suggest that neither CML13 nor CML14 interacts with ATM2-IQ1 (Figs 4B, 6B, D, F). Similar to ATM1 assays, the motility of ATM2 was fastest when tested using combinations of CaM and CML13 or CML14 (Fig. 7B).
Fig. 7.
Actin sliding assays indicate that CaM, CML13, and CML14 can function as light chains for ATM1 and ATM2. (A) Actin sliding velocity of ATM1 in the presence of CaM, CML13, and CML14 alone or in different combinations of two putative MLCs. The concentrations used for CaM, CML13, and CML14 were 10, 30, and 30 µM, respectively. (B) Actin sliding velocity of ATM2 in the presence of CaM, CML13, and CML14 alone or in different combinations of two putative MLCs. The concentrations used for CaM, CML13, and CML14 were each 30 µM. (C) The Ca2+ sensitivity of ATM1 in the presence of CaM and CML13. The concentrations used for CaM and CML13 were 10 µM and 30 µM, respectively. (D) The Ca2+ sensitivity of ATM2 in the presence of CaM and CML13. The concentrations used for CaM and CML13 were 10 µM and 30 µM, respectively. Motility values are the mean ±SD and were determined by measuring the displacements of actin filaments that were smoothly moving for distances >10 μm as described in the Materials and methods.
The Ca2+ sensitivity of ATM1 and ATM2 was tested using CaM and CML13 as representative MLCs under conditions of increasing concentrations of free Ca2+ (Fig. 7C, D). ATM1 displayed reduced motility only at high Ca2+ concentrations (i.e. >1 mM), whereas ATM2 activity was inhibited by much lower Ca2+ concentrations, with a marked reduction in motility observed at 30 µM free Ca2+ (Fig. 7D). Taken together, our actin motility assays clearly indicate that along with CaM, both CML13 and CML14 can function as MLCs.
Dexamethasone-inducible CML13/14 hpRNAi lines phenocopy myosin VIII mutants
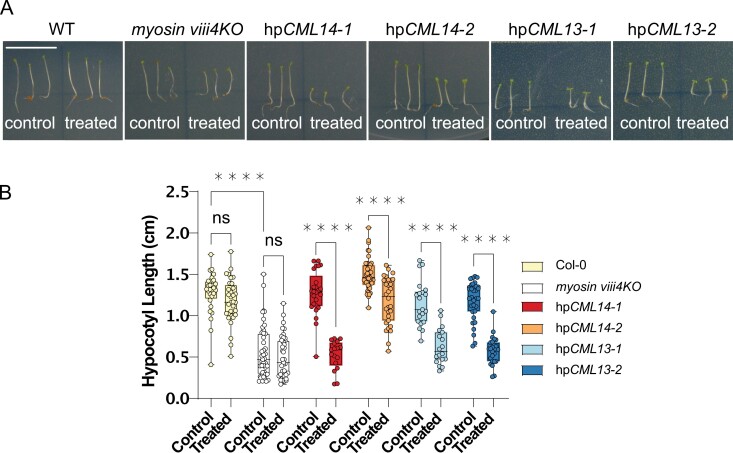
We previously showed that suppression of CML13 or CML14 expression results in strong, aberrant phenotypes throughout development (Symonds et al., 2023). An earlier report examined the phenotypes of myosin mutants and found that a myosin VIII mutant, atm1, and myosin XI triple mutants (X1-1, -2, -K), xi3KO, exhibited reduced hypocotyl elongation relative to wild-type plants (Olantunji and Kelly, 2020). Thus, we compared hypocotyl lengths in our hpCML13 and hpCML14 RNAi plants with wild-type (WT) and myosin VIII mutants under control and 5 μM Dex-treated conditions (Fig. 8). We used the quadruple myosin VIII mutant, myosin viii4 (Talts et al., 2016), and observed a clear reduction in hypocotyl length compared with WT plants (Fig. 8). In the absence of Dex, hypocotyl length in hpCML13 and hpCML14 RNAi lines resembled that of WT seedlings. In contrast, treatment with Dex caused a marked reduction in hypocotyl length in these hpRNAi lines, resulting in a phenotype comparable with that of the myosin viii4KO mutant (Fig. 8). Dex treatment had no discernible effect on either WT or myosin viii4KO seedlings.
Fig. 8.
Hypocotyl lengths of Col-0, myosin viii4KO, and Dex-inducible CML13,14 hpRNAi lines. Seeds were sown onto 0.5× MS agar plates with (treatment) or without (control) 5 µM Dex and cold stratified in the dark at 4 °C for 48 h. Plates were then moved to the growth chamber, still in the dark, and the hypocotyls were allowed to elongate for 5 d before imaging (A) and measuring the hypocotyl lengths with ImageJ (B). Boxes show the mean ±SD of each line on each of the control or treatment plates, whiskers portray the range of the data acquired, and each point is a single biological replicate (one-way ANOVA with Tukey’s test for multiple comparisons, with 20–45 biological replicates, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001). White bar in (A)=1 cm, and all images are of the same magnification.
Discussion
Despite the importance of MLCs for myosin function, relatively little is known about the identity and properties of plant MLCs. While previous reports have indicated that CaM probably functions as an MLC in plants (Yokota and Shimmen, 1994; Haraguchi et al., 2014), evidence to support speculation that plants also use non-CaM MLCs has been limited (Haraguchi et al., 2018). Multiple, independent lines of analysis in our study support our hypothesis that Arabidopsis CML13 and CML14 function as MLCs given that: (i) they bind specifically to all four myosin VIIIs in planta and show some degree of preference in binding to different IQ domains (Figs 2, 4); (ii) they interact at physiological concentrations (i.e. 200 nM) with IQ domains from myosin VIIIs in vitro (Fig. 5; Supplementary Fig. S6); (iii) they co-localize with myosin VIIIs when co-expressed in plant cells (Fig. 3); (iv) they support in vitro myosin activity (Fig. 7); and (v) suppression of CML13 or CML14 expression by RNAi results in an impaired hypocotyl-extension phenotype comparable with that of a mutant lacking all four myosin VIII isoforms (Fig. 8).
The mutiple IQ domains of a given myosin allow MLCs to bind and confer structural integrity and, in some cases, impact activity (Heissler and Sellers, 2014). Different MLCs often possess specificity for distinct IQ domains within a given myosin (Heissler and Sellers, 2014). While our data suggest that CaM, CML13, and CML14 show preference for distinct IQ domains on plant myosin, in general, designations of MLCs as ‘essential’ or ‘regulatory’ should be avoided for plant MLCs given that the plant-specific myosin VIII and XI families are unconventional myosins.
Although the IQ motif is broadly defined according to the IQXXXRGXXXR consensus, there is considerable variation among IQ domains (Bähler and Rhoads, 2002; Vetter and Leclerc, 2003). The canonical CaM-binding IQ motif has a large hydrophobic residue immediately upstream of the first consensus Arg residue and another following the conserved Gly and second Arg: IQXXΦRGΦXXRXXΦ, where Φ refers to large hydrophobic residues (Houdusse et al., 2006). It is noteworthy that the first IQ domain (IQ1) for all four Arabidopsis myosin VIII isoforms adheres to this pattern (Fig. 1A). In contrast, although the IQ2 domains of these myosin VIIIs are highly conserved, they possess smaller hydrophobic residues (e.g. Val and Ile) preceding the Arg–Gly consensus, followed by a Glu (Fig. 1A). Negatively charged residues within IQ domains, and other CaM-binding domains, have been reported to impede interaction with CaM (Vetter and Leclerc, 2003; Slaughter et al., 2005). These notable differences in primary sequence suggest that the IQ1 and IQ2 domains of Arabidopsis myosin VIIIs may be occupied by non-CaM MLCs, consistent with our observations that CML13 and CML14 are MLCs. Structurally, how the sequence variation among these IQ domains and the peripheral regions impacts MLC specificity, binding affinity, and myosin VIII function remains an open question.
Using the SL in planta protein interaction system we observed some differences in MLC preference among the IQ motifs of ATM1 and ATM2 as representative myosin VIII isoforms (Fig. 4). Despite the divergence of ATM1-IQ3 from the canonical IQ consensus motif, it bound CaM, CML13, and CML14 with comparable signal strength (Fig. 4A), indicating that all essential residues for interaction with different MLCs are present. Given the adherence of ATM1-IQ4, and ATM2-IQ3 to the IQ consensus, it is not surprising that they bound CaM, but these data also indicate that CML13 and CML14 can bind to both conserved and divergent IQ domains (Fig. 4A, B). In contrast, we observed specificity of ATM2-IQ2 with CML13 and CML14, and of ATM1-IQ2 with CML13 alone (Fig. 4A, B), suggesting that CML13 and CML14 probably act as the specific MLCs for these myosins. We speculate that the conserved Glu at IQ domain position 8 within the consensus motif of IQ2 in myosin VIIIs may decrease CaM affinity without impacting CML13/14-IQ interaction. Furthermore, the specificity differences between ATM1-IQ2 and ATM2-IQ2 may result from the Lys/Arg residues at position 9 and/or the Glu/Leu residues at position 13, respectively (Houdusse et al., 2006). However, analysis of solved structures will be needed in the future to fully elucidate the binding properties of IQ/CML complexes.
Collectively, our study indicates that CaM, CML13, and CML14 interact with myosin VIII IQ domains, but our data from in planta and in vitro assays did not align in all cases, a phenomenon previously reported for CaM binding to IQ domains in plant proteins (Bürstenbinder et al., 2013). For example, ATM2-IQ1 did not interact with any of the MLCs tested in the SL assay (Fig. 4B), whereas CaM showed strong interaction with GST–ATM2-IQ1 + 2 and an ATM2-IQ1 synthetic peptide in vitro as judged by overlay assays (Fig. 5) or D-CaM binding spectrophotometry (Fig. 6), respectively. These differences may reflect the cellular conditions of the SL system or the simplicity of the in vitro assays where there is an absence of CaM/CML binding competitors. Regardless, as CaM alone was able to support ATM2 motility to a greater level than either CML13 or CML14 alone (Fig. 7), this represents strong evidence that it can serve as an MLC for ATM2. We are unaware of any previous in vitro studies on the motility of ATM2 or the identity of its MLCs. Thus, when considered together with our binding and motility data (Figs 5–7), the collective evidence suggests that CaM is likely to be an MLC for ATM1, ATM2, and myosin VIIIs in general.
It is also interesting that we observed weak signals for ATM1-IQ2 interaction with each of the MLCs tested using the in planta SL system, with only CML13 emerging as a putative interactor (Fig. 4). Despite this, in vitro motility of ATM1 was supported by a combination of CaM and either CML13 or CML14 (Fig. 7A). In actin sliding (motility) assays, it is thought that all IQ domains need to be occupied for smooth actin movement (Haraguchi et al., 2018). Given that we observed such smooth ATM1 activity with CaM and CML14 even in the absence of CML13, this suggests that they can function as MLCs at ATM1-IQ2, at least under in vitro conditions. However, as CML13, but not CML14 or CaM, interacted with ATM1-IQ2 in the in planta SL interaction assays (Fig. 4), this suggests that CML13 might be the preferred MLC in vivo. We used the well-established High Five insect cell expression system (Vaughn et al., 1977; Haraguchi et al., 2014) to prepare myosin for activity assays, and one caveat of this method is that other proteins, including conserved CaM, might co-purify with myosin preparations. However, SDS–PAGE analysis indicated that our recombinant ATM1 and ATM2 samples were very pure and free of contamination by native CaM or other proteins (Supplementary Fig. S7). Moreover, as CMLs are unique to plants, and their presence increased ATM1 and ATM2 in vitro activity, our data suggest that High Five cells do not possess proteins that can substitute for CML13 or CML14 as MLCs (Haraguchi et al., 2018).
We also corroborated earlier work that suggested that CML24 is a putative interactor with some myosin VIII isoforms (Abu-Abied et al., 2006). The fact that cml24 point mutants display actin and cortical microtubule defects led to speculation that CML24 may interact with myosins during mechanical stress to aid in the organization of the cytoskeleton (Abu-Abied et al., 2006; Wang et al., 2011). Although we showed the in planta interactions of CML24 with the neck domains of several myosin VIII isoforms (Fig. 2C), it generally exhibited weaker SL signals than CaM, CML13, or CML14. Thus, we restricted our co-localization, in vitro binding, and myosin motility assays to the latter group. As such, further studies will be needed to assess whether CML24 serves as a bona fide MLC.
Myosin VIIIs have been suggested to function in the organization of the cytoskeleton at the plasma membrane (Golomb et al., 2008; Bar-Sinai et al., 2022). The punctate localization pattern that we observed (Fig. 3) using myosin VIIIs comprised of neck and tail regions is consistent with earlier studies and is thought to reflect the location of myosin VIII cargo to which the tail domains bind (Golomb et al., 2008; Haraguchi et al 2014). Our FRET-FLIM and co-localization data indicate that myosin VIIIs recruit CML13 and CML14 as MLCs to the plasma membrane as components of the acto-myosin cytoskeleton (Fig. 3). Myosin VIII-A and VIII-B typically displayed a more diffuse localization compared with ATM1 and ATM2, consistent with previous reports (Avisar et al. 2009). Interestingly, FRET-FLIM suggested that while CML13 and CML14 bind better to ATM1 and VIIIA than to ATM2 and VIIIB, the preference of CaM binding was to ATM2. Although we did not test all 50 Arabidopsis CMLs, our SL analysis indicates clear specificity among the CML family for myosin VIII interaction (Fig. 2). However, this does not exclude the possibility that other CMLs or non-CML proteins might also function as MLCs. Although a weak ATM1–CaM binding was demonstrated by FRET-FLIM, our SL analyses, in vitro binding assays, and motility tests all demonstrate that CaM can function as an MLC with ATM1, supporting a previous report (Haraguchi et al., 2014).
Myosin VIIIs associate with the actin network and localize to plasma membranes, plasmodesmata, endosomes, and other internal structures, suggesting a breadth of roles within cells (Golomb et al., 2008; Haraguchi et al., 2014; Nebenführ and Dixit, 2018; Bar-Sinai et al., 2022). However, insight into their functions has been hindered by an apparent genetic redundancy. Although no obvious phenotype was initially observed in an Arabidopsis mutant lacking all four myosin VIIIs (Talts et al., 2016), a subsequent study reported a subtle phenotype for atm1 single knockouts, a reduced hypocotyl elongation that was recoverable by sucrose supplementation (Olatunji and Kelley, 2020). We performed similar assays using the quadruple myosin VIII knockout (myosin viii4KO) and observed an even stronger suppression of hypocotyl extension in comparison with WT plants than reported for the single atm1 KO, supporting a role for myosin VIIIs in hypocotyl growth (Fig. 8). Given that our hpCML13 and hpCML14 RNAi plants showed a phenotype comparable with the viii4KO mutant only when treated with Dex, we speculate that CML13 and CML14 participate in myosin VIII function during hypocotyl elongation. Recently, ATM1 was also implicated in root apical meristem organization (Olatunji et al., 2023), and myosin VIIIs were reported to function in Agrobacterial transformation of Arabidopsis cells (Liu et al., 2023, Preprint). The use of Physcomitrella patens as a model demonstrated that the loss of all five myosin VIIIs resulted in a pleiotropic phenotype that included reduced size and growth rate of gametophytes, suggesting roles in cell expansion and hormone homeostasis (Wu et al., 2011). Thus, going forward, further exploration of correlative phenotypes between CML and myosin mutants is warranted.
In general, our data provide strong evidence that CaM, CML13, and CML14 are bona fide Arabidopsis MLCs. The importance of Ca2+ in this interaction is unclear and may vary depending on cellular conditions. While we observed reduced motility in both ATM1 and ATM2 assays in the presence of CaM and CML13 at high Ca2+ levels (Fig. 7C, D), we did not test CML14 across a range of Ca2+ concentrations, and thus it remains possible that the response to Ca2+ may vary among light chains. In the yeast or mammalian class V myosin model, CaM is thought to be the MLC responsible for the Ca2+ regulation of the myosin V holoenzyme by dissociating from IQ2 in the presence of Ca2+ (Batters and Veigel, 2016). However, the physiological relevance of this Ca2+-induced dissociation remains unclear given that under in vitro conditions levels of ≥100 µM Ca2+ are often required to dissociate CaM from the IQ domains (Yokata et al., 1999; Manceva et al., 2007). Indeed it has been speculated that the myosin V paradigm is not applicable to plant myosins (Batters and Veigel, 2016). The in vivo picture is thus likely to be quite complex, and the role of Ca2+ may vary among MLCs and specific IQ domains within a given myosin. As CML13 and CML14 are mainly Ca2+ insensitive (Vallone et al., 2016; Teresinski et al., 2023), one possibility is that a conformational change or an increase in the rate of dissociation of Ca2+–CaM at IQ1 regulates Ca2+ sensitivity as has been reported previously for myosin Va and Ic, respectively (Manceva et al., 2007; Shen et al., 2016). Our in vitro binding data (Fig. 6E, F), showing a Ca2+-induced conformational change in CaM when bound to ATM1-IQ1 and ATM2-IQ1, are consistent with this speculation. An alternative hypothesis considers the impact of competition for CaM between myosins and other CaM-binding proteins under conditions of elevated Ca2+. CML13, CML14, and possibly other MLCs may occupy some IQ domains of plant myosins and afford them structural stability when Ca2+ levels rise in response to stimuli and free CaM levels are limiting due to CaM interaction with various targets.
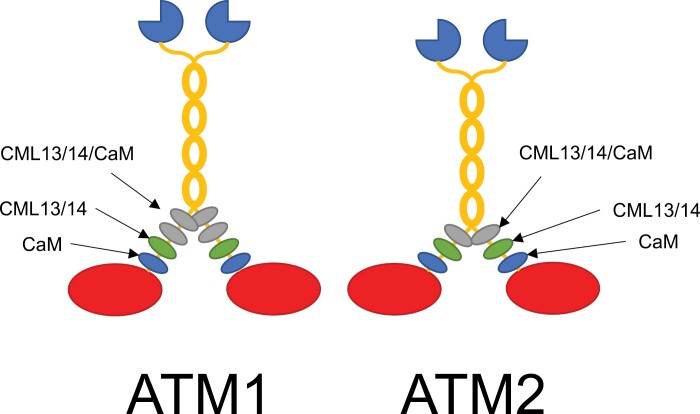
Our discovery of CML13/14 as novel MLCs may explain why several Arabidopsis myosin XI isoforms lacked motility when tested with CaM as the sole MLC (Haraguchi et al., 2018). A working model of CaM, CML13, and CML14 as MLCs is presented in Fig. 9. Our motility assays indicate that both CaM and another MLC such as CML13 or CML14 are required for maximal activity. Split-luciferase in planta and D-CaM/CML in vitro binding assays point to a preference for CaM at the first IQ position and CML13 or CML14 at the second IQ position. We did not observe a clear binding preference at the third IQ position. It will be important in future studies to see if binding specificity extends to in vivo associations and whether conditions, such as biotic or abiotic stress, impact the interaction of CaM/CML with particular IQ domains.
Fig. 9.
Working model of CaM, CML13, and CML14 as light chains for Arabidopsis myosins ATM1 and ATM2. Actin-binding, catalytic myosin head domains are presented in red, the neck regions are decorated with light chains represented as blue (CaM), green (CML13/14), or gray (any of CaM/CML13/14) ovals, the coiled-coil dimerization regions are in yellow, and the cargo-binding tail domains are dark blue, three-quarter circles. Light chains bind to the IQ domains within the neck region of myosins to provide the leverage and structural integrity needed for the power stroke. We speculate that CaM is the preferred light chain at the IQ1 position (nearest the head) for both ATM1 and ATM2, whereas CML13 or CML14 are preferred light chains at the IQ2 position. Other IQ domains did not show a clear preference in our tests and may be occupied by CaM, CML13, CML14, or possibly other light chains.
A major challenge in elucidating specific roles for CML13 and CML14 concerns their potential number of targets, including CAMTAs, IQD proteins, and myosins (Teresinski et al., 2023). In Arabidopsis, these putative targets represent 56 different proteins, each with multiple IQ domains, and thus, like CaM, the number and variety of cellular processes that CML13/14 may participate in is expansive. Both CML13 and CML14 are important in plant growth throughout development, as suppression of either leads to a pleiotropic phenotype (Symonds et al., 2023). Moreover, CML13 and CML14 function in abiotic stress responses, and a recent report found that cml13 mutants exhibit enhanced germination under salinity stress (Hau et al., 2023). Assessing which of these physiological events are due to CML13/14 operating as MLCs should be an area of future exploration.
Speculation about the presence of non-CaM MLCs in plants was raised several decades ago but, to our knowledge, the present study is the first to empirically demonstrate that specific CMLs can function as MLCs (Vahey et al., 1982; Ma and Yen, 1989). The identification of CML13 and CML14 as novel MLCs should help accelerate myosin research. Among the key, unresolved questions to be addressed are whether CML13/14 function as class XI MLCs, how the specificity of CaM or CML-IQ interaction is achieved, and whether the phosphorylation of CML13 and CML14 impacts myosin activity/function.
Supplementary data
The following supplementary data are available at JXB online.
Table S1. List of oligonucleotide primers used in this study.
Table S2. Description of plasmid constructs used in this study.
Fig. S1. Alignment and phylogenetic tree of Arabidopsis CML13 and CML14 with predicted orthologs from various plant species.
Fig. S2. Alignment and phylogenetic tree of Arabidopsis CML13 and CML14 with known MLCs from various organisms.
Fig. S3. Anti-C-Luciferase immunoblot of CLuc–CML fusion proteins from split-luciferase assays.
Fig. S4. A representative, whole leaf image of split-luciferase protein interaction assays of CaM, CML13, CML14, and CML42 (negative control) with ATM1.
Fig. S5. Representative phasor plots of the FRET-FLIM analysis presented in Fig. 3.
Fig. S6. In vitro dot-blot fluorescent overlay assays of CaM/CML interaction with myosin VIII IQ domains.
Fig. S7. SDS–PAGE analysis of purified ATM1 and ATM2 from High Five insect cells without co-expression of CaM.
Acknowledgements
We thank Dr Zongchao Jia (Queen’s University) for the kind gift of plasmid pET28b-GB1.
Contributor Information
Kyle Symonds, Department of Biology, Queen’s University, Kingston, ON, Canada.
Howard J Teresinski, Department of Biology, Queen’s University, Kingston, ON, Canada.
Bryan Hau, Department of Biology, Queen’s University, Kingston, ON, Canada.
Vikas Dwivedi, Institute of Plant Sciences, Volcani Institute, ARO, Rishon LeZion 7528809, Israel.
Eduard Belausov, Institute of Plant Sciences, Volcani Institute, ARO, Rishon LeZion 7528809, Israel.
Sefi Bar-Sinai, Institute of Plant Sciences, Volcani Institute, ARO, Rishon LeZion 7528809, Israel.
Motoki Tominaga, Faculty of Education and Integrated Arts and Sciences, Waseda University, 2-2 Wakamatsu-cho, Shinjuku-ku, Tokyo 162-8480, Japan; Graduate School of Science and Engineering, Waseda University, 2-2 Wakamatsu-cho, Shinjuku-ku, Tokyo 162-8480, Japan.
Takeshi Haraguchi, Department of Biology, Graduate School of Science, Chiba University, Inage-ku, Chiba 263-8522, Japan.
Einat Sadot, Institute of Plant Sciences, Volcani Institute, ARO, Rishon LeZion 7528809, Israel.
Kohji Ito, Department of Biology, Graduate School of Science, Chiba University, Inage-ku, Chiba 263-8522, Japan.
Wayne A Snedden, Department of Biology, Queen’s University, Kingston, ON, Canada.
Peter Bozhkov, Swedish University of Agricultural Sciences, Sweden.
Author contributions
WS, KS, KI, ES, and MT: conceptualization; KS, BH, HJT, VD, EB, SB-S, and TH: performing experiments and collecting data. All authors contributed to data interpretation and manuscript writing.
Conflict of interest
The authors have no conflicts to declare.
Funding
Funding was provided by a Natural Sciences and Engineering Council (NSERC) Discovery grant (2018-04928 to WS), the Israel Science Foundation (grant 776/14 to ES), by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JP 20001009 and JP 23770060 to MT; JP 17K07436, JP 26440131, and JP 21570159 to KI), and a grant from the Hamaguchi Foundation for the Advancement of Biochemistry (JP 22K20623 to TH).
Data availability
All data supporting the findings of this study are available within the paper and its supplementary data published online.
References
- Abu-Abied M, Belausov E, Hagay S, Peremyslov V, Dolja V, Sadot E. 2018. Myosin XI-K is involved in root organogenesis, polar auxin transport, and cell division. Journal of Experimental Botany 69, 2869–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Abied M, Golomb L, Belausov E, Huang S, Geiger B, Kam Z, Staiger CJ, Sadot E. 2006. Identification of plant cytoskeleton-interacting proteins by screening for actin stress fiber association in mammalian fibroblasts. The Plant Journal 48, 367–379. [DOI] [PubMed] [Google Scholar]
- Alaimo A, Malo C, Areso P, Aloria K, Millet O, Villarroel A. 2013. The use of dansyl-calmodulin to study interactions with channels and other proteins. Methods in Molecular Biology 998, 217–231. [DOI] [PubMed] [Google Scholar]
- Altman D. 2013. Myosin: fundamental properties and structure. In: Roberts GCK, ed. Encyclopaedia of biophysics. Berlin, Heidelberg: Springer, 1664–1671. [Google Scholar]
- Avisar D, Abu-Abied M, Belausov E, Sadot E, Hawes C, Sparkes IA. 2009. A comparative study of the involvement of 17 Arabidopsis myosin family members on the motility of Golgi and other organelles. Plant Physiology 150, 700–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler M, Rhoads A. 2002. Calmodulin signaling via the IQ motif. FEBS Letters 513, 107–113. [DOI] [PubMed] [Google Scholar]
- Baluška F, Cvrčková F, Kendrick-Jones J, Volkmann D. 2001. Sink plasmodesmata as gateways for phloem unloading. Myosin VIII and calreticulin as molecular determinants of sink strength? Plant Physiology 126, 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Sinai S, Belausov E, Dwivedi V, Sadot E. 2022. Collisions of cortical microtubules with membrane associated myosin VIII tail. Cells 11, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashandy H, Jalkanen S, Teeri TH. 2015. Within leaf variation is the largest source of variation in agroinfiltration of Nicotiana benthamiana. Plant Methods 11, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batters C, Veigel C. 2016. Mechanics and activation of unconventional myosins. Traffic 17, 860–871. [DOI] [PubMed] [Google Scholar]
- Belausov E, Dwivedi V, Yechezkel S, Bar-Sinai S, Sadot E. 2023. Documentation of microtubule collisions with myosin VIII ATM1 containing membrane-associated structures. Methods in Molecular Biology 2604, 77–88. [DOI] [PubMed] [Google Scholar]
- Buchnik L, Abu-Abied M, Sadot E. 2015. Role of plant myosins in motile organelles: is a direct interaction required? Journal of Integrative Plant Biology 57, 23–30. [DOI] [PubMed] [Google Scholar]
- Bürstenbinder K, Savchenko T, Müller J, Adamson AW, Stamm G, Kwong R, Zipp BJ, Dinesh DC, Abel S. 2013. Arabidopsis calmodulin-binding protein IQ67-domain 1 localizes to microtubules and interacts with kinesin light chain-related protein-1. Journal of Biological Chemistry 288, 1871–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. 2008. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiology 146, 368-376. [DOI] [PMC free article] [PubMed]
- Cheng Y, Patel DJ. 2004. An efficient system for small protein expression and refolding. Biochemical and Biophysical Research Communications 317, 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham DE. 2007. Calcium signaling. Cell 131, 1047–1058. [DOI] [PubMed] [Google Scholar]
- DeFalco TA, Bender KW, Snedden WA. 2009. Breaking the code: Ca2+ sensors in plant signaling. The Biochemical Journal 425, 27–40. [DOI] [PubMed] [Google Scholar]
- DeFalco TA, Chiasson D, Munro K, Kaiser BN, Snedden WA. 2010. Characterization of GmCaMK1, a member of a soybean calmodulin-binding receptor-like kinase family. FEBS Letters 584, 4717–4724. [DOI] [PubMed] [Google Scholar]
- Duan Z, Tominaga M. 2018. Actin–myosin XI: an intracellular control network in plants. Biochemical and Biophysical Research Communications 506, 403–408. [DOI] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert T-M, Werner S, Jones JDG, Patron NJ, Marillonnet S. 2014. A golden gate modular cloning toolbox for plants. ACS Synthetic Biology 3, 839–843. [DOI] [PubMed] [Google Scholar]
- Gifford JL, Walsh MP, Vogel HJ. 2007. Structures and metal-ion-binding properties of the Ca2+-binding helix–loop–helix EF-hand motifs. The Biochemical Journal 405, 199–221. [DOI] [PubMed] [Google Scholar]
- Goldstein D. 1979. Calculation of the concentrations of free cations and cation–ligand complexes in solutions containing multiple divalent cations and ligands. The Biophysical Journal 26, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb L, Abu-Abied M, Belausov E, Sadot E. 2008. Different subcellular localizations and functions of Arabidopsis myosin VIII. BMC Plant Biology 8, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Tannier E, Comte N, Parsons DP. 2021. Seaview Version 5: a multiplatform software for multiple sequence alignment, molecular phylogenetic analyses, and tree reconciliation. Methods in Molecular Biology 2231, 241–260. [DOI] [PubMed] [Google Scholar]
- Haraguchi T, Ito K, Duan Z, Rula S, Takahashi K, Shibuya Y, Hagino N, Miyatake Y, Nakano A, Tominaga M. 2018. Functional diversity of class XI myosins in Arabidopsis thaliana. Plant and Cell Physiology 59, 2268–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T, Tamanaha M, Suzuki K, Yoshimura K, Imi T, Tominaga M, Sakayama H, Nishiyama T, Murata T, Ito K. 2022. Discovery of ultrafast myosin, its amino acid sequence, and structural features. Proceedings of the National Academy of Sciences, USA 119, e2120962119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T, Tominaga M, Matsumoto R, Sato K, Nakano A, Yamamoto K, Ito K. 2014. Molecular characterization and subcellular localization of Arabidopsis class VIII myosin, ATM1. Journal of Biological Chemistry 289, 12343–12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman MA, Finan D, Sivaramakrishnan S, Spudich JA. 2011. Principles of unconventional myosin function and targeting. Annual Review of Cell and Developmental Biology 27, 133–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hau B, Symonds K, Teresinski H, Janssen A, Duff L, Smith M, Benidickson K, Plaxton W, Snedden WA. 2023. Arabidopsis calmodulin-like proteins CML13 and CML14 interact with calmodulin-binding transcriptional activators and function in salinity stress response. Plant and Cell Physiology, pcad152. [DOI] [PubMed] [Google Scholar]
- Heissler SM, Sellers JR. 2014. Myosin light chains: teaching old dogs new tricks. Bioarchitecture 4, 169–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn A, Sadot E. 2014. The unique enzymatic and mechanistic properties of plant myosins. Current Opinion in Plant Biology 22, 65–70. [DOI] [PubMed] [Google Scholar]
- Houdusse A, Gaucher J-F, Krementsova E, Mui S, Trybus KM, Cohen C. 2006. Crystal structure of apo-calmodulin bound to the first two IQ motifs of myosin V reveals essential recognition features. Proceedings of the National Academy of Sciences, USA 103, 19326–19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Ikebe M, Kashiyama T, Mogami T, Kon T, Yamamoto K. 2007. Kinetic mechanism of the fastest motor protein, Chara myosin. Journal of Biological Chemistry 282, 19534–19545. [DOI] [PubMed] [Google Scholar]
- Ito K, Yamaguchi Y, Yanase K, Ichikawa Y, Yamamoto K. 2009. Unique charge distribution in surface loops confers high velocity on the fast motor protein Chara myosin. Proceedings of the National Academy of Sciences, USA 106, 21585–21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakei T, Sumiyoshi H, Higashi-Fujime S. 2012. Characteristics of light chains of Chara myosin revealed by immunological investigation. Proceedings of the Japan Academy Series B Physical and Biological Sciences 88, 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmar M, Mühlhausen S. 2017. Myosin repertoire expansion coincides with eukaryotic diversification in the Mesoproterozoic era. BMC Evolutionary Biology 17, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari P, Dahiya P, Livanos P, et al. 2021. IQ67 DOMAIN proteins facilitate preprophase band formation and division-plane orientation. Nature Plants 7, 739–747. [DOI] [PubMed] [Google Scholar]
- Kurth EG, Peremyslov VV, Turner HL, Makarova KS, Iranzo J, Mekhedov SL, Koonin EV, Dolja VV. 2017. Myosin-driven transport network in plants. Proceedings of the National Academy of Sciences, USA 114, E1385–E1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Verde V, Dominici P, Astegno A. 2018. Towards understanding plant calcium signaling through calmodulin-like proteins: a biochemical and structural perspective. International Journal of Molecular Sciences 19, 1331–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Lee L-Y, Hsu F-Y, Yu Y, Rao P, Gelvin SB. 2023. Myosin VIII and XI isoforms interact with Agrobacterium VirE2 protein and help direct transport from the plasma membrane to the perinuclear region during plant transformation. bioRxiv 06.531343. [Preprint].
- Ma YZ, Yen LF. 1989. Actin and myosin in pea tendrils. Plant Physiology 89, 586–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceva S, Lin T, Pham H, Lewis JH, Goldman YE, Ostap EM. 2007. Calcium regulation of calmodulin binding to and dissociation from the myo1c regulatory domain. Biochemistry 46, 11718–11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack E, Braam J. 2003. Calmodulins and related potential calcium sensors of Arabidopsis. New Phytologist 159, 585–598. [DOI] [PubMed] [Google Scholar]
- Meijering E, Dzyubachyk O, Smal I. 2012. Methods for cell and particle tracking. Methods in Enzymology 504, 183–200. [DOI] [PubMed] [Google Scholar]
- Nebenführ A, Dixit R. 2018. Kinesins and myosins: molecular motors that coordinate cellular functions in plants. Annual Review of Plant Biology 69, 329–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olatunji D, Clark NM, Kelley DR. 2023. The class VIII myosin ATM1 is required for root apical meristem function. Development 150, dev201762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olatunji D, Kelley DR. 2020. A role for Arabidopsis myosins in sugar-induced hypocotyl elongation. MicroPublication Biology 2020, doi: 10.17912/micropub.biology.000276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paysan-Lafosse T, Blum M, Chuguransky S, et al. 2023. InterPro in 2022. Nucleic Acids Research 51, D418–D427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Morgun EA, Kurth EG, Makarova KS, Koonin EV, Dolja VV. 2013. Identification of myosin XI receptors in Arabidopsis defines a distinct class of transport vesicles. The Plant Cell 25, 3022–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Prokhnevsky AI, Dolja VV. 2010. Class XI myosins are required for development, cell expansion, and F-actin organization in Arabidopsis. The Plant Cell 22, 1883–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzalis N, Heinlein M. 2017. The roles of membranes and associated cytoskeleton in plant virus replication and cell-to-cell movement. Journal of Experimental Botany 69, 117–132. [DOI] [PubMed] [Google Scholar]
- Powell CJ, Jenkins ML, Parker ML, Ramaswamy R, Kelsen A, Warshaw DM, Ward GE, Burke JE, Boulanger MJ. 2017. Dissecting the molecular assembly of the Toxoplasma gondii MyoA motility complex. Journal of Biological Chemistry 292, 19469–19477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW. 2017. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rula S, Suwa T, Kijima ST, Haraguchi T, Wakatsuki S, Sato N, Duan Z, Tominaga M, Uyeda TQP, Ito K. 2018. Measurement of enzymatic and motile activities of Arabidopsis myosins by using Arabidopsis actins. Biochemical and Biophysical Research Communications 495, 2145–2151. [DOI] [PubMed] [Google Scholar]
- Sattarzadeh A, Franzen R, Schmelzer E. 2008. The Arabidopsis class VIII myosin ATM2 is involved in endocytosis. Cell Motility and the Cytoskeleton 65, 457–468. [DOI] [PubMed] [Google Scholar]
- Shen M, Zhang N, Zheng S, Zhang W-B, Zhang H-M, Lu Z, Su QP, Sun Y, Ye K, Li X-D. 2016. Calmodulin in complex with the first IQ motif of myosin-5a functions as an intact calcium sensor. Proceedings of the National Academy of Sciences, USA 113, E5812–E5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Higgins DG. 2014. Clustal omega. Current Protocols in Bioinformatics 48, 3.13.1–3.13.16. [DOI] [PubMed] [Google Scholar]
- Slaughter BD, Unruh JR, Allen MW, Bieber Urbauer RJ, Johnson CK. 2005. Conformational substates of calmodulin revealed by single-pair fluorescence resonance energy transfer: influence of solution conditions and oxidative modification. Biochemistry 44, 3694–3707. [DOI] [PubMed] [Google Scholar]
- Symonds K, Teresinski H, Hau B, Chiasson D, Benidickson K, Plaxton W, Snedden WA. 2023. Arabidopsis CML13 and CML14 have essential and overlapping roles in plant development. Plant and Cell Physiology doi: 10.1093/pcp/pcad142. [DOI] [PubMed] [Google Scholar]
- Talts K, Ilau B, Ojangu EL, Tanner K, Peremyslov VV, Dolja VV, Truve E, Paves H. 2016. Arabidopsis myosins XI1, XI2, and XIK are crucial for gravity-induced bending of inflorescence stems. Frontiers in Plant Science 7, 1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teresinski HJ, Hau B, Symonds K, Kilburn R, Munro KA, Doner NM, Mullen R, Li VH, Snedden WA. 2023. Arabidopsis calmodulin-like proteins CML13 and CML14 interact with proteins that have IQ domains. Plant, Cell & Environment 46, 2470–2491. [DOI] [PubMed] [Google Scholar]
- Terrak M, Rebowski G, Lu RC, Grabarek Z, Dominguez R. 2005. Structure of the light chain-binding domain of myosin V. Proceedings of the National Academy of Sciences, USA 102, 12718–12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga M, Nakano A. 2012. Plant-specific myosin XI, a molecular perspective. Frontiers in Plant Science 3, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahey M, Titus M, Trautwein R, Scordilis S. 1982. Tomato actin and myosin: contractile proteins from a higher land plant. Cell Motility 2, 131–147. [Google Scholar]
- Vallone R, La Verde V, D’Onofrio M, Giorgetti A, Dominici P, Astegno A. 2016. Metal binding affinity and structural properties of calmodulin-like protein 14 from Arabidopsis thaliana. Protein Science 25, 1461–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Eeckhout D, Gadeyne A, et al. 2022. Mapping of the plant SnRK1 kinase signaling network reveals a key regulatory role for the class II T6P synthase-like proteins. Nature Plants 8, 1245–1261. [DOI] [PubMed] [Google Scholar]
- Vaughn JL, Goodwin RH, Tompkins GJ, McCawley P. 1977. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera; Noctuidae). In Vitro 13, 213–217. [DOI] [PubMed] [Google Scholar]
- Vetter SW, Leclerc E. 2003. Novel aspects of calmodulin target recognition and activation. European Journal of Biochemistry 270, 404–414. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang B, Gilroy S, Wassim Chehab E, Braam J. 2011. CML24 is involved in root mechanoresponses and cortical microtubule orientation in Arabidopsis. Journal of Plant Growth Regulation 30, 467–479. [Google Scholar]
- Wu S-Z, Bezanilla M, Bezanilla M. 2014. Myosin VIII associates with microtubule ends and together with actin plays a role in guiding plant cell division. eLife 3, e03498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S-Z, Ritchie JA, Pan A-H, Quatrano RS, Bezanilla M. 2011. Myosin VIII regulates protonemal patterning and developmental timing in the moss Physcomitrella patens. Molecular Plant 4, 909–921. [DOI] [PubMed] [Google Scholar]
- Yokota E, Muto S, Shimmen T. 1999. Inhibitory regulation of higher-plant myosin by Ca2+ ions. Plant Physiology 119, 231–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota E, Shimmen T. 1994. Isolation and characterization of plant myosin from pollen tubes of lily. Protoplasma 177, 153–162. [Google Scholar]
- Zeng H, Zhu Q, Yuan P, Yan Y, Yi K, Du L. 2023. Calmodulin and calmodulin-like protein-mediated plant responses to biotic stresses. Plant, Cell & Environment 46, 3680–3703. [DOI] [PubMed] [Google Scholar]
- Zhu X, Dunand C, Snedden W, Galaud J-P. 2015. CaM and CML emergence in the green lineage. Trends in Plant Science 20, 483–489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and its supplementary data published online.