Abstract
INTRODUCTION:
Celiac disease (CD) and type 1 diabetes (T1D) often co-occur and share genetic components in the human leukocyte antigen (HLA) class II region. We aimed to study the usefulness of HLA genotyping in predicting the risk of developing T1D in patients with CD and the temporal relationship between these diseases.
METHODS:
A cohort of 1,886 Sardinian patients, including 822 with CD, 1,064 with T1D, and 627 controls, underwent HLA class II typing. Seventy-six of 822 patients with CD were also affected by T1D (CD-T1D), and their HLA genotypes were analyzed for specific HLA associations with CD, T1D, and controls.
RESULTS:
High-risk HLA-DQ genotypes, including HLA-DQ2.5/DQ8, -DQ2.5/DQ2.5, and -DQ2.5/DQ2.3, were strongly associated with CD-T1D with frequencies of 34.5%, 15.9%, and 18.8%, respectively. Conversely, certain HLA genotypes associated with CD seemed to confer protection against T1D development. Therefore, HLA genotyping allows for the identification of those patients with CD who might develop T1D. The frequency of patients with CD preceding T1D is higher in younger children than older ones, with implications for the early childhood approach to diabetes prevention.
DISCUSSION:
CD is a condition for future T1D development, and specific HLA genotypes can predict this risk. Early screening for celiac autoimmunity and subsequent HLA typing in CD children could help identify those at high risk of T1D, allowing for proactive interventions and immunotherapies to preserve β-cell function. These findings may support the re-evaluation of HLA typing in children with CD.
KEYWORDS: type 1 diabetes (T1D), celiac disease (CD), human leukocyte antigen (HLA), genotyping
INTRODUCTION
Celiac disease (CD) is an immune-mediated systemic disorder elicited by gluten and related prolamins in genetically susceptible individuals (1). It is characterized by a variable combination of gluten-dependent clinical manifestations, production of CD-specific antibodies, strong association with HLA-DQ2 or HLA-DQ8 haplotypes, and enteropathy (2).
Patients with CD also have an increased risk of developing other autoimmune diseases in comparison with the healthy population (3,4), and a frequency up to 15% (5) of such associations has been observed. One of the most commonly reported is with type 1 diabetes (T1D). Indeed, in patients with T1D, the prevalence of CD is about 8% (6–8). However, the development of T1D in patients with CD is less frequently described, and these studies are mostly limited to either cross-sectional studies or studies of diabetes-related autoantibodies in patients with CD (9).
The connection of both diseases with the HLA-DR3 and -DR4 haplotypes has been considered as a major responsible for their association (9), and the HLA-DR3-DQ2/DR3-DQ2 and HLA-DR3-DQ2/DR4-DQ8 genotypes have been linked to a higher risk of association of T1D and CD (10,11). In addition, other studies, besides HLA-DR3/DR4, have related the increased risk of association with history of familial T1D and with SH2B3 polymorphisms, although the overlap of the 2 diseases is not fully explained by these risk factors (12).
Therefore, the temporal co-occurrence of CD and T1D (CD-T1D) and the HLA association need to be further analyzed in different populations.
Recently, it has been described that monitoring for pancreatic islet autoantibodies, combined with HLA genetic risk assessment, can allow for the identification of patients with high risk to develop T1D while still having a normal glucose concentration in the blood (13,14). Moreover, progression of T1D in diabetic patients' relatives, who did not have diabetes but had a high risk to clinically develop the disease, was delayed by about 2 years using teplizumab, an anti-CD3 antibody showing weak agonistic activity on the T-cell receptor-CD3 signaling complex (15). In the past few years, clinical trials in patients with new-onset (stage 3) T1D highlighted several other immunological agents that may be able to preserve β-cell function (16–18). Therefore, nowadays, T1D scientific community not only has the tools to identify the individuals who will develop the disease in the future but also a paraphernalia of low-risk immunotherapies able to preserve endogenous β-cell function and make metabolic control substantially easier (17).
Children with CD (19) represent a category of patients with significantly higher risk to subsequently develop T1D. However, not all these patients carry the same risk of T1D (9), and therefore, the separation of those with higher risk from those with a neglectable risk using HLA typing and islet-specific autoantibodies may offer the possibility to intervene even before the clinical diagnosis of T1D (stage 2 or 3), to maximize the preservation of β-cells (16) from autoimmunity.
Hence, the goal of this study was to identify the HLA class II genotypes that predispose Sardinian children with CD to the future development of T1D.
METHODS
The study was conducted with a cohort of 1,886 Sardinian patients. Among them, 822 were diagnosed with CD, with 76 of these individuals also being affected by T1D. In addition, there were 1,064 patients solely diagnosed with T1D, whereas 627 healthy subjects were included as controls. The HLA class II typing for patients with CD, T1D, and the control group had already been performed as part of previous studies (20–24) (Table 1). The patients with CD-T1D were collected in the period from 2011 to 2019. Among the 76 patients with CD-T1D, 54 developed T1D before or at the same time as CD diagnosis, whereas in 22, T1D was diagnosed after CD onset. Diagnosis of T1D, in patients with CD-T1D and T1D, was obtained by measuring fasting blood sugar, random blood sugar test, glycated hemoglobin test, pancreatic autoantibodies, and HLA typing. Diagnosis of CD was performed according to European Society for Paediatric Gastroenterology Hepatology and Nutrition (ESPGHAN) guidelines (2,25,26). An ethics committee approval was obtained for this study (authorization no. 1046 obtained on July 15, 2016).
Table 1.
Patients with CD-T1D, CD, and T1D and controls participating in the study
| Diseases | No. of patients | Females | Males | Ratio of females/males |
| CD-T1D | 76 | 43 | 33 | 1.3 |
| CD | 746 | 497 | 249 | 2 |
| T1D | 1,064 | 416 | 648 | 0.64 |
| Controls | 627 | 308 | 319 | 0.97 |
CD-T1D, celiac disease type 1 diabetes.
IgA anti–tissue transglutaminase antibody (TtG-IgA) and IgA antiendomysial antibody (EMA)
TtG-IgA was determined using commercial kit (ImmunoCAP; Phadia, Milan, Italy) after serum dilution, and EMA was detected by immunofluorescence (Delta Biologicals, Rome, Italy).
HLA class II typing and haplotypes' analysis
HLA-DRB1, -DQA1, and -DQB1 genotypes were determined by polymerase chain reaction with sequence-specific primers (SSP) using Olerup SSP typing kits (Olerup SSP AB, Stockholm, Sweden). The identification of the different HLA-DRB1, -DQA1, and -DQB1 haplotypes was obtained by following the segregation of HLA types in families.
Small bowel intestinal histopathology
For each patient, 2 or more biopsies were taken from the second/third portion of the duodenum (yielding at least 4 samples), and at least 1 or more biopsies were taken from the duodenal bulb (2,25,27). Intestinal villous atrophy has been graded according to the Marsh classification (28), modified by Oberhuber in types 3c, 3b, 3a, 2, 1, and 0 (29), always by the same board-certified pathologists.
Statistical analysis
χ2 test and odds ratio (OR) were used to compare the frequencies of the HLA class II genotypes among CD-T1D, CD, T1D, and controls. In the case of sample size lower than 5, a 2-tailed Fisher exact test was used. A P value lower than 0.05 was considered statistically significant. The Student t test was used as a statistical method to assess the difference between the mean age of the study groups.
RESULTS
This retrospective cross-sectional study was aimed at identifying the HLA-DQ genotypes in patients affected by CD and T1D. First, to calculate the different ORs, the HLA class II distribution of patients with CD-T1D, CD, and T1D was compared with the HLA frequencies of controls. Next, when appropriate, the ORs were compared with one another.
As expected, in the group of patients with CD, the HLA-DQ2.5/DQ2.5 and -DQ2.5/DQ2.2 molecules were found to be the most frequently associated and showed the highest OR. Also, the frequencies of HLA-DQ2.5/DQ8 and HLA-DQ2.5/x molecules were significantly increased, but with a lower OR (Table 2). Interestingly, besides the strong association with the HLA-DQ2.5/DQ2.2, the HLA-DQ2.2 alone showed a lower frequency vs controls (Table 2). A slightly significant increment of the HLA-DQ2.5 molecule encoded in trans was observed in patients with CD. In addition to the HLA-DQ7/DQ2.2 or HLA-DQ7/DQ2.3 genotype (Table 2), also the Sardinian DRB1*04:05, DQA1*05, DQB1*03:01 haplotype in combination with the DR7 haplotype contributed to the generation of the HLA-DQ2.5 molecule encoded in trans. Finally, the remaining HLA genotypes not significantly associated with CD were found in 47.7% of controls and in only 9.9% of patients with CD (Table 2).
Table 2.
Comparison of frequencies of HLA genotypes between controls and each patient groups
| HLA genotypesa | CD-T1D (n = 76) | T1D (n = 1,064) | CD (n = 746) | Controls (n = 627) | |
| DQ2.5/DQ8 | No. (%) | 30 (39.5) | 316 (29.7) | 57 (7.6) | 21 (3.3) |
| P value | 2 × 10−30 | 3.2 × 10−39 | 6.2 × 10−4 | / | |
| OR (CI) | 18.8 (10–35.4) | 12.2 (7.7–19.2) | 2.4 (1.4–4) | / | |
| DQ2.5/DQ2.5 | No. (%) | 29 (38.2) | 329 (30.9) | 153 (20.5) | 33 (5.3) |
| P value | 1.3 × 10−21 | 1.9 × 10−35 | 2 × 10−16 | / | |
| OR (CI) | 11.1 (6.2–19.8) | 8.1 (5.5–11.7) | 4.6 (3.1–6.9) | / | |
| DQ2.5/DQ2.3 | No. (%) | 3 (3.9) | 26 (2.4) | 13 (1.7) | 4 (0.6) |
| P value | 1.6 × 10−2 | 1.2 × 10−2 | Ns | / | |
| OR (CI) | 6.4 (1.4–29.1) | 3.9 (1.4–11.2) | — | / | |
| DQ8/DQ8 | No. (%) | 1 (1.3) | 24 (2.3) | 10 (1.3) | 2 (0.3) |
| P value | ns | 9 × 10−4 | Ns | / | |
| OR (CI) | / | 7.2 (1.7–30.6) | — | / | |
| DQ2.5/DQ2.2 | No. (%) | 0 | 14 (1.3) | 111 (14.9) | 17 (2.7) |
| P value | ns | 3.9 × 10−2 | 1.1 × 10−14 | / | |
| OR (CI) | / | 0.5 (0.2–1) | 6.3 (3.7–10.6) | / | |
| DQ7/DQ2.2 or DQ7/DQ2.3 | No. (%) | 0 | 2 (0.2) | 41 (5.5) | 20 (3.2) |
| P value | ns | 1 × 10−4 | 3.9 × 10−2 | / | |
| OR (CI) | / | 0.1 (0.1–0.2) | 1.8 (1–3) | / | |
| DQ2.5/x | No. (%) | 6 (7.9) | 208 (19.5) | 271 (36.3) | 165 (26.3) |
| P value | 2 × 10−2 | 1.2 × 10−3 | 7.2 × 10−5 | / | |
| OR (CI) | 0.2 (0.1–0.6) | 0.1 (0.5–0.9) | 1.6 (1.3–2.) | / | |
| DQ2.2/x | No. (%) | 0 | 4 (0.4) | 17 (2.3) | 28 (4.5) |
| P value | ns | 1 × 10−4 | 2.3 × 10−2 | / | |
| OR (CI) | / | 0.1 (0.1–0.2) | 0.5 (0.3–0.9) | / | |
| DQ8/x | No. (%) | 2 (2.6) | 48 (4.5) | 15 (2) | 38 (6.1) |
| P value | ns | ns | 1 × 10−4 | / | |
| OR (CI) | — | — | 0.3 (0.2–0.6) | / | |
| Others | No. (%) | 5 (6.6) | 93 (8.7) | 74 (9.9) | 299 (47.7) |
| P value | 9.1 × 10−12 | 4.6 × 10−75 | 1.7 × 10−52 | / | |
| OR (CI) | 0.1 (0.1–0.2) | 0.1 (0.1–0.2) | 0.1 (0.1–0.2) | / |
CD-T1D, celiac disease type 1 diabetes; CI, confidence interval; HLA, human leukocyte antigen; OR, odds ratio.
The HLA genotypes analyzed are those that confer greater susceptibility for CD or T1D.
Analysis of patients with T1D showed a highly significant association with the HLA-DQ2.5/DQ2.5 and HLA-DQ2.5/DQ8 molecules, even stronger than that observed in CD (Table 2). Other significant associations were found for HLA-DQ8/DQ8 and HLA-DQ2.5/DQ2.3 molecules (Table 2). These 2 genotypes were observed also in patients with CD but did not reach any significant association.
By contrast, the presence of the HLA-DQ2.5/DQ2.2 genotype was significantly diminished by up to tenfold when compared with patients with CD, as well as in comparison with the control group (Table 2).
In patients with T1D, a reduced association was also found for HLA-DQ genotypes producing the HLA-DQ2.5 molecule in trans, as with the HLA-DQ2.2 or HLA-DQ2.3 in combination with the HLA-DQ7 (Table 2). The HLA-DQ2.5/x genotypes, positively associated with CD, and the HLA-DQ2.2/x molecules were not significantly related with T1D. Finally, the remaining HLA genotypes not significantly associated with T1D were found in 47.7% of controls and in only 8.7% of patients with T1D (Table 2).
A distinctive HLA association was identified in patients with CD-T1D: The HLA molecules that were most frequently observed were HLA-DQ2.5/HLA-DQ8 and HLA-DQ2.5/HLA-DQ2.5, exhibiting a higher frequency compared with both patients with CD and T1D (Table 2).
The next more frequent HLA genotype was HLA-DQ2.5/HLA-DQ2.3, which was found to be associated also with T1D. Interestingly, the HLA-DQ2.5/HLA-DQ2.2 strongly associated with CD was not found at all in CD-T1D (Table 2). Also, the HLA-DQ2.2 or HLA-DQ2.3 in combination with the HLA-DQ7 haplotype, producing the HLA-DQ2.5 molecule in trans and significantly associated with CD, was absent in CD-T1D (Table 2). Finally, the remaining HLA genotypes not significantly associated with CD-T1D were found in 47.7% of controls and in only 6.6% of patients with CD-T1D (Table 2).
In short, these data show a strong association with HLA-DQ2.5/HLA-DQ8 and HLA-DQ2.5/HLA-DQ2.5 genotypes in all the 3 groups of patients (T1D, CD, and CD-T1D), whereas other HLA molecules, associated with CD (HLA-DQ2.5/HLA-DQ2.2 and HLA-DQ2.2 or HLA-DQ2.3 in combination with the HLA-DQ7), were not associated with T1D development and therefore also with CD-T1D (Table 2).
Overall, within our cohort of 822 patients with CD, we identified 76 cases of T1D (9.2%). However, in patients carrying the HLA-DQ2.5/HLA-DQ8, HLA-DQ2.5/HLA-DQ2.5, or HLA-DQ2.5/HLA-DQ2.3 molecules, the frequency of T1D significantly rose to 34.5%, 15.9%, and 18.8%, respectively. These findings confirm the distinct association of these 3 HLA genotypes with CD-T1D (Table 3).
Table 3.
Frequencies of low- and high-risk HLA genotypes associated with the development of T1D in patients with CD
| High-risk and low-risk HLA genotypes | HLA genotypes | CD (n = 746) | CD-T1D (n = 76) | Percentage of T1D in CD | T1D onset before or at the same time of CDb | T1D onset after CDb |
| High-riska | DQ2.5/DQ8 | 57 | 30 | 34.5% | 22 | 8 |
| DQ2.5/DQ2.5 | 153 | 29 | 15.9% | 20 | 9 | |
| DQ2.5/DQ2.3 | 13 | 3 | 18.8% | 2 | 1 | |
| Low-risk | Other HLA genotypes | 523 | 14 | 2.6% | 10 | 4 |
CD-T1D, celiac disease type 1 diabetes; HLA, human leukocyte antigen.
The group of high-risk HLA genotypes was further subdivided by each individual susceptibility HLA genotype for T1D.
The last 2 columns report the number of patients who developed T1D before and after CD.
Interestingly, in patients with CD-T1D, 71.1% developed T1D before or at the same time of CD diagnosis, whereas in the remaining, 28.9% T1D appeared after the CD diagnosis (Table 3). The mean age of CD was 8.16 (range: 1–20) and for T1D was 11.05 (range: 1–23). The 2 diseases varied significantly for the age of onset with a P = 0.0002. In CD-T1D, the mean age of CD onset was 6.53 (range: 1–13), whereas for T1D, the mean age of CD onset was 8.5 (range: 1–18); P = 0.04.
We also subdivided the patients with CD-T1D into 5 age classes according to CD onset, observing in younger children an increased percentage of patients who developed T1D after CD (Table 4).
Table 4.
Distribution of patients with CD-T1D according to the age of onset of CD
| Age of onset of CD expressed in yr | All ages | <10.64a | <8.16a | <5.67a | <5.09a |
| CD-T1D | 76 | 70 | 54 | 30 | 21 |
| T1D onset after CD | 22 | 19 | 19 | 12 | 10 |
| Percentage of T1D onset after CD | 28.9% | 27.1% | 35.2% | 40.0% | 47.6% |
CD-T1D, celiac disease type 1 diabetes.
For each age group, the total number of patients with CD-T1D, the number, and the percentage of those who developed T1D after CD onset are reported.
The high-risk HLA-DQ2.5/HLA-DQ8 and HLA-DQ2.5/HLA-DQ2.5 genotypes did not show a preferential association with the age of onset in patients with T1D, CD, and CD-T1D (data not shown).
The female/male ratio was maximum in CD and minimum in T1D, whereas in patients with CD-T1D, an intermediate ratio of 1.3 was observed (Table 1).
Finally, no difference was observed in the severity of intestinal damage between patients with CD and CD-T1D (data not shown).
DISCUSSION
Patients with CD have an increased risk of developing other autoimmune diseases such as T1D. Indeed, in comparison with the prevalence of T1D observed in Sardinian general population (0.366%) (30), the frequency of T1D in patients with CD (9.2%) was 25.1 times higher. HLA class II typing may offer the opportunity to identify potential patients susceptible to T1D.
In our cohort of 822 patients with CD, 76 were also affected by T1D, and in 28.9% of these patients with CD-T1D, the onset of T1D was observed after the diagnosis of CD. This may have practical implications for T1D prevention because nowadays, there are therapies able to slow down the autoimmune process that finally results in β-cell destruction and insulin deficiency.
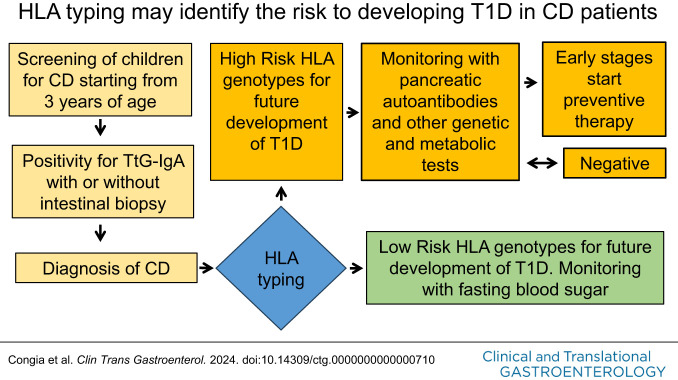
Therefore, in this study, we sought and identified the HLA class II genotypes that are associated with high-risk of future development of T1D in Sardinian children with CD.
First, we have subdivided the HLA genotypes in CD-T1D according to the OR (Table 2). The HLA-DQ2.5/HLA-DQ8, HLA-DQ2.5/HLA-DQ2.5, and HLA-DQ2.5/HLA-DQ2.3 genotypes conferred the highest risk (Table 2). Indeed, the frequency of T1D observed in patients with CD carrying these HLA genotypes was 34.5%, 15.9%, and 18.7%, respectively, and considering them all together, a frequency of T1D of 21.8% was observed, whereas patients with CD with low-risk HLA genotypes had a much lower T1D prevalence of only 2.7% (Table 3).
As the frequency of these HLA genotypes was higher in patients with CD-T1D compared with what was observed in CD and T1D alone, it is conceivable that they may be specific to patients with CD-T1D.
Interestingly, in the 76 patients with CD-T1D, these 3 HLA genotypes explained almost all the HLA variability reaching a frequency of 81.6% among all the other HLA genotypes (Table 3).
Conversely, several HLA genotypes frequently observed in CD showed a very low association or were absent in CD-T1D and T1D. For instance, the HLA-DQ2.5/HLA-DQ2.2 genotype, conferring the highest OR in CD, was rarely observed in T1D and was absent in patients with CD-T1D as well as the HLA-DQ2.5 molecule encoded in trans. Therefore, these HLA genotypes, which are strongly associated with CD, seem to be not predisposing for the future development of T1D.
In addition, the HLA-DQ2.5/x genotypes were observed in CD-T1D but with an association lower than that observed in controls, thus conferring a low risk of T1D susceptibility.
Altogether, these data confirm that CD-T1D shows with HLA class II genes a peculiar and different association from that reported in CD and T1D.
Another finding emerging from our data is a significantly different age of onset between CD and T1D. We have found that CD begins at around 8.16 (range 1–20) years, whereas T1D has an average of onset at 11.05 (range 1–23). More importantly, we observed in younger CD children, an increased percentage of patients who developed T1D after CD (Table 4). Therefore, the discovery that CD precedes the onset of T1D and that in younger CD children T1D starts after CD onset could have practical implications. In other terms, our data suggest that an earlier CD diagnosis followed by HLA typing may allow for identification of a larger number of patients with CD with high risk of future T1D development at a very early stage (phase 1) of disease.
It is important to note that in the last ESPGHAN-2020 guidelines, the HLA typing has been removed from the no-biopsy approach to diagnosis of CD. The reasons adopted were that HLA typing has been demonstrated to have a weak positive predictivity and that it is not universally available and quite costly in some countries.
Obviously, the guidelines have been designed for CD diagnosis without taking into account that the HLA typing, as shown in this article, allows to establish in CD children also the risk of future T1D development. In light of our results, it is worth considering that HLA typing in children with CD could be re-evaluated, given the recent decision of the Italian parliament to start a screening campaign for CD and T1D among children and also taking into account the numerous mass screening initiatives for T1D (31,32).
In conclusion, CD represents a high-risk condition for the future development of T1D. In this study, we uncovered significant variations in T1D risk based on HLA genotypes.
Our data may suggest that to prevent the future development of T1D in CD children, a population screening program for celiac autoimmunity with antitransglutaminase IgA (TtG-IgA) antibody starting from 3 years of age should be instituted. Children who test positive for TtG-IgA autoantibodies should undergo HLA typing to identify a greater number of patients with CD who are at high risk of T1D.
This strategy may detect patients with CD at the very early stages of T1D (phase 1) who should undergo periodic pancreatic autoantibody monitoring, further immune, genetic, and metabolic tests (14), and eventually low-risk immunotherapies able to preserve endogenous β-cell function.
CONFLICTS OF INTEREST
Guarantor of the article: Mauro Congia, MD.
Specific author contributions: E.S., M.C., C.S., and M.R.R.: contributed to the conception and design of the work; the acquisition, analysis, and the interpretation of data for the work. R.R., R.D.J., D.B., M.C., and S.M.: contributed to drafting the work or reviewing it critically for important intellectual content. F.C., M.C., and C.R.: final approval of the version to be published. M.C. and E.S.: accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Financial support: This research did not receive any specific grant from funding agencies in the public or commercial sectors.
Potential competing interests: None to report.
Study Highlights.
WHAT IS KNOWN
✓ Type 1 diabetes (T1D) and celiac disease frequently coexist in the same patient.
✓ T1D and celiac disease share human leukocyte antigen (HLA) genetic susceptibility.
WHAT IS NEW HERE
✓ In celiac disease (CD), only a fraction of HLA genotypes is permissive for T1D development.
✓ Some HLA genotypes conferring high risk of CD may protect from the future development of T1D.
✓ All HLA genotypes associated with T1D confer susceptibility to CD.
✓ HLA typing in CD may predict the risk of future development of T1D.
ACKNOWLEDGMENTS
We thank the not-for-profit organization Diabete Zero ODV for the study design and Fondazione Sardegna (prot. U44.2021/AI.40.PL prat. 2020.2284) for supporting D.D. in an unrelated work. The article was written in the memory of Caterina Chessa.
Contributor Information
Enrico Schirru, Email: schirrue@unica.it.
Rossano Rossino, Email: rossino40@gmail.com.
Daniela Diana, Email: dani.diana2@gmail.com.
Rita D. Jores, Email: rdjores@yahoo.com.
Davide Baldera, Email: d.baldera19@gmail.com.
Sandro Muntoni, Email: smuntoni@unica.it.
Claudia Spiga, Email: claudia.spiga87@gmail.com.
Carlo Ripoli, Email: carlo.ripoli@aob.it.
Maria R. Ricciardi, Email: rossella.ricciardi@tiscali.it.
Francesco Cucca, Email: fcucca@uniss.it.
REFERENCES
- 1.Rubin JE, Crowe SE. Celiac disease. Ann Intern Med 2020;172(1):ITC1–ITC16. [DOI] [PubMed] [Google Scholar]
- 2.Husby S Koletzko S Korponay-Szabo IR, et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J Pediatr Gastroenterol Nutr. 2012;54(1):136–60. [DOI] [PubMed] [Google Scholar]
- 3.Lauret E, Rodrigo L. Celiac disease and autoimmune-associated conditions. Biomed Res Int 2013;2013:127589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flores Monar GV, Islam H, Puttagunta SM, et al. Association between type 1 diabetes mellitus and celiac disease: Autoimmune disorders with a shared genetic background. Cureus 2022;14(3):e22912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neuhausen SL, Steele L, Ryan S, et al. Co-occurrence of celiac disease and other autoimmune diseases in celiacs and their first-degree relatives. J Autoimmun 2008;31(2):160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohn A, Sofia AM, Kupfer SS. Type 1 diabetes and celiac disease: Clinical overlap and new insights into disease pathogenesis. Curr Diab Rep 2014;14(8):517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dube C, Rostom A, Sy R, et al. The prevalence of celiac disease in average-risk and at-risk western European populations: A systematic review. Gastroenterology 2005;128(4 Suppl 1):S57–67. [DOI] [PubMed] [Google Scholar]
- 8.Volta U, Tovoli F, Caio G. Clinical and immunological features of celiac disease in patients with Type 1 diabetes mellitus. Expert Rev Gastroenterol Hepatol 2011;5(4):479–87. [DOI] [PubMed] [Google Scholar]
- 9.Smigoc Schweiger D, Mendez A, Kunilo Jamnik S, et al. High-risk genotypes HLA-DR3-DQ2/DR3-DQ2 and DR3-DQ2/DR4-DQ8 in co-occurrence of type 1 diabetes and celiac disease. Autoimmunity 2016;49(4):240–7. [DOI] [PubMed] [Google Scholar]
- 10.Zubkiewicz-Kucharska A, Jamer T, Chrzanowska J, et al. Prevalence of haplotype DQ2/DQ8 and celiac disease in children with type 1 diabetes. Diabetol Metab Syndr 2022;14(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vidan-Jeras B. When type 1 diabetes meets celiac disease. HLA 2018;92(Suppl 2):64–6. [DOI] [PubMed] [Google Scholar]
- 12.Hagopian W, Lee HS, Liu E, et al. Co-occurrence of type 1 diabetes and celiac disease autoimmunity. Pediatrics 2017;140(5):e20171305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anand V, Li Y, Liu B, et al. Islet autoimmunity and HLA markers of presymptomatic and clinical type 1 diabetes: Joint analyses of prospective cohort studies in Finland, Germany, Sweden, and the U.S. Diabetes Care 2021;44(10):2269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bluestone JA, Buckner JH, Herold KC. Immunotherapy: Building a bridge to a cure for type 1 diabetes. Science 2021;373(6554):510–6. [DOI] [PubMed] [Google Scholar]
- 15.Herold KC, Bundy BN, Long SA, et al. An anti-CD3 antibody, teplizumab, in relatives at risk for type 1 diabetes. N Engl J Med 2019;381(7):603–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dayan CM, Besser REJ, Oram RA, et al. Preventing type 1 diabetes in childhood. Science 2021;373(6554):506–10. [DOI] [PubMed] [Google Scholar]
- 17.Tatovic D, Dayan CM. Replacing insulin with immunotherapy: Time for a paradigm change in type 1 diabetes. Diabet Med 2021;38(12):e14696. [DOI] [PubMed] [Google Scholar]
- 18.Gregory JW, Carter K, Cheung WY, et al. Phase II multicentre, double-blind, randomised trial of ustekinumab in adolescents with new-onset type 1 diabetes (USTEK1D): Trial protocol. BMJ Open 2021;11(10):e049595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ludvigsson JF, Ludvigsson J, Ekbom A, et al. Celiac disease and risk of subsequent type 1 diabetes: A general population cohort study of children and adolescents. Diabetes Care 2006;29(11):2483–8. [DOI] [PubMed] [Google Scholar]
- 20.Lampis R, Morelli L, De Virgiliis S, et al. The distribution of HLA class II haplotypes reveals that the Sardinian population is genetically differentiated from the other Caucasian populations. Tissue Antigens 2000;56(6):515–21. [DOI] [PubMed] [Google Scholar]
- 21.Jores RD, Frau F, Cucca F, et al. HLA-DQB1*0201 homozygosis predisposes to severe intestinal damage in celiac disease. Scand J Gastroenterol 2007;42(1):48–53. [DOI] [PubMed] [Google Scholar]
- 22.Schirru E, Jores RD, Cicotto L, et al. High frequency of low-risk human leukocyte antigen class II genotypes in latent celiac disease. Hum Immunol 2011;72(2):179–82. [DOI] [PubMed] [Google Scholar]
- 23.Schirru E, Jores RD, Rossino R, et al. Low-risk human leukocyte antigen genes and mild villous atrophy typify celiac disease with immunoglobulin A deficiency. J Pediatr Gastroenterol Nutr 2021;72(6):889–93. [DOI] [PubMed] [Google Scholar]
- 24.Zoledziewska M, Perra C, Orru V, et al. Further evidence of a primary, causal association of the PTPN22 620W variant with type 1 diabetes. Diabetes 2008;57(1):229–34. [DOI] [PubMed] [Google Scholar]
- 25.Husby S Koletzko S Korponay-Szabo I, et al. European Society Paediatric Gastroenterology, Hepatology and Nutrition guidelines for diagnosing coeliac disease 2020. J Pediatr Gastroenterol Nutr. 2020;70(1):141–56. [DOI] [PubMed] [Google Scholar]
- 26.Walker-Smith J Guandalini S Schmitz J, et al. Revised criteria for diagnosis of coeliac disease. Report of working group of European Society of Paediatric Gastroenterology and Nutrition. Arch Dis Child. 1990;65(8):909–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pais WP, Duerksen DR, Pettigrew NM, et al. How many duodenal biopsy specimens are required to make a diagnosis of celiac disease? Gastrointest Endosc 2008;67(7):1082–7. [DOI] [PubMed] [Google Scholar]
- 28.Marsh MN, Crowe PT. Morphology of the mucosal lesion in gluten sensitivity. Baillieres Clin Gastroenterol 1995;9(2):273–93. [DOI] [PubMed] [Google Scholar]
- 29.Oberhuber G. Histopathology of celiac disease. Biomed Pharmacother 2000;54(7):368–72. [DOI] [PubMed] [Google Scholar]
- 30.Songini M, Lombardo C. The Sardinian way to type 1 diabetes. J Diabetes Sci Technol 2010;4(5):1248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Couzin-Frankel J. Efforts to screen kids for type 1 diabetes multiply. Science 2024;383(6688):1164–5. [DOI] [PubMed] [Google Scholar]
- 32.Bosi E, Catassi C. Screening type 1 diabetes and celiac disease by law. Lancet Diabetes Endocrinol. 2024;12(1):12–4. [DOI] [PubMed] [Google Scholar]