Abstract
Cellular activation is critical for the propagation of human immunodeficiency virus type 1 (HIV-1) infection. It has been suggested that truly naive CD4+ T cells are resistant to productive HIV-1 infection because of their constitutive resting state. Memory and naive CD4+ T-cell subsets from 11 HIV-1-infected individuals were isolated ex vivo by a combination of magnetic bead depletion and fluorescence-activated cell sorting techniques with stringent criteria of combined expression of CD45RA and CD62L to identify naive CD4+ T-cell subsets. In all patients HIV-1 provirus could be detected within naive CD45RA+/CD62L+ CD4+ T cells; in addition, replication-competent HIV-1 was isolated from these cells upon CD4+ T-cell stimulation in tissue cultures. Memory CD4+ T cells had a median of fourfold more replication-competent virus and a median of sixfold more provirus than naive CD4+ T cells. Overall, there was a median of 16-fold more integrated provirus identified in memory CD4+ T cells than in naive CD4+ T cells within a given patient. Interestingly, there was a trend toward equalization of viral loads in memory and naive CD4+ T-cell subsets in those patients who harbored CXCR4-using (syncytium-inducing) viruses. Within any given patient, there was no selective usage of a particular coreceptor by virus isolated from memory versus naive CD4+ T cells. Our findings suggest that naive CD4+ T cells may be a significant viral reservoir for HIV, particularly in those patients harboring CXCR4-using viruses.
Human immunodeficiency virus type 1 (HIV-1) replication is sustained by continuous rounds of de novo infection of a pool of rapidly turning over CD4+ T cells in vivo (20, 21, 27, 35). It is believed that activated CD4+ T cells which have recently encountered antigen contribute to this pool (6, 17, 21, 35). T cells can be subdivided phenotypically into memory and naive subsets. Naive T cells have not yet encountered antigen, coexpress CD45RA and CD62L, and lack markers of cellular activation (1, 16, 22, 25, 28, 31, 37). Thus, it is assumed that the pool of rapidly turning over CD4+ T cells reflects only the memory phenotype. This is supported by previous observations demonstrating preferential HIV-1 isolation and accumulation of provirus within the memory versus naive subsets of CD4+ T cells obtained from HIV-1-infected individuals (4, 5, 8, 32).
Recent studies have shown that although memory (CD45RO+) and naive (CD45RA+) CD4+ T cells are equally susceptible to acute infection by laboratory-adapted (T cell line-tropic) strains of HIV-1, memory CD4+ T cells have a greater capacity to produce infectious virus in tissue cultures after activation by various physiologic stimuli, such as interleukin 2 (IL-2), or anti-CD3 plus anti-CD28 costimulation (5, 30, 34, 36). In particular, Roederer et al. (30) isolated highly purified populations of naive cells identified by the dual expression of both CD45RA and CD62L from normal HIV-uninfected blood donors and showed that these cells were inherently resistant to productive infection by the LAI strain of HIV-1 after anti-CD3 and anti-CD28 costimulation. These findings have questioned the importance of naive CD4+ T cells as a potential reservoir for HIV-1 and suggest that naive CD4+ T cells in vivo should be devoid of HIV-1. Previous studies that have identified culturable HIV-1 from naive CD4+ T cells in HIV-1-infected individuals only used a single marker, i.e., CD45RA or CD45RO expression, to separate naive from memory T cells. The increased expression of CD45RA on memory T cells in HIV-1-infected individuals may have confounded these analyses due to the contamination of the naive T-cell compartment with a CD45RA+ population of memory T cells (30). It has thus been suggested that future studies examining naive CD4+ T-cell subsets in HIV-1-infected individuals should utilize cells with dual expression of CD45RA and CD62L in order to define purer populations of naive cells.
The surface expression of the HIV-1 coreceptors CCR5 and CXCR4 on CD4+ T cells is differentially expressed on memory versus naive T cells. Specifically, CCR5, the coreceptor for macrophage-tropic viruses (2, 7, 12–15), is largely restricted to a CD26high subset of memory T cells. CXCR4, the dominant coreceptor for T cell line-tropic viruses (11, 18, 33), is expressed on both memory and naive cells, although more so on naive T cells (3, 23, 34, and unpublished observations). It is unknown how this differential expression of the major HIV-1 coreceptors correlates with the susceptibility of these subsets of cells to viruses of various phenotypes (CCR5-using versus CXCR4-using virus) in vivo.
In the present study, we sought to determine whether naive T cells from HIV-1-infected individuals are truly infected with HIV-1 by using more stringent criteria to identify naive CD4+ T cells based on the combined phenotype of CD4, CD45RA, and CD62L with an approach involving quantitative micrococulture and PCR. In addition, we looked for evidence of phenotypic and genotypic compartmentalization of viruses within memory and naive T-cell subsets by examining coreceptor usage and heteroduplex analysis of virus isolates obtained from respective cell subsets. We confirm that naive T cells are indeed infected in vivo in HIV-1-infected individuals at all stages of disease, although the majority of culturable virus and provirus resides within memory T cells, especially in patients who harbor only macrophage-tropic (non-syncytium-inducing [NSI]) viruses. We also demonstrate that naive CD4+ T cells may be an important viral reservoir in those patients who harbor CXCR4-using (syncytium-inducing [SI]) viruses.
MATERIALS AND METHODS
Antibodies and reagents.
The following antibodies and reagents were obtained from PharMingen (San Diego, Calif.): CD62L-fluorescein isothiocyanate and CD45RA-phycoerythrin. CD4-perdinin chlorophyll protein was obtained from Becton Dickinson (San Jose, Calif.). MT2 cells were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. U87MG (microglia) cells expressing the human CD4 gene and various chemokine receptors were kindly provided by Dan Littman. RPMI 1640 (BioWhittaker, Walkersville, Md.) was supplemented with glutamine (2 mM) (Biofluids, Rockville, Md.), penicillin-streptomycin (Biofluids), HEPES (15 mM) (Biofluids), and 10% fetal calf serum (FCS). IL-2 was obtained from Boehringer (Mannheim, Germany).
Patients.
Eleven HIV-1-infected adults (Table 1) were subjected to apheresis to obtain peripheral blood mononuclear cells (PBMCs) according to a protocol approved by the National Institute of Allergy and Infectious Diseases institutional review board.
TABLE 1.
Baseline clinical profiles of patients in this study
| Patient | Baseline CD4 count/μl | Therapy | Duration of therapy | Viremia (RNA copies/ml) | Naive CD4 (%)b | Memory CD4 (%) | Virus phenotype |
|---|---|---|---|---|---|---|---|
| 1 | 997 | None | <500 | 50.9 | 49.1 | NSI | |
| 2 | 422 | HAARTa | 2 yr | 1,439 | 32.4 | 67.6 | NSI |
| 3 | 586 | None | 27,140 | 32.3 | 67.7 | NSI | |
| 4 | 189 | HAART | 2 yr | 27,680 | 26.8 | 73.2 | SI |
| 5 | 565 | None | 10,900 | 62.7 | 37.3 | NSI | |
| 6 | 144 | HAART | 1 yr | 1,085 | 31.1 | 68.9 | NSI |
| 7 | 279 | HAART | 1 yr | 66,520 | 30.5 | 69.5 | SI |
| 8 | 587 | HAART | 0.5 yr | <500 | 47.3 | 52.7 | SI |
| 9 | 304 | HAART | 2 wk | <500 | 40 | 60 | NSI |
| 10 | 547 | HAART | 1.5 yr | <500 | 48 | 52 | SI |
| 11 | 663 | None | <500 | 25.4 | 74.6 | NSI |
HAART, highly active antiretroviral therapy; usually one protease inhibitor and two nucleoside reverse transcriptase inhibitors.
Based on percentages of CD4 cells in PBMC.
Isolation of memory and naive CD4+ T-cell populations.
PBMCs were Ficoll separated (lymphocyte separation medium [LSM]; Organon Teknika, Durham, N.C.) and washed in phosphate-buffered saline (BioWhittaker)–1% FCS (HyClone, Ogden, Utah). Prior to sorting, the CD4+ T-cell population was enriched by depletion of CD14+ and CD8+ cells with magnetic beads according to the manufacturer’s instructions (Dynal, Lake Success, N.Y.). Cells were stained with CD62L-fluorescein isothiocyanate, CD45RA-phycoerythrin, and CD4-perdinin chlorophyll protein at room temperature, washed, and then sorted on an Elite (Coulter, Hialeah, Fla.) cell sorter. In general, 5 to 10 million cells of a particular phenotype were sorted.
Micrococulture assay.
In order to determine the frequency of sorted memory and naive cells from patients carrying replication-competent HIV-1, micrococulture assays were carried out as previously described (10). Briefly, duplicate fivefold serial dilutions were performed in an IL-2-containing medium (20 U/ml). To induce activation, anti-CD3 antibody (mouse ascites fluid at a 1/4,000 dilution) and irradiated autologous PBMCs were added to the culture. The following day, 3-day-old CD3-stimulated, CD8-depleted PBMCs from an HIV-negative donor were added to amplify infectious virus. Cultures were restimulated with CD3-blasts on day 7. Supernatants were collected on day 14 and analyzed for p24 antigen by enzyme-linked immunosorbent assay (Coulter).
Assays for integrated and total HIV-1 DNA.
Genomic DNA from purified memory and naive CD4+ T cells was prepared with the Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minn.). The total copy number of HIV-1 DNA was determined and quantitated by a single-round PCR as previously described (33), with primers RU5-5′ (5′-GGTCTCTCTGGTTAGACCAGAT-3′) and RU5-3′ (5′-CTGCTAGAGATTTTCCACACTG-3′). The assay was performed in duplicate when possible, with an input DNA of 1 to 2 μg. PCR products were analyzed by gel electrophoresis followed by liquid hybridization with a 32P-end-labeled probe RU5 (5′-AGTAGTGTGTGCCCGTCTGT-3′). After liquid hybridization, bands were quantified by PhosphorImager analysis (Molecular Dynamics, Sunnyvale, Calif.) by using a standard curve based on a PCR of known copy numbers of serially diluted ACH-2 DNA. For the determination of integrated DNA, cellular DNA was serially diluted, in duplicate, into tubes containing 200,000, 40,000, 8,000, 1,600, 320, 16, or 2.4 cell equivalents and then subjected to a nested PCR, in which first-round primers consisted of conserved sequences of human Alu (Alu-long terminal repeat [LTR] 5′) and HIV-1 LTR (Alu-LTR 3′) and second-round primers consisted of a portion of the LTR region of HIV-1 DNA (NI-2 5′ and NI-2 3′) as previously described (33). PCR products were analyzed by gel electrophoresis and then by Southern hybridization with a 32P-end-labeled probe (NI). The frequency of cells carrying integrated HIV-1 DNA was determined from the limiting dilution PCR data by the statistical method of Myers et al. (24).
Virus phenotyping and determination of coreceptor usage.
Cultured virus was phenotyped by using an MT2 cell assay in which 100 μl of culture supernatant (>5,000 reverse transcriptase counts/μl) was added to MT2 cells and monitored for the presence of syncytia over a 3-week period. The coreceptor usage of virus isolates was determined by using U87MG cells expressing the human CD4 gene and one of the following receptors: CXCR4, CCR5, CCR1, CCR2b, and CCR3. U87MG cells were maintained in Dulbecco’s modified Eagle medium (BioWhittaker) with 10% FCS, puromycin (1 μg/ml), and G418 (500 μg/ml). The medium for U87MG cells expressing CD4 alone did not contain G418. A 100-μl volume of culture supernatant was added to plated U87MG cells (4 × 104 cells per well) in 24-well plates, and cultures were maintained in 2 ml of Dulbecco’s modified Eagle medium supplemented as described above. Cultures were examined daily for syncytia, and supernatants were tested for reverse transcriptase activity on days 7 and 14 as previously described (26).
HTA.
cDNA was synthesized from RNA extracted from plasma and viral culture supernatants as previously described (26). Heteroduplex mobility shift assays (HMA) and heteroduplex tracking assays (HTA) were performed by using nested PCR products of the V3-V5 region of HIV-1 envelope amplified from cDNA from patient plasma samples obtained on the day of sorting, from DNA from ex vivo-sorted memory and naive cells, and from cDNA from viral culture supernatants of memory and naive cells. Nested PCR of the V3-V5 region was performed by using primers ED5 and ED12 in the first round and DR7 and DR8 in the second round as previously described (26). 32P-labeled single-stranded probes were generated from DR7-DR8-derived PCR products of cDNA from viral culture supernatants of memory and naive cells (26). Driver sequences consisted of envelope PCR products from cDNA of plasma, from DNA derived from freshly sorted memory and naive cells, and from an unrelated sample derived from ACH-2 cells. Probes were mixed with driver sequences at a ratio of 1:100 in an annealing buffer, denatured at 94°C for 3 min, placed on ice for 5 min, and heated to 55°C for 5 min to form heteroduplexes (26). The resulting reaction mixtures were electrophoresed on 5% polyacrylamide gels (acrylamide/bis ratio, 37.5:1) at 250 V for 3 h, stained with ethidium bromide to assure the even migration of bands, dried, and scanned using a phosphorimager as previously described (26).
Statistical analysis.
Comparisons between memory and naive CD4+ T-cell subsets were made using a one-sided Wilcoxon signed rank test. Data are shown as median values in order to better represent the central tendency. P values were corrected for multiple comparisons by the Bonferroni method. In comparisons between memory and naive CD4+ T cells within the subgroup of patients with SI viruses, P values are not reported because of the small number of patients (three to four) in the comparison.
RESULTS
Characterization of HIV-1 viral load in memory and naive CD4+ T-cell subsets.
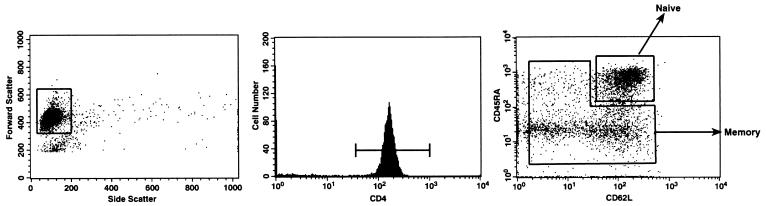
We isolated CD4+ T-cell subsets from 11 HIV-1-infected individuals who presented at various disease stages (Table 1) by using a combination of magnetic bead depletion and flow cytometric sorting techniques. As previously described (16, 22, 25, 28, 29, 31), we defined naive CD4+ T cells as those which dually expressed CD45RA and CD62L and all other cells as memory (i.e., CD45RA−/CD62L−, CD45RA+/CD62L−, or CD45RA−/CD62L+). It should be noted that the memory compartment consisted of activated effector cells as well as resting memory cells (Fig. 1). The purity of sorted populations was ≥96% (data not shown).
FIG. 1.
Fluorescence-activated cell sorter analysis and sorting of CD4+ T cells. CD4+-enriched T cells were obtained by negative bead selection and subsequently stained for the expression of CD62L, CD45RA, and CD4. Sorting gates are shown for lymphocytes (left panel), CD4-positive T cells (middle panel), and naive and memory cells (right panel). Cell purities were always assessed immediately and 36 h postsorting and were >95%.
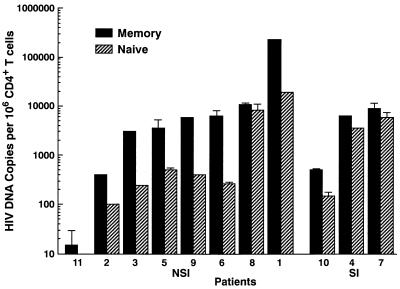
The amount of replication-competent HIV-1 within CD4+ T-cell subsets was determined by using a sensitive quantitative micrococulture assay. In this assay, serially diluted freshly sorted memory or naive CD4+ T cells were activated in vitro with anti-CD3 and IL-2 as previously described (8), and supernatants from each culture were collected on day 14 for the determination of HIV-1 p24 by enzyme-linked immunosorbent assay. Both integrated and unintegrated replication-competent proviruses can be induced in this assay (9). Infectious virus was detected from both stimulated memory and naive CD4+ T-cell subsets in all 11 patients studied (Fig. 2). Within a given patient, the frequency of cells carrying infectious HIV-1 in memory CD4+ T-cell subsets was a median of fourfold greater than that in naive CD4+ T-cell subsets (range, 0.5- to 19-fold; P < 0.01).
FIG. 2.
Quantitative virus isolation from memory and naive CD4+ T-cell subsets. Frequencies of memory and naive CD4+ T cells carrying replication-competent HIV-1 DNA were determined by activating sorted CD4+ T-cell memory and naive subsets on day 0. For each assay, a statistical method developed by Myers et al. (24) was used to calculate copy numbers or infectious units per million cells. Patients are grouped according to the presence of NSI or SI virus isolated in tissue cultures based on MT2 cell assays. Note the log scale of the y axis.
HIV-1 proviral DNA was detected in both memory and naive CD4+ T-cell subsets in all patients studied (Fig. 3) except in the naive CD4+ T-cell population in patient 11. The naive CD4+ T cells in this patient presumably contained provirus at levels below the limit of detection of the assay (10 copies/million cells), since small amounts of virus could be induced in tissue cultures (Fig. 2) in this subset. Within a given patient there was a median of sixfold greater total HIV-1 provirus in memory cells than in naive cells (range, 1.3- to 24-fold; P < 0.01).
FIG. 3.
Quantitation of total proviral HIV-1 DNA in memory and naive CD4+ T-cell subsets. The number of copies of total HIV-1 proviral DNA was determined in freshly sorted memory and naive CD4+ T-cell subsets. Patients are grouped according to the presence of NSI or SI virus isolated in tissue cultures based on MT2 cell assays. Note the log scale of the y axis.
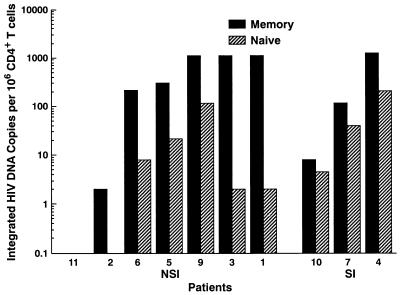
The amount of integrated HIV-1 DNA was determined by utilizing a previously described Alu-LTR nested PCR method that has a sensitivity at the single copy level (10). The integrated form of HIV-1 DNA was detected in both memory and naive subsets in the majority of patients studied (Fig. 4). In patient 2, integrated provirus was undetected in the naive CD4+ T-cell subset, and in patient 11 integrated provirus was undetected in both subsets. Of note, when comparing the amounts of integrated HIV-1 DNA found within memory and naive CD4+ T-cell subsets we observed even greater differences than that observed with total provirus, with a median of 16-fold more integrated HIV-1 DNA in memory CD4+ T cells (range, 1.8- to 561-fold; P < 0.01).
FIG. 4.
Quantitation of integrated HIV-1 DNA in memory and naive CD4+ T-cell subsets. The number of copies of integrated HIV-1 proviral DNA was determined in freshly sorted memory and naive CD4+ T-cell subsets. Integrated provirus was undetectable in patient 11 in either CD4+ T-cell subset with an input DNA of 2 μg in the assay. Data for patient 8 were not determined. Patients are grouped according to the presence of NSI or SI virus isolated in tissue cultures based on MT2 cell assays. Note the log scale of the y axis.
There was a trend toward equalization of the amounts of virus in memory and naive subsets in those patients who harbored SI viruses (4, 7, 8, and 10), even though the average CD4+ T-cell counts of both groups were fairly comparable, i.e., 526 versus 400 cells/μl, respectively (P = 0.5). For example, there was a median of 10-fold more replication-competent HIV-1 isolated in memory versus naive CD4+ T-cell subsets in those patients with NSI viruses (P = 0.05) but only threefold more in those patients with SI viruses. Similarly, there was a median of 12-fold more total HIV-1 provirus in memory versus naive subsets in patients with NSI viruses (P = 0.05) but only 1.7-fold more in patients with SI viruses; there was a median of 23-fold more integrated HIV-1 DNA in memory versus naive cells in patients harboring NSI viruses (P = 0.05) but only threefold more in patients with SI viruses.
Spectrum of coreceptor usage by HIV-1 isolated from memory and naive CD4+ T-cell subsets.
Since the major HIV-1 coreceptors CCR5 and CXCR4 are differentially expressed on memory and naive CD4+ T-cell subsets, we looked for evidence of phenotypic compartmentalization of viruses isolated from memory and naive subsets within individual patients. In 4 of 11 patients, SI viruses were isolated from both memory and naive subsets (patients 4, 7, 8, and 10) by MT2 cell assay. In order to determine which coreceptors were used by viruses induced from sorted memory and naive subsets, we infected U87MG/CD4 cells stably expressing each of the chemokine receptors CCR1, CCR2b, CCR3, CCR5, and CXCR4 with virus isolated from either memory or naive CD4+ T cells from each patient. Table 2 shows coreceptor usage of bulk culture supernatants isolated from respective CD4+ T-cell subsets. Within a given patient, coreceptor usage by viruses isolated from memory and naive subsets was identical, indicating a lack of strict phenotypic compartmentalization of viruses between these CD4+ T-cell subsets. The usage of CXCR4 of all HIV-1 isolates, as determined with U87MG/CD4 cells, directly correlated with an SI phenotype.
TABLE 2.
Phenotypic characteristics of viral isolates
| Patient | Coreceptor usage of HIV-1 isolates
|
|
|---|---|---|
| Memory CD4+ T cell | Naive CD4+ T cell | |
| 1 | CCR5, CCR1 | CCR5, CCR1 |
| 2 | CCR5 | CCR5 |
| 3 | CCR5 | CCR5 |
| 4 | CCR5, CXCR4 | CCR5, CXCR4 |
| 5 | CCR5 | CCR5 |
| 6 | CCR5, CCR1 | CCR5, CCR1 |
| 7 | CCR5, CXCR4 | CCR5, CXCR4 |
| 8 | CXCR4 | CXCR4 |
| 9 | CCR5 | CCR5 |
| 10 | CCR1, CXCR4, CCR3, CCR5 | CCR1, CXCR4, CCR3, CCR5 |
| 11 | NDa | ND |
ND, not detected.
Genotypic characterization of HIV-1 in memory and naive CD4+ T-cell subsets.
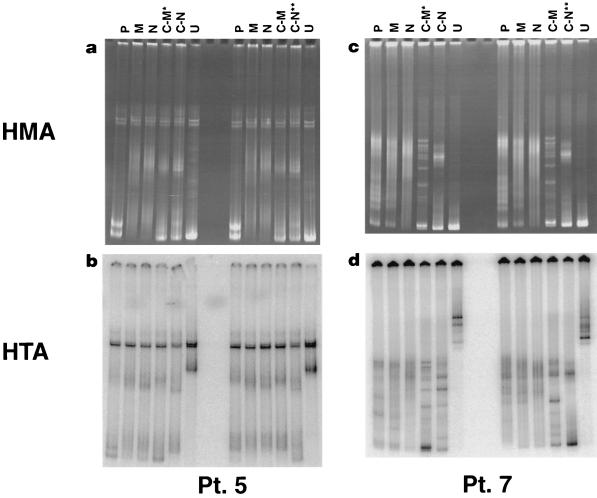
We also looked for evidence of genotypic compartmentalization of viruses within these subsets in a given patient. HTA were performed on samples from three patients by using nested PCR products of the V3-V5 envelope region of viruses derived from patient plasma, ex vivo memory and naive CD4+ T cells, and their respective cultures, in order to determine the genetic relatedness of viruses derived from these samples. Data for two representative patients are presented in Fig. 5. We could not demonstrate consistent genetic differences between viruses derived from cultures of memory versus naive CD4+ T cells. For example, although viruses derived from cultures of naive CD4+ T cells were genetically distinct by HTA in patient 5, identical viruses could be identified in both cultures, respectively, in patient 7 (see the legend to Fig. 5). Of interest, in all three patients studied, envelope sequences amplified from patient plasma migrated more closely with envelope sequences obtained from memory cells than with those from naive cells, indicating that viruses within plasma are genetically more similar to those derived from memory cells in these patients.
FIG. 5.
HTA and accompanying HMA of amplified envelopes obtained from cDNA in plasma (P), DNA in ex vivo-sorted memory (M) and naive (N) CD4+ T cells, and supernatant viral culture cDNA of memory (C-M) and naive (C-N) CD4+ T cells in two representative patients. U, unrelated sample. For each patient, two HTA were performed, with a probe made from virus cultured from memory CD4+ T cells (C-M*) (left side of gel) and a probe made from virus cultured from naive CD4+ T cells (C-N**) (right side of gel). In patient 5 (a and b), the migration patterns between envelopes from cultured memory and naive CD4+ T cells differed, indicating genetic differences between viruses cultured from these two subsets; however, in patient 7 (c and d) migration occurred equally along the length of the gel, indicating the presence of viruses with genetically identical envelopes. In both patients, envelope PCR products from cultured memory CD4+ T cells migrated equally with those from plasma virus (left side of gel), whereas there was a retardation of migration in the plasma viruses compared to viruses from naive CD4+ T cells with the probe of virus cultured from naive CD4+ T cells (C-N**) (right side of gel), thus indicating the closer genetic similarity of viruses in plasma and memory cells than in plasma and naive cells.
DISCUSSION
The present study has demonstrated the presence of inducible virus and provirus in naive CD4+ T cells from HIV-1-infected individuals by using specific criteria for identifying naive CD4+ T cells, namely, CD45RA+/CD62L+ cells. Overall, memory CD4+ T cells contained about fourfold more replication-competent virus and sixfold more total provirus than naive CD4+ T cells. This quantitative difference in provirus levels is similar to previous studies that depended upon the expression of CD45RA alone as a marker for the naive phenotype (5, 9, 32). Of note was the fact that memory CD4+ T cells contained a median of 16-fold more integrated provirus than naive CD4+ T cells in the cohort of 11 patients who were studied. This observation is consistent with the concept that HIV-1 can enter naive CD4+ T cells in vivo and initiate reverse transcription but is unable to integrate into the genome as efficiently as in memory CD4+ T cells (8). Recent in vitro studies have also demonstrated that CD45RA+ CD4+ T cells can be acutely infected with HIV-1 but are less efficient in replicating virus upon physiologic stimulation (8, 30, 34, 36). The mechanisms responsible for these differences in viral replicative capacity have not been clearly defined, and possible explanations include lower levels of deoxynucleoside triphosphate substrates and a deficiency of other cellular factors in naive cells that are necessary for completion of a viral replicative cycle (19, 34).
Of note, we observed a tendency for equalization in the amounts of HIV-1 within naive and memory CD4+ T-cell subsets in those patients with SI viruses. Given the low-level to absent CCR5 expression and high-level CXCR4 expression in naive CD4+ T cells, it is probable that CXCR4-using viruses are able to more readily infect naive CD4+ T cells than viruses that use CCR5 alone. There was considerably more virus measured by proviral DNA and infectious units in memory than in naive CD4+ T cells when the isolated virus was NSI. However, these differences between memory and naive cells were much smaller when the isolated virus was SI. These smaller differences suggest that naive CD4+ T cells may represent a significant viral reservoir in patients harboring SI viruses. In addition, absolute reductions in the naive CD4+ T-cell population have been observed with progression of HIV-1 disease (19, 29, 34). Whether the onset of decline of naive CD4+ T cells in HIV-1-infected individuals correlates with the emergence of CXCR4-using (SI) viruses will be an avenue for future study.
We asked whether there was evidence for a phenotypic or genotypic compartmentalization of viruses isolated from memory and naive CD4+ T-cell subsets, given the differing levels of chemokine receptor expression and migration patterns these cells display in vivo (16, 22, 25, 28, 29, 31). Phenotypically, all virus isolates within a given patient were identical, whether isolated from memory or naive subsets. It should be noted, however, that viral phenotypic analysis was performed on bulk cultures isolated from respective memory and naive subsets and thus differences in coreceptor usage of individual virus variants isolated from memory and naive cells could not be ruled out. Heteroduplex analysis also did not show clear genotypic differences between viruses isolated from these two subsets in the three patients whom we studied. These findings indicate that there is likely a considerable mixing of viruses in vivo within these two cell subsets. It was of interest that viruses isolated from memory cells were genotypically more closely related to those found in the plasma by heteroduplex analysis in the three patients studied. This is consistent with the notion that activated effector CD4+ cells, which also have a memory phenotype, are likely responsible for the majority of virus produced within the plasma.
We have thus shown that HIV-1 can be identified within naive CD4+ subsets displaying dual expression of CD45RA and CD62L and that this cell population may represent a significant viral reservoir of virus in those patients harboring SI viruses.
ACKNOWLEDGMENT
Mario A. Ostrowski and Tae-Wook Chun contributed equally to this work.
REFERENCES
- 1.Akbar A, Terry L, Timms A, Beverley P, Janossy G. Loss of CD45R and gain of UCHL1 reactivity is a feature of primed T cells. J Immunol. 1988;140:2171–2178. [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Bleul C C, Wu L, Hoxie J A, Springer T A, Mackay C R. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cayota A, Vuillier F, Scott-Algara D, Dighiera G. Preferential replication of HIV-1 in memory CD4+ subpopulation. Lancet. 1990;336:941–942. doi: 10.1016/0140-6736(90)92311-5. [DOI] [PubMed] [Google Scholar]
- 5.Cayota A, Vuillier F, Scott-Algara D, Feuillie V, Dighiero G. Differential requirements for HIV-1 replication in naive and memory CD4 T cells from asymptomatic HIV-1 seropositive carriers and AIDS patients. Clin Exp Immunol. 1993;91:241–248. doi: 10.1111/j.1365-2249.1993.tb05890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheynier R, Henrichwark S, Hadida F, Pelletier E, Oksenhendler E, Autran B, Wain-Hobson S. HIV and T cell expansion in splenic white pulps is accompanied by infiltration of HIV-specific cytotoxic T lymphocytes. Cell. 1994;78:373–387. doi: 10.1016/0092-8674(94)90417-0. [DOI] [PubMed] [Google Scholar]
- 7.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 8.Chun T, Carruth L, Finzi D, Shen X, DiGiuseppe A, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn T C, Kuo Y, Brookmeyer R, Zeiger M A, Barditch-Crovo P, Siliciano R F. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 9.Chun T, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano R F. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. 1995;1:1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 10.Chun T W, Stuyver L, Mizell S B, Ehler L A, Mican J A, Baseler M, Lloyd A L, Nowak M A, Fauci A S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di M P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 13.Doms R, Peiper S C. Unwelcomed guests with master keys: how HIV uses chemokine receptors for cellular entry. Virology. 1997;235:179–190. doi: 10.1006/viro.1997.8703. [DOI] [PubMed] [Google Scholar]
- 14.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 15.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 16.Dutton R W, Bradley L M, Swain S L. T cell memory. Annu Rev Immunol. 1998;16:201–223. doi: 10.1146/annurev.immunol.16.1.201. [DOI] [PubMed] [Google Scholar]
- 17.Fauci A S. Host factors and the pathogenesis of HIV-induced disease. Nature. 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 18.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 19.Gao W-Y, Cara A, Gallo R C, Lori F. Low levels of deoxynucleotides in peripheral blood lymphocytes: a strategy to inhibit human immunodeficiency virus type 1 replication. Proc Natl Acad Sci USA. 1993;90:8925–8928. doi: 10.1073/pnas.90.19.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho D D. HIV-1 dynamics in vivo. Philos Trans R Soc Lond B Biol Sci. 1995;349:33–40. [Google Scholar]
- 21.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 22.MacKay C. Migration pathways and immunologic memory among T lymphocytes. Semin Immunol. 1992;4:51–58. [PubMed] [Google Scholar]
- 23.Mo H, Monard S, Pollack H, Ip J, Rochford G, Wu L, Hoxie J, Borkowsky W, Ho D D. Expression patterns of the HIV type 1 co-receptors CCR5 and CXCR4 on CD4+ T cells and monocytes from cord and adult blood. AIDS Res Hum Retroviruses. 1998;14:607–617. doi: 10.1089/aid.1998.14.607. [DOI] [PubMed] [Google Scholar]
- 24.Myers L E, McQuay L J, Hollinger F B. Dilution assay statistics. J Clin Microbiol. 1994;32:732–739. doi: 10.1128/jcm.32.3.732-739.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okumura M, Fujii Y, Takeuchi Y, Inada K, Nakahara K, Matsuda H. Age-related accumulation of LFA-1high cells in a CD8+ CD45RAhigh T cell population. Eur J Immunol. 1993;23:1057–1063. doi: 10.1002/eji.1830230512. [DOI] [PubMed] [Google Scholar]
- 26.Ostrowski M, Krakauer D, Li Y, Justement S, Learn G, Ehler L, Stanley S, Nowak M, Fauci A. Effect of immune activation on the dynamics of human immunodeficiency virus replication and on the distribution of viral quasispecies. J Virol. 1998;72:7772–7784. doi: 10.1128/jvi.72.10.7772-7784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 28.Picker L, Treer J, Ferguson D, Collins P, Buck D, Terstappen L. Control of lymphocyte recirculation in man. I. Differential regulation of the peripheral lymph node homing receptor l-selectin on T cells during the virgin to memory cell transition. J Immunol. 1993;150:1105–1121. [PubMed] [Google Scholar]
- 29.Roederer M, Dubs J G, Anderson M T, Raju P A, Herzenberg L A, Herzenberg L A. CD8 naive T cell counts decrease progressively in HIV-infected adults. J Clin Investig. 1995;95:2061–2066. doi: 10.1172/JCI117892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roederer M, Raju P A, Dipendra K M, Herzenberg L A, Herzenberg L A. HIV does not replicate in naive CD4 T cells stimulated with CD3/CD28. J Clin Investig. 1997;99:1555–1564. doi: 10.1172/JCI119318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanders M, Makgoba M, Dhse D. Human naive and memory T cells. Immunol Today. 1988;9:195–199. doi: 10.1016/0167-5699(88)91212-1. [DOI] [PubMed] [Google Scholar]
- 32.Schnittman S M, Lane H C, Greenhouse J, Justement J S, Baseler M, Fauci A S. Preferential infection of CD4+ memory T cells by human immunodeficiency virus type 1: evidence for a role in the selective T-cell functional defects observed in infected individuals. Proc Natl Acad Sci USA. 1990;87:6058–6062. doi: 10.1073/pnas.87.16.6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spina C A, Prince H E, Richman D D. Preferential replication of HIV-1 in the CD45RO memory cell subset of primary CD4 lymphocytes in vitro. J Clin Investig. 1997;99:1774–1785. doi: 10.1172/JCI119342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini I A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 36.Woods T C, Roberts B D, Butera S T, Folks T M. Loss of inducible virus in CD45RA naive cells after human immunodeficiency virus-1 entry accounts for preferential viral replication in CD45RO memory cells. Blood. 1997;89:1635–1641. [PubMed] [Google Scholar]
- 37.Young A, Hay J, MacKay C. Lymphocyte recirculation and life span in vivo. Curr Top Microbiol Immunol. 1993;184:161–173. doi: 10.1007/978-3-642-78253-4_13. [DOI] [PubMed] [Google Scholar]