Abstract
The HTLV-1 singly spliced open reading frame I protein, p12I, is highly unstable and appears to be necessary for persistent infection in rabbits. Here we demonstrate that p12I forms dimers through two putative leucine zipper domains and that its stability is augmented by specific proteasome inhibitors. p12I is ubiquitylated, and mutations of its unique carboxy-terminus lysine residue to an arginine greatly enhance its stability. Interestingly, analysis of 53 independent HTLV-1 strains revealed that the natural p12I alleles found in ex vivo samples of tropical spastic paraparesis-HTLV-1-associated myelopathy patients contain a Lys at position 88 in some cases, whereas arginine is consistently found at position 88 in HTLV-1 strains from all adult T-cell leukemia-lymphoma (ATLL) cases and healthy carriers studied. This apparent segregation of different alleles in tropical spastic paraparesis-HTLV-associated myelopathy and ATLL or healthy carriers may be relevant in vivo, since p12I binds the interleukin-2 receptor β and γc chains, raising the possibility that the two natural alleles might affect differently the regulation of these molecules.
The human T-cell lymphotropic/leukemia virus type 1 (HTLV-1) genome spans approximately 9 kb and encodes the structural (gag and env) and enzymatic (reverse transcriptase, protease, and integrase) proteins (29) and, at the 3′ region, contains four different open reading frames (ORF; ORFsI to IV): ORFIII and ORFIV encode the viral transactivator p40 Tax, the p27Rex protein, a posttranscriptional regulator of RNA expression, and p21Rex, a protein with unknown function (8, 30). Singly and doubly spliced mRNA from the ORFI and the ORFII have been found in cultured T cells (1, 3, 10, 15) and macrophages infected by HTLV-1 (16), as well as in ex vivo samples of HTLV-1-infected individuals (1, 15). The singly spliced mRNA from the ORFI encodes a membrane-associated hydrophobic protein of 12 kDa (99 amino acids) (18) with two potential transmembrane regions, TM-1 and TM-2, and at least four putative SH3 binding motifs (PXXP) (8). To date, it has been difficult to assess ORFI protein(s) expression in infected cells; however, indirect evidence suggest its importance, since the ablation of the acceptor splice site for p12I-protein expression interferes with the ability of a biologically active HTLV-1 molecular clone to establish a persistent infection in the rabbit model (4).
The p12I protein shares regions of genetic similarity with the E5 oncoprotein encoded by the bovine papillomavirus type 1 (27), enhances the E5 transforming ability (9) and, like E5, binds to the 16-kDa subunit of the vacuolar ATPase (9, 17). E5 forms dimers and binds to cellular receptors such as platelet-derived growth factor β, epithelial growth factors, and colony-stimulating factor receptors (6, 20, 22, 24). p12I specifically binds to the immature forms of both β and γc chains of the interleukin-2 receptor (IL-2R) and decreases their surface expression in transfected cells (21), presumably by retarding the translocation of the receptor chains from the endoplasmic reticulum compartment to the Golgi and subsequently to the plasma membrane. Since p12I interacts with both the β and γc chains of the IL-2R, we postulated that p12I may form dimers. Here, we demonstrate that p12I indeed forms dimers, providing a rational explanation for how the same region of p12I may interact with both IL-2R chains. In addition, we provide evidence that the p12I metabolic instability is mediated in part by ubiquitylation at a single Lys residue at position 88 and subsequent proteasomal degradation, as well as by destabilizing residues at its amino terminus. Finally, analysis of natural alleles of p12I in 53 cases of HTLV-1 infection suggests that the presence of a p12I allele carrying a lysine at position 88 could be important in tropical spastic paraparesis-HTLV-1-associated myelopathy (TSP-HAM).
MATERIALS AND METHODS
Expression plasmids.
The pME18s plasmid was used to express the p12I cDNA, tagged with the AU1 or the HA1 epitopes, as well as all p12I mutants tagged with the AU1 epitope (15, 17).
The lysine-to-arginine mutation at position 88 in the p12IHA1 plasmid was generated by PCR with the upstream primer 5′-CTTTCTCCCCTGGAGGGC-3′ and the downstream primer 5′-CTGCTCTAGACGGTTTGCTATCC-3′. This 100-bp fragment was used in a subsequent PCR with an upstream primer 5′-ATTCTCGAGCACCTCGCCTTCC-3′, and the resultant 450-bp product was cloned into the XhoI and XbaI sites of a modified pME18s vector missing the XhoI stuffer fragment. The introduction of the genetic mutation was verified by DNA sequencing.
DNA transfection and protein detection.
One million (HeLa-TAT) or 1.2 × 106 (293T) cells were plated in a 100-mm3 dish and transfected the next day with 10 μg each of plasmid by the calcium phosphate method (11). At 24 h after transfection, the cells were harvested, washed twice with 1× phosphate-buffered saline (PBS), and lysed with 1× RIPA buffer (1% deoxycholic acid, 0.1% sodium dodecyl sulfate [SDS], 1% Triton X-100, 0.15 M NaCl, 50 mM Tris-Cl [pH 7.5]), containing 20 μg of leupeptin and aprotinin per ml and 10 μg of trypsin inhibitor per ml, 1 mM sodium orthovanadate, and 1 mM AEBSF. In some cases, HeLa-TAT cells were metabolically labeled for 4 h with 200 μCi of EXPRE35S, as previously described (21). Cell lysates were precleared for 2 h with normal rabbit serum and protein A-agarose beads (Boehringer Mannheim); beads were then pelleted, and the supernatant was reacted overnight at 4°C with specific antibody (Ab). Immunocomplexes, bound with protein A-agarose beads, were extensively washed with cold 1× RIPA buffer and boiled in 1× SDS-Laemmli buffer (Novex, San Diego, Calif.). Proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) on 10, 16, and 18% polyacrylamide gels and then transferred to nitrocellulose membranes and detected by Western blot analysis. Labeled proteins were analyzed in SDS–15% PAGE gel and visualized on X-Omat film (Kodak, Eastman, N.Y.). The Ab against AU1 (BAbCO; Berkeley Antibody, Richmond, Calif.) and HA1 (12CA5; Boehringer Mannheim, Indianapolis, Ind.) epitopes were used to immunoprecipitate the tagged wild-type p12I and mutants.
Proteasome inhibitors’ treatment of transfected cells and p12I ubiquitylation.
HeLa-TAT or 293T cells were transfected as described previously and treated with proteasome inhibitors 20 h after transfection: cells were incubated with either 10 μM lactacystin for 8 h or 30 μM MG115 with or without 50 μM MG132 for 2 h each (Calbiochem, La Jolla, Calif.). After lysis, the amount of total protein was measured by Bradford assay (Bio-Rad, Hercules, Calif.); 30 μg of total cell protein was loaded in a 16% SDS gel, transferred to a nitrocellulose membrane, and analyzed by Western blotting with the appropriate Ab. Anti-ubiquitin serum (Sigma, St. Louis, Mo.) was used in p12I-ubiquitylation experiments for immunoprecipitation and Western blot analysis.
Cycloheximide treatments of transfected cells.
293T cells were plated at 4 × 105 cells/well in six-well plates, and the following morning they were transfected by the calcium phosphate method. Two micrograms of the p12I expression plasmid was used for the transfection, and the amount of DNA transfected was normalized to 4 μg with pME18s. Approximately 6 h after transfection, cells were washed two times in 1× PBS, and fresh medium was added (Dulbecco modified Eagle medium, 10% fetal calf serum, 1% penicillin-streptomycin [GIBCO BRL, Grand Island, N.Y.]). Eighteen hours later, the cells were treated with 10 μg of cycloheximide (Sigma) per ml, and cells were harvested at the indicated time points in lysis buffer containing aprotinin (20 μg/ml), AEBSF (1 mM), dithiothreitol (0.5 mM), and leupeptin (20 μg/ml). The protein concentration was determined by using the Bradford assay (Bio-Rad). Equivalent amounts of each sample were prepared in SDS loading buffer (Novex) with 5% β-mercaptoethanol, heated at 95°C for 5 min, and electrophoresed on an SDS–16% PAGE gel (Novex), followed by transfer to nitrocellulose membrane (Schleicher & Schuell, Keene, N.H.) by using a Bio-Rad Transblot cell at 100 V for 2 h at 4°C. p12I stability assays were performed three to four times with 15 to 50 μg of protein.
Western blot analysis.
Membranes were blocked in 3% bovine serum albumin (Sigma) with 0.2% Tween 20 in 1× PBS for 1 h at room temperature, followed by an overnight incubation at 4°C in a 1:1,000 dilution of either anti-HA1 (Boehringer Mannheim) or anti-AU1 Ab (BAbCO) in blocking buffer. After this washing, blots were incubated at room temperature for 1 h in biotin-conjugated donkey anti-mouse immunoglobulin G (Jackson Immunoresearch, West Grove, Pa.) at a 1:5,000 dilution, and a streptavidin-horseradish peroxidase conjugate was used for the final 1-h incubation. Detection of blotted proteins was performed by use of enhanced chemiluminescence (Amersham, Arlington Heights, Ill.) according to the manufacturer’s instructions.
PCR amplification and restriction-enzyme analysis of the HTLV-1 ORFI.
First, 100 ng of plasmid DNA or 1 μg of chromosomal DNA was amplified in 50 μl containing 10 mM Tris-HCl–1.5 mM MgCl2–50 mM KCl (pH 8.3)–200 μM deoxynucleoside triphosphates–2.5 U of Taq DNA polymerase (Boehringer Mannheim) and 50 pmol of primers ORFI (6768 to 6785; 5′-CACCTCGCCTTCCAACTG-3′) and PX1AS (7160 to 7142, 5′-GCTGTGCTTGACGGTTTGC-3′). Amplification was carried out in a Thermal Cycler (Perkin-Elmer Cetus, Norwalk, Conn.) for 30 cycles (30 s at 94°C, 15 s at 58°C, and 30 s at 72°C). PCR products were run on a 1.5% low-melt agarose gel and purified with QIAEXII or purified directly by using the QIAquick PCR purification kit (Qiagen, Valencia, Calif.). Then, 10 to 50 ng of purified PCR product was cleaved with 10 U of ApaI (GIBCO BRL) at 30°C for 1 h. Cleaved and uncleaved products were electrophoresed in a 1.5 to 2% agarose gel and transferred to nylon membrane (Hybond-N Plus; Amersham International, Buckingham, United Kingdom). Next, 120 ng of a whole HTLV-1 provirus was labeled by random primer biotinylation according to the manufacturer’s instructions (Kirkegaard & Perry Laboratories, Gaithersburg, Md.). Prehybridization was performed in 0.25 M Na2HPO4 (pH 7.2)–7% SDS for 1 h at 65°C. Filters were hybridized with 50 ng of biotinylated probe per ml in 0.25 M Na2HPO4 (pH 7.2)–7% SDS for 8 h at 65°C and washed twice in 20 mM Na2HPO4 (pH 7.2)–5% SDS for 10 min at 65°C and in 20 mM Na2HPO4 (pH 7.2)–1% SDS for 15 min at 65°C. Finally, chemiluminescent detection was performed by using a DNADetector kit (Kirkegaard & Perry Laboratories) as described in the supplier’s manual.
RESULTS
Analysis of p12I motifs.
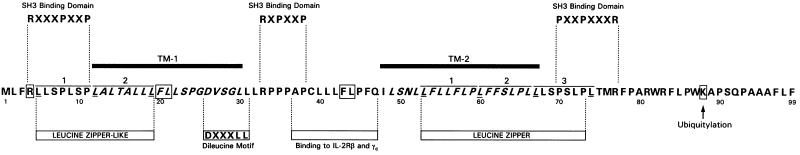
The analysis of the p12I amino acid sequence revealed the presence of a leucine zipper-like motif and a leucine zipper motif in the first and in the second transmembrane (TM) regions, respectively (Fig. 1). Leucine zipper domains are known to mediate noncovalent interactions between opposing zipper regions to form homo- or hetero-oligodimers in vitro and in vivo (13). Interestingly, the p12I leucine zipper motif is highly conserved among the envelope TM proteins of other retroviruses (8), as well as in the equivalent ORFI of STLV-1 (26).
FIG. 1.
Amino-acid sequence of the singly spliced ORFI putative p12I protein. The amino acid single code is used. TM-1 and TM-2 stand for the putative TM regions of the protein. The two amino acids (FL) are boxed because they correspond to possible destabilizing residues according to the N degron rule.
TM-1 and TM-2 contribute to p12I intermolecular association.
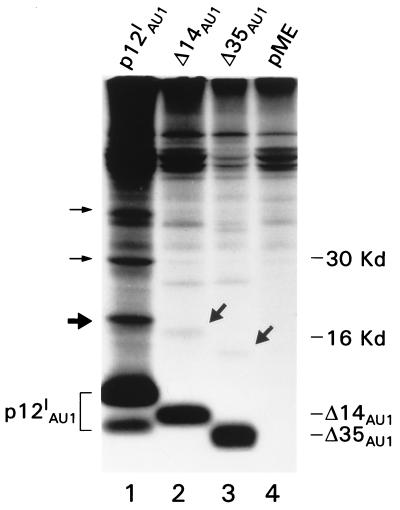
Immunoprecipitation of p12I from transfected, metabolically labeled HeLa-TAT cells resulted in the detection of the p12I doublet and at least three additional protein bands of ca. 20, 30, and 40 kDa (Fig. 2 [see arrows in lane 1]). A number of possibilities exist as to the identity of these additional bands: (i) p12I forms dimers and multimers, (ii) p12I undergoes posttranslational modification such as ubiquitylation, or (iii) p12I binds to a number of cellular proteins. The expression of two p12IAU1 deletion mutants (p12IΔ14 and p12IΔ35), lacking the amino-terminal 14 and 35 amino acids, respectively (17), revealed coprecipitated proteins with respectively lower molecular weights (Fig. 2 [see arrows in lanes 2 and 3]) as well, making the possibility of coprecipitating cellular proteins less likely. Despite the fact that the cell lysates were reconstituted in denaturing condition (1× RIPA buffer) and that removal of mercaptoethanol in the Laemmli buffer did not alter the size of the coprecipitated bands (data not shown), the possibility of dimer formation could not be definitively discounted. Because of the complexity of bands coprecipitated with p12I, we chose to demonstrate directly that p12I forms dimers by coexpressing p12I tagged with two different epitopes (p12IAU1 and p12IHA1) in HeLa-TAT cells. Interaction between p12I molecules was demonstrated by immunoprecipitation with the α-AU1 Ab and Western blot with α-HA1 (Fig. 3A, lane 8). Similarly, immunoprecipitation with the α-HA1 Ab followed by Western blotting with the AU1 Ab demonstrated binding of the two tagged p12I molecules (Fig. 3A). Controls for the specificity of Ab recognition and the expression of each tagged p12I protein are shown in the remaining lanes of panel A (1 to 4, 6 and 7, and 9 to 12).
FIG. 2.
High-molecular-weight proteins detected in immunoprecipitation of wild-type and p12I mutants. Radio immunoprecipitation was performed by using anti-AU1 Ab in cells transfected with p12IAU1 (lane 1), vector only (lane 4), and the Δ14 and Δ35 mutants (lanes 2 and 3, respectively). The thick and thin arrows indicate the positions of extra bands in the p12I wild-type transfectants (lane 1) and p12I mutant transfectants (lanes 2 and 3).
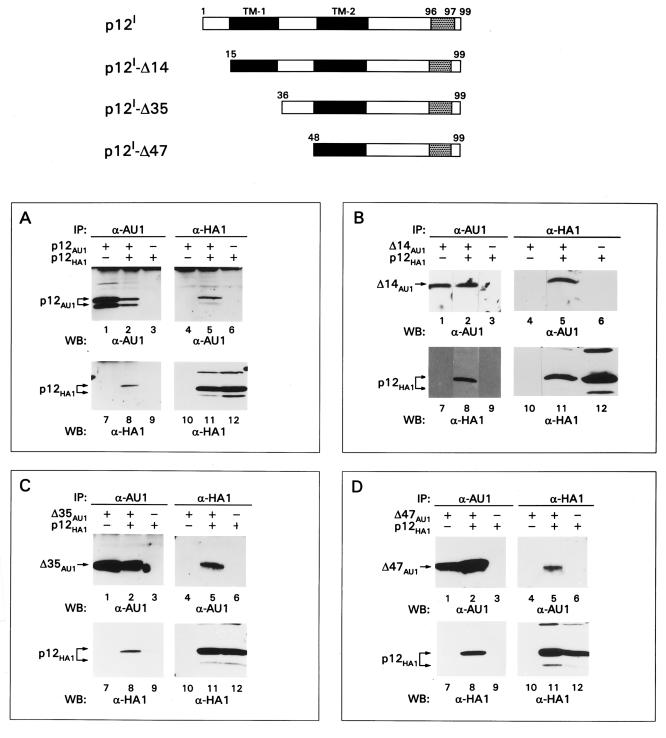
FIG. 3.
p12I regions involved in intermolecular interaction of p12I. The top part of the figure is a graphical representation of p12I deletion mutants. The numbers refer to the amino acid position within p12I. The black bars encompass the putative TM domains of p12I. The gray areas stand for the epitope location. IP, immunoprecipitation; WB, Western blot.
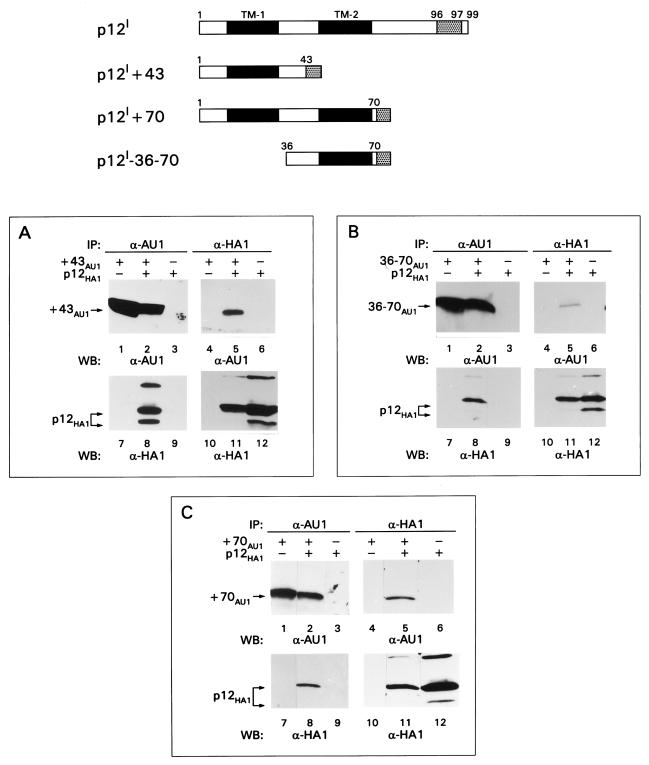
To identify the region(s) of p12I involved in the intermolecular interaction, the wild-type p12IHA1 was coexpressed with AU1-tagged p12I deletion mutants (17) lacking portions of the amino terminus (Δ14, Δ35, and Δ47), carboxy terminus (+43, +70), or a combination of both (36 to 70) (Fig. 3 and 4). p12I mutants lacking up to the first 47 amino acids retained their ability to bind the wild-type p12IHA1, suggesting that a binding site could be present between amino acids 48 and 99 (lanes 5 and 8 of Fig. 3B, C, and D). Surprisingly, however, the p12I+43 mutant lacking the carboxy-terminal 55 amino acids maintained its ability to bind to the wild-type p12IHA1 (Fig. 4A, lanes 5 and 8), indicating that an additional binding site was present at the amino terminus of p12I. The interaction of mutant p12I-36–70 with wild-type p12I HA1 suggested that one binding site was present between amino acids 36 and 70 (Fig. 4B, lanes 5 and 8) and, as expected, the p12I+70 mutant interacted with wild-type p12I (Fig. 4C, lanes 5 and 8). Controls for the specificity of Ab recognition and the expression of p12I wild-type and mutant proteins are shown in the remaining lanes (1 to 4, 6 and 7, and 9 to 12) of Fig. 3B, C, and D and Fig. 4.
FIG. 4.
p12I regions involved in intermolecular interaction of p12I (continued). See the legend to Fig. 3 for an explanation of panels, symbols, and abbreviations.
All together, these results indicate the existence of at least two interactive sites within p12I, one within the first 43 amino acids at the amino terminus of p12I and the second within (but not limited to) the 36-to-70-amino-acid-stretch of p12I.
These findings suggest that both leucine zipper motifs, the first overlapping with TM-1 and the second located within TM-2 (Fig. 1), may be involved in the dimerization of p12I and raise the interesting question as to whether the TM-1 regions of two p12I molecules interact either with each other or with TM-2.
p12I is ubiquitylated and degraded in the proteasome.
As demonstrated in Fig. 2, additional bands are consistently detected in the p12I immunoprecipitate. This finding raised the possibility that p12I could form multimers and/or that posttranslational modifications such as ubiquitylation may induce high-molecular-weight complexes. The latter hypothesis appeared possible in light of the difficulty of expressing p12I in mammalian cells (unpublished observation) and given the presence of a lysine residue at the carboxy terminus that could serve as a substrate for ubiquitin. Since ubiquitylated proteins are degraded by the proteolytic machinery of the proteasome (12, 32), we assessed the effect of proteasome inhibitors (7, 19) on the p12I steady-state level. Lactacystin treatment significantly increased the steady-state level of p12I (Fig. 5A) as did treatment with MG115 or MG132 (data not shown).
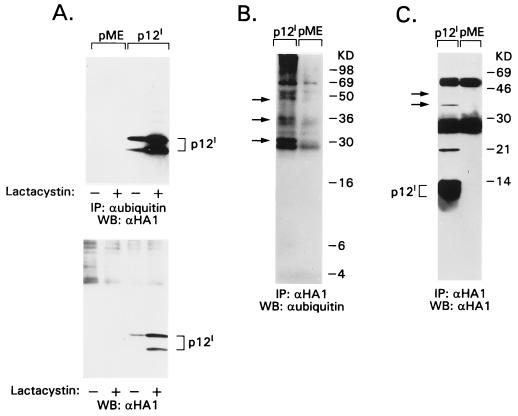
FIG. 5.
p12I is ubiquitylated and stabilized by proteasome inhibitors. 293T cells were transiently transfected with p12I HA1 expression plasmid or pME vector control and lysed in 1× RIPA buffer 24 h later. (A) An antiubiquitin immunoprecipitate of cell lysates is shown in the top panel. The bottom panel shows results with 50 μg of total cell lysate. An anti-HA1 Western blot was performed for both membranes. (B and C) An anti-HA1 immunoprecipitate of cell lysates was performed, and the immunoprecipitates were split into duplicate membranes. (B) Antiubiquitin Western blot. (C) Blot probed with α-HA1 Ab. Arrows indicate superimposable bands present on both blots. + and −, presence or absence, respectively, of drugs.
To assess whether p12I is ubiquitylated, cell lysates containing p12I were immunoprecipitated with Ab against ubiquitin and immunoblotted with the α-HA1 Ab. The p12I doublet was readily detected, whereas ubiquitylated forms of p12I were only weakly detectable (Fig. 5A, upper panel). However, when immunoprecipitation with the α-HA1 Ab was followed by immunoblotting with α-ubiquitin Ab, only bands larger than 30 kDa were detected (Fig. 5B). These same bands were identified as p12I, as demonstrated by the immunoblotting of a duplicate membrane with the α-HA1 Ab (Fig. 5C).
Thus, p12I is ubiquitylated and degraded in the proteasome, although it appears that the majority of p12I in the cells is ubiquitin-free, as suggested by the finding that ubiquitin-free p12I molecules outnumber the ubiquitylated p12I molecules (as is evident from Fig. 5A). It is possible that multimers of ubiquitin-free p12I are able to bind to ubiquitylated p12I molecules and/or that ubiquitin-free p12I molecules bind to other ubiquitylated proteins. This interpretation would explain the preponderance of ubiquitin-free molecules immunoprecipitated by the α-ubiquitin Ab (Fig. 5A) or by the α-HA1 Ab (Fig. 5C), as well as the fact that the seemingly ubiquitin-free p12I is stabilized by proteasome inhibitors (Fig. 5A).
p12I is ubiquitylated at the carboxy-terminus Lys residue 88.
To investigate further the contribution of different portions of p12I to its instability, some of the p12I deletion mutants described in Fig. 3 and 4 were expressed in the presence or absence of proteasome inhibitors. p12I mutants that retained the carboxy terminus (Δ35 and Δ47) were stabilized by proteasome inhibitors, whereas the steady-state level of the +70 and 36–70 mutants was not altered in the presence of proteasome inhibitors (Fig. 6A), suggesting the presence of a ubiquitylation site at the carboxy terminus of the protein.
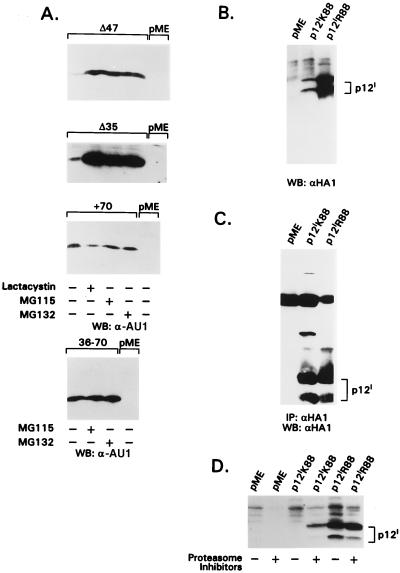
FIG. 6.
Sensitivity of p12I mutants and p12IK88 or p12IR88 to proteasome inhibitors. Panels A, B, and C show the results of Western blot assays with α-HA1 or α-AU1 Ab in transiently transfected 293T cells. In panel C, a Western blot was preceded by α-HA1 immunoprecipitation. p12IR88 is a mutant that encodes an R instead of a K in codon 88. (A) p12I mutants Δ47 and Δ35 are stabilized by proteasome inhibitors, while +70 and 36–70 mutants lacking the Lys 88 residue are not. (B) The steady-state level of the isogene mutant p12IR88 is greatly increased compared to p12IK88. (C) High-molecular-weight forms of p12I are clearly present in cells transfected with p12IK88 but are absent in those expressing p12IR88. (D) The steady-state levels of p12IK88 and p12IR88 in the presence of all three proteasome inhibitors at the concentration indicated in the Materials and Methods are shown.
The p12I cDNA used in this study was cloned from the HTLV-1 LAF cell line, established from a patient with TSP-HAM (18), and contains a unique Lys amino acid residue at position 88. Of interest, this amino acid residue is an arginine in most viral strains obtained from ex vivo samples of HTLV-1-infected individuals (8). Since lysine is a target for covalent binding of ubiquitin (31), it was plausible that ubiquitylation of Lys 88 would render p12I sensitive to proteasome degradation. To prove this hypothesis and to assess the importance of this amino acid in the natural alleles of p12I, a point mutation was introduced in the p12I cDNA to generate a K-to-R change at position 88 (p12IR88 mutant). The steady-state levels of both p12IR88 and p12IK88 were assessed in a Western blot assay and, as demonstrated in Fig. 6B, the steady-state level of p12IR88 was significantly higher (ca. 10-fold) than p12IK88. Consistent with this finding, p12IR immunoprecipitates did not contain the high-molecular-weight complexes present in the p12IK88 immunoprecipitates (Fig. 6C) and the steady-state level of expression of p12IR88 was not significantly increased by proteasome inhibitors (Fig. 6D).
To define better the differential stability of the p12IR88 and p12IK88 alleles, the expression of both proteins was analyzed after cycloheximide treatment of cells transfected with either p12IK88 or p12IR88 cDNAs. As demonstrated in Fig. 7, the p12IK88 protein was undetectable by Western blotting after 8 h of cycloheximide treatment, whereas p12IR88 was still detectable up to 24 h after treatment. Taken together, these results indicate that ubiquitylation of the carboxy-terminal lysine residue appears to target p12IK88 to degradation in the proteasome complex and that the stability of p12I natural alleles that carry an arginine at position 88 is much greater than the stability of those alleles containing a lysine at residue 88. Ubiquitylation, however, does not appear to be the only mechanism that contributes to p12I instability. The removal of the amino-terminal 47 amino acids greatly increased the steady-state level of the truncated protein, regardless of the continued presence of the unique lysine (see, for example, reference 17 and Fig. 3D). These results are consistent with the existence of destabilizing amino acids at the amino terminus of the protein (31).
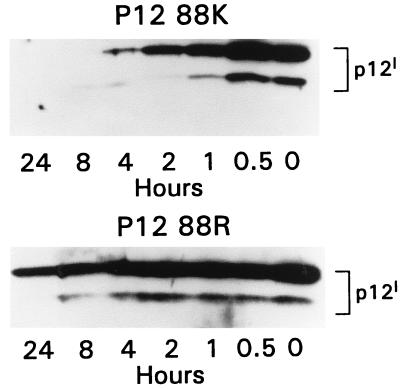
FIG. 7.
Differential stability of p12IK88 and p12IR88. Western blot analysis with anti-HA1 Ab of total proteins from transfected cells after treatment with 10 μg of cycloheximide per ml for the indicated time intervals. The single substitution of an Arg for a Lys at position 88 significantly increased the half-life of the protein. The experiment was repeated three to four times for each protein.
Distribution of the natural p12I alleles in TSP-HAM and ATLL patients and healthy carriers.
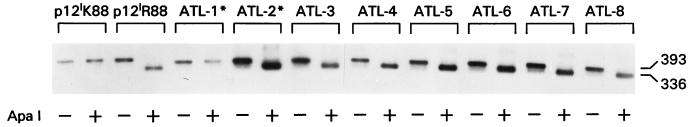
Analysis of the p12I ORF in 21 HTLV-1 strains from different geographical areas (8) demonstrated that the p12I putative amino acid sequence is highly conserved and that, although Lys at position 88 is rare, it is found exclusively in TSP-HAM patients. Because this amino acid change (R⇆K) leads to significant differences in the stability of p12I and possibly in its biological effects, we extended our study to an additional 32 ex vivo samples from healthy carriers, TSP-HAM or adult T-cell leukemia-lymphoma (ATLL) patients, as well as to families in which both diseases occur. To do so, we took advantage of the finding that, although Arg can be encoded by six different codons and only two of them would reconstitute an ApaI site, the arginine AGG codon (which reconstitutes the GGGCCC ApaI site) is preferentially found in HTLV-1 strains (Fig. 1). The absence of the ApaI site does not prove the presence of a lysine at position 88; nevertheless, because ApaI is unique within the ORFI, ApaI cleavage represents a rapid method to screen for p12I alleles carrying an arginine. With this assay, we analyzed PCR products from an additional 21 cases of ATLL, 2 of whom had concomitant TSP-HAM, 9 of whom were solely TSP-HAM cases, and 2 of whom were healthy carriers. A representative result from some of these samples is presented in Fig. 8. Collectively, the data obtained in this and the previous study (8) indicate that, in ATLL or healthy carriers, an Arg at position 88 is consistently found regardless of the geographical origin of the patients (Table 1). In fact, the ATLL samples were from different geographical regions, including the Caribbean, South America (Brazil, Paraguay, and Columbia), North America (Alaska), Africa, Europe, and Japan. Similarly, the five TSP-HAM cases that carried a lysine at position 88 (Table 1) did not cluster geographically. Thus, lysine 88 was found only in 5 of 17 TSP-HAM cases. Lysine at position 88 was found in one ATLL patient who also had TSP-HAM (ATL-1* in Fig. 8). However, in this patient’s sample, DNA sequencing revealed an in-frame termination codon immediately preceding the lysine. The significance of these findings is unclear at present, but it is likely that selective pressure in the host rather than random mutation in the virus explains this observation.
FIG. 8.
Natural allelic mutation at position 88 in samples obtained from HTLV-1-infected patients. Southern blot analysis of PCR product from the HTLV-I ORFI before and after ApaI cleavage. ATL-1* and ATL-2* indicate patients with both TSP-HAM and ATLL.
TABLE 1.
Naturally occurring alleles of p12I that carry a Lys or an Arg at position 88a
| p12 type | No. of naturally occurring alleles in patients with:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| ATLL
|
TSP-HAM
|
ATLL plus TSP-HAM
|
HC
|
|||||
| This study | Reference 8 | This study | Reference 8 | This study | Reference 8 | This study | Reference 8 | |
| Total | 19 | 6 | 9 | 8 | 2 | 0 | 2 | 7 |
| p12K | 0 | 0 | 0 | 5 | 1b | 0 | 0 | 0 |
| p12R | 19 | 6 | 9 | 3 | 1 | 0 | 2 | 7 |
The table presents a summary of all cases analyzed in this study (left column for each clinical diagnosis). The amino acid sequence of p12I obtained from ex vivo samples was presented in Franchini (8 [Fig. 6]) and is reported in the right column of each patient’s group. HC, healthy carrier.
In this case, the Lys was preceded by an in-frame termination codon.
DISCUSSION
Intracellular protein degradation is an important mechanism for the modulation of specific proteins and the elimination of damaged proteins. The ubiquitin-proteasome pathway appears to be the major system for selective protein degradation in eukaryotic cells (32). Multiple molecules of ubiquitin, a 76-amino-acid protein, are attached to the target protein making a polyubiquitylated substrate that is rapidly degraded by the 26S proteasome, an ATP-dependent complex of proteases (32). Proteolysis by the ubiquitin system regulates a variety of cell functions, including cell cycle, various signal-transduction pathways, and embryogenesis (12, 23). Recently, it has been demonstrated that the proteasome proteolysis pathway is involved in the cytoplasmic degradation of proteins retained in the endoplasmic reticulum and functions as the “quality control” of the endoplasmic reticulum, degrading abnormal, misfolded, or unassembled proteins (2, 14).
In this study we have demonstrated that the HTLV-1 p12I protein dimerizes and is a substrate of ubiquitylation and proteasome degradation. Specific inhibitors of the proteasome (7, 19) increased the steady-state level of p12I as well as of the high-molecular-weight products of polyubiquitylated p12I. To prove the p12I ubiquitylation directly, the lysine residue at position 88 (p12IK) was substituted with an arginine (p12IR), since an internal lysine is thought to be one of the two determinants of protein metabolic instability in eukaryotes (31). Indeed p12IR was not recognized as a target for ubiquitylation since polyubiquitylated complexes were not detected and p12IR was significantly more stable than p12IK88.
The proteasome destabilization of incoming viral proteins has been postulated to be an intracellular defense mechanism against viral infection (28). In fact, in the case of human immunodeficiency virus type 1, an early block of intracellular proteasome activity by MG132 increased the efficiency of human immunodeficiency virus infection and the cellular amount of p17, p24, and p66 gag products (28). Relevant to this concept, we provide evidence that the HTLV-1 p12I protein is also targeted and degraded by the ubiquitin proteasome system.
Only two natural allelic variants of p12I were found in ex vivo samples from HTLV-1-infected individuals: at position 88, the more frequent one carries an Arg (p12IR) and the less frequent one a Lys (p12IK) and the latter was found only in some TSP-HAM cases. The full appreciation of these findings awaits a better understanding of p12I function in regard to its binding to the IL-2R β and γc chains. Interestingly, HTLV-1 p12I expression in the course of HTLV-1 infection of human lymphocytes in vitro does not appear to be important (5, 25), whereas it appears to be essential for viral infectivity in vivo (4), raising the possibility that the culture conditions used (phytohemagglutinin stimulation and IL-2 addition) may override the requirement for p12I expression in vitro. A better definition of the complex interactions of the natural alleles of p12I with the components of signaling pathways will help in understanding its role in the infection of T lymphocytes in vitro and in vivo.
ACKNOWLEDGMENTS
We thank A. Gessain and S. Kamihira for supplying DNA material. We thank Steven Snodgrass for editorial assistance.
R. Trovato was supported by a fellowship from Istituto Superiore di Sanità, Rome, Italy.
REFERENCES
- 1.Berneman Z N, Gartenhaus R B, Reitz M S, Jr, Blattner W A, Manns A, Hanchard B, Ikehara O, Gallo R C, Klotman M E. Expression of alternatively spliced human T-lymphotropic virus type 1 (HTLV-I) pX mRNA in infected cell lines and in primary uncultured cells from patients with adult T-cell leukemia/lymphoma and healthy carriers. Proc Natl Acad Sci USA. 1992;89:3005–3009. doi: 10.1073/pnas.89.7.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonifacino J S. Reversal of fortune for nascent proteins. Nature. 1996;384:405–406. doi: 10.1038/384405a0. [DOI] [PubMed] [Google Scholar]
- 3.Ciminale V, Pavlakis G N, Derse D, Cunningham C P, Felber B K. Complex splicing in the human T-cell leukemia virus (HTLV) family of retroviruses: novel mRNAs and proteins produced by HTLV-I. J Virol. 1992;66:1737–1745. doi: 10.1128/jvi.66.3.1737-1745.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins N D, Newbound G C, Albrecht B, Beard J L, Ratner L, Lairmore M D. Selective ablation of human T-cell lymphotropic virus type 1 p12I reduces viral infectivity in vivo. Blood. 1998;91:4701–4707. [PubMed] [Google Scholar]
- 5.Derse D, Mikovits J, Ruscetti F. X-I and X-II open reading frames of HTLV-I are not required for virus replication or for immortalization of primary T-cells in vitro. Virology. 1997;237:123–128. doi: 10.1006/viro.1997.8781. [DOI] [PubMed] [Google Scholar]
- 6.DiMaio D, Lai C-C, Klein O. Virocrine transformation: the intersection between viral transforming proteins and cellular signal transduction pathways. Annu Rev Microbiol. 1998;52:397–421. doi: 10.1146/annurev.micro.52.1.397. [DOI] [PubMed] [Google Scholar]
- 7.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 8.Franchini G. Molecular mechanisms of human T-cell leukemia/lymphotropic virus type 1 infection. Blood. 1995;86:3619–3639. [PubMed] [Google Scholar]
- 9.Franchini G, Mulloy J C, Koralnik I J, Lo Monico A, Sparkowski J J, Andresson T, Goldstein D J, Schlegel R. The human T-cell leukemia/lymphotropic virus type I p12I protein cooperates with the E5 oncoprotein of bovine papillomavirus in cell transformation and binds the 16-kilodalton subunit of the vacuolar H+ ATPase. J Virol. 1993;67:7701–7704. doi: 10.1128/jvi.67.12.7701-7704.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furukawa K, Furukowa K, Shiku H. Alternatively spliced mRNA of the pX region of human T-lymphotropic virus type 1 proviral genome. FEBS Lett. 1991;295:141–145. doi: 10.1016/0014-5793(91)81404-v. [DOI] [PubMed] [Google Scholar]
- 11.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 12.Hochstrasser M. Ubiquitin, proteasomes and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 13.Hodge D R, Chen Y M, Samuel K P. Oligomerization of the HIV type 2 Nef protein: mutational analysis of the heptad leucine repeat motif and cysteine residues. AIDS Res Hum Retroviruses. 1995;11:65–79. doi: 10.1089/aid.1995.11.65. [DOI] [PubMed] [Google Scholar]
- 14.Kopito R R. ER quality control: the cytoplasmic connection. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- 15.Koralnik I, Gessain A, Klotman M E, Lo Monico A, Berneman Z N, Franchini G. Protien isoforms encoded by the pX region of the human T-cell leukemia/lymphotropic virus type I. Proc Natl Acad Sci USA. 1992;89:8813–8817. doi: 10.1073/pnas.89.18.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koralnik I, Lemp J F, Jr, Gallo R C, Franchini G. In vitro infection of human macrophages by human T-cell leukemia/lymphotropic virus type I (HTLV-I) AIDS Res Hum Retroviruses. 1992;8:1845–1849. doi: 10.1089/aid.1992.8.1845. [DOI] [PubMed] [Google Scholar]
- 17.Koralnik I, Mulloy J C, Andresson T, Fullen J, Franchini G. Mapping of the intermolecular association of the human T-cell leukemia/lymphotropic virus type 1 p12I and the vacuolar H+ ATPase 16 kDa subunit protein. J Gen Virol. 1995;76:1909–1916. doi: 10.1099/0022-1317-76-8-1909. [DOI] [PubMed] [Google Scholar]
- 18.Koralnik I J, Fullen J, Franchini G. The p12I, p13II, and p30II proteins encoded by human T-cell leukemia/lymphotropic virus type I open reading frames I and II are localized in three different cellular compartments. J Virol. 1993;67:2360–2366. doi: 10.1128/jvi.67.4.2360-2366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee D H, Goldberg A L. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J Biol Chem. 1996;271:27280–27284. doi: 10.1074/jbc.271.44.27280. [DOI] [PubMed] [Google Scholar]
- 20.Martin P, Vass W C, Schiller J T, Lowry D R, Velu T J. The bovine papillomavirus E5 transforming protein can stimulate the transforming activity of EGF and CSF1 receptors. Cell. 1998;59:21–32. doi: 10.1016/0092-8674(89)90866-0. [DOI] [PubMed] [Google Scholar]
- 21.Mulloy J C, Crowley R W, Fullen J, Leonard W J, Franchini G. The human T-cell leukemia/lymphotropic virus type I p12I protein binds the interleukin-2 receptor β and γc chains and affects their expression on the cell surface. J Virol. 1996;70:3599–3605. doi: 10.1128/jvi.70.6.3599-3605.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilson L A, Di Maio D. Platelet-derived growth factor receptor can mediate tumorigenic transformation by the bovine papilloma E5 protein. Mol Cell Biol. 1993;13:4137–4145. doi: 10.1128/mcb.13.7.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pagano M, Tam S W, Theodoras A M, Beer-Romano P, Del Sal G, Chau V, Yew P R, Draetta G F, Rolfe M. Role of the ubiquitin-proteasome pathway in regulating the abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 24.Petti L, Di Maio D. Stable association between the bovine papillomavirus E5 transforming protein and activated platelet-derived growth factor receptor in transformed mouse cells. Proc Natl Acad Sci USA. 1992;89:6736–6740. doi: 10.1073/pnas.89.15.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robek M D, Wong F-H, Ratner L. Human T-cell leukemia virus type 1 pX-I and pX-II open reading frames are dispensable for the immortalization of primary lymphocytes. J Virol. 1998;72:4458–4462. doi: 10.1128/jvi.72.5.4458-4462.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saksena N K, Srinivasan A, Ge Y C, Xiang S-H, Azad A, Bolton W, Herve V, Reddy S, Diop O, Miranda-Saksena M, Rawlinson W D, Vandamme A-M, Barre-Sinoussi F. Simian T-cell leukemia virus type I from naturally infected feral monkeys from central and west Africa encodes a 91-amino acid p12 (orf-I) protein as opposed to a 99-amino acid protein encoded by HTLV type I from humans. AIDS Res Hum Retroviruses. 1997;13:425–432. doi: 10.1089/aid.1997.13.425. [DOI] [PubMed] [Google Scholar]
- 27.Schlegel R, Wade-Glass M, Rabson M S, Yang Y C. The E5 transforming gene of bovine papillomavirus encodes a small, hydrophobic polypeptide. Science. 1986;233:464–467. doi: 10.1126/science.3014660. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz O, Marechal V, Friguet B, Arenzana-Seisdedos F, Heard J-M. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. J Virol. 1998;72:3845–3850. doi: 10.1128/jvi.72.5.3845-3850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seiki M, Hattori S, Hirayama Y, Yoshida M. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci USA. 1983;80:3618–3622. doi: 10.1073/pnas.80.12.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uchiyama T. Human T cell leukemia virus type I (HTLV-I) and human diseases. Annu Rev Immunol. 1997;15:15–37. doi: 10.1146/annurev.immunol.15.1.15. [DOI] [PubMed] [Google Scholar]
- 31.Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]