Abstract
The Sendai virus P protein is an essential component of the viral RNA polymerase (P-L complex) required for RNA synthesis. To identify amino acids important for P-L binding, site-directed mutagenesis of the P gene changed 17 charged amino acids, singly or in groups, and two serines to alanine within the L binding domain from amino acids 408 to 479. Each of the 10 mutants was wild type for P-L and P-P protein interactions and for binding of the P-L complex to the nucleocapsid template, yet six showed a significant inhibition of in vitro mRNA and leader RNA synthesis. To determine if binding was instead hydrophobic in nature, five conserved hydrophobic amino acids in this region were also mutated. Each of these P mutants also retained the ability to bind to L, to itself, and to the template, but two gave a severe decrease in mRNA and leader RNA synthesis. Since all of the mutants still bound L, the data suggest that L binding occurs on a surface of P with a complex tertiary structure. Wild-type biological activity could be restored for defective polymerase complexes containing two P mutants by the addition of wild-type P protein alone, while the activity of two others could not be rescued. Gradient sedimentation analyses showed that rescue was not due to exchange of the wild-type and mutant P proteins within the P-L complex. Mutants which gave a defective RNA synthesis phenotype and could not be rescued by P establish an as-yet-unknown role for P within the polymerase complex, while the mutants which could be rescued define regions required for a P protein function independent of polymerase function.
Sendai virus, a paramyxovirus, is an enveloped virus with a single-stranded, negative-sense, nonsegmented RNA genome of about 15 kb (for reviews, see references 25 and 27). The genome RNA is completely encapsidated by the nucleocapsid protein, NP, which renders the RNA nuclease resistant. The RNA-dependent RNA polymerases of negative-strand RNA viruses are unique in that the helical ribonucleoprotein complex or nucleocapsid (NC), not the RNA alone, serves as the template for mRNA synthesis and genome replication. The viral RNA polymerase is composed of the phosphoprotein (P, 568 amino acids [aa]) and the large protein (L, 2,228 aa) and has been shown to be packaged within the virion with ca. 300 molecules of the P protein and 30 molecules of the L protein (30). Transcription by the viral polymerase initiates at the precise 3′ end of the encapsidated genome, yielding first positive-strand leader RNA (le+, 55 nucleotides [nt]), which is followed by the sequential synthesis of the six major mRNAs in the gene order of NP, P/C/V, M, F, HN, and L mRNAs (26–28). These mRNAs, but not leader RNA, are modified by the viral polymerase by capping and methylation at the 5′ end and by polyadenylation at the 3′ end. The exact mechanism of mRNA synthesis has yet to be defined, but the L protein is believed to contain most of the catalytic activities necessary for viral RNA synthesis and processing, although presently there is little direct experimental data on this point.
The P protein is involved in multiple protein-protein interactions that are required for RNA synthesis. First, P complexes with L to form the RNA polymerase; however, the precise function of the P protein within the complex is unknown. P appears to be important for the proper folding of the L protein, since L must be coexpressed with P to be stable (20, 21). Through deletion analysis, the L binding domain within the P protein has been mapped to aa 412 to 479 (12, 34). Binding of the P-L complex to the NC template occurs through the P protein subunit of the polymerase (23, 31) between two noncontiguous NC binding sites (32, 33) in the C-terminal half of the P protein.
The P protein has also been shown to oligomerize forming a homotrimeric complex, where the oligomerization domain was mapped to aa 344 to 411 (9, 11, 24). Computer analysis predicts that this domain has a coiled-coil structure which is important for the P-P interaction. This oligomerization domain overlaps the most amino terminal NC binding domain, and it has been proposed that the different protein-protein interactions necessary to form the P-NC and P-P interactions occur on different faces of the coiled-coil. However, the data are also consistent with a model where P oligomerization is required for the P-NC interaction, as has been shown for the vesicular stomatitis virus (VSV) P protein (17, 18). Another possible role for P protein in viral RNA synthesis has recently been described. Curran (8, 9) provides evidence that while part of the NC associated P protein is found in the P-L complex, the rest is bound to the template independently of L, and this latter form of P is also essential for mRNA synthesis.
It was our goal to characterize the P-L interaction with respect to the specific P amino acids which are required for binding L protein. We initially selected clustered charge-to-alanine mutagenesis as our approach because of the likelihood of targeting residues residing on the surface of the protein and of producing stable mutants capable of exhibiting altered phenotypes (1, 2, 7, 36). This approach was complimented by the mutagenesis of conserved hydrophobic amino acids in the L binding domain. We present evidence that while all of these mutants maintained their ability to complex with itself and L protein, a significant number proved to be defective in viral RNA synthesis. Our studies were able to define specific residues within the L binding domain which are required for an independent function of P, but not for P-L complex activity.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Sendai virus was propagated in embryonic chicken eggs and polymerase-free wild-type (wt) NCs were prepared from purified virus by banding on CsCl gradients (4). Recombinant vaccinia virus (VV) containing the gene for the phage T7 RNA polymerase (VVT7) (15) was a gift of E. Niles (State University of New York, Buffalo) and was grown in Vero cells. Infections and transfections were performed in human lung carcinoma A549 cells from the American Type Culture Collection. The Sendai plasmids pGEM-P/C and pGEM-L, and pTM-GST-P with each gene cloned downstream of the T7 promoter were as described by Curran et al. (10) and Chandrika et al. (5), respectively. Rabbit anti-Sendai virus antibody (α-SV), rabbit anti-L antibody (α-L) (34), rabbit polyclonal antibody against P peptides corresponding to aa 274 to 298 and aa 453 to 477 (α-P, provided by K. Gupta, Chicago, Ill.) (3), and rabbit anti-glutathione S-transferase antibody (α-GST) (5) have all been described.
Deletion and mutagenesis.
Charge-to-alanine and other alanine substitution mutagenesis targeted the putative L binding domain of the Sendai virus P protein from aa 408 to 479 (12, 34), generating mutants containing one to four alanine substitutions. The mutants P2, P4, P5, P408/9, and S426A were constructed with the Sculptor oligonucleotide-directed in vitro Mutagenesis System Version 2.1 Kit (Amersham) according to the manufacturer’s protocol. The mutagenic oligonucleotides used are shown in Table 1 with the introduced new silent restriction enzyme sites (underlined) to screen putative mutants. An EcoRI-NdeI-digested fragment from nt 1,130 to 1,717 containing the mutation site(s) was then subcloned into pGEM-P/C at the same restriction sites, where a second EcoRI site located in the multiple cloning region upstream of the P gene in the original vector had been removed. The constructs were confirmed by sequencing. PCR-directed mutagenesis (19) was used to generate mutants P1, S419A, K453A, P455/6, and all the hydrophobic-to-alanine mutations. Site-specific mutations were constructed by introducing mismatches into the oligonucleotides used to prime the PCR amplification. This technique required two amplification steps which made use of a mutagenic oligonucleotide primer and its complement (not shown) along with two outside oligonucleotide primers (Table 1). The primary PCR amplification paired each outside primer with one mutagenic primer generating 5′ and 3′ arms. A secondary PCR product from the overlapping arms with the outside primers was then ligated into the pCR II vector (TA Cloning Kit; Invitrogen Corp.). Colonies were screened by PCR, and the mutations in positive clones were verified by digestion at the silent restriction site. The EcoRI-NdeI-digested DNA fragment containing the mutation site(s) was subcloned into pGEM-P/C at the same restriction sites and confirmed by sequencing. The mutant 2S447 was constructed by linearizing pGEM-P/C with PpuMI. The overhangs were filled in with T4 DNA polymerase and ligated by using T4 DNA ligase. This resulted in a 3-bp insertion coding for a serine residue.
TABLE 1.
Oligonucleotide primers for P mutations
| Primer group and mutant | Primer sequencea | Enzyme |
|---|---|---|
| Mutagenic primer group | ||
| P408/9 | (+)-CGGATATCTACGCGGCATTCTCTGAG | EcoRV |
| P1 | (+)-CTCTGCGTATCAGGCCGCGCAGAACTC | BsoFI |
| S419A | (+)-GAACAGAACGCATTGCTCATGTCCAACC | NlaIII |
| S426A | (+)-CCAACCTAGCTACGCTGCATATCATCAC | BsoFI |
| P2 | (+)-CATCACAGCTGCAGGTGGCGCGACTGACAAC | BsoFI |
| P3 | (+)-CCTCCGTTTTTGCAGCATCAGCGGCCACCGCGACTAAGGC | BsoFI |
| K453A | (+)-CCTCCGTTTTTGCAGCATCAAAAGAGAACAAGACTAAGGC | BsoFI |
| P455/6 | (+)-CCTCCGTTTTTGCAAAATCAGCAGCGAACAAGACTAAGGC | BsoFI |
| P4 | (+)-CAAGACTGCGGCTACCGCGTTTGCCCCATC | BsoFI |
| P5 | (+)-CCATCTATGGCCACCCTAGCCGCGATGAAGTACAAACCG | BsoFI |
| L421A | (+)-CAGAACTCATTGGCCATGTCCAACCTATC | NlaIII |
| L425A | (+)-GCTGATGTCCAACGCTAGCACACTTCATATCATCAC | NheI |
| L428A | (+)-GATGTCCAACCTAAGTACTGCTCATATCATCACAG | ScaI |
| I430A | (+)-CAACCTATCTACACTGCACGCCATCACAGATAGAG | BsgI |
| G436A | (+)-CACAGATAGAGGCGCCAAGACTGACAAC | HaeII |
| Outside primer group | ||
| Primer 1 | (+)-ATCTACACAGGATGAGC | |
| Primer 2 | (−)-GATCTCGAGCCCGGGATCTAGTTG | XhoI |
The oligonucleotide sequences are written 5′ to 3′. The plus symbol refers to the messenger sense (+) of the oligonucleotide. The underlined sequence indicates the introduced silent restriction enzyme site shown on the right.
Protein analysis.
To measure various protein-protein interactions, A549 cells (35-mm dishes) were infected with VVT7 at a multiplicity of infection of 2.5 PFU/cell for 1 h at 37°C. The cells were then transfected with the wt or mutant pGEM-P/C (1.67 μg), pGEM-L (1.67 μg), and/or pTM-GST-P (0.2 μg) by using Lipofectin (Life Technologies) in Opti-MEM medium (GIBCO) as described in each experiment. At 5 h posttransfection (p.t.), the cells were labeled with Tran35S-label (100 μCi/ml) in Dulbecco modified Eagle medium with 0.1× cysteine and methionine from 4 to 18 h p.t. at 37°C. Cytoplasmic cell extracts were then prepared by lysolecithin permeabilization in 300 μl of Sendai virus reaction mix salts (RM salts; 100 mM HEPES [pH 8.5], 50 mM NH4Cl, 7 mM KCl, 4.5 mM magnesium acetate, 1 mM dithiothreitol, 1 mM spermidine) with 0.25% Nonidet P-40, and the lysates were clarified at 13,000 rpm for 25 min at 4°C.
For immunoprecipitation, samples of the 35S-labeled supernatants (50 μl) were incubated with 1 μl each of α-SV, α-L, α-P, and/or α-GST antibodies as described in the figure legends, and the antigen-antibody complex was collected with inactivated Staphylococcus aureus (Cowan) as described previously (4, 20). For bead-binding with glutathione-Sepharose beads, 50 μl was brought up to a volume of 450 μl with RM salts. The Sepharose beads (15 μl per reaction), preblocked in RM salts containing 0.1% Nonidet P-40, 0.5% nonfat dry milk, and 10 mg of bovine serum albumin per ml and equilibrated in RM salts, were added and incubated for 15 min at 4°C. The beads were washed, and the proteins were separated by sodium dodecyl sulfate (SDS)–7.5% polyacrylamide gel electrophoresis (PAGE) and visualized by autoradiography. For analysis of P-L complex binding to NC, the wt or mutant P proteins (5 μg) and the wt L protein (5 μg) were coexpressed in VVT7-infected cells (100-mm dishes), and lysolecithin extracts were prepared in RM salts in the absence of detergent, but with 1 mM ATP. Equal samples were incubated in the absence or presence of the wt Sendai polymerase-free nucleocapsids (wt RNA-NP) (2.5 μg) for 30 min at 30°C. The samples were fractioned on step gradients containing 2.5 ml of 30 and 50% (vol/vol) glycerol in 10 mM HEPES (pH 8.5) and 1 mM EDTA at 50,000 rpm for 1.4 h at 4°C in an SW55 rotor. The pellets were resuspended, immunoprecipitated with α-SV and α-L antibodies and analyzed by SDS–7.5% PAGE and autoradiography. For glycerol gradient analysis of the exchange assay, cells were either transfected with wt P plasmid in the presence or absence of wt L plasmid and labeled with Tran35S-label or transfected with plasmids to express the wt or mutant polymerases, wt P+L, L421A+L, or K453A+L, which were unlabeled. Cytoplasmic extracts were prepared in complete RM, and samples of 35S-labeled wt P were mixed with each unlabeled polymerase extract and incubated for 30 min at 30°C. The samples were fractionated by centrifugation on a 5 to 20% (vol/vol) glycerol gradient in the SW41 rotor at 29,000 rpm for 46 h as described previously (20). Gradient fractions (1 ml) were collected, immunoprecipitated (0.5 ml) with the α-P antibody, and analyzed by SDS-PAGE. The P protein was quantitated on a phosphorimager and plotted in arbitrary units.
In vitro RNA synthesis.
wt or mutant P (1.5 μg) and wt L (0.5 μg) plasmids were cotransfected into VVT7-infected cells (60-mm dish), along with a negative control containing no plasmid DNA. At 18 h p.t., lysolecithin cytoplasmic cell extracts were prepared, and the supernatant was treated with micrococcal nuclease as described previously (5). Samples of 10 and 90 μl were used for immunoblot analysis and in vitro transcription, respectively. For mRNA synthesis, [α-32P]CTP and 1 μg of Sendai virus polymerase-free wt RNA-NP template were added and incubated at 30°C for 2 h. Total RNA was isolated with the Qiagen RNEasy Total RNA Kit and analyzed directly by electrophoresis on a 1.5% agarose-acid–6 M urea gel and autoradiography. The products were quantitated on a phosphorimager. For leader RNA analysis, the extracts were incubated with CTP at 1 mM in the absence of radiolabeled nucleotide and wt RNA-NP. Total RNA was phenol-chloroform extracted, separated by electrophoresis on a 8% polyacrylamide–8 M urea gel, and electroblotted onto Hybond N nitrocellulose. le+ RNA was detected by Northern analysis as described previously (22) with the complementary 32P-end-labeled oligonucleotide, 5′-AAATCCTGTA TAACTTCATT ACATATCCCA TACATGTTTT TTCTCTTGTT TGGT-3′. The blots were exposed to Kodak X-Omat film, and the RNA quantitated on the phosphorimager.
For the rescue experiments, VVT7-infected cells (60-mm dishes) were transfected with no plasmids (Mock) or with wt or mutant P plasmids alone (5 μg), and separate dishes were cotransfected in duplicate with the same P plasmids and wt L plasmid at a 1:1 ratio (3 μg each). It has been shown previously that this ratio of P to L is suboptimal for in vitro transcription (∼25% [20]). To maintain about the same final amount of viral polymerase used in regular in vitro transcription experiments, the amount of P plasmid was doubled from 1.5 to 3 μg, since only 60% of the extract was used for the reaction. Cell extracts (110 μl) were prepared, duplicate dishes were pooled, and nuclei were pelleted as described above. Cytoplasmic extracts with the wt or mutant P-L complexes were then divided into three 60-μl aliquots and to these were added 30 μl of a cell extract which contained either mock, wt P, or mutant P protein, where the P proteins were expressed alone. In vitro transcription with radiolabeled nucleotide and template was performed and analyzed as described above. For analysis of P protein expression, samples of cell extracts were separated by SDS–7.5% PAGE and electroblotted onto nitrocellulose (Schleicher & Schuell) by using the Mini-Protean electrophoresis system (Bio-Rad). Proteins were detected with α-P antibody and the enhanced chemiluminescence protein identification system (Amersham Life Science).
RESULTS
Mutant P proteins defective in in vitro transcription.
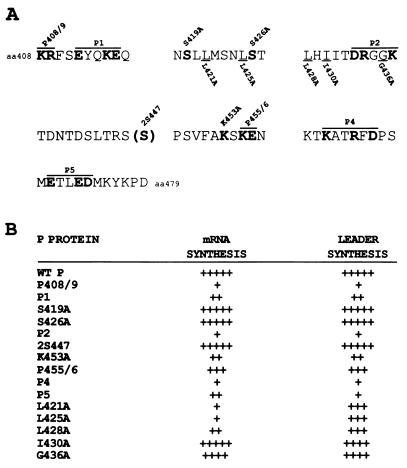
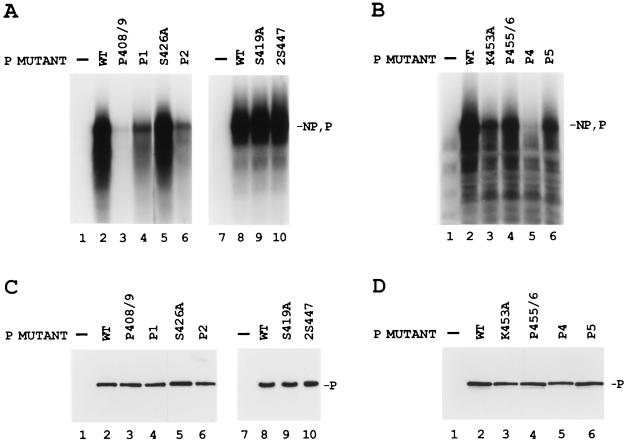
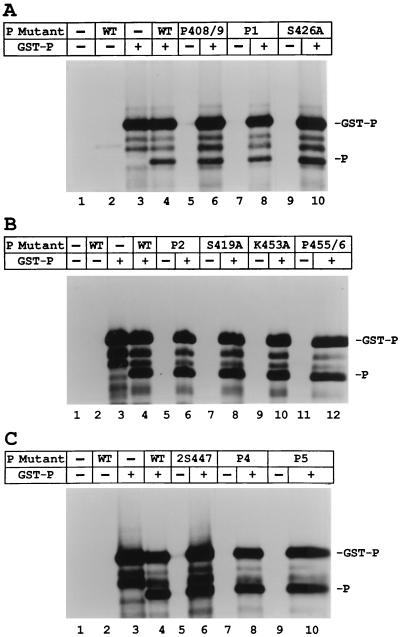
Ten charge-to-alanine or serine-to-alanine mutants were constructed in the L binding domain of the P protein as shown in Fig. 1A. To test for the biological activity of the mutant P proteins, VVT7-infected A549 cells were cotransfected with the wt or mutant P plasmid together with L plasmid and incubated overnight, and cytoplasmic extracts were then prepared. Polymerase-free Sendai virus RNA-NP template and radiolabeled nucleotide were added, and transcription in vitro was carried out as described in Materials and Methods. The wt P and L proteins were active in the synthesis of full-length NP and P mRNA products (Fig. 2A, lanes 2 and 8, and Fig. 2B, lane 2), whereas no synthesis of mRNAs was detected in the absence of viral proteins (Fig. 2A, lanes 1 and 7, and Fig. 2B, lane 1). The mutants S419A, S426A, and 2S447 retained wild-type levels of transcriptional activity (Fig. 2A, lanes 5, 9, and 10), while P455/6 gave about half the activity (Fig. 2B, lane 4). The mutants P1, P2, K453A, and P5 showed a significant reduction (60 to 90%) in activity (Fig. 2A, lanes 4 and 6, and 2B, lanes 3 and 6, respectively), while P408/9 and P4 were the most impaired, yielding very little in the way of mRNA products (Fig. 2A, lane 3, and Fig. 2B, lane 5, respectively). Where mRNA synthesis occurred, it was not determined whether the ratio of NP to P mRNAs was altered by any of the mutations. In some experiments there were background products in the mock-transfected samples (Fig. 2B, lane 1), which probably resulted from residual vaccinia virus or T7 polymerase activity due to incomplete nuclease treatment of the cell extracts prior to the start of transcription. Immunoblot analysis of samples of the cell extracts showed that the wt and mutant P proteins were nearly equally expressed (Fig. 2C and D), so the differences were in the activities of the proteins. Radiolabeling of cells expressing L with each of the mutant P protein showed that L protein was stably expressed in each case (see Fig. 4 and 7). Thus, mutagenesis of P amino acids throughout the L binding domain produced a spectrum of mRNA synthesis phenotypes which are summarized in Fig. 1B.
FIG. 1.
Summary of the in vitro transcription data of the mutant P proteins. (A) A schematic of the P protein and the amino acid sequences from aa 408 to 479 of the P mutants. Charge-to-alanine mutants named above are highlighted by boldface lettering and are indicated by an overline. Hydrophobic-to-alanine mutants named below are underlined. 2S447 indicates the insertion of a serine. (B) In vitro transcription of the mutant P proteins from Fig. 2, 3, and 6. The plus signs refer to the amount of transcription in multiple experiments (3 to 6) compared to wt P, as follows: +++++, >80%; ++++, 60 to 80%; +++, 40 to 60%; ++, 20 to 40%; and +, <20%.
FIG. 2.
In vitro transcription of the wt RNA-NP with the charge-to-alanine mutant P proteins. (A and B) VVT7-infected A549 cells transfected with no plasmids (−) or cotransfected with wt or mutant P plasmids together with L plasmid. Cytoplasmic cell extracts were incubated with wt RNA-NP in the presence of [α-32P]CTP. The transcripts were purified and analyzed by gel electrophoresis as described in Materials and Methods. The positions of the NP and P transcripts are indicated. (C and D) Immunoblot analysis of samples of the cytoplasmic extracts with α-P antibody, where the position of the P protein is indicated. Panels C and D correspond to the samples in panels A and B, respectively.
FIG. 4.
Binding of the charge-to-alanine mutant polymerase complexes to NCs. (A to C) VVT7-infected A549 cells were incubated alone or cotransfected with wt or mutant P and wt L plasmids as indicated in different sets of experiments, each with a control for VV infection alone to measure nonspecific background for that set of samples. Cells were labeled with Tran35S-label, and cytoplasmic cell extracts were prepared, incubated in the absence or presence of polymerase-free wt RNA-NP, and pelleted through glycerol as described in Materials and Methods. The pellets were immunoprecipitated with α-SV and α-P antibodies and analyzed by SDS-PAGE. The positions of the P and L proteins are indicated.
FIG. 7.
Binding of the hydrophobic-to-alanine mutant polymerase complexes to NCs. VVT7-infected A549 cells were cotransfected with wt P or mutant P plasmids and wt L plasmid as indicated. Cells were labeled with Tran35S-label, and cell extracts were prepared as described in Materials and Methods. The extract was divided in half, incubated in the absence or presence of polymerase-free wt RNA-NP, and pelleted through glycerol as described in Materials and Methods. The pellets were immunoprecipitated with α-SV and α-P antibodies and analyzed by SDS-PAGE. The positions of the P and L proteins are indicated.
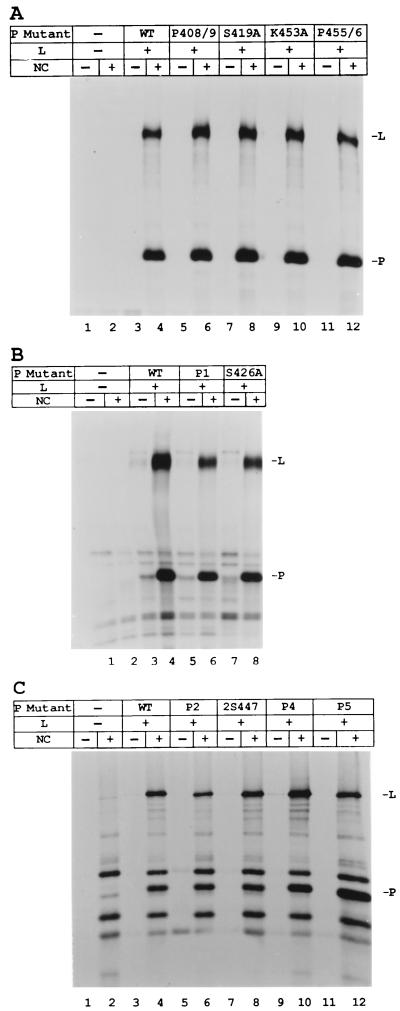
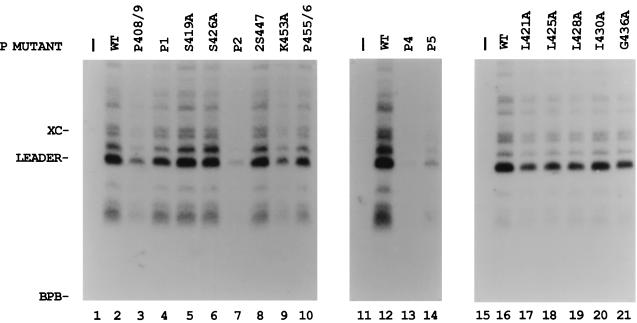
We also tested whether the P proteins could synthesize the first transcription product, the 55-nt le+ RNA from the wt RNA-NP template. The infected, transfected extracts were used for the synthesis of unlabeled product RNAs. The small RNA products were separated by acrylamide-urea gel electrophoresis and analyzed by Northern blotting with an le+-specific probe as described in Materials and Methods. The wt P-L complex synthesized le+ RNA of 55 nt, as well as longer products, due to readthrough of the leader-NP gene boundary (Fig. 3, lanes 2, 12, and 16) as reported earlier (5, 35), while no le+ RNA product was detected in the mock samples in which no viral proteins were expressed (Fig. 3, lanes 1, 11, and 15). S419A, S426A, and 2S447, which showed no reduction of mRNA synthesis, also synthesized le+ RNA at or near wt levels (Fig. 3, lanes 5, 6, and 8). Mutants P408/9, P1, P2, K453A, P455/6, and P4 were reduced in their ability to synthesize le+ RNA (Fig. 3, lanes 3, 4, 7, 9, 10, and 13) by amounts similar to their reduction in mRNA synthesis. In contrast, P5 synthesized mRNA at 25% of wt levels, but only synthesized le+ RNA at 6% of wt P (Fig. 3, lane 14). Since le+ RNA and mRNA synthesis are decreased proportionally in most of the mutants, these data indicate that the defect appears to be in the initiation of RNA synthesis and that, once started, elongation is not affected.
FIG. 3.
le+ RNA synthesis with the mutant P proteins. VVT7-infected A549 cells were transfected with no plasmids (−) or the wt P or mutant P plasmid and wt L plasmid as indicated. Cytoplasmic extracts were prepared as described for in vitro transcription in Materials and Methods and incubated with the wt RNA-NP. The total RNA products were analyzed by Northern blot with a 32P-end-labeled oligonucleotide complementary to le+ RNA as described in Materials and Methods. The 55-nt le+ (leader) RNA and the positions of the xylene cyanol (XC) and bromophenol blue (BPB) dyes are indicated.
The mutant P proteins form P-L complexes and assemble onto the NC template.
Since these P mutations reside in the L binding domain, perhaps those that were defective in RNA synthesis were not capable of forming the viral polymerase. We, therefore, utilized a NC binding assay which measures both P-L complex formation and the assembly of the polymerase onto the NC template. VVT7-infected cells were transfected with no plasmids (−) or with wt or mutant P plasmids, together with L plasmid, and labeled with Tran35S-label. Cytoplasmic extracts were divided, incubated in the presence or absence of NCs, and pelleted through glycerol as described in Materials and Methods. Since the polymerase binds NC through the P subunit (23), L will sediment with NC only when the P-L complex is formed. When the wt or mutant P proteins were coexpressed with L and pelleted in the absence of NC, little or no P and L were present in the pellet (Fig. 4A, B and C, odd-numbered lanes). In the presence of NC, both the P and the L proteins were in the pellets for wt P and all the mutants, and thus each mutant P bound both L and NC (Fig. 4A, B and C, even-numbered lanes). These data also show that L protein is stably expressed with each of the mutant P proteins. The other bands in two sets of experiments (Fig. 4B and C) are vaccinia virus proteins that nonspecifically pelleted (lanes 2). These data suggest that the defects in RNA synthesis with some P mutants are not due to the inability of P and L to form a polymerase complex and to assemble this complex onto the nucleocapsid template.
The mutant P proteins are capable of oligomerization.
The P oligomerization site is just upstream of the L binding domain and was postulated to extend into the L binding domain (11). To test whether the P-P interaction was altered in the mutants, radiolabeled wt or mutant P proteins were expressed in cells together with GST-P fusion protein, and the cobinding of P with GST-P to glutathione-Sepharose beads was used as a measure of complex formation. GST-P protein bound to the beads as expected (Fig. 5A, B and C, lanes 3). wt P protein expressed alone did not bind to the beads (lanes 2); however, when P protein was expressed in the presence of GST-P, P cobound with GST-P to the beads showing complex formation (lanes 4). Each of the P mutants also cobound to beads when expressed in the presence of the GST-P protein (Fig. 5A, B, and C, even-numbered lanes), but not when expressed alone (Fig. 5A, B, and C, odd-numbered lanes). These data show that all of the P mutants are capable of oligomerization.
FIG. 5.
Cobinding of the charge-to-alanine mutant P proteins and the GST-P fusion protein to glutathione-Sepharose beads. (A to C) VVT7-infected A549 cells were transfected with the wt or mutant P plasmids alone or together with GST-P plasmid as indicated. The cells were labeled with Tran35S-label, and cell extracts prepared and incubated with glutathione-Sepharose beads; the bound proteins were then analyzed by SDS-PAGE as described in Materials and Methods. The positions of the P and GST-P proteins are indicated.
Hydrophobic mutant P proteins are defective in in vitro transcription.
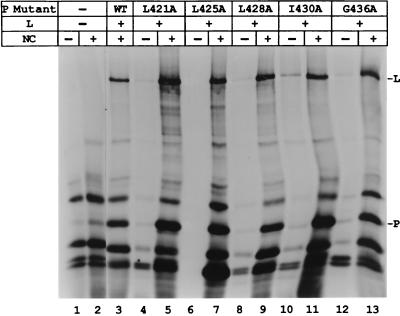
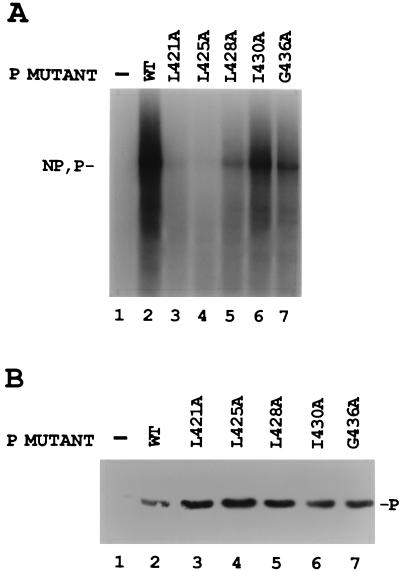
Since individual changes of the charged amino acids and serines in the L binding domain did not affect P-L complex formation, conserved hydrophobic amino acids in this region of P were also changed to alanine (Fig. 1A) and tested for their biological activity. Compared to the wt P positive control, only mutant I430A synthesized NP and P mRNAs (Fig. 6A, lane 6) at levels near that of the wt. Mutants L421A and L425A gave little or no mRNA synthesis (Fig. 6A, lanes 3 and 4), while L428A and G436A gave intermediate activity (lanes 5 and 7). Immunoblot analysis showed that the wt and mutant P proteins were similarly expressed (Fig. 6B). Thus, mutagenesis of hydrophobic P amino acids within the L binding domain produced two mutants which were basically inactive, as summarized in Fig. 1B. Analysis of le+ RNA synthesis by the hydrophobic mutants showed that I430A and G436A synthesized significant le+ in the same proportion as the mRNA products (Fig. 3, lanes 20 and 21, and Fig. 6). Surprisingly, L421A, L425A, and L428A also synthesized le+ RNA at about half the level of wt P (Fig. 3, lanes 17 to 19), although their synthesis of mRNA was much decreased (Fig. 6, lanes 3 to 5, and Fig. 1B). This increased amount of le+ RNA relative to mRNA suggested that these mutants may be defective in their ability to transcribe past the leader-NP boundary.
FIG. 6.
In vitro transcription with the hydrophobic-to-alanine mutant P proteins. (A) VVT7-infected A549 cells were transfected with no plasmids (−) or cotransfected with wt or mutant P plasmids together with L plasmid. Cell extracts were prepared and incubated with wt RNA-NP in the presence of [α-32P]CTP. The transcripts were purified and analyzed by gel electrophoresis. The relative positions of the NP and P transcripts are indicated. (B) Immunoblot analysis on samples of the cytoplasmic extracts with α-P peptide antibody, where the position of the P protein is indicated. One-half of the wt sample (lane 2) was lost.
The hydrophobic mutant P proteins form P-L, P-NC, and P-P interactions.
P-L complex formation with the hydrophobic P mutants was analyzed by utilizing the NC binding assay. When each of the radiolabeled mutant P proteins was coexpressed with L and incubated in the absence of NC, little or no P or L was present in the pellet after centrifugation (Fig. 7, even-numbered lanes), while in the presence of NC both the P and L proteins were in the pellet, showing the P-L complex bound NCs (Fig. 7, odd-numbered lanes). These data show that all of these mutant P proteins are able to form a polymerase complex and to bind to the template. We also showed that all of the hydrophobic P mutants were capable of oligomerization, since each cobound to beads when expressed in the presence, but not the absence, of the GST-P protein (data not shown). The hydrophobic, like the charge-to-alanine, P mutants are thus able to form all of the known protein-protein interactions necessary for transcription, yet the biological data suggest that two of these residues are essential for polymerase activity.
Transcriptional activity is rescued by wt P protein for some but not all P mutants.
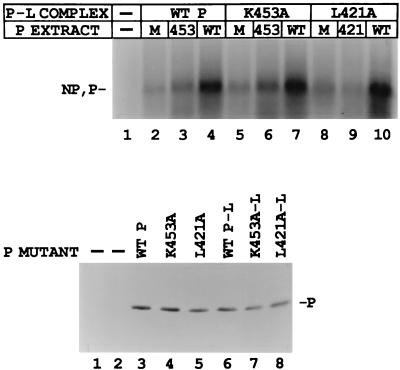
Another possible role for P protein in transcription, aside from its activity as part of the polymerase complex, has recently been described. Curran (8, 9) provides evidence that P alone is bound to the template independently of L and that this form of P is also essential for mRNA synthesis. The exact mechanism for this supplemental function of P has yet to be delineated. To test whether the defect in the activity of any of the mutant P proteins could be polymerase independent, we designed an experiment to determine whether wt P protein could rescue the defective polymerases. We first tested K453A and L421A, whose activities were decreased by ∼70 and 95%, respectively, for both le+ RNA and mRNA synthesis (Fig. 1B). The wt P-L or mutant (K453A-L or L421A-L) polymerase complexes were expressed in one extract by utilizing a suboptimal ratio of P and L plasmids which gave reduced (20 to 25%) transcription with the addition of a mock-transfected extract (Fig. 8, top panel, lanes 2, 5, and 8). When a wt P extract, where P protein was expressed alone as described in Materials and Methods, was added to the suboptimal wt P-L complex, the transcriptional activity was significantly stimulated (Fig. 8, top panel, lanes 4, 7, and 10), up to the level observed when P and L were coexpressed at the optimum ratio of plasmids (data not shown). The addition of the P K453A extract to wt P-L, however, was only able to partially rescue activity (∼30%, lanes 2 and 3, respectively), a result consistent with its limited overall activity. Addition of the mutant L421A extract to wt P-L was not able to rescue the activity of the wt polymerase at all (data not shown). The K453A-L and L421A-L polymerases could both also be rescued to full activity by the addition of wt P (lanes 7 and 10, respectively), while neither mutant protein restored the activity of its respective polymerase (lanes 6 and 9). Immunoblot analysis showed that the P proteins were all expressed at similar levels (Fig. 8, bottom panel).
FIG. 8.
Rescue of in vitro transcription of the K453A-L and L421A-L polymerases by wt P protein. (Top panel) VVT7-infected A549 cells were transfected with no plasmids or wt or mutant P plasmids alone and separate dishes cotransfected with the same P plasmids and wt L plasmid. Cell extracts containing the viral polymerases were then divided into three aliquots, and to these were added cell extract containing either mock, wt P, or mutant P protein, where the P proteins were expressed alone as described in Materials and Methods. The samples were incubated with wt RNA-NP in the presence of [α-32P]CTP. The transcripts were purified and analyzed by gel electrophoresis. The relative positions of the NP and P transcripts are indicated. (Bottom panel) Immunoblot analysis with α-P peptide antibody, where the two mock lanes are for the P-L and P samples. The position of the P protein is indicated.
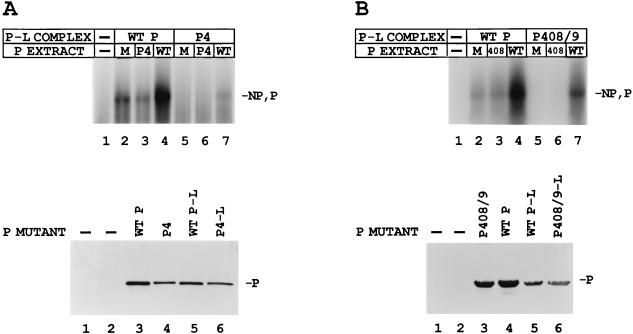
Two other mutants, P408/9 and P4, which showed more severe defects in transcription, yielding little mRNA or le+ RNA synthesis (Fig. 1B), were also tested for their ability to be rescued by the addition of wt P. Although wt P gave significant mRNA synthesis when added to the wt polymerase (Fig. 9A and B, top panel, lanes 4). wt P did not restore the P4 or P408/9 polymerase complexes to full activity (lanes 7). For P4 only a small amount of rescue could be seen (<10%); however, P408/9 was partially rescued (∼50 to 60%). Likewise, neither the mutant P408/9 nor P4 alone was successful in restoring full activity to the wt polymerase (Fig. 9A and B Top, lanes 3) or to its respective mutant polymerase (lanes 6). Immunoblot analysis showed that the proteins in each set, polymerase or P alone, were similarly expressed (Fig. 9A and B, bottom).
FIG. 9.
Rescue of in vitro transcription of the P4-L (A) and P408/9-L (B) polymerases by the wt P protein. (Top panels) VVT7-infected cells were transfected with no plasmids (−) or wt or mutant P plasmids alone, and separate dishes were cotransfected with the same P plasmids and wt L plasmid. Cell extracts containing the wt or mutant viral polymerases were divided into three aliquots, and to these were added cell extract containing either mock, wt P, or mutant P protein, where the P proteins were expressed alone. The samples were incubated with wt RNA-NP in the presence of [α-32P]CTP. The transcripts were purified and analyzed by gel electrophoresis. The relative positions of the NP and P transcripts are indicated. (Bottom panels) Immunoblot analysis with α-P peptide antibody of each set of samples with mock lanes (−) for both P-L and P samples. The position of the P protein is indicated.
Lack of exchange between P alone and the P-L complex.
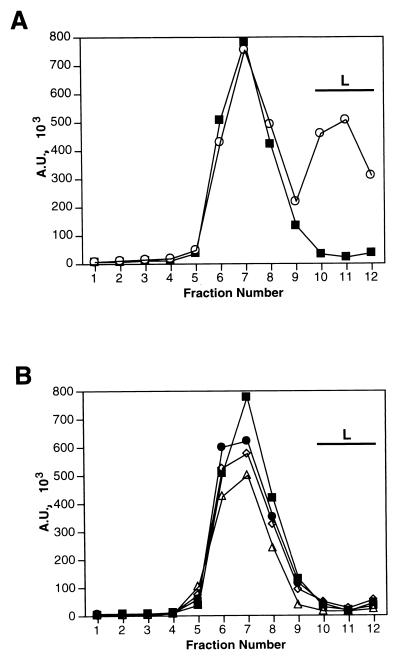
We wanted to test whether the mechanism of rescue of transcription is that the added P protein exchanges with the P within the P-L polymerase complex. This was assayed by sedimentation analysis of 35S-labeled wt P protein on glycerol gradients either alone, after coexpression with L, or after incubation with unlabeled wt or mutant P-L extracts under transcription conditions. As the control for complex formation, glycerol gradient analysis showed that P protein expressed alone sedimented, as a trimer (9), primarily in fractions 6 to 8 (Fig. 10A). However, when the P protein was expressed with L, a portion of P now cosedimented with L in fractions 10 to 12 (Fig. 10A), indicative of polymerase complex formation. Unlabeled polymerase complexes were prepared with wt P-L and the mutants L421A-L and K453A-L, all of which could be rescued with wt P. After incubation of radiolabeled wt P with the unlabeled wt and mutant polymerase complexes, in each case P was still found in sedimentation analysis only in fractions 6 to 8, profiles which were identical to the sedimentation profile of P alone (Fig. 10B). Since there was no shift of the added P to sediment with L (Fig. 10B), the data suggest that exchange between a free pool of P protein and P associated with the L protein did not occur during rescue.
FIG. 10.
Analysis of exchange between unlabeled wt or mutant polymerase complexes and radiolabeled wt P protein. (A) VVT7-infected A549 cells were transfected with the wt P plasmid alone (■) or together with the L plasmid (○) and then labeled with Tran35S-label. Cytoplasmic extracts were fractioned on a glycerol gradient, and the proteins were immunoprecipitated with α-SV and α-L antibodies and analyzed by SDS-PAGE. The P band was quantitated on the phosphorimager, and plotted in arbitrary units (A.U.) as described in Materials and Methods. (B) VVT7-infected cells were transfected with wt P plasmid alone (■) and labeled with Tran-35S-label or with wt L plasmid together with either wt P (◊), K453A (●), or L421A (▵) plasmids, which were unlabeled. Cytoplasmic extracts containing the unlabeled polymerase complexes were prepared, incubated with an equal volume of the 35S-labeled wt P cell extract, and analyzed as described above. The position of sedimentation of the L protein is indicated by the bar.
DISCUSSION
The modular P subunit of the Sendai virus RNA-dependent RNA polymerase performs multiple functions in viral RNA synthesis. The P protein oligomerizes and forms the polymerase complex with L protein and mediates binding of the polymerase to NCs (9, 11, 12, 23, 33, 34). In an effort to identify the amino acids within P that are important for complex formation with L and for function, seven charged-to-alanine and five hydrophobic-to-alanine mutants were constructed in the region spanning the L binding site. We assessed the ability of each of the mutants to transcribe viral mRNA and le+ RNA and found a spectrum of wt and defective phenotypes as summarized in Fig. 1B.
The mutants P455/6, L428A, and G436A gave intermediate mRNA synthesis relative to wt P, and the mutants P1, P2, K453A, P5, L421A, and L425A were significantly reduced in mRNA synthesis (Fig. 2 and 6). The mutants P408/9 and P4 were the most impaired, yielding little or no mRNA products. Thus, sites essential for activity are interspersed throughout the region. This was unexpected since Curran et al. (12) proposed that only the residues from 412 to 445 were necessary for L binding and P function. Clearly, these data show that the distal portion of this domain from aa 446 to 479 is also critical for polymerase function. Another interesting feature of this set of mutants is that P2 was temperature sensitive. The P2-L complex showed significant defects in both mRNA and leader RNA synthesis (>85% inhibition) at 37°C (Fig. 2 and 3). However, when the P2-L complex was synthesized at 32°C instead of 37°C, it was able to synthesize mRNA at 75% of wt L levels (data not shown). In other studies (1, 2, 36) charge-to-alanine mutagenesis yielded ca. 30% temperature-sensitive mutants, while the present study yielded 14%.
In general, the mutant P proteins showed comparable defects in both the synthesis of the first transcript, the le+ RNA, and the downstream mRNAs (Fig. 1B), which suggests that these mutants are defective in the initiation of RNA synthesis, which equally affects all downstream RNA synthesis. However, there were a few interesting exceptions. P5 synthesized mRNA at 25% of wt levels, but only synthesized le+ RNA at 6% of wt levels. This mutant polymerase is thus defective in initiation at the 3′ end of the genome but is now apparently able to initiate at the internal NP gene start site. A VSV mutant with one change in the VSV N protein was also previously shown to initiate more frequently at the start of the N gene rather than at the le+ gene (6). In contrast, the hydrophobic mutants L421A, L425A, and L428A synthesized le+ RNA in increased amounts relative to mRNA synthesis (Fig. 3 and 6). This indicates that these mutants may be unable to properly transverse the leader-NP boundary and that P is necessary for moving the polymerase into the next transcriptional unit. Also of interest was that conserved serines, S419 and S426, within the L binding domain proved to be nonessential for Sendai virus P function in transcription.
To determine whether the defective phenotypes were a consequence of a defect in any of the required protein-protein interactions, NC binding and P oligomerization assays were employed. Each of the mutants, in fact, retained each of these protein-protein interactions which are necessary for biological activity. These data suggest that the mutations did not introduce global changes in P protein conformation. Thus, we propose that the L binding domain may be folded in such a way that the residues important for P-L complex formation are spaced throughout a complex tertiary structure where mutation of no single site abolishes binding, although deletion of multiple sites does abolish binding (12, 34). Another possibility for the defects of some of the P mutants is that P may be binding a cell protein required for viral RNA synthesis and the mutations may have altered this interaction. Further work needs to be done to address this hypothesis, although there is currently no evidence that P binds cellular proteins.
Since protein-protein interactions did occur, we next sought to determine whether the defects in RNA synthesis were due to abrogation of polymerase function or possibly to a defect in a supplemental role of the P protein which was recently identified by Curran (8, 9). Earlier studies had suggested that there was more P required for viral mRNA synthesis than was accounted for by assuming P functioned only in the P-L complex. Cell extracts optimized for Sendai virus or VSV mRNA synthesis contained an excess of P protein (5, 13), and immunoelectron microscopy of intracellular Sendai virus NCs showed that P was bound to the template both independently of and together with L, whereas L was only found associated with P (31). Curran (8) demonstrated that α-L antibody-immunoselected P-L complexes were surprisingly inactive in transcription, but that that activity could be completely restored by the addition of P alone. P proteins containing deletions of either the L binding domain or the C-terminal NC binding domain were unable to rescue transcription, suggesting that P somehow interacts with both L in the bound polymerase complex and the template.
To test whether the supplemental function of P was defective in the mutants, we expressed P and L with suboptimal levels of P plasmid and then sought to determine whether added wt P could restore (rescue) full transcriptional activity to the polymerase. Our data showed that rescue by wt P occurred for the polymerase complexes containing the K453A and L421A mutant P proteins but not for those with P408/9 or P4 (Fig. 8 and 9). Interestingly, none of the mutant P proteins could rescue the wt polymerase. These results suggest that mutants which are rescued are defective in the supplemental role of P, while polymerase function remains intact. Mutants which cannot be rescued by wt P are defective in both supplemental and polymerase functions.
Two models may be proposed for the rescue of transcription by wt P. First, exchange may occur between the P protein in the polymerase complex and free P creating a wt polymerase. Second, the supplemental role of the P protein might be independent of the polymerase complex, where P binding somehow alters the nucleocapsid aiding in the movement and/or activity of the polymerase on the template. To address the first model for rescue, there are several lines of evidence which suggest that no exchange occurs. The sedimentation analyses shown here (Fig. 10) showed no incorporation of wt P into preformed wt or mutant P-L complexes when rescue did occur and two of the P mutants could not be rescued. In addition, previous data showed that the P and L proteins must be coexpressed in order to form the polymerase complex and an active polymerase (20). Likewise, it has also been demonstrated that different forms of the P protein required coexpression for oligomerization (11, 24). One caveat to these experiments, however, was that each of the assays used to determine exchange were done with the proteins in the absence of the viral template. Thus, it cannot be ruled out that exchange occurs only during RNA synthesis while the polymerase and excess P protein are associated with the template.
We favor a supplemental role of P in RNA synthesis that is independent of the polymerase. One mechanism might be that P binds to the template to increase the accessibility of the polymerase. By electron microscopy the Sendai virus NCs in infected cells have been observed to have both a relatively tight pitch of 5.3 or 6.8 nm and a more extended conformation with a pitch of 37.5 nm (14, 29). Thus, binding of P could be functioning in the transition between these pitch states to open the NC helix for RNA synthesis. Alternatively, or at the same time, P may function to transiently remove NP from the NC to expose the RNA (8). This model would predict that the P mutants might not bind NCs with the same affinity as wt P. While our assay for binding suggests that they all do bind NC, subtle differences may not be distinguishable. Certain characteristics of the P oligomer may facilitate the supplemental function. Curran (9) suggests a model whereby the P protein trimer can “walk” down the template through the formation and breakage of multiple weak contacts. In addition, Gao and Lenard (18) provided evidence that the VSV P-L complex is bound more tightly than P alone to NCs. If this is analogous for Sendai virus, then it would lend further evidence that P alone functions to increase the processivity of the polymerase, as it would be easier for the supplemental P to move on the template.
Two of the P mutants, P408/9 and particularly P4, have defects in the activity of the polymerase complex. The reason for this is unknown, but the data would suggest that either P itself contributes to the catalytic activity of the polymerase or that these incorrect contacts with L protein during formation of the complex cause folding alterations in L that inactivate the catalytic activity of the protein. These alterations must be subtle since L protein is stable when expressed with each mutant P and is not degraded as it is when synthesized in the absence of P. In summary, the L binding domain is important both for polymerase activity and for the unique supplemental role of P alone in transcription. Furthermore, in some cases residues important for the catalytic activity of the P-L complex can be genetically separated from those for the supplemental function.
ACKNOWLEDGMENTS
We thank Joyce Feller for assistance with the leader RNA analysis.
This work was supported by NIH grant AI14594 (to S.A.M.).
REFERENCES
- 1.Bass S H, Mulkerrin M G, Wells J A. A systematic mutational analysis of hormone-binding determinants in the human growth hormone receptor. Proc Natl Acad Sci USA. 1991;88:4498–4502. doi: 10.1073/pnas.88.10.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett W F, Paoni N F, Keyt B A, Botstein D, Jones A J S, Presta L, Wurm F M, Zoller M J. High resolution analysis of functional determinants on human tissue-type plasminogen activator. J Biol Chem. 1991;266:5191–5201. [PubMed] [Google Scholar]
- 3.Byrappa S, Hendricks S, Pan Y-B, Seyer J M, Gupta K G. Intracellular phosphorylation of the Sendai virus P protein. Virology. 1995;208:408–413. doi: 10.1006/viro.1995.1169. [DOI] [PubMed] [Google Scholar]
- 4.Carlsen S R, Peluso R W, Moyer S A. In vitro replication of Sendai virus wild-type and defective interfering particle genome RNAs. J Virol. 1985;54:493–500. doi: 10.1128/jvi.54.2.493-500.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandrika R, Horikami S M, Smallwood S, Moyer S A. Mutations in conserved domain I of the Sendai virus L polymerase protein uncouple transcription and replication. Virology. 1995;213:352–363. doi: 10.1006/viro.1995.0008. [DOI] [PubMed] [Google Scholar]
- 6.Chuang J L, Perrault J. Initiation of vesicular stomatitis virus mutant polR1 transcription internally at the N gene in vitro. J Virol. 1997;71:1466–1475. doi: 10.1128/jvi.71.2.1466-1475.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham B C, Wells J A. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 8.Curran J. Reexamination of the Sendai virus P protein domains required for RNA synthesis. A possible supplemental role for the P protein. Virology. 1996;221:130–140. doi: 10.1006/viro.1996.0359. [DOI] [PubMed] [Google Scholar]
- 9.Curran J. A role for the Sendai virus P protein trimer in RNA synthesis. J Virol. 1998;72:4274–4280. doi: 10.1128/jvi.72.5.4274-4280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curran J, Boeck R, Kolakofsky D. The Sendai virus P gene expresses both an essential protein and an inhibitor of RNA synthesis by shuffling modules via mRNA editing. EMBO J. 1991;10:3079–3085. doi: 10.1002/j.1460-2075.1991.tb07860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curran J, Boeck R, Lin-Marq N, Lupas A, Kolakofsky D. Paramyxovirus phosphoproteins form homotrimers as determined by an epitope dilution assay, via predicted coiled coils. Virology. 1995;214:139–149. doi: 10.1006/viro.1995.9946. [DOI] [PubMed] [Google Scholar]
- 12.Curran J, Pelet T, Kolakofsky D. An acidic activation-like domain of the Sendai virus P protein is required for RNA synthesis and encapsidation. Virology. 1994;202:875–884. doi: 10.1006/viro.1994.1409. [DOI] [PubMed] [Google Scholar]
- 13.De B P, Banerjee A K. Requirements and functions of vesicular stomatitis virus L and NS proteins in the transcription process in vitro. Biochem Biophys Res Commun. 1985;126:40–49. doi: 10.1016/0006-291x(85)90568-6. [DOI] [PubMed] [Google Scholar]
- 14.Egelman E H, Wu S-S, Amrein M, Portner A, Murti G. The Sendai virus nucleocapsid exists in at least four different helical states. J Virol. 1989;63:2233–2243. doi: 10.1128/jvi.63.5.2233-2243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Y, Greenfield N J, Cleverley D Z, Lenard J. The transcriptional form of the phosphoprotein of vesicular stomatitis virus is a trimer: structure and stability. Biochemistry. 1996;35:14569–14573. doi: 10.1021/bi9613133. [DOI] [PubMed] [Google Scholar]
- 17.Gao Y, Lenard J. Multimerization and transcriptional activation of the phosphoprotein (P) of vesicular stomatitis virus by casein kinase-II. EMBO J. 1995;14:1240–1247. doi: 10.1002/j.1460-2075.1995.tb07107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Y, Lenard J. Cooperative binding of the multimeric phosphoprotein (P) of vesicular stomatitis virus to polymerase (L) and template: pathways of assembly. J Virol. 1995;69:7718–7723. doi: 10.1128/jvi.69.12.7718-7723.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higuchi R, Krummel B, Saiki R K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horikami S M, Curran J, Kolakofsky D, Moyer S A. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J Virol. 1992;66:4901–4908. doi: 10.1128/jvi.66.8.4901-4908.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horikami S M, Hector R E, Smallwood S, Moyer S A. The Sendai virus C protein binds L polymerase protein to inhibit viral RNA synthesis. Virology. 1997;235:261–270. doi: 10.1006/viro.1997.8702. [DOI] [PubMed] [Google Scholar]
- 22.Horikami S M, Moyer S A. Synthesis of leader RNA and editing of the P mRNA during transcription by purified measles virus. J Virol. 1991;65:5342–5347. doi: 10.1128/jvi.65.10.5342-5347.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horikami S M, Moyer S A. Alternative amino acids at a single site in the Sendai virus L protein produce multiple defects in RNA synthesis in vitro. Virology. 1995;211:577–582. doi: 10.1006/viro.1995.1440. [DOI] [PubMed] [Google Scholar]
- 24.Horikami S M, Smallwood S, Moyer S A. The Sendai virus V protein interacts with the NP protein to regulate viral genome RNA replication. Virology. 1996;222:383–390. doi: 10.1006/viro.1996.0435. [DOI] [PubMed] [Google Scholar]
- 25.Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum Press, Inc.; 1991. [Google Scholar]
- 26.Kolakofsky D, Vidal S, Curran J. Paramyxovirus RNA synthesis and P gene expression. In: Kingsbury D, editor. The paramyxoviruses. New York, N.Y: Plenum Press, Inc.; 1991. pp. 215–233. [Google Scholar]
- 27.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, et al., editors. Virology. New York, N.Y: Raven Press; 1996. pp. 1177–1204. [Google Scholar]
- 28.Moyer S A, Horikami S M. The role of viral and host cell proteins in paramyxovirus transcription and replication. In: Kingsbury D, editor. The paramyxoviruses. New York, N.Y: Plenum Press; 1991. pp. 249–274. [Google Scholar]
- 29.Pelet T, Delenda C, Gubbay O, Garcin D, Kolakofsky D. Partial characterization of a Sendai virus replication promoter and the rule of six. Virology. 1996;224:405–414. doi: 10.1006/viro.1996.0547. [DOI] [PubMed] [Google Scholar]
- 30.Portner A, Murti K G. Localization of P, NP and M proteins on Sendai virus nucleocapsids using immunogold labeling. Virology. 1986;150:469–478. doi: 10.1016/0042-6822(86)90311-9. [DOI] [PubMed] [Google Scholar]
- 31.Portner A, Murti K G, Morgan E M, Kingsbury D W. Antibodies against Sendai virus L protein: distribution of the protein in nucleocapsids revealed by immunoelectron microscopy. Virology. 1988;163:236–239. doi: 10.1016/0042-6822(88)90257-7. [DOI] [PubMed] [Google Scholar]
- 32.Ryan K W, Kingsbury D W. Carboxyl-terminal region of Sendai virus P protein is required for binding to viral nucleocapsids. Virology. 1988;167:106–112. doi: 10.1016/0042-6822(88)90059-1. [DOI] [PubMed] [Google Scholar]
- 33.Ryan K W, Portner A. Separate domains of Sendai virus P protein are required for binding to viral nucleocapsids. Virology. 1990;174:515–521. doi: 10.1016/0042-6822(90)90105-z. [DOI] [PubMed] [Google Scholar]
- 34.Smallwood S, Ryan K W, Moyer S A. Deletion analysis defines a carboxyl-proximal region of Sendai virus P protein that binds to the polymerase L protein. Virology. 1994;202:154–163. doi: 10.1006/viro.1994.1331. [DOI] [PubMed] [Google Scholar]
- 35.Vidal S, Kolakofsky D. Modified model for the switch from Sendai virus transcription to replication. J Virol. 1989;63:1951–1958. doi: 10.1128/jvi.63.5.1951-1958.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wertman K F, Drubin D G, Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]