Abstract
Domestic cats infected with the horizontally transmitted feline leukemia virus subgroup A (FeLV-A) often produce mutants (termed FeLV-C) that bind to a distinct cell surface receptor and cause severe aplastic anemia in vivo and erythroblast destruction in bone marrow cultures. The major determinant for FeLV-C-induced anemia has been mapped to a small region of the surface envelope glycoprotein that is responsible for its receptor binding specificity. Thus, erythroblast destruction may directly or indirectly result from FeLV-C binding to its receptor. To address these issues, we functionally cloned a putative cell surface receptor for FeLV-C (FLVCR) by using a human T-lymphocyte cDNA library in a retroviral vector. Expression of the 2.0-kbp FLVCR cDNA in naturally resistant Swiss mouse fibroblasts and Chinese hamster ovary cells caused substantial susceptibility to FeLV-C but no change in susceptibilities to FeLV-B and other retroviruses. The predicted FLVCR protein contains 555 amino acids and 12 hydrophobic potential membrane-spanning sequences. Database searches indicated that FLVCR is a member of the major-facilitator superfamily of transporters and implied that it may transport an organic anion. RNA blot analyses showed that FLVCR mRNA is expressed in multiple hematopoietic lineages rather than specifically in erythroblasts. These results suggest that the targeted destruction of erythroblasts by FeLV-C may derive from their greater sensitivity to this virus rather than from a preferential susceptibility to infection.
Feline leukemia viruses (FeLVs) cause prevalent contagious infections of domestic cats that result in proliferative, degenerative, and immunosuppressive disorders (16, 17, 32). The viruses recovered from infected cats often contain mixtures of components that utilize distinct cell surface receptors and have been classified into the A, B, and C interference groups (44). FeLV-A strains are horizontally transmitted in saliva, preferentially infect cat cells, and often cause a slowly developing immunodeficiency and T-cell lymphoma (16, 17, 32, 36). In contrast, FeLV subgroups B and C have broader host ranges (17, 21, 32) and are formed from FeLV-A in infected cats, with FeLV-B being formed by recombination with endogenously inherited retroviral sequences (32, 37, 45, 46) and FeLV-C apparently being formed by mutations (32, 42). Although the receptor genes for FeLV-A and FeLV-C have not been cloned or unambiguously identified, FeLV-A has been shown to bind to a 70-kDa protein that has been proposed to be the cell surface receptor (13). The receptor for FeLV-B, however, has been identified as the sodium-dependent phosphate symporter Pit1 (50), which is also used by gibbon ape leukemia virus (33) and 10A1 murine leukemia virus (30, 54).
FeLV-C formation in cats has been tightly associated with a fatal pure erythrocyte aplasia, also known as erythroid aplasia or aplastic anemia (1, 10, 20, 32, 35, 40–42). This disease, which is specific for the erythroid lineage and is similar to human aplastic anemia (9, 23), involves depletion of erythroid CFU (CFU-E) and burst-forming units (BFU-E) in vivo, and it can be induced in bone marrow cultures by FeLV-C infections (1, 35, 43, 51). Studies of cats infected with FeLV-A/FeLV-C chimeras and site-directed mutants have demonstrated that aplastic anemia is determined predominantly by a small variable region, vr1, in the envelope glycoprotein of FeLV-C that also determines the receptor binding specificity of the glycoprotein (4, 42). These findings suggest that the aplastic anemia may result from FeLV-C binding to its receptor, either on erythroblasts or on other cells that control the bone marrow microenvironment (1, 27, 28). By analogy to other retroviruses (4, 19, 22, 52), the FeLV-C envelope glycoprotein might perturb the normal function of its receptor, resulting in cell killing or in changes in cytokine production. The observation that FeLV-C can infect other hematopoietic cells, including myeloid and lymphoid cells (7), implies that erythroblasts may be either exceptionally sensitive to secondary sequelae of infection or critically dependent on the normal function of the FeLV-C receptor.
We have addressed these issues by functionally cloning and characterizing a putative cell surface receptor for FeLV-C (FLVCR). This was done with a human T-lymphocyte cDNA library in a retroviral vector by procedures that have recently been used to clone cDNAs that encode coreceptors for simian immunodeficiency viruses (8) and receptors for xenotropic and polytropic murine leukemia viruses (3, 48, 55) and for simian type D retroviruses (39, 49). This method has major advantages, in part because full library representation occurs within relatively small cultures of naturally resistant murine NIH 3T3 fibroblasts and because each cell in the culture expresses only a small number (usually one or fewer) of human cDNAs. The cells that expressed the FeLV-C receptor were selected by infection, and the FLVCR cDNA was cloned from the cellular DNA by PCR. Our results suggest that FLVCR is a member of a large superfamily of transporters and that it is most closely related to evolutionary branches of this superfamily that function to transport the organic anion(s). These results have important implications for our understanding of FeLV-induced diseases.
MATERIALS AND METHODS
Cells and viruses.
Mouse NIH 3T3, human TE671, and Chinese hamster ovary (CHO) cells were used as target cells for infection. TECeB15, TELCeB6 (6), and Phoenix-Eco (provided by Garry Nolan, Stanford University, Stanford, Calif.) are packaging cell lines that produce replication-defective retrovirus. TECeB15 and TELCeB6 cells do not contain retroviral envelope genes and therefore produce noninfectious virus. CHO cells were maintained in Dulbecco’s modified alpha medium supplemented with 10% fetal bovine serum (FBS), and Phoenix-Eco cells were maintained in Dulbecco’s minimal essential medium supplemented with high glucose and 10% FBS. All other cell lines were maintained in Dulbecco’s minimal essential medium supplemented with low glucose and 10% FBS.
lacZ(FeLV-B), lacZ(A-MLV), and lacZ(RD114) producer cells were provided by Yasuhiro Takeuchi (Institute of Cancer Research, London, United Kingdom). lacZ(FeLV-C) pseudotype virus was generated by transfection (calcium phosphate precipitation [Stratagene]) of TELCeB6 cells with the FBCsalf vector (FeLV-C Sarma envelope gene [41] cloned into the FBsalf retroviral expression vector [6]). Transfectants were selected with phleomycin (50 μg/ml), and resistant colonies were pooled 2 weeks after the start of selection. Infection of target cells with lacZ pseudotype virus was carried out as previously described (47). Replication-defective FeLV-C pseudotype virus carrying the puromycin resistance gene, puro(FeLV-C), was generated by transducing TECeB15 cells with replication-defective puro(RD114) pseudotype virus to introduce the puromycin resistance gene [the puro(RD114) pseudotype virus was produced from FLYRD18 cells (6) transfected with pBabe-puro retroviral expression vector (31)]. Transduced cells were selected with puromycin (1 μg/ml), and pooled resistant colonies were then transfected with FBCsalf envelope expression vector. Transfected cells were selected with phleomycin, and puro(FeLV-C) pseudotype virus was harvested from a pooled population of resistant cells and used for infection studies.
Receptor cloning.
A human T-lymphocyte cDNA library, cloned into the retroviral vector pBabe-X (25), was generously provided by R. Sutton (Baylor College of Medicine, Houston, Tex.). Approximately 10 μg of retroviral plasmid library DNA was transfected into Phoenix-Eco packaging cells (2 × 106 cells in a 100-mm-diameter tissue culture plate) with SuperFect transfection reagent (Qiagen, Valencia, Calif.). Two days after transfection, the viral supernatant was filtered and added with Polybrene (8 μg/ml) to 10 100-mm tissue culture dishes, each containing 5 × 105 NIH 3T3 cells. After 16 h of incubation, the viral supernatant was replaced with fresh medium. The following day, the transduced NIH 3T3 cells were transferred to 150-mm tissue culture plates and incubated with 10 ml of puro(FeLV-C) supernatant for 16 h. The cells were then incubated with another 10 ml of puro(FeLV-C) supernatant for a further 4 h, after which the medium was replaced. The next day, the cells in each plate were transferred to two 150-mm plates and puromycin was added at 5 μg/ml. Selection medium was replaced every 2 days until resistant colonies had appeared. Resistant colonies were then tested for susceptibility to the lacZ(FeLV-C) pseudotype.
Isolation of receptor cDNA and expression in mammalian cells.
The transduced cDNA was recovered by subjecting 250 ng of genomic DNA, isolated from lacZ(FeLV-C)-sensitive NIH 3T3 clones, to PCR amplification with the Expand PCR kit from Boehringer Mannheim (Indianapolis, Ind.). The 2.0-kb FLVCR cDNA C10 was amplified with primers complementary to pBabe-X vector sequences flanking the cDNA insert (upstream primer, 5′-GATCCCAGTGTGCTGGAAAG-3′; downstream primer, 5′-GGTGGGGTCTTTCATTCC-3′). The PCR was run for 30 cycles with annealing at 54°C for 1.5 min and extension at 68°C for 7 min. The amplified DNA was cloned into the pCDNA3.1V5HisTOPO vector (Invitrogen, Carlsbad, Calif.) and was named pCD3.1VHC10. The DNA sequence was determined at the Microbiology and Molecular Immunology Core Facility on the PE/ABD 377 sequencer by using dye terminator cycle-sequencing chemistry (Applied Biosystems, Foster City, Calif.).
CHO cells expressing human FLVCR were generated by transfection of the pCD3.1VHC10 expression vector. Transfectants were selected with G418 (1 mg/ml), and resistant cells were analyzed for sensitivity to lacZ(FeLV-C).
Northern blots.
Multiple-tissue Northern blots containing approximately 2 μg of poly(A)+ RNA from various human tissues were obtained from Clontech (Palo Alto, Calif.). The blots were probed with full-length FLVCR cDNA that was labeled with 32P by using the random-primer extension system from NEN Life Science Products (Boston, Mass.).
Nucleotide sequence accession number.
The human FeLV-C receptor DNA and protein sequences has been assigned Genbank accession no. AF118637.
RESULTS
Isolation of a putative human FLVCR cDNA.
To isolate the FLVCR cDNA, we used a human T-lymphocyte cDNA library subcloned into the pBabe-X retroviral vector (see Materials and Methods). NIH 3T3 murine fibroblasts, which are naturally resistant to FeLV-C infections, were transduced with the retroviral library. Transduced cells were challenged with a replication-defective FeLV-C pseudotype virus that encodes the dominant selectable marker for puromycin resistance. Puromycin selection yielded 16 resistant clones, 1 of which (termed C10) was reproducibly susceptible to infection by lacZ(FeLV-C) pseudotype virus. The putative receptor cDNA was isolated by subjecting genomic DNA isolated from C10 cells to PCR amplification with primers specific for the pBabe-X vector. A DNA product of 2.0 kbp was amplified and cloned into a mammalian expression vector (see Materials and Methods) to generate the expression plasmid pCD3.1VHC10. NIH 3T3 fibroblasts and CHO cells transiently transfected with pCD3.1VHC10 were highly susceptible to lacZ(FeLV-C) infection, whereas cells transfected with vector alone were resistant (data not shown). We then isolated a CHO cell clone, CC1011, that stably expressed the FLVCR. CC1011 cells were highly susceptible to lacZ(FeLV-C), whereas no infections were observed in control CHO cells (Table 1). In contrast, the CC1011 cells were completely resistant to FeLV-B, amphotropic murine leukemia virus, and RD114 feline endogenous retrovirus (Table 1). These results strongly suggest that the 2.0-kbp C10 DNA encodes a protein that specifically mediates FeLV-C infections.
TABLE 1.
CHO cells expressing FLVCR are susceptible to FeLV-C infection
| Target cella | Titer of lacZ pseudotype (cfu/ml)
|
|||
|---|---|---|---|---|
| FeLV-Cb | FeLV-B | A-MLVc | RD114 | |
| TE671 | 3.6 × 105 | 1.0 × 106 | 5.0 × 106 | 2.1 × 106 |
| CHO | 0 | 0 | 0 | 0 |
| CC1011 | 2.3 × 104 | 0 | 0 | 0 |
TE671 are human rhabdomyosarcoma cells, and CC1011 is a G418-resistant CHO clone transfected with human FLVCR cDNA.
The titers of lacZ(FeLV-C) are average values from three infection studies.
A-MLV, amphotropic murine leukemia virus.
The FLVCR protein.
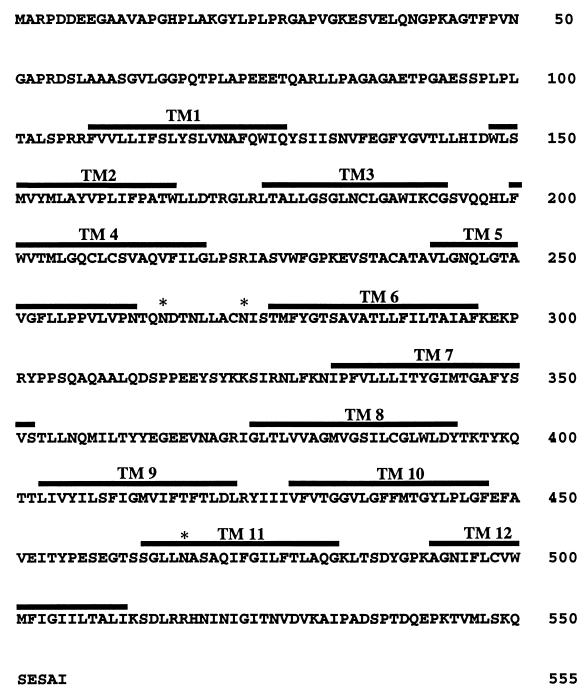
Figure 1 shows the predicted amino acid sequence and major structural features of the human FLVCR. The open reading frame encodes a protein of 555 amino acids that has three NX(S/T) sites for potential N-linked glycosylation. However, the first of these sites (NDT) may be inefficiently glycosylated because it contains an acidic amino acid in the X position (reviewed in reference 18). The Kyte-Doolittle (26) plot of FLVCR suggests the presence of 12 hydrophobic potential transmembrane (TM) sequences, consistent with its possible presence in cellular membranes (see Fig. 2).
FIG. 1.
Amino acid sequence and major structural features of the human FLVCR protein. The FLVCR protein contains 555 amino acids with 12 hydrophobic potential TM sequences. TM sequences were identified by the Kyte-Doolittle algorithm (26) and are indicated by bold lines over the amino acid sequence. Potential N-linked glycosylation sites are shown by asterisks. The protein contains several dileucines, which may be required for endocytosis (15).
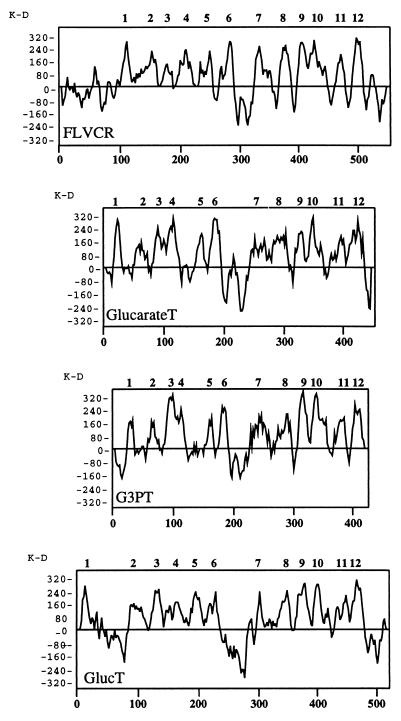
FIG. 2.
Hydrophobicity plot of human FLVCR and comparison to plots of several members of the MFS transporters. Hydrophobicity plots were generated by the Kyte-Doolittle algorithm (26). The human FLVCR has a similar hydrophobicity plot to that of bacterial glucarate and glycerol-3-phosphate transporters (GlucarateT and G3PT, respectively) and to that of the type 2 glucose transporter (GlucT) from human liver (12). The results suggest that FLVCR contains 12 TM domains, which are numbered above the hydrophobicity plot. This is a general characteristic of MFS transporters, which typically contain 12 TM region with a large hydrophilic region between TM6 and TM7.
BLAST (2) comparisons with sequences in the databases showed 99% identity to an expressed sequence tag (EST176269) derived from human colon carcinoma Caco-2 cells. FLVCR also showed strong homology (45% identity) to a Caenorhabditis elegans protein (P = 1 × 10−113) of unknown function and weaker homologies (21% identity) to a bacterial glycerol-3-phosphate transporter (P = 1 × 10−5) and to a bacterial glucarate transporter (P = 2 × 10−5). Interestingly, the bacterial transporters are both members of the ancient major-facilitator superfamily (MFS) of transporters, which also generally contain 12 hydrophobic TM sequences (38). The MFS transporters have been classified into 17 evolutionary branches or subgroups that transport distinct categories of solutes (38). FLVCR is most closely related to the organic-phosphate antiporter (OPA) and anion/cation symporter (ACS) subfamilies, which both transport organic anions. As shown in Fig. 2, FLVCR has a size and hydrophobicity profile that is closely similar to those of various MFS transporters, including the type 2 glucose transporter from human liver and the bacterial glucarate and glycerol-3-phosphate transporters. Like the MFS transporters, FLVCR has a relatively large hydrophilic sequence between TM6 and TM7.
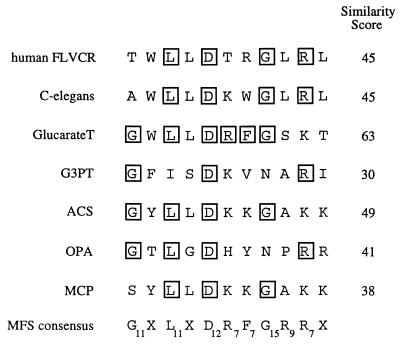
Further evidence that FLVCR is a member of the MFS superfamily is shown in Fig. 3. Specifically, it has been found that the most highly conserved sequence in the MFS transporters occurs in the hydrophilic sequence of defined length that separates TM2 and TM3 (38). The FLVCR sequence in this region is compared in Fig. 3 with the corresponding sequences of representative transporters in the MFS superfamily and with the consensus sequence for all 17 evolutionarily distinct subfamilies of the MFS transporters (38). The subscripts at the consensus amino acid positions indicate the numbers of the 17 MFS subfamilies that adhere to the consensus at that site. Thus, the most highly conserved amino acids are glycine at position 1 (in 11 of the 17 subfamilies), leucine at position 3 (in 11 of the 17 subfamilies), aspartic acid at position 5 (in 12 of the 17 subfamilies), and glycine at position 8 (in 15 of the 17 subfamilies). The similarity scores indicated in Fig. 3 are the sums of the subscripts for all positions of identity of the query sequence and the consensus sequence. In contrast to the scores indicated for these MFS transporters, random peptides of this length would have an average similarity score of only 4.0. Thus, FLVCR closely matches this consensus in both its length and its sequence.
FIG. 3.
Signature sequence for MFS transporters compared with the corresponding sequence of the human FLVCR. The MFS signature sequence, which occurs in the hydrophilic loop that separates TM2 and TM3, is the most highly conserved sequence in this transporter superfamily. The sequence for FLVCR is shown at the top, and residues identical to the consensus are boxed. Other MFS transporter sequences are also shown for C. elegans (accession no. AF002196) and for the bacterial glucarate transporter (GlucarateT) (accession no. P42237) and glycerol-3-phosphate transporter (G3PT) (accession no. AJ235270). In addition, the consensus sequences are shown for the ACS, OPA, and monocarboxylate porter (MCP) subfamilies of the MFS transporters (38). The bottom line shows the overall consensus sequence for all MFS transporters as compiled by Pao et al. (38). In this line, each amino acid is indicated by a subscript, which indicates its frequency of occurrence in the consensus sequences of the 17 distinct lineages of MFS transporters. For example, glycine (G) at position 1 occurs in 11 of the 17 subfamilies whereas G at position 8 occurs in 15 of the 17 subfamilies. The similarity score for each sequence is the sum of these subscript score for each corresponding position. Random sequences would have very low average similarity scores.
FLVCR expression in human tissues.
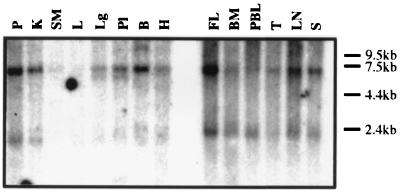
We analyzed the tissue distribution of FLVCR expression by Northern blot analysis with the 2.0-kb C10 cDNA as a probe (Fig. 4). A 2.0-kb RNA transcript was present in all hematopoietic tissues including peripheral blood lymphocytes, and was most abundant in the fetal liver. However, relatively little of this RNA transcript was present in nonhematopoietic tissues except the pancreas and kidneys. In addition, a 7.5-kb RNA was detected in most tissues except the liver and skeletal muscle.
FIG. 4.
FLVCR RNA expression in human tissues. The multiple tissue Northern blots containing poly(A)+ RNA from various human tissues were probed with 32P-labeled C10 cDNA. FLVCR RNA transcript was present in all hematopoietic tissues and also in the pancreas and kidney. Relatively little RNA expression was present in other tissues. An additional transcript of 7.5 kb was present in most tissues. P, pancreas; K, kidney; SM, skeletal muscle; L, liver; Lg, lung; Pl, placenta; B, brain; H, heart; FL, fetal liver; BM, bone marrow; PBL, peripheral blood lymphocyte; T, thymus; LN, lymph node; S, spleen.
DISCUSSION
Functional cloning of human FLVCR.
We report the functional cloning and characterization of a 2.0-kbp human cDNA from human T lymphocytes that encodes a putative cell surface receptor (FLVCR) that mediates FeLV-C infections. This cloning was done with a representative cDNA library in a retroviral vector, by procedures that have recently been successfully used to clone coreceptors for simian immunodeficiency viruses (8) and receptors for xenotropic and polytropic murine leukemia viruses (3, 48, 55), and for type D primate retroviruses (39, 49). Expression of FLVCR cDNA in NIH 3T3 fibroblasts (data not shown) and in CHO cells (Table 1), which are both resistant to FeLV-C, resulted in substantial susceptibility of the cells to this virus. In contrast, FLVCR did not confer susceptibility to infections of FeLV-A (data not shown), FeLV-B, amphotropic murine leukemia viruses, or RD114 feline endogenous retrovirus (Table 1), suggesting that FLVCR is highly specific for FeLV-C. Northern blot analyses indicated expression of a 2.0-kb FLVCR mRNA in diverse hematopoietic tissues including peripheral blood lymphocytes, with relatively little expression in other tissues. These results are in accordance with a report that FeLV-C can infect diverse hematopoietic cells in vivo (7) and with our isolation of the FLVCR cDNA from a T-lymphocyte library.
Our evidence does not exclude the possibility that additional or alternative FeLV-C receptors occur in certain cells. The FLVCR that we have identified is a member of a large and ancient MFS transporter superfamily that has many related members. Moreover, FeLV-C is closely related to FeLV-A and apparently is formed from FeLV-A within infected cats by a small number of mutations that modify the vr1 domain of the envelope glycoprotein (32, 42). These mutations evidently change the receptor-binding specificity of the virus (4, 42) and are critical for the onset of aplastic anemia (1, 10, 20, 32, 35, 40–42). The close precursor-progeny and sequence similarities of FeLV-A and FeLV-C imply that FeLV-A might use a receptor that is related to FLVCR. In accordance with this hypothesis, a putative receptor for FeLV-A that was identified by biochemical methods has an apparent Mr 70,000 (13), consistent with the expected size of a glycosylated form of FLVCR (Fig. 1) and with the sizes of MFS transporters (38). Furthermore, it is conceivable that some FeLV-C isolates might use FLVCR or closely related MFS transporters with differing efficiencies. These relationships appear strikingly similar to those for human immunodeficiency virus type 1, which is transmitted in a form that employs the CCR5 coreceptor but progresses by in vivo mutations that modify the small variable region V3 in the surface envelope glycoprotein to form derivatives that use related coreceptors such as CXCR4 (5, 8, 11, 14, 29). All of the immunodeficiency virus coreceptors are members of a common protein superfamily, and the different forms of human immunodeficiency virus also have different cellular tropisms and pathogenic effects (11, 29).
The FLVCR protein.
The FLVCR protein contains 555 amino acids with three NX(S/T) sites for potential N-linked glycosylation and with 12 hydrophobic potential TM sequences. The FLVCR protein is closely related (45% identity) to a C. elegans protein of unknown function and is less closely related (21% identity) to bacterial glycerol-3-phosphate and glucarate transporters, which are both members of the MFS superfamily of transporters (38). This large superfamily, which is ancient, contains at least 17 evolutionarily distinct branches, most of which separated from the trunk shortly after the origin of the superfamily approximately 2 billion years ago (38). As a consequence of their ancient divergences, most MFS transporters are only distantly related in their sequences (38). Moreover, each branch or subfamily has specialized in the transport of a particular category of solutes. The glycerol-3-phosphate and glucarate transporters occur in the OPA and ACS subfamilies of MFS transporters, which have both specialized for transport of different types of organic anions. These relationships suggest that FLVCR is also a transporter and that it may transport an organic anion.
The identification of FLVCR as a member of the MFS transporter superfamily is strongly supported by additional considerations. Like FLVCR, MFS transporters typically contain 12 TM sequences with a relatively large hydrophilic loop between TM6 and TM7. These similarities are illustrated in Fig. 2, which compares the sizes and hydrophobicity profiles of FLVCR with those of several members of the MFS family including the widely studied mammalian facilitated glucose transporters. In addition, FLVCR has a conserved signature sequence in the hydrophilic region between TM2 and TM3 that is highly conserved throughout the MFS superfamily (Fig. 3). These features strongly suggest that FLVCR is a member of the MFS transporter superfamily and imply that its amino and carboxyl termini are in the cytosol and that its TM domains each traverse the membrane. Accordingly, the regions most likely to interact with FeLV-C occur in the hydrophilic extracellular loops between TM1 and TM2, TM3 and TM4, TM5 and TM6, TM7 and TM8, TM9 and TM10, and TM11 and TM12. As expected, this implies that the potential sites of N-linked glycosylation between TM5 and TM6 are in the lumen of vesicles or facing the extracellular milieu.
General implications.
Our results strongly suggest that FLVCR is expressed in diverse hematopoietic cells including peripheral blood lymphocytes and T cells but is only weakly or negligibly expressed in many other tissues (Fig. 4). This is consistent with evidence that FeLV-C preferentially infects different lineages of hematopoietic cells in vivo (7). Thus, although the major determinant of FeLV-C-induced aplastic anemia has been mapped to the vr1 region, which controls the receptor-binding specificity of the envelope glycoprotein (42), it is clear that FLVCR is not restricted to erythroblasts. In addition, our results suggest that FLVCR is likely to be a facilitative transporter for organic anion(s) (see above), and we propose that it transports an important nutrient or regulator that is critical for erythroblast survival in vivo and in stroma-dependent bone marrow cultures. By analogy to other retroviral receptors (22, 24, 34, 53), it is likely that FLVCR transport function would be inhibited or down-modulated following infection with FeLV-C, and it is conceivable that this transport function is more critical for erythroblasts than for other hematopoietic cells. Alternatively, this transport activity might be necessary for function or signaling by stromal or nurse cells in the erythropoietic microenvironment (1, 27, 28). Our present results should facilitate further investigations and elucidation of the normal FLVCR function. In addition, these results raise the possibility that FeLV-A, which is a closely related progenitor of FeLV-C, may also use an MFS transporter for infection. Such a result would confirm a strong similarity to human immunodeficiency virus type 1, which also evolves by env gene mutations in vivo to form divergent progeny that utilize distinct members of a receptor superfamily and that have different cellular tropisms and pathogenic effects (5, 11, 14, 29).
ACKNOWLEDGMENTS
We are extremely grateful to Richard Sutton for providing the human retroviral cDNA library, to Yasuhiro Takeuchi and Garry Nolan for providing the packaging cells, and to Susan Kozak for helping with the Northern blot analysis. We are also grateful to our coworkers Emily Platt, Mariana Marin, Navid Madani, Shawn Kuhmann, and Ali Nouri for helpful suggestions.
This work was supported by NIH grant CA25810.
REFERENCES
- 1.Abkowitz J L, Holly R D, Grant C K. Retrovirus-induced feline pure red cell aplasia. Hematopoietic progenitors are infected with feline leukemia virus and erythroid burst-forming cells are uniquely sensitive to heterologous complement. J Clin Investig. 1987;80:1056–1063. doi: 10.1172/JCI113160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battini J L, Rasko J E, Miller A D. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc Natl Acad Sci USA. 1999;96:1385–1390. doi: 10.1073/pnas.96.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brojatsch J, Kristal B S, Viglianti G A, Khiroya R, Hoover E A, Mullins J I. Feline leukemia virus subgroup C phenotype evolves through distinct alterations near the N terminus of the envelope surface glycoprotein. Proc Natl Acad Sci USA. 1992;89:8457–8461. doi: 10.1073/pnas.89.18.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1 infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cosset F L, Takeuchi Y, Battini J L, Weiss R A, Collins M K L. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dean G A, Groshek P M, Mullins J I, Hoover E A. Hematopoietic target cells of anemogenic subgroup C versus nonanemogenic subgroup A feline leukemia virus. J Virol. 1992;66:5561–5568. doi: 10.1128/jvi.66.9.5561-5568.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng H, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature (London) 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 9.Doney K C, Torok-Storb B, Dahlberg S, Buckner C D, Martin P, Hansen J A, Thomas E D, Storb R. Immunosuppressive therapy of severe aplastic anemia. Prog Clin Biol Res. 1984;148:259–270. [PubMed] [Google Scholar]
- 10.Dornsife R E, Gasper P W, Mullins J I, Hoover E A. Induction of aplastic anemia by intra-bone marrow inoculation of a molecularly cloned feline retrovirus. Leuk Res. 1989;13:745–755. doi: 10.1016/0145-2126(89)90087-8. [DOI] [PubMed] [Google Scholar]
- 11.D’Souza M P, Harden V A. Chemokines and HIV-1 second receptors. Confluence of two fields generates optimism in AIDS research. Nat Med. 1996;2:1293–1300. doi: 10.1038/nm1296-1293. [DOI] [PubMed] [Google Scholar]
- 12.Fukumoto H, Seino S, Imura H, Seino Y, Eddy R L, Fukushima Y, Byers M G, Shows T B, Bell G I. Sequence, tissue distribution, and chromosomal localization of mRNA encoding a human glucose transporter-like protein. Proc Natl Acad Sci USA. 1998;85:5434–5438. doi: 10.1073/pnas.85.15.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh A K, Bachmann M H, Hoover E A, Mullins J I. Identification of a putative receptor for subgroup A feline leukemia virus on feline T cells. J Virol. 1992;66:3707–3714. doi: 10.1128/jvi.66.6.3707-3714.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glushakova S, Grivel J C, Fitzgerald W, Sylwester A, Zimmerberg J, Margolis L B. Evidence for the HIV-1 phenotype switch as a causal factor in acquired immunodeficiency. Nat Med. 1998;4:346–349. doi: 10.1038/nm0398-346. [DOI] [PubMed] [Google Scholar]
- 15.Hamer I, Haft C R, Paccaud J P, Maeder C, Taylor S, Carpentier J L. Dual role of a dileucine motif in insulin receptor endocytosis. J Biol Chem. 1997;272:21685–21691. doi: 10.1074/jbc.272.35.21685. [DOI] [PubMed] [Google Scholar]
- 16.Hardy W D. Feline oncoretroviruses. In: Levy J A, editor. The Retroviridae. Vol. 2. New York, N.Y: Plenum Press; 1993. pp. 109–180. [Google Scholar]
- 17.Hoover E A, Mullins J I. Feline leukemia virus infection and diseases. J Am Vet Med Assoc. 1991;199:1287–1297. [PubMed] [Google Scholar]
- 18.Hubbard S C, Ivatt R J. Synthesis and processing of asparagine-linked oligosaccharides. Annu Rev Biochem. 1981;50:555–583. doi: 10.1146/annurev.bi.50.070181.003011. [DOI] [PubMed] [Google Scholar]
- 19.Hunter E. Viral entry and receptors. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 71–120. [PubMed] [Google Scholar]
- 20.Jarrett O, Golder M C, Toth S, Onions D E, Stewart M F. Interaction between feline leukaemia virus subgroups in the pathogenesis of erythroid hypoplasia. Int J Cancer. 1984;34:283–288. doi: 10.1002/ijc.2910340222. [DOI] [PubMed] [Google Scholar]
- 21.Jarrett O, Laird H M, Hay D. Determinants of the host range of feline leukaemia viruses. J Gen Virol. 1973;20:169–175. doi: 10.1099/0022-1317-20-2-169. [DOI] [PubMed] [Google Scholar]
- 22.Kavanaugh M P, Miller D G, Zhang W, Law W, Kozak S L, Kabat D, Miller A D. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc Natl Acad Sci USA. 1994;91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelleher J F, Luban N L, Mortimer P P, Kamimura T. Human serum “parvovirus”: a specific cause of aplastic crisis in children with hereditary spherocytosis. J Pediatr. 1983;102:720–722. doi: 10.1016/s0022-3476(83)80243-1. [DOI] [PubMed] [Google Scholar]
- 24.Kim J W, Closs E I, Albritton L M, Cunningham J M. Transport of cationic amino acids by the mouse ecotropic receptor. Nature (London) 1991;352:725–728. doi: 10.1038/352725a0. [DOI] [PubMed] [Google Scholar]
- 25.Kitamura T, Onishi M, Kinoshita S, Shibuya A, Miyajima A, Nolan G P. Efficient screening of retroviral cDNA expression libraries. Proc Natl Acad Sci USA. 1995;92:9146–9150. doi: 10.1073/pnas.92.20.9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 27.Linenberger M L, Abkowitz J L. Haematological disorders associated with feline retrovirus infections. Baillieres Clin Haematol. 1995;8:73–112. doi: 10.1016/S0950-3536(05)80233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linenberger M L, Abkowitz J L. In vivo infection of marrow stromal fibroblasts by feline leukemia virus. Exp Hematol. 1992;20:1022–1027. [PubMed] [Google Scholar]
- 29.Littman D R. Chemokine receptors: keys to AIDS pathogenesis. Cell. 1998;93:677–680. doi: 10.1016/s0092-8674(00)81429-4. [DOI] [PubMed] [Google Scholar]
- 30.Miller D G, Miller A D. A family of retroviruses that utilize related phosphate transporters for cell entry. J Virol. 1994;68:8270–8276. doi: 10.1128/jvi.68.12.8270-8276.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neil J C, Fulton R, Rigby M, Stewart M. Feline leukaemia virus: generation of pathogenic and oncogenic variants. Curr Top Microbiol Immunol. 1991;171:67–93. doi: 10.1007/978-3-642-76524-7_4. [DOI] [PubMed] [Google Scholar]
- 33.O’Hara B, Johann S V, Klinger H P, Blair D G, Rubinson H, Dunn K J, Sass P, Vitek S M, Robbins T. Characterization of the human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1990;1:119–127. [PubMed] [Google Scholar]
- 34.Olah Z, Lehel C, Anderson W B, Eiden M V, Wilson C A. The cellular receptor for gibbon ape leukemia virus is a novel high affinity sodium-dependent phosphate transporter. J Biol Chem. 1994;269:25426–25431. [PubMed] [Google Scholar]
- 35.Onions D, Jarrett O, Testa N, Frassoni F, Toth S. Selective effect of feline leukemia virus on early erythroid precursors. Nature (London) 1982;296:156–158. doi: 10.1038/296156a0. [DOI] [PubMed] [Google Scholar]
- 36.Overbaugh J, Donahue P R, Quackenbush S L, Hoover E A, Mullins J I. Molecular cloning of a feline leukemia virus that induces fatal immunodeficiency disease in cats. Science. 1988;239:906–910. doi: 10.1126/science.2893454. [DOI] [PubMed] [Google Scholar]
- 37.Overbaugh J, Riedel N, Hoover E A, Mullins J I. Transduction of endogenous envelope genes by feline leukaemia virus in vitro. Nature (London) 1988;332:731–734. doi: 10.1038/332731a0. [DOI] [PubMed] [Google Scholar]
- 38.Pao S S, Paulsen I T, Saier M H., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rasko J E, Battini J L, Gottschalk R J, Mazo I, Miller A D. The RD114/simian type D retrovirus receptor is a neutral amino acid transporter. Proc Natl Acad Sci USA. 1999;96:2129–2134. doi: 10.1073/pnas.96.5.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riedel N, Hoover E A, Dornsife R E, Mullins J I. Pathogenic and host range determinants of the feline aplastic anemia retrovirus. Proc Natl Acad Sci USA. 1988;85:2758–2762. doi: 10.1073/pnas.85.8.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riedel N, Hoover E A, Gasper P W, Nicholson M O, Mullins J I. Molecular analysis and pathogenesis of the feline aplastic anemia retrovirus, feline leukemia virus C-Sarma. J Virol. 1986;60:242–250. doi: 10.1128/jvi.60.1.242-250.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rigby M A, Rojko J L, Stewart M A, Kociba G J, Cheney C M, Rezanka L J, Mathes L E, Hartke J R, Jarrett O, Neil J C. Partial dissociation of subgroup C phenotype and in vivo behaviour in feline leukaemia viruses with chimeric envelope genes. J Gen Virol. 1992;73:2839–2847. doi: 10.1099/0022-1317-73-11-2839. [DOI] [PubMed] [Google Scholar]
- 43.Rojko J L, Cheney C M, Gasper P W, Hamilton K L, Hoover E A, Mathes L E, Kociba G J. Infectious feline leukaemia virus is erythrosuppressive in vitro. Leuk Res. 1986;10:1193–1199. doi: 10.1016/0145-2126(86)90237-7. [DOI] [PubMed] [Google Scholar]
- 44.Sarma P, Log T. Subgroup classification of feline leukemia and sarcoma viruses by viral interference and neutralisation tests. Virology. 1973;54:160–169. doi: 10.1016/0042-6822(73)90125-6. [DOI] [PubMed] [Google Scholar]
- 45.Sheets R L, Pandey R, Jen W, Burman P R. Recombinant feline leukemia virus genes detected in naturally occurring feline lymphosarcomas. J Virol. 1993;67:3118–3125. doi: 10.1128/jvi.67.6.3118-3125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart M A, Warnock M, Wheeler A, Wilkie N, Mullins J I, Onions D E, Neil J C. Nucleotide sequence of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J Virol. 1986;66:1219–1222. doi: 10.1128/jvi.58.3.825-834.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tailor C S, Kabat D. Variable regions A and B in the envelope glycoproteins of feline leukemia virus subgroup B and amphotropic murine leukemia virus interact with discrete receptor domains. J Virol. 1997;71:9383–9391. doi: 10.1128/jvi.71.12.9383-9391.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tailor C S, Nouri A, Lee C G, Kozak C, Kabat K. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc Natl Acad Sci USA. 1999;96:927–932. doi: 10.1073/pnas.96.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tailor C S, Nouri A, Zhao Y, Takeuchi Y, Kabat D. A sodium-dependent neutral-amino-acid transporter mediates infections of feline and baboon endogenous retroviruses and simian type D retroviruses. J Virol. 1999;73:4470–4474. doi: 10.1128/jvi.73.5.4470-4474.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeuchi Y, Vile R G, Simpson G, O’Hara B, Collins M K L, Weiss R A. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J Virol. 1992;66:1219–1222. doi: 10.1128/jvi.66.2.1219-1222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Testa N G, Onions D, Jarrett O, Frassoni F, Eliason J F. Haemopoietic colony formation (BFU-E, GM-CFC) during the development of pure red cell hypoplasia induced in the cat by feline leukaemia virus. Leuk Res. 1983;7:103–116. doi: 10.1016/0145-2126(83)90001-2. [DOI] [PubMed] [Google Scholar]
- 52.Wang H, Dechant E, Kavanaugh M, North R A, Kabat D. Effects of ecotropic murine retroviruses on the dual-function cell surface receptor/basic amino acid transporter. J Biol Chem. 1992;267:23617–23624. [PubMed] [Google Scholar]
- 53.Wang H, Kavanaugh M P, North R A, Kabat D. Cell surface receptor for ecotropic murine retroviruses is a basic amino acid transporter. Nature (London) 1991;352:729–731. doi: 10.1038/352729a0. [DOI] [PubMed] [Google Scholar]
- 54.Wilson C A, Farrell K B, Eiden M V. Properties of a unique form of the murine amphotropic leukemia virus receptor expressed on hamster cells. J Virol. 1994;68:7697–7703. doi: 10.1128/jvi.68.12.7697-7703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Y L, Guo L, Xu S, Holland C A, Kitamura T, Hunter K, Cunningham J M. Receptors for polytropic and xenotropic mouse leukaemia viruses encoded by a single gene at Rmc1. Nat Genet. 1999;21:216–219. doi: 10.1038/6005. [DOI] [PubMed] [Google Scholar]