Abstract
Hepatitis C virus (HCV) is prevalent worldwide and has become a major cause of liver dysfunction and hepatocellular carcinoma. The high prevalence of HCV reflects the persistent nature of infection and the large frequency of cases that resist the current interferon (IFN)-based anti-HCV therapeutic regimens. HCV resistance to IFN has been attributed, in part, to the function of the viral nonstructural 5A (NS5A) protein. NS5A from IFN-resistant strains of HCV can repress the PKR protein kinase, a mediator of the IFN-induced antiviral and apoptotic responses of the host cell and a tumor suppressor. Here we examined the relationship between HCV persistence and resistance to IFN therapy. When expressed in mammalian cells, NS5A from IFN-resistant HCV conferred IFN resistance to vesicular stomatitis virus (VSV), which normally is sensitive to the antiviral actions of IFN. NS5A blocked viral double-stranded RNA (dsRNA)-induced PKR activation and phosphorylation of eIF-2α in IFN-treated cells, resulting in high levels of VSV mRNA translation. Mutations within the PKR-binding domain of NS5A restored PKR function and the IFN-induced block to viral mRNA translation. The effects due to NS5A inhibition of PKR were not limited to the rescue of viral mRNA translation but also included a block in PKR-dependent host signaling pathways. Cells expressing NS5A exhibited defective PKR signaling and were refractory to apoptosis induced by exogenous dsRNA. Resistance to apoptosis was attributed to an NS5A-mediated block in eIF-2α phosphorylation. Moreover, cells expressing NS5A exhibited a transformed phenotype and formed solid tumors in vivo. Disruption of apoptosis and tumorogenesis required the PKR-binding function of NS5A, demonstrating that these properties may be linked to the IFN-resistant phenotype of HCV.
Eukaryotic viruses establish persistent infection by avoiding the innate defenses of the host cell, escaping acquired immunity, and blocking host-mediated programmed cell death (20, 22, 64). Hepatitis C virus (HCV), a hepacivirus and member of the Flaviviridae (16, 38), mediates persistent infection within a majority of infected individuals. Viral persistence is a major factor contributing to the accumulating prevalence of HCV, which now exceeds 2% of the world population (2). Persistent HCV infection often leads to chronic hepatitis and liver cirrhosis and is strongly associated with the development of hepatocellular carcinoma and lymphoproliferative disorders (65, 66, 86). The molecular mechanisms of HCV persistence and pathogenesis are poorly understood, although these processes clearly involve avoidance of the host immune response through the evolution of viral quasispecies (12, 20, 22, 52) and alteration of host signaling pathways by interaction with specific viral proteins (60, 62).
Of central importance to these problems is the high level of viral resistance to alpha interferon (IFN-α) therapeutic regimens for the treatment of HCV infection. It is now clear that IFN therapies are effective in only approximately 30% of treated patients, though response rates differ between HCV genotypes (36, 37, 43). The recent introduction of IFN with ribavirin combination therapeutic regimens has moderately improved the response rate to anti-HCV therapy (55). However, overcoming IFN resistance remains a major challenge for effective IFN-based therapy and future management of the HCV pandemic. Problematically, resistance to IFN and development of persistent infection are major features of the most widespread HCV genotypes, 1A and 1B (53). Thus, pathogenesis due to HCV may be more severe in individuals infected with HCV genotype 1. Indeed, in independent studies, genotype 1 infection was the single factor consistently associated with IFN resistance, development of persistent infection, and severe liver pathology (3, 11, 23, 25, 77). These features support the hypothesis that that HCV persistence and pathogenesis may be linked to the IFN-resistant phenotype.
We have recently demonstrated that the nonstructural 5A (NS5A) protein from IFN-resistant strains of HCV genotypes 1A and 1B can repress the actions of the IFN-induced protein kinase PKR, an immediate-early effector of the cellular antiviral response induced by IFN (29, 31, 32). PKR mediates the antiviral actions of IFN, in part by phosphorylating the alpha subunit of eukaryotic initiation factor 2 (eIF-2α), resulting in acute inhibition of mRNA translation and a concomitant block in viral replication (56, 57; reviewed in references 17, 30, and 76). In addition, PKR facilitates IFN-induced transcriptional programs by participating in the activation of nuclear factor kappa B (NF-κB) and IFN-regulatory factor 1 (IRF-1) (46). Along with its antiviral properties, PKR has been defined as a tumor suppressor (58), and it is an important regulator of cellular pathways that control gene expression and specific apoptotic programs within dividing cells (17). Our results suggest that HCV represses PKR function through the actions of the viral NS5A protein, which binds and inhibits PKR in vivo (29, 32). Importantly, mutations within a discrete region of the PKR-binding domain of NS5A (previously termed the IFN-sensitivity-determining region [ISDR] [Fig. 1]), which were identified in IFN-sensitive strains of HCV (14, 23, 24, 47), rendered NS5A unable to bind PKR and inhibit PKR catalytic activity (29, 32). In the present report, we demonstrate that expression of NS5A in mammalian cells provides viral resistance to IFN by removing the IFN-induced, PKR-imposed block on mRNA translation during virus infection. NS5A repression of PKR similarly blocked PKR-dependent eIF-2α phosphorylation and the initiation of host apoptotic programs induced by double-stranded RNA (dsRNA). Our results suggest that disruption of PKR-dependent translational control and apoptotic programs may confer oncogenic potential to HCV.
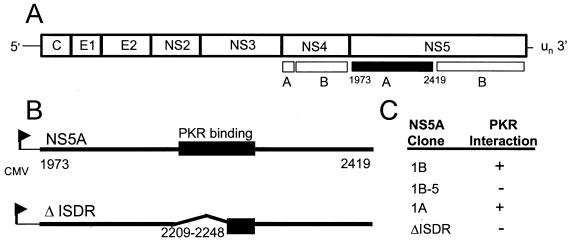
FIG. 1.
Structural representations of the HCV polyprotein and functional characteristics of the NS5A expression constructs used in this study. (A) The HCV polyprotein and NS5A cleavage product (filled region). (B) Structural representations of full-length NS5A representing HCV 1A and HCV 1B isolates (upper) and the ΔISDR NS5A deletion construct (lower) are shown. The 64-aa PKR-binding domain is indicated (29, 32). CMV, cytomegalovirus promoter. (C) Functional properties of the NS5A expression constructs. NS5A 1B and 1B-5 are isogenic except for four amino acid substitutions in the ISDR of NS5A 1B-5 that are associated with IFN sensitivity (23, 24) and confer loss of function (29). The ΔISDR NS5A construct is isogenic to NS5A 1A except that it lacks aa 2209 to 2248, which correspond to the entire ISDR. We have previously determined that this region is required for interaction with PKR and inhibition of protein kinase activity (29, 32). The PKR-binding property of each construct is indicated.
MATERIALS AND METHODS
Construction of NS5A expression plasmids.
The NS5A constructs used in these studies are depicted in Fig. 1. NS5A 1A and NS5A 1B were independently cloned from HCV RNA isolated from separate patients infected with HCV genotypes 1A and 1B, respectively (32). In each case, the patient had failed to respond to IFN therapy, and the resulting viral isolates were labeled as IFN resistant. NS5A 1B-5 is isogenic to NS5A 1B, except for four amino acid mutations within the ISDR (29) corresponding to IFN-sensitive isolates of HCV (23, 24, 47). Previous studies have determined that the NS5A 1B-5 protein does not bind PKR and does not repress PKR activity (29). The ΔISDR NS5A construct encodes a nonfunctional deletion mutant of NS5A 1A in which the first 39 amino acids (aa) (aa 2209 to 2248) of the PKR-binding domain, corresponding to the HCV ISDR, have been removed (32). Deletion of aa 2209 to 2248 disrupts the ability of NS5A to bind and inhibit PKR, thereby rendering the protein inactive (32). For expression of NS5A 1B in mammalian cells, the entire 1.4-kb NS5A coding region (32) was fused at the N terminus to the 8-aa FLAG epitope tag and placed under control a tetracycline (Tet)-regulated minimal cytomegalovirus promoter in the pTRE response plasmid (Clontech) to yield pTRE-NS5A 1B. Expression of NS5A 1B-5 in mammalian cells was facilitated by cloning the 1.4-kb insert from pFLAG NS5A 1B-5 (29) into the HindIII site of pTRE to yield pTRE-NS5A 1B-5. pNeo-NS5A 1A and pNeo-ΔISDR were constructed by cloning the 1.4-kb HindIII/XbaI insert of pYES2-NS5A 1A-wt and pcDNA3.1/His-ΔISDR, respectively (32), into the corresponding sites of pcDNA1Neo (Invitrogen).
Cell culture and transfection.
The Tet-Off gene expression system and HeLa S3 Tet-Off cells (Clontech) were used to establish the HeLa 1B and HeLa 1B-5 cell lines harboring pTRE-NS5A 1B and pTRE-NS5A 1B-5, respectively. In this system, expression of NS5A is induced by removal of Tet from the culture medium. HeLa S3 Tet-Off cells (Clontech) were transfected with pTRE-NS5A 1B or pTRE-NS5A 1B-5 and selected in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS), l-glutamine (200 μg/ml), G418 (100 μg/ml), hygomycin B (100 μg/ml), and Tet (2 μg/ml). Clonal lines of HeLa 1B and HeLa 1B-5 were generated by limiting dilution cloning, expanded, and maintained in selective DMEM as described by the system manufacturer. NIH 3T3 cells (American Type Culture Collection) were grown in DMEM containing 10% FBS as described previously (84). NIH 3T3 cell lines stably expressing the PKR inhibitory protein P58IPK have been described previously (6). Vector control NIH 3T3 cell lines (Neo) and those expressing NS5A 1A or the ΔISDR NS5A mutant were derived by transfecting cells via the DEAE-dextran–chloroquine method (84) with 3 to 10 μg of pcDNA1neo, pNeoNS5A 1A, and pNeoΔISDR, respectively. Transfected cells were selected by growth in DMEM containing 10% FBS and 600 μg of the neomycin analog G418 per ml. Drug-resistant clones were isolated, expanded, and tested for stable transgene expression. By this method, we isolated several clones expressing high or low levels of wild-type or mutant NS5A. Except for the tumorigenicity studies (below), Neo control clone 5-2, ΔISDR clone 3C2, and NS5A 1A clone 5C6 were used in all analyses.
Cell growth analysis and tumorigenicity assays.
Growth characteristics of stable NIH 3T3 cell lines constitutively expressing NS5A or P58IPK (control) were determined as described elsewhere (6). For determination of culture saturation density, cells were seeded at 105 cells/55-mm-diameter culture dish in selective DMEM containing 10% FBS and 400 μg of G418 per ml. Cells were counted every 24 h, and saturation density was determined by measuring the number of total cells in culture 4 days after reaching confluency. To assess cloning efficiency, 5 × 102, 5 × 103, or 104 cells were suspended in 0.35% agar–DMEM solution with 20% FBS and overlaid in duplicate onto six-well culture dishes containing 0.7% agar–DMEM with 10% FBS. Cloning efficiency was determined 14 days later and is presented in Table 1 as a percentage of the number of colonies observed/total number of cells plated. Colonies were defined as an isolated cluster of four or more cells. Determination of oncogenic potential was made by injecting 4- to 6-week old athymic nude mice (nu/nu; Charles River) subcutaneously near the left hind limb with 2 × 106 cells in phosphate-buffered saline (PBS). Mice were housed in microisolator cages in a pathogen-free facility and observed for up to 50 days for the formation of solid tumors. For analysis of tumor phenotypes, tumors were excised from nu/nu mice under sterile conditions, washed in PBS, and minced into 1- to 5-mm fragments. Tumor fragments were homogenized by first incubating in 1% collagenase–0.1% Dispase (Gibco BRL) in PBS and then blending in a Dounce vessel homogenizer. Homogenized tumors were incubated at 37°C for 2 h, centrifuged at 600 × g for 10 min, and resuspended in fresh DMEM with 10% FCS. Tumor cell suspensions were seeded into multiwell plates for the generation of tumor-derived clonal cell lines. Cell growth characteristics are shown in Table 1 and are representative of four experiments from each of four independent clones within the groups examined.
TABLE 1.
Growth properties of cells expressing wild-type and mutant NS5A
| Cell linea | Doubling time (h) | Saturation density (105 cells/cm2) | Cloning efficiency (%) | Mice with tumors/5 tested | Latency (days)b |
|---|---|---|---|---|---|
| Neo 5-10 | 27.6 ± 0.1 | 1.6 ± 0.1 | 0 | 0 | |
| P58-20 | 17.3 ± 1.5 | 3.7 ± 0.2 | 15 | 5 | 12–20 |
| NS5A 1A | |||||
| Clone 5C6 | 17.7 ± 0.6 | 4.0 ± 0.2 | 14 | 5 | 17–25 |
| Clone 4A1 | 22.4 ± 2.1 | 3.3 ± 0.1 | 4 | 4 | 22–44 |
| ΔISDR clone 3C2d | 17.5 ± 1.3 | 3.8 ± 0.3 | 3.9 | 0 |
Neo 5-10 and P58-20 are negative and positive control cell lines, respectively, and data for each represent results from five independent clonal cell lines. NS5A 1A clone 5C6 cells express NS5A to levels approximately fivefold higher than those expressed by NS5A 1A clone 4A1 cells. Results are representative of four independent cell lines from each group of high- and low-expressing NS5A clones and four ΔISDR clones. Values shown are averages from four experiments.
Defined as the time, in days, to produce 3-mm-diameter tumors.
Protein analysis.
Unless otherwise noted, cell extracts were prepared in buffer I (50 mM KCl, 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 20% glycerol, 0.5% Triton X-100, 100 U of aprotinin per ml, 1 mM phenylmethylsulfonyl fluoride, 20 mM Tris [pH 7.5]) exactly as described elsewhere (84). Extracts were clarified by 4°C centrifugation at 12,000 × g; supernatants were collected and stored at −70°C. Cell extract protein concentration was determined by the Bio-Rad Bradford assay as described by the manufacturer. Determination of protein expression was carried out by immunoblot analyses of 25 to 50 μg of detergent-soluble protein from cell extracts as previously described (28). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. Bound proteins were detected by probing the membranes with a primary monoclonal antibody (MAb) specific to NS5A (a generous gift from T. Imagawa, Osaka University), human PKR (48) (generously provided by A. Hovanessian, Pasteur Institute), murine PKR (Santa Cruz Biotechnology), or mammalian eIF-2α (generously provided by Scot Kimball, Pennsylvania State University). Proteins were visualized by enhanced chemiluminescence and autoradiography.
For protein biosynthetic labeling, cultured cell monolayers were rinsed three times with ice-cold PBS and then incubated for 5 h in methionine- and cysteine-deficient medium containing 50 μCi of [35S]methionine-cysteine (Dupont) per ml. Labeled cells were rinsed three times with ice-cold PBS and subjected to extract preparation as described above. Radiolabel incorporation was quantitated by scintillation counting of trichloroacetic acid-precipitated cell extracts. Labeled proteins were separated by electrophoresis through 12.5% polyacrylamide gels. Proteins were visualized by autoradiography of the dried gel.
For isoelectric focusing of eIF-2α, cell extracts were prepared by first rinsing cell monolayers three times with ice-cold PBS containing 86 mM NaF, 10 mM 2-aminopurine, and 17.2 mM each β-glycerolphosphate and Na2MoO4. Rinsed monolayers were lysed by incubation for 2 min in ice-cold isoelectric focusing lysis buffer as described elsewhere (75). Extracts were clarified by centrifugation at 12,000 × g at 4°C for 10 min; the detergent-soluble fraction was collected and stored at −70°C until used. Extracts (20 μg) were separated by single-dimension isoelectric focusing essentially as described elsewhere (75). After transfer to nitrocellulose membrane, the positions of eIF-2α were detected by immunoblot analyses using the anti-eIF-2α MAb. This procedure allows for the discrimination of basally phosphorylated eIF-2α from the more acidic serine 51-hyperphosphorylated eIF-2α species. In some experiments, a mixture of hemin-treated and untreated rabbit reticulocyte lysate was included in the analysis for positive identification of serine 51-phosphorylated eIF-2α (phosphorylated by the reticulocyte-specific HRI protein kinase [15]).
Virus infection.
For viral infection, HeLa 1B or HeLa 1B-5 cells were cultured at a density of 4 × 105 cells/60-mm-diameter dish in Tet-deficient (Tet−) selective medium for 10 h at 37°C to facilitate induction of NS5A expression. Parallel control cultures were similarly incubated in Tet+ medium. NIH 3T3-derived cell lines were cultured at a density of 2 × 104 cells/60-mm-diameter dish and incubated in selective medium for 24 h. Prior to infection of cultures, the medium was replaced with fresh selective medium alone, or selective medium containing murine or human IFN-α, and incubated for a further 16 h at 37°C. HeLa 1B, HeLa 1B-5, and NIH 3T3-derived cell lines were infected with vesicular stomatitis virus (VSV; strain HR-W+; a kind gift from Phillip Marcus and Margaret Sekellick) at a multiplicity of infection (HeLA cells) of 10 or 20 (NIH 3T3 cells). Virion attachment was facilitated by a 45-min incubation at 4°C and was followed by a 5-h incubation at 37°C. For each experiment, mock-infected control cultures were similarly incubated in the absence of added virus. Viral protein synthesis was determined by biosynthetic pulse-labeling and SDS-PAGE analyses.
Determination of apoptosis.
Detection of apoptotic cells was facilitated using the terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) procedure (Boehringer Mannheim). Cell monolayers were incubated (in the absence of exogenous IFN) for 16 h in medium containing 50 ng of actinomycin D per ml in the presence or absence of the specified concentrations of the synthetic dsRNA, poly(riboinosine-ribocytosine) (pIC; Sigma). After trypsin detachment of monolayers, cells were processed for TUNEL analyses as described by the kit manufacturer. The frequency of apoptotic nuclei (green fluorescence) within a given culture was quantitated by flow cytometric analysis using a FACScan cytometer (Becton Dickinson Immunocytometry Systems). Data are presented as histograms of relative fluorescence intensity or plotted as a pIC dose-response curve based on histogram-derived data.
RESULTS
NS5A from IFN-resistant HCV provides viral resistance to IFN.
Previous work demonstrated that NS5A could directly inhibit the translational regulatory properties of PKR in vivo (29, 32). These results suggested that HCV might mediate resistance to IFN, at least in part, through NS5A repression of PKR. Thus, it was essential to determine if NS5A could overcome the IFN-induced block on viral mRNA translation when expressed during viral infection. Since HCV does not replicate efficiently in cell culture, we developed a system based on VSV infection of stable cell lines expressing NS5A from IFN-resistant and IFN-sensitive HCV (Fig. 1). VSV replication involves dsRNA intermediates, which are potent activators of PKR. We chose the VSV model because unlike many eukaryotic viruses, VSV does not encode a mechanism to inhibit the antiviral properties of PKR that are activated during infection (79). Hence, VSV is sensitive to the antiviral actions of IFN mediated through PKR phosphorylation of eIF-2α (49, 79).
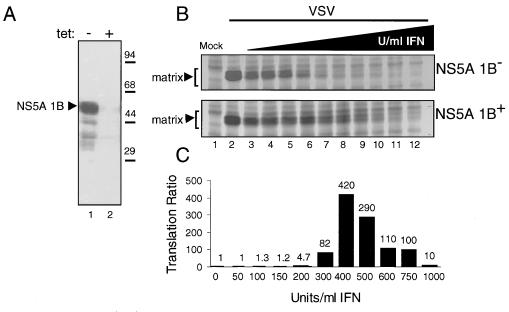
We prepared clonal HeLa S3 cell lines (HeLa 1B) that express NS5A, isolated from IFN-resistant HCV-1B (32) from a Tet-regulated promoter (Fig. 1). Removal of Tet from the culture medium induced the stable expression of NS5A 1B in HeLa 1B cell lines (Fig. 2A), and the level of NS5A 1B was discretely regulated by titrating Tet back into the culture medium (not shown). We used HeLa 1B cells to determine if expression of NS5A could prevent the IFN-induced block on viral mRNA translation imposed by PKR during VSV infection. HeLa 1B cells were infected with VSV in the presence or absence of Tet and increasing amounts of IFN. As revealed by [35S]methionine-cysteine pulse-labeling and quantitation of VSV matrix protein synthesis, IFN treatment significantly reduced viral mRNA translation in Tet+ HeLa 1B cultures not expressing NS5A 1B, and a complete block in viral mRNA translation was achieved at 300 U of IFN per ml (NS5A 1B [Fig. 2B, upper panel]). These results are consistent with the PKR-mediated antiviral actions of IFN (30, 40). In contrast, induction of NS5A 1B expression (NS5A 1B+) in Tet− cultures supported viral mRNA translation in the presence of increasing concentrations of IFN (Fig. 2B, lower panel). As shown in Fig. 2C, determination of the ratio of VSV matrix protein translation in NS5A 1B+ to that in NS5A 1B− cultures revealed an apparent rescue of VSV mRNA translation in NS5A 1B cultures, beginning at 300 U of IFN per ml. Though the rescue of VSV mRNA translation extended over the range of IFN concentrations, the strongest rescue effect was clearly seen at IFN concentrations of 400 and 500 U/ml (Fig. 2C). These studies provide evidence that NS5A from IFN-resistant HCV can mediate viral resistance to IFN. Importantly, our results suggest that NS5A may provide IFN resistance by preventing the PKR-imposed block on viral mRNA translation.
FIG. 2.
Inducible expression of NS5A reduces IFN sensitivity of viral mRNA translation. (A) Inducible expression of NS5A 1B in HeLa 1B cells. Extracts (50 μg) prepared from Tet− (lane 1) and Tet+ (lane 2) cultures of HeLa 1B cells were analyzed by immunoblotting using a MAb specific to NS5A. The arrowhead denotes the position of NS5A 1B. We confirmed that PKR was efficiently expressed in cells from Tet− and Tet+ cultures (not shown). Positions of molecular mass standards are indicated in kilodaltons. (B) Expression of NS5A supports viral mRNA translation in IFN-treated HeLa 1B cells infected with VSV. Viral protein synthesis in cells treated with increasing concentrations of IFN was determined by biosynthetic labeling and autoradiography as described in Materials and Methods. The level of each viral protein was quantitated from autoradiograms by using a Bio-Rad GS700 imaging densitometer and computer software supplied by the manufacturer. Panels show the biosynthesis of the 29-kDa VSV matrix protein (denoted by arrowheads) in the presence (NS5A 1B+; lower panel) and absence (NS5A 1B−; upper panel) of NS5A 1B expression. The far-left lane of each panel shows an extract prepared from uninfected, untreated control cultures. Lanes represent IFN concentrations of 0 (lanes 1 and 2), 50 (lane 3), 100 (lane 4), 150 (lane 5), 200 (lane 6), 300 (lane 7), 400 (lane 8), 500 (lane 9), 600 (lane 10), 750 (lane 11), and 1,000 (lane 12) U/ml. (C) The relative level of matrix protein translation for each sample was determined by first subtracting the optical density (within the region indicated for mock-infected extracts by brackets in panel B) from that obtained for each subsequent lane. Data are presented as a ratio of VSV matrix protein translation in cells expressing NS5A to that observed in cells not expressing NS5A, for each concentration of IFN. The value of each ratio is shown above the corresponding bar.
IFN sensitivity is conferred by ISDR mutations within the PKR-binding domain of NS5A.
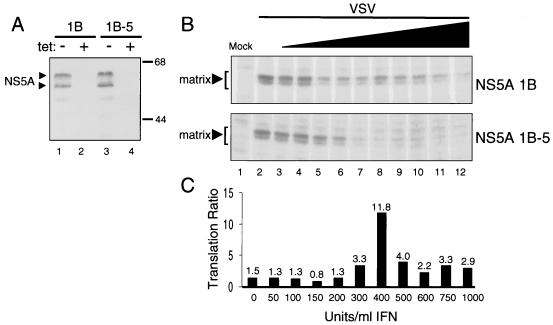
To determine if NS5A rescue of VSV mRNA translation in IFN-treated cells was dependent on the PKR-regulatory function of NS5A, we similarly prepared a HeLa cell line expressing the NS5A 1B-5 mutant from a Tet-regulated promoter. The NS5A 1B-5 construct is isogenic to NS5A 1B but harbors four amino acid substitutions within the ISDR, corresponding to IFN-sensitive HCV (23, 24, 29; reviewed in reference 34). The ISDR mutations in NS5A 1B-5 map to within the PKR-binding domain and render the protein nonfunctional and unable to bind or regulate PKR (29) (Fig. 1). We examined the ability of NS5A 1B and NS5A 1B-5 to rescue VSV mRNA translation upon parallel infection of the respective IFN-treated cell lines (Fig. 3). Removal of Tet from the culture medium induced expression of NS5A 1B and NS5A 1B-5 to approximately equal levels (Fig. 3A). Interestingly, each protein migrated as a 55/58-kDa dimer when separated by high-resolution SDS-PAGE, consistent with isoforms representing physiological levels of NS5A phosphorylation (reviewed in reference 60). Each cell line was infected with VSV in the absence of Tet and increasing amounts of IFN. Analysis of protein synthetic rates demonstrated an acute sensitivity of VSV mRNA translation to IFN in HeLa 1B-5 cells. As shown in Fig. 3B (lower panel), synthesis of the viral matrix protein was completely abolished in HeLa 1B-5 cells treated with an IFN concentration of 400 U/ml. In contrast, viral protein synthesis was sustained in HeLa 1B cells throughout the range of IFN concentrations (Fig. 3B, upper panel), supporting our previous observations (Fig. 2). Determination of the viral matrix protein translation ratio demonstrated that expression of NS5A 1B-5 was not sufficient to rescue viral protein synthesis from the antiviral actions of IFN (Fig. 3C). Expression of NS5A 1B conferred nearly a 12-fold increase in the level of viral protein synthesis compared to cells expressing the nonfunctional NS5A 1B-5 mutant. Importantly, these results suggest that ISDR mutations (corresponding to IFN-sensitive HCV [23, 24, 29]) that abrogate NS5A function confer IFN sensitivity to viral replication. The loss of PKR-regulatory function associated with mutations within the NS5A ISDR suggests that IFN sensitivity in this system may be due in part to the inability of NS5A 1B-5 to repress the antiviral actions of PKR. Similar to the results shown in Fig. 2C, we noted a rescue curve in which the highest level of rescue over VSV protein synthesis occurred between 300 and 500 U of IFN per ml. These seemingly narrow rescue curves may simply reflect differential levels of PKR induction at the various IFN concentrations and/or the more pleiotropic actions of IFN upon viral and cellular metabolism (76).
FIG. 3.
ISDR mutations within the PKR-binding domain of NS5A confer IFN sensitivity to viral mRNA translation. (A) Inducible expression of NS5A 1B and NS5A 1B-5 in HeLa S3 cells. Extracts (50 μg) prepared from Tet− (lanes 1 and 3) and Tet+ (lane 2 and 4) cultures of HeLa 1B (lanes 1 and 2) and HeLa 1B-5 (lanes 3 and 4) cell lines were analyzed by immunoblotting using a MAb specific to NS5A. Arrowheads denote the positions NS5A, which migrates on SDS-PAGE as hypo- and hyperphosphorylated isoforms (60). We confirmed that PKR was efficiently expressed in both cell lines in the presence and absence of Tet (not shown). Positions of molecular mass standards are indicated in kilodaltons. (B) Expression of NS5A 1B, but not NS5A 1B-5, supports viral mRNA translation in IFN-treated HeLa cell lines infected with VSV. Viral protein synthesis in HeLa 1B (upper panel) and HeLa 1B-5 (lower panel) cell lines, cultured in the absence of Tet and treated with increasing concentrations of IFN, was determined by biosynthetic labeling and autoradiography as described in Materials and Methods. The level of each viral protein was quantitated as described for Fig. 2. Panels show the biosynthesis of the 29-kDa VSV matrix protein (denoted by arrowheads) in the presence NS5A 1B and NS5A 1B-5 (upper and lower panels, respectively). The far-left lane of each panel shows an extract prepared from uninfected, untreated control cultures. Lanes represent IFN concentrations of 0 (lanes 1 and 2), 50 (lane 3), 100 (lane 4), 150 (lane 5), 200 (lane 6), 300 (lane 7), 400 (lane 8), 500 (lane 9), 600 (lane 10), 750 (lane 11), and 1,000 (lane 12) U/ml. (C) Translation ratios. The relative level of matrix protein translation for each sample was determined as for Fig. 2. Data are presented as a ratio of VSV matrix protein translation in cells expressing NS5A 1B to that observed in cells expressing NS5A 1B-5, for each concentration of IFN. The value of each ratio is shown above the corresponding bar.
IFN resistance and rescue of viral mRNA translation requires the PKR-regulatory function of NS5A and occurs through disruption of eIF-2α phosphorylation.
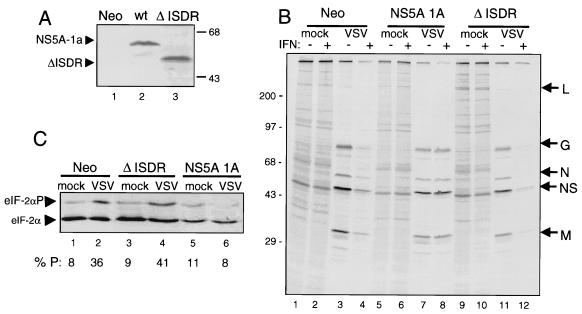
To confirm that NS5A could provide viral resistance to IFN and determine the molecular mechanisms by which NS5A may rescue viral mRNA translation, we prepared NIH 3T3 cell lines that constitutively express NS5A from IFN-resistant HCV 1A (NS5A 1A) (32). As controls, we also prepared stable NIH 3T3 cell lines that harbor the empty transfection vector (Neo) or that constitutively express a nonfunctional deletion mutant of NS5A 1A (ΔISDR) lacking the first 39 aa of the PKR-binding domain (32). This 39-aa region of NS5A has been previously defined as the HCV ISDR (14, 23, 24, 47) (Fig. 1). As shown in Fig. 4A, immunoblot analysis demonstrated that NS5A 1A and the ΔISDR mutant were efficiently expressed in stable cell lines. We confirmed that PKR was expressed to similar levels in all NIH 3T3-derived cell lines (not shown).
FIG. 4.
The PKR-regulatory function of NS5A is required for rescue of viral mRNA translation and ablation of virus-induced eIF-2α phosphorylation in IFN-treated cells. (A) Constitutive expression of NS5A 1A and the ΔISDR construct in NIH 3T3 cell lines. Immunoblots of extracts prepared from Neo control (lane 1), NS5A 1A (wild type [wt]; lane 2), and ΔISDR (lane 3) cells were probed with a MAb specific to NS5A. Arrowheads denote the positions of NS5A 1A and the ΔISDR proteins. (B) Removal of the IFN-induced block on viral mRNA translation requires the PKR-regulatory function of NS5A. NIH 3T3 cell lines were mock infected or infected with VSV in the presence (+) or absence (−) of 100 U of IFN per ml, as shown above each lane. Proteins were pulse-labeled with [35S]methionine-cysteine, separated by SDS-PAGE, and visualized by autoradiography. Shown are representative analyses of Neo control (lanes 1 to 4), NS5A 1A (lanes 5 to 8), and ΔISDR (lanes 9 to 12) cell lines. Positions of molecular mass standards are indicated in kilodaltons. Arrows at right show the positions of the five VSV proteins. (C) NS5A prevents virus-induced eIF-2α phosphorylation in IFN-treated, VSV-infected cells. Soluble extracts were prepared from mock-infected (lanes 1, 3, and 5) or VSV-infected (lanes 2, 4, and 6), IFN-treated cells and subjected to single-dimension isoelectric focusing. Proteins were transferred to nitrocellulose and subjected to immunoblot analysis with a MAb specific to eIF-2α. Arrowheads point to the positions of basally phosphorylated eIF-2α (lower band) and eIF-2α phosphorylated on serine 51, the site phosphorylated by PKR (upper band) (75). Serine 51-phosphorylated eIF-2α as a percentage of the total eIF-2α present in each sample was quantitated by scanning densitometry and is shown below each lane as % P.
Neo control cells and those expressing NS5A 1A or the nonfunctional ΔISDR NS5A mutant were infected with VSV in the presence or absence of 100 U of IFN per ml. Without IFN, VSV RNA was efficiently translated at similar rates in all three cell lines (Fig. 4B). IFN treatment significantly limited viral polypeptide synthesis in Neo control and ΔISDR cell lines. However, viral mRNA translation in cells expressing NS5A 1A remained unaltered by IFN treatment (Fig. 4B; compare lanes 4, 8, and 12). Examination of eIF-2α phosphorylation in IFN-treated NIH 3T3 cell lines demonstrated that VSV induced nearly a fivefold increase in the level of phosphorylated eIF-2α in both Neo and ΔISDR cell lines, consistent with the activation of PKR by viral dsRNA. In contrast, virus-induced eIF-2α phosphorylation was completely abolished in IFN-treated cells expressing NS5A 1A. Importantly, the concomitant rescue of viral mRNA translation and inhibition of virus-induced eIF-2α phosphorylation was dependent on the ability of NS5A to bind and inhibit PKR. Viral mRNA expression was not affected by IFN treatment and remained comparable throughout all experiments (not shown). Our results demonstrate that NS5A can provide viral resistance to IFN and, importantly, provide a molecular mechanism by which HCV mediates resistance to IFN therapy.
Expression of NS5A blocks eIF-2α phosphorylation and apoptosis induced by dsRNA PKR agonists.
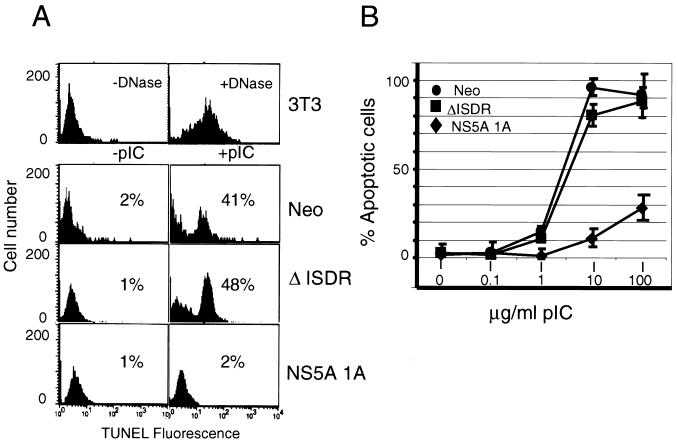
Recent studies have defined PKR function (4, 18) and phosphorylation of eIF-2α (81) as requisite components of apoptotic signaling induced by dsRNA (reviewed in reference 82). Our results suggested that by inhibiting PKR, NS5A may disrupt PKR-dependent dsRNA signaling and apoptotic programs during HCV infection. The ability to disrupt or delay host apoptotic programs induced by dsRNA is a common determinant shared by viruses which, like HCV, mediate persistent infection, including human immunodeficiency virus type 1, herpesviruses, poliovirus, and the DNA tumor viruses (64, 82, 85). To determine if NS5A can confer a similar antiapoptotic potential to HCV, we examined the ability of NS5A cell lines to undergo apoptosis induced by dsRNA. Treatment with the dsRNA pIC readily induced apoptosis in Neo control and ΔISDR cell lines (Fig. 5A), suggesting that dsRNA signaling pathways remained in tact in these cells. In contrast, cells expressing NS5A were resistant to dsRNA-induced apoptosis and retained this resistant phenotype even when exposed to high levels of pIC (Fig. 5). Thus, resistance to dsRNA-induced apoptosis required the intact PKR-binding domain of NS5A. Taken together, these results demonstrate that NS5A from IFN-resistant HCV can block PKR-dependent apoptotic signaling induced by dsRNA.
FIG. 5.
NS5A provides resistance to apoptosis induced by dsRNA PKR agonists. (A) Flow cytometric analysis of TUNEL fluorescence. Neo control, ΔISDR, and NS5A 1A cell lines were cultured for 16 h in medium containing actinomycin D (50 ng/ml) with (right column) or without (left column) 1 μg of the synthetic dsRNA pIC per ml. The number of apoptotic cells was determined by quantitation of green fluorescence intensity (TUNEL fluorescence). As a control, NIH 3T3 cells were treated with DNase prior to the TUNEL reaction (upper panels). Results shown are representative of three experiments done independently of those shown in panel B. (B) Resistance to dsRNA-induced apoptosis requires the PKR-regulatory function of NS5A. Neo control, ΔISDR, and NS5A 1A cells were cultured as described for panel A and incubated in the presence of increasing concentrations of pIC. TUNEL fluorescence from three independent experiments was quantitated as for panel A and plotted as the average percentage of apoptotic cells for each concentration of pIC. Error bars indicate the standard error from the mean for each titration point.
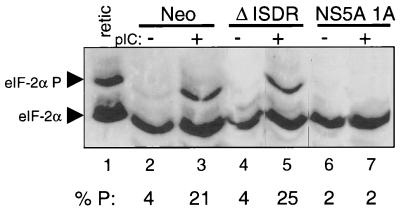
Phosphorylation of eIF-2α serine 51 by PKR is essential for dsRNA-induced apoptosis (81). To determine if the block in apoptosis imposed by NS5A could be attributed to inhibition of eIF-2α phosphorylation, we examined the levels and extent of phosphorylated eIF-2α after exposure of cells to dsRNA. As seen in Fig. 6, pIC induced a greater than fivefold increase in the level of serine 51-phosphorylated eIF-2α in Neo control cells, consistent with the dsRNA-dependent actions of PKR. In contrast, cells expressing NS5A 1A were refractory to dsRNA-dependent signaling, and the induction of serine 51 phosphorylation was completely abolished (Fig. 6, lanes 6 and 7). Importantly, the loss of PKR-binding activity of the ΔISDR mutant restored serine 51 phosphorylation induced by pIC, and these cells exhibited an approximate sixfold increase of phosphorylated eIF-2α. Thus, constitutive expression of NS5A 1A from IFN-resistant HCV blocks serine 51 phosphorylation of eIF-2α induced by dsRNA. Together, these results suggest that NS5A may block PKR-dependent signaling events during HCV infection, including the activation of PKR by viral dsRNA. Importantly, inhibition of PKR provides a molecular link by which HCV can both resist the antiviral actions of IFN and avoid host apoptotic programs induced by dsRNA.
FIG. 6.
NS5A blocks eIF-2α phosphorylation induced by dsRNA. Extracts were prepared from Neo control (lanes 2 and 3), ΔISDR (lanes 4 and 5), and NS5A 1A cells (lanes 6 and 7) that were incubated for 16 h in the presence or absence of pIC (1 μg/ml). Proteins (20 μg) were subjected to single-dimension isoelectric focusing and anti-eIF-2α immunoblot analyses. The upper and lower arrowheads denote the positions of serine 51-phosphorylated eIF-2α and basally phosphorylated eIF-2α, respectively. As a control, hemin-treated and untreated rabbit reticulocyte lysate mixture (retic) was included in the analysis (lane 1). The percentage of serine 51-phosphorylated eIF-2α from the total was determined by scanning laser densitometry and is indicated below each lane as % P.
PKR inhibition confers oncogenic potential to NS5A from IFN-resistant HCV.
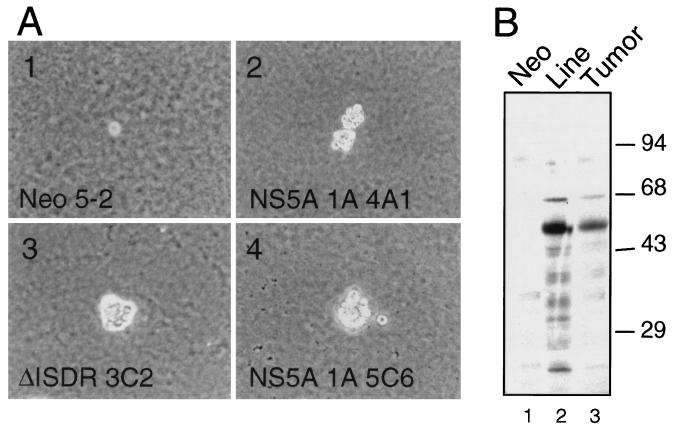
The translational control and antiproliferative properties of PKR have defined this protein kinase as a tumor suppressor (17). We therefore hypothesized that by blocking PKR function, NS5A might alter the growth properties of HCV-infected cells. To begin to examine this hypothesis, we characterized the growth properties of cells expressing NS5A 1A or the nonfunctional ΔISDR NS5A mutant. As shown in Table 1, comparison with Neo control and P58-20 cells (which overexpresses the cellular PKR inhibitory protein P58IPK [6]) indicated that both NS5A 1A and ΔISDR cells exhibited a growth-stimulatory phenotype with a characteristic reduction in doubling time and an increase in culture saturation density. Moreover, constitutive expression of NS5A 1A or the ΔISDR mutant supported colony formation of cells cultured on soft-agar medium (Table 1 and Fig. 7A). In contrast, cells expressing NS5A 1A, but not those expressing the ΔISDR nonfunctional NS5A mutant, generated solid tumors after injection into athymic mice. As noted previously, those mice receiving control P58-20 cells exhibited aggressive tumor growth (6). NS5A expression was confirmed in tumor-derived cells prepared from those tumors recovered from cells expressing NS5A 1A (Fig. 7B). This study demonstrates that constitutive expression of NS5A 1A from IFN-resistant HCV can induce malignant transformation of immortalized cells. The anchorage-independent growth observed in the ΔISDR cell lines suggests that NS5A can potentiate the immortalized phenotype of NIH 3T3 cells and stimulate cell growth through PKR-independent pathways. However, perturbation of these pathways themselves is not sufficient to induce oncogenic transformation. Importantly, our studies demonstrate that NS5A oncogenicity, defined by the ability to form tumors in vivo, required the PKR-regulatory function of NS5A.
FIG. 7.
The PKR-regulatory function of NS5A is required for solid tumor growth in vivo but not for cell growth on soft-agar medium. (A) Soft-agar colony formation of Neo control (clone 5-2; image 1), NS5A 1A (clones 4A1 and 5C6; images 2 and 4, respectively), and ΔISDR cell lines (clone 3C2; image 3). For reference, panel 1 shows a single Neo control cell. Magnification is ×100. (B) NS5A is expressed in solid tumors generated from NS5A 1A cells. Extracts prepared from Neo control (lane 1) and NS5A 1A cells (lane 2), or solid tumor-derived cells recovered from mice inoculated with the NS5A 1A 5C6 clone (lane 3), were subjected to immunoblot analysis with anti-NS5A MAb. Positions of molecular mass standards are shown in kilodaltons.
DISCUSSION
Inhibition of PKR function and rescue of viral mRNA translation in IFN-treated cells: a molecular mechanism of HCV IFN resistance.
Our results demonstrate that the NS5A protein from HCV can provide viral resistance to IFN by removing the IFN-induced, PKR-imposed block on mRNA translation during an actual viral infection. The following evidence allows us to attribute the rescue of viral mRNA translation to NS5A-mediated PKR repression: (i) NS5A from IFN-resistant HCV can physically bind PKR to repress protein kinase activity and the phosphorylation of eIF-2α (29, 32), (ii) NS5A expression resulted in a block in eIF-2α phosphorylation induced by viral infection or exogenous dsRNA, and (iii) the block in eIF-2α phosphorylation and rescue of viral mRNA translation required the PKR-binding function of NS5A and was disrupted by ISDR mutations that correspond to IFN-sensitive HCV (29, 32) (Fig. 2 to 4 and 6). These observations support a recent study, which demonstrated that the inducible expression of NS5A could provide IFN resistance to encephalomyocarditis virus and VSV, as determined by significant increases in viral titer (68). We note that in addition to the processes described here, HCV may mediate IFN resistance through other mechanisms, including those that are independent of NS5A or PKR. In this regard, HCV infection may block the PKR-dependent activation of NF-κB and IRF-1 induced by IFN, resulting in an attenuated IFN response (42, 45, 46). Recent results from our laboratory suggest that NS5A may disrupt virus-induced activation of the ERK protein kinases to possibly block induction of STAT activity and the transcription of IFN-stimulated genes (83). Moreover, the HCV core protein has been shown to suppress the production of IFN and other immunomodulatory cytokines during infection, possibly contributing to HCV IFN resistance and suppression of the virus-specific cytotoxic T-lymphocyte response.
Maintaining the translational competence of the host cell is critical for HCV, which replicates at an extremely high rate that can average in excess of 1012 virion particles/day/ml of blood examined (63). Coupled with the high rate of virion production, the pressure exerted by administration of therapeutic doses of IFN is a major factor in the generation of HCV quasispecies diversity and viral fitness. Compared to those viral quasispecies that failed to respond to IFN therapy, IFN-sensitive quasispecies of HCV 1B have been shown to harbor mutations within the ISDR, an important region of the PKR-binding domain of NS5A (23, 29). Though the majority of these studies have been conducted within Japanese patient populations (where this correlation remains strong [34]), it should be noted that these observations may be controversial, as the correlation between ISDR mutations and IFN sensitivity has not been reliably reproduced in patient populations outside Japan (35, 41, 67, 90). However, our results suggest that ISDR mutations may confer IFN sensitivity to viral replication by rendering NS5A unable to repress PKR (Fig. 3 and 4). Moreover, previous analyses of NS5A, representing a limited subset of IFN-sensitive HCV quasispecies, revealed that ISDR and PKR-binding domain mutations or deletions abolish the ability of NS5A to bind and inhibit PKR (29, 32). It is noteworthy that recent analyses of HCV dynamics during IFN therapy indicated that IFN functions to block de novo virion production of IFN-sensitive quasispecies (63), consistent with the antiviral actions of PKR. As demonstrated here with VSV, our studies indicate that the level of HCV mRNA translation, and hence viral persistence, would be severely compromised during IFN therapy by quasispecies mutations that abolish NS5A function.
Disruption of PKR-dependent apoptosis is associated with the IFN-resistant phenotype of HCV: implications for viral persistence.
Evasion of host apoptosis is an important element by which viruses maintain persistent infection. Here we have shown that NS5A from IFN-resistant HCV can disrupt dsRNA-induced host apoptotic signaling by inhibiting PKR. Disruption of host dsRNA signaling may be critical for HCV, which has the potential to activate PKR through interactions with stem-loop dsRNA structures located within the 5′ and 3′ untranslated regions of the HCV genome (10, 50). NS5A inhibition of PKR and the resulting block in eIF-2α phosphorylation may therefore allow HCV to avoid host apoptosis induced by viral dsRNA.
Activation of PKR and suppression of cellular mRNA translation are necessary for initiation of apoptotic programs induced by dsRNA and such proinflammatory mediators as bacterial endotoxin and tumor necrosis factor alpha (9, 18, 81, 87). In combination with the transcriptional regulation of apoptotic effector genes, such as Fas/Apo1, FADD, and BAX, eIF-2α phosphorylation is thought to promote apoptosis by limiting mRNA expression and the synthesis of protective, anti-apoptotic gene products (4, 18, 81). Recent evidence indicates that PKR can signal apoptosis through FADD-dependent mechanisms (4), suggesting that NS5A may additionally allow HCV to avoid dsRNA-independent mechanisms of apoptosis by blocking death receptor signaling cascades. It is important to note that the HCV core protein has been shown to potentiate death receptor signaling and apoptosis in response to tumor necrosis factor alpha (88, 91). Inhibition of PKR function by NS5A may counteract the apoptotic potential of both HCV dsRNA and the viral core protein, thereby blocking the initiation of PKR-dependent host antiviral programs during HCV infection. The ability to block dsRNA apoptotic signaling required an intact PKR-binding domain on NS5A (Fig. 5 and 6), confirming that NS5A blocks dsRNA-induced apoptosis at the level of PKR activity. Taken together, our data provide evidence that HCV evasion of dsRNA-induced host apoptosis may be limited to those viral quasispecies that can inhibit PKR. We propose that NS5A inhibition of PKR links IFN resistance with the ability of HCV to evade host apoptosis and thereby establish persistent infection. This idea is supported by the observations that viral persistence is not a common feature in infections with HCV genotypes 2 to 6 (53), which collectively exhibit a higher response rate to IFN therapy (3, 59). Accordingly, IFN resistance may now define a major determinant in the progression from acute to persistent HCV infection (51, 59).
Disruption of PKR function by NS5A links IFN resistance with the oncogenic potential of HCV.
HCV RNA is present in a high frequency of liver tumors found in patients with chronic HCV infection (33, 73), and recent studies have identified HCV sequences in non-Hodgkin’s lymphoma B cells of HCV carriers (61). These studies implicate HCV in the etiology of virus-related malignancy (19). However, the molecular mechanisms underlying the oncogenic potential of HCV remain unclear. Others have shown that the NS3 and core proteins of HCV have oncogenic potential when overexpressed in NIH 3T3 cells (13, 70, 74). Work by Ray et al. (69, 71) suggests that the oncogenic potential of the HCV core protein may reside within its ability to repress transcription from the p53 and p21WAF1/Cip1/Sid1 promoters. However, evidence that the viral core protein can potentiate Fas-induced apoptosis (72) and enhance cell death signaling though interactions with members of the tumor necrosis factor receptor family suggests that this viral protein may also have antiproliferative properties (54, 88, 91). Here we provide evidence supportive of a role for NS5A in HCV-related cellular proliferative disorders. We have demonstrated that expression of NS5A from IFN-resistant HCV, and constitutive inhibition of PKR, can induce a transformed phenotype in murine NIH 3T3 cell lines (Table 1). This is consistent with previous work from our laboratory and others demonstrating that disruption of eIF-2α phosphorylation through expression of an S51A eIF-2α mutant, dominant-negative PKR, or cellular PKR inhibitors could induce malignant transformation of NIH 3T3 cells (6–8, 44). Taken together, these studies indicate that PKR exerts its antiproliferative effects, at least in part, by phosphorylating serine 51 of eIF-2α and limiting mRNA translation.
Recent work suggests that PKR activity is strictly regulated during the cell division cycle (27, 89). Moreover, Aktas et al. (1) have demonstrated that PKR-mediated eIF-2α phosphorylation is required for the control of cyclin D1 translation and G1 cell cycle arrest that occur in response to intracellular calcium depletion and activation of PKR (80). In accordance with these observations, Balachandran and colleagues (4) demonstrated that inducible expression of PKR results in altered cell cycle kinetics, accumulation of cells in G1, and potentiation of apoptosis induced by dsRNA. In contrast, loss of PKR function or abrogation of eIF-2α phosphorylation induced oncogenic transformation (5, 6, 8, 21) and rendered cells refractory to apoptosis induced by PKR agonists, including dsRNA (4, 18, 81). Thus, the PKR pathway may regulate cell growth, in part, by enforcing a translational control or apoptotic checkpoint on cell proliferation. By this model, oncogenic potential is conferred to NS5A through disruption of the PKR checkpoint. In addition, a recent analysis of liver tissue from HCV-infected patients has documented an aberrantly low level of apopototic nuclei within infected hepatocellular tumors, though PKR levels remained relatively high (78). Taken together, these results suggest that PKR-dependent antitumor programs may be blocked in HCV-infected cells. We cannot formally exclude the possibility that NS5A transforms cells through pathways that are independent of eIF-2α or PKR. Indeed, our analyses revealed that NS5A has growth-stimulatory potential that is independent of the ability to regulate PKR (Table 1). It is thus interesting that the carboxyl-terminal region of NS5A may function as a viral transactivator of transcription (26, 39). Though the relevance of NS5A transactivation function remains unclear, the possibility remains that it affects cell growth processes to stimulate cell proliferation. Importantly, however, we emphasize that the oncogenic potential of NS5A, defined by tumor induction in vivo, resides in its ability to repress PKR-mediated eIF-2α phosphorylation. Thus, our results provide evidence to link the oncogenic potential of HCV with viral persistence and resistance to IFN therapy, through NS5A-mediated inhibition of PKR.
ACKNOWLEDGMENTS
We are grateful to Dagma Daniel for excellent administrative support. We thank T. Imagawa (Osaka University) for antibody to NS5A, M. Wambach and N. Tang for outstanding technical assistance, P. Marcus and M. Sekellick for providing VSV stock, and S. Polyak for initially providing the NS5A 1B clone. We thank M. Korth and C. Blakely for helpful discussions and critical review of the manuscript.
This work was supported in part by National Institutes of Health grants AI22646, RR00166, and AI41629 (M.G.K.) and the Helen Hay Whitney Foundation (M.G.).
REFERENCES
- 1.Aktas H, Fluckiger R, Acosta J, Savage J, Palakurthi S, Halperin J. Depletion of intracellular calcium stores, phosphorylation of eIF-2α, and sustained inhibition of translation mediate the anticancer effects of clotrimazole. Proc Natl Acad Sci USA. 1998;95:8280–8285. doi: 10.1073/pnas.95.14.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter M J H. Epidemiology of hepatitis C. Hepatology. 1997;26:62–65. doi: 10.1002/hep.510260711. [DOI] [PubMed] [Google Scholar]
- 3.Amoroso P, Rapicetta M, Tosti M E M A, Spada E, Bounocore S, Lettieri G, Peirri P, Chionne P, Ciccaglione A R, Sagliocca L. Correlation between virus genotype and chronicity rate in acute hepatitis C. J Hepatol. 1998;28:939–944. doi: 10.1016/s0168-8278(98)80340-1. [DOI] [PubMed] [Google Scholar]
- 4.Balachandran S, Kim C N, Yeh W-C, Mak T W, Barber G N. Activation of the dsRNA-dependent protein kinase, PKR, induces apoptosis through FADD-mediated death signaling. EMBO J. 1998;17:6888–6902. doi: 10.1093/emboj/17.23.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barber G N, Jagus R, Meurs E F, Hovanessian A G, Katze M G. Molecular mechanisms responsible for malignant transformation by regulatory and catalytic domain variants of the interferon-induced enzyme RNA-dependent protein kinase. J Biol Chem. 1995;270:17423–17428. doi: 10.1074/jbc.270.29.17423. [DOI] [PubMed] [Google Scholar]
- 6.Barber G N, Thompson S, Lee T G, Strom T, Jagus R, Darveau A, Katze M G. The 58-kilodalton inhibitor of the interferon-induced double-stranded RNA-activated protein kinase is a tetratricopeptide repeat protein with oncogenic properties. Proc Natl Acad Sci USA. 1994;91:4278–4282. doi: 10.1073/pnas.91.10.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barber G N, Wambach M, Thompson S, Jagus R, Katze M G. Mutants of the RNA-dependent protein kinase (PKR) lacking double-stranded RNA binding domain I can act as transdominant inhibitors and induce malignant transformation. Mol Cell Biol. 1995;15:3138–3146. doi: 10.1128/mcb.15.6.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benkirane M, Neuveut C, Chun R F, Smith S M, Samuel C E, Gatignol A, Jeang K-T. Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J. 1997;16:611–624. doi: 10.1093/emboj/16.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beyaert R, Fiers W. Molecular mechanisms of tumor necrosis factor-induced cytotoxicity. What we do understand and what we do not. FEBS Lett. 1994;340:9–16. doi: 10.1016/0014-5793(94)80163-0. [DOI] [PubMed] [Google Scholar]
- 10.Blight K J, Rice C M. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1997;71:7345–7352. doi: 10.1128/jvi.71.10.7345-7352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruno S, Silini E, Crosignani A, Borzio F, Leandro G, Bono F, Asti M, Rossi S, Larghi A, Cerino A, Podda M, Mondelli M U. Hepatitis C virus genotypes and risk of hepatocellular carcinoma in cirrhosis: a prospective study. Hepatology. 1997;25:754–758. doi: 10.1002/hep.510250344. [DOI] [PubMed] [Google Scholar]
- 12.Bukh J, Miller R, Purcell R. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin Liver Dis. 1995;15:41–63. doi: 10.1055/s-2007-1007262. [DOI] [PubMed] [Google Scholar]
- 13.Chang J, Yang S-H, Cho Y-G, Hwang S B, Hahn Y S, Sung Y C. Hepatitis C virus core from two different genotypes has an oncogenic potential but is not sufficient for transforming primary rat embryo fibroblasts in cooperation with the H-ras oncogene. J Virol. 1998;72:3060–3065. doi: 10.1128/jvi.72.4.3060-3065.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chayama K, Tsubota A, Kobayashi M, Okamoto K, Hashimoto M, Miyano Y, Koike H, Koida I, Arase Y, Saitoh S, Suzuki Y, Murashima N, Ikeda K, Kumada H. Pretreatment virus load and multiple amino acid substitutions in the interferon sensitivity-determining region predict the outcome of interferon treatment in patients with chronic genotype 1b hepatitis C virus infection. Hepatology. 1997;25:745–749. doi: 10.1002/hep.510250342. [DOI] [PubMed] [Google Scholar]
- 15.Chen J-J, London I M. Regulation of protein synthesis by heme-regulated eIF-2α kinase. Trends Biochem Sci. 1995;20:105–108. doi: 10.1016/s0968-0004(00)88975-6. [DOI] [PubMed] [Google Scholar]
- 16.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 17.Clemens M J, Elia A. The double-stranded RNA-dependent protein kinase PKR: structure and function. J Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 18.Der S D, Yang Y-L, Weissman C, Williams B R G. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:3279–3283. doi: 10.1073/pnas.94.7.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Bisceglie A M. Hepatitis C and hepatocellular carcinoma. Semin Liver Dis. 1995;15:64–69. doi: 10.1055/s-2007-1007263. [DOI] [PubMed] [Google Scholar]
- 20.Domingo E, Baranowski E, Ruiz-Jarabo C M, Martin-Hernandez A M, Saiz J C, Escarmis C. Quasispecies structure and persistence of RNA viruses. Emerging Infect Dis. 1998;4:521–527. doi: 10.3201/eid0404.980402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donzé O, Jagus R, Koromilas A E, Hershey J W B, Sonenberg N. Abrogation of translation initiation factor eIF-2 phosphorylation causes malignant transformation of NIH 3T3 cells. EMBO J. 1995;14:3828–3834. doi: 10.1002/j.1460-2075.1995.tb00052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duarte E A, Novella I S, Weaver S C, Domingo E, Wain-Hobson S, Clarke D K, Moya A, Elena S F, de la Torre J C, Holland J J. RNA virus quasispecies: significance for viral disease and epidemiology. Infect Agents Dis. 1994;3:201–214. [PubMed] [Google Scholar]
- 23.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Maruno F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 24.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murankami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. J Clin Investig. 1995;96:224–230. doi: 10.1172/JCI118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernadez I, Castellano G, Domingo M J, Fuertes A, Colina F, Canga F, DelaCruz F J, DelaCamara A G, Solis J A. Influence of viral genotype and the level of viremia on the severity of liver injury and the response to interferon therapy. Scand J Gastroenterol. 1997;32:7–76. doi: 10.3109/00365529709025066. [DOI] [PubMed] [Google Scholar]
- 26.Fukuma T, Enomoto N, Marumo F, Sato C. Mutations in the interferon-sensitivity determining region of hepatitis C virus and transcriptional activity of the nonstructural region 5A protein. Hepatology. 1998;28:1147–1153. doi: 10.1002/hep.510280433. [DOI] [PubMed] [Google Scholar]
- 27.Gale, M., Jr., C. Zhou, E. J. Firpo, M. G. Katze, A. Rudendsky, and B. R. Franza, Jr. 1999. Unpublished observations.
- 28.Gale M, Jr, Blakely C M, Hopkins D A, Melville M W, Wambach M, Romano P R, Katze M G. Regulation of interferon-induced protein kinase PKR: modulation of P58IPK inhibitory function by a novel protein, P52rIPK. Mol Cell Biol. 1998;18:859–871. doi: 10.1128/mcb.18.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gale M, Jr, Blakely C M, Kwieciszewski B, Tan S-L, Dossett M, Korth M J, Polyak S J, Gretch D R, Katze M G. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gale M, Jr, Katze M G. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol Ther. 1998;78:29–46. doi: 10.1016/s0163-7258(97)00165-4. [DOI] [PubMed] [Google Scholar]
- 31.Gale M, Jr, Korth M J, Katze M G. Repression of the PKR protein kinase by hepatitis C virus: a potential mechanism for interferon resistance. Clin Diagn Virol. 1998;10:157–162. doi: 10.1016/s0928-0197(98)00034-8. [DOI] [PubMed] [Google Scholar]
- 32.Gale M, Jr, Korth M J, Tang N M, Tan S-L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 33.Haruna Y, Hayashi N, Kamada T, Hytiroglou P, Thung S N, Gerber M A. Expression of hepatitis C virus in hepatocellular carcinoma. Cancer. 1994;73:2253–2258. doi: 10.1002/1097-0142(19940501)73:9<2253::aid-cncr2820730904>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 34.Herion D, Hoofnagle J. The interferon sensitivity determining region: all hepatitis C virus isolates are not the same. Hepatology. 1997;25:769–771. doi: 10.1002/hep.510250346. [DOI] [PubMed] [Google Scholar]
- 35.Hofgärtner W T, Polyak S J, Sullivan D, Carithers Jr RL, Gretch D R. Mutations in the NS5A gene of hepatitis C virus in North American patients infected with HCV genotype 1a or 1b. J Med Virol. 1997;53:118–126. doi: 10.1002/(sici)1096-9071(199710)53:2<118::aid-jmv3>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 36.Hoofnagle J H. Therapy of acute and chronic viral hepatitis. Adv Intern Med. 1994;39:241–275. [PubMed] [Google Scholar]
- 37.Iino S, Hino K, Yasuda K. Current state of interferon therapy for chronic hepatitis C. Intervirology. 1994;37:87–100. doi: 10.1159/000150362. [DOI] [PubMed] [Google Scholar]
- 38.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato N, Lan K H, Ono-Nita S K, Shiratori Y, Omata M. Hepatitis C virus nonstructural region 5A protein is a potent transcriptional activator. J Virol. 1997;71:8856–8859. doi: 10.1128/jvi.71.11.8856-8859.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katze M G. Regulation of the interferon-induced PKR: can viruses cope? Trends Microbiol. 1995;3:75–78. doi: 10.1016/s0966-842x(00)88880-0. [DOI] [PubMed] [Google Scholar]
- 41.Khorsi H, Castelain S, Wyseur A, Izopet J, Canva V, Rombout A, Capron D, Capron J-P, Lunel F, Stuyver L, Duverlie G. Mutations of hepatitis C virus 1b NS5A 2209-2248 amino acid sequence do not predict the response to recombinant interferon-alpha therapy in French patients. J Hepatol. 1997;27:72–77. doi: 10.1016/s0168-8278(97)80282-6. [DOI] [PubMed] [Google Scholar]
- 42.Kirchhoff S, Koromilas A E, Schaper F, Grashof M, Sonenberg N, Hauser H. IRF-1 induced cell growth inhibition and interferon induction requires the activity of the protein kinase PKR. Oncogene. 1995;11:439–445. [PubMed] [Google Scholar]
- 43.Koff R S. Therapy in chronic hepatitis C: say goodbye to the 6-month interferon regimen. Am J Gastroenterol. 1997;91:2072–2074. [PubMed] [Google Scholar]
- 44.Koromilas A E, Roy S, Barber G N, Katze M G, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 45.Kumar A, Haque J, Lacoste J, Hiscott J, Williams B R G. Double-stranded RNA-dependent protein kinase activates transcription factor NF-κB by phosphorylating IκB. Proc Natl Acad Sci USA. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar A, Yang Y-L, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, Williams B R G. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-κB. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurosaki M, Enomoto N, Murakami T, Sakuma I, Asahina Y, Yamamoto C, Ikeda T, Tozuka S, Izumi N, Marumo F, Sato C. Analysis of genotypes and amino acid residues 2209 to 2248 of the NS5A region of hepatitis C virus in relation to the response to interferon-β therapy. Hepatology. 1997;25:750–753. doi: 10.1002/hep.510250343. [DOI] [PubMed] [Google Scholar]
- 48.Laurent A G, Krust B, Galabru J, Svab J, Hovanessian A G. Monoclonal antibodies to interferon induced 68,000 Mr protein and their use for the detection of double-stranded RNA dependent protein kinase in human cells. Proc Natl Acad Sci USA. 1985;82:4341–4345. doi: 10.1073/pnas.82.13.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee S B, Bablanian R, Esteban M. Regulated expression of the interferon-induced protein kinase p68 (PKR) by vaccinia virus recombinants inhibits the replication of vesicular stomatitis virus but not that of poliovirus. J Interferon Cytokine Res. 1996;16:1073–1078. doi: 10.1089/jir.1996.16.1073. [DOI] [PubMed] [Google Scholar]
- 50.Lee S Y, Lui W M, Maizel J V. Phylogenetic evidence for the improved RNA higher-order structure in internal ribosome entry sequences of HCV and pestiviruses. Virus Genes. 1998;17:279–295. doi: 10.1023/a:1008073905920. [DOI] [PubMed] [Google Scholar]
- 51.Lee W M, Reddy K R, Tong M J, Black M, van Leeuwen D J, Hooinger F B, Mullem K D, Pimstone N, Albert D, Gardner S the Consensus Interferon Study Group. Early hepatitis C virus-RNA responses predict interferon treatment outcomes in chronic hepatitis C. Hepatology. 1998;28:1411–1415. doi: 10.1002/hep.510280533. [DOI] [PubMed] [Google Scholar]
- 52.Martell M, Esteban J I, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gomez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinot-Peignoux M, Marcellin P, Pouteau M, Castelnau C, Boyer N, Poliquin M, Degott C, Descombes I, Le Breton V, Milotova V, Benhamou J P, Erlinger S. Pretreatment serum hepatitis C virus RNA levels and hepatitis C virus genotype are the main and independent prognostic factors of sustained response to interferon alfa therapy in chronic hepatitis C. Hepatology. 1995;22:1050–1056. [PubMed] [Google Scholar]
- 54.Matsumoto M, Hsieh T-Y, Zhu N, VanArsdale T, Hwang S B, Gorbalenya A E, Lo S-Y, Ou J-H, Ware C F, Lai M C. Hepatitis C virus core protein interacts with the cytoplasmic tail of the lymphotoxin-β receptor. J Virol. 1997;71:1301–1309. doi: 10.1128/jvi.71.2.1301-1309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McHutchison J G, Gordon S C, Schiff E R, Shiffman M L, Lee W M, Goodman Z D, Ling M H, Cort S, Albrecht J K for the Interferon Interventional Therapy Group. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N Engl J Med. 1998;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- 56.Merrick W C, Hershey J W B. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J, Mathews M, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 31–70. [Google Scholar]
- 57.Meurs E, Chong K L, Galabru J, Thomas N, Kerr I, Williams B R G, Hovanessian A G. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 58.Meurs E F, Galabru J, Barber G N, Katze M G, Hovanessian A G. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1993;90:232–236. doi: 10.1073/pnas.90.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morisco F, Tuccillo C, Sessa G, Brunasso G, Caporaso N. Chronic hepatitis C long-term responders to human leukocyte interferon-alpha therapy: persistence of a sustained biochemical and virological response during 5 years of surveillance. Eur J Gastroenterol Hepatol. 1998;10:399–403. doi: 10.1097/00042737-199805000-00008. [DOI] [PubMed] [Google Scholar]
- 60.Neddermann P, Tomei L, Steinkühler C, Gallinari P, Tramontano A, De Francesco R. The nonstructural proteins of the hepatitis C virus: structure and functions. Biol Chem. 1997;378:469–476. [PubMed] [Google Scholar]
- 61.Negro F, Levrero M. Does hepatitis C virus replicate in cells of the hematopoetic lineage? Hepatology. 1998;28:261–264. doi: 10.1002/hep.510280134. [DOI] [PubMed] [Google Scholar]
- 62.Nelson D R, Lau J Y N. Pathogenesis of chronic hepatitis C virus infection. Antiviral Ther. 1999;3:25–35. [PubMed] [Google Scholar]
- 63.Neumann A U, Lam N P, Dahari H, Gretch D R, Wiley T E, Layden T J, Perelson A S. Hepatitis C virus dynamic in vivo and the antiviral efficacy of interferon-α therapy. Science. 1999;282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 64.O’Brien V. Viruses and apoptosis. J Gen Virol. 1998;79:1833–1845. doi: 10.1099/0022-1317-79-8-1833. [DOI] [PubMed] [Google Scholar]
- 65.Ohsawa M, Shingu N, Miwa H, Yoshihara H, Kubo M, Tsukuma H, Teshima H, Hashimoto M, Aozasa K. Risk of non-Hodgkin’s lymphoma in patients with hepatitis C virus infection. Int J Cancer. 1999;80:237–239. doi: 10.1002/(sici)1097-0215(19990118)80:2<237::aid-ijc12>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 66.Okuda K. Hepatitis C and hepatocellular carcinoma. J Gastroenterol Hepatol. 1998;13:294–298. doi: 10.1111/j.1440-1746.1998.tb01896.x. [DOI] [PubMed] [Google Scholar]
- 67.Pawlotsky J-M, Germanidis G, Neumann A U, Pellerin M, Frainais P-O, Dhumeaux D. Interferon resistance of hepatitis C virus genotype 1b: relationship to nonstructural 5A gene quasispecies mutations. J Virol. 1998;72:2795–2805. doi: 10.1128/jvi.72.4.2795-2805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Polyak S J, Paschal D, McArdla S, Gale M, Jr, Moradpour D, Gretch D R. Characterization of the effects of hepatitis C virus nonstructural 5A protein expression in human cell lines and on interferon-sensitive virus replication. Hepatology. 1999;29:1262–1270. doi: 10.1002/hep.510290438. [DOI] [PubMed] [Google Scholar]
- 69.Ray R B, Steele R, Meyer K, Ray R. Hepatitis C virus core protein represses p21(WAF1/Cip1/Sid1) promoter activity. Gene. 1998;208:331–336. doi: 10.1016/s0378-1119(98)00030-4. [DOI] [PubMed] [Google Scholar]
- 70.Ray R B, Lagging L M, Meyer K, Ray R. Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J Virol. 1996;70:4438–4443. doi: 10.1128/jvi.70.7.4438-4443.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ray R B, Steele R, Meyer K, Ray R. Transcriptional repression of p53 promoter by hepatitis C virus core protein. J Biol Chem. 1997;272:10983–10986. doi: 10.1074/jbc.272.17.10983. [DOI] [PubMed] [Google Scholar]
- 72.Ruggieri A, Harada T, Matsuura Y, Miyamura T. Sensitization to Fas-mediated apoptosis by hepatitis C virus core protein. Virology. 1997;229:68–76. doi: 10.1006/viro.1996.8420. [DOI] [PubMed] [Google Scholar]
- 73.Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, et al. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sakamuro D, Furukawa T, Takegami T. Hepatitis C virus nonstructural protein NS3 transforms NIH 3T3 cells. J Virol. 1995;69:3893–3896. doi: 10.1128/jvi.69.6.3893-3896.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Savinova O, Jagus R. Use of vertical slab isoelectric focusing and immunoblotting to evaluate steady-state phosphorylation of eIF-2α in cultured cells. Methods Companion Methods Enzymol. 1997;11:419–425. doi: 10.1006/meth.1996.0438. [DOI] [PubMed] [Google Scholar]
- 76.Sen G C, Ransohoff R M. Interferon-induced antiviral actions and their regulation. Adv Virus Res. 1993;42:57–102. doi: 10.1016/s0065-3527(08)60083-4. [DOI] [PubMed] [Google Scholar]
- 77.Sharara A I, Hunt C M, Hamilton J D. Hepatitis C. Ann Intern Med. 1996;125:658–668. doi: 10.7326/0003-4819-125-8-199610150-00006. [DOI] [PubMed] [Google Scholar]
- 78.Shimada A, Shiota G, Miyatya H, Kamahora T, Kawasaki H, Shiraki K, Hino S, Terada T. Aberrant expression of double-stranded RNA-dependent protein kinase in hepatocytes of chronic hepatitis and differentiated hepatocellular carcinoma. Cancer Res. 1998;58:4434–4438. [PubMed] [Google Scholar]
- 79.Shors S T, Beattie E, Paoletti K, Tartaglia J, Jacobs B L. Role of the vaccinia virus E3L and K3L gene products in rescue of VSV and EMCV from the effects of IFN-alpha. J Interferon Cytokine Res. 1998;18:721–729. doi: 10.1089/jir.1998.18.721. [DOI] [PubMed] [Google Scholar]
- 80.Srivastava S P, Davies M V, Kaufman R J. Calcium depletion from the endoplasmic reticulum activates the double-stranded RNA-dependent protein kinase (PKR) to inhibit protein synthesis. J Biol Chem. 1995;270:16619–16624. doi: 10.1074/jbc.270.28.16619. [DOI] [PubMed] [Google Scholar]
- 81.Srivastava S P, Kumar K U, Kaufman R J. Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J Biol Chem. 1998;273:2416–2423. doi: 10.1074/jbc.273.4.2416. [DOI] [PubMed] [Google Scholar]
- 82.Tan, S.-L., and M. G. Katze. The emerging role of the interferon-induced PKR protein kinase as an apoptotic effector: a new face of death? J. Interferon Cytokine Res., in press. [DOI] [PubMed]
- 83.Tan S-L, Nakao H, He Y, Vijaysri V, Neddermann P, Jacobs B L, Mayer B J, Katze M G. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc Natl Acad Sci USA. 1999;96:5533–5538. doi: 10.1073/pnas.96.10.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang N M, Ho C Y, Katze M G. The 58-kDa cellular inhibitor of the double stranded RNA-dependent protein kinase requires the tetratricopeptide repeat 6 and DnaJ motifs to stimulate protein synthesis in vivo. J Biol Chem. 1996;271:28660–28666. doi: 10.1074/jbc.271.45.28660. [DOI] [PubMed] [Google Scholar]
- 85.Teodoro J G, Branton P E. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Trepo C, Berthillon P, Vitvitski L. HCV and lymphoproliferative diseases. Ann Oncol. 1998;9:469–470. doi: 10.1023/a:1008298228620. [DOI] [PubMed] [Google Scholar]
- 87.Yang Y-L, Reis L F L, Pavlovic J, Aguzzi A, Schäfer R, Kumar A, Williams B R G, Aguet M, Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.You L R, Chen C M. Hepatitis C virus core protein enhances NF-κB signal pathway triggering by lymphotoxin-beta receptor ligand and tumor necrosis factor alpha. J Virol. 1999;73:1672–1681. doi: 10.1128/jvi.73.2.1672-1681.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zamanian-Daryoush M, Der S D, Williams B R G. Cell cycle regulation of the double stranded RNA activated protein kinase, PKR. Oncogene. 1999;17:315–326. doi: 10.1038/sj.onc.1202293. [DOI] [PubMed] [Google Scholar]
- 90.Zeuzem S, Lee J-H, Roth W K. Mutations in the nonstructural 5A gene of European hepatitis C virus isolates and response to interferon alfa. Hepatology. 1997;25:740–744. doi: 10.1002/hep.510250341. [DOI] [PubMed] [Google Scholar]
- 91.Zhu N, Khoshan A, Schneider R, Matsumoto M, Dennert G, Ware C F, Lai M C. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J Virol. 1998;72:3691–3697. doi: 10.1128/jvi.72.5.3691-3697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]