Abstract
The avian adenovirus CELO can, like the human adenoviruses, transform several mammalian cell types, yet it lacks sequence homology with the transforming, early regions of human adenoviruses. In an attempt to identify how CELO virus activates the E2F-dependent gene expression important for S phase in the host cell, we have identified two CELO virus open reading frames that cooperate in activating an E2F-inducible reporter system. The encoded proteins, GAM-1 and Orf22, were both found to interact with the retinoblastoma protein (pRb), with Orf22 binding to the pocket domain of pRb, similar to other DNA tumor virus proteins, and GAM-1 interacting with pRb regions outside the pocket domain. The motif in Orf22 responsible for the pRb interaction is essential for Orf22-mediated E2F activation, yet it is remarkably unlike the E1A LxCxD and may represent a novel form of pRb-binding peptide.
Avian adenoviruses resemble human adenoviruses in many respects (37). Both adenovirus types form icosahedral capsids of 70 to 75 nm, with hexons and pentons as the major subunits (12, 13, 24, 32, 35, 38, 43, 47). The viral capsids contain a linear double-stranded DNA molecule that is associated with viral core proteins (34). Replication and viral assembly occur in the nucleus of the infected cell (8, 36, 46).
The chicken embryo lethal orphan (CELO) virus is representative of the type 1 fowl adenoviruses. Characterization of CELO virus is of importance both for the potential application of the virus as a vaccine vector and for its use as a novel adenovirus serotype for gene transfer. CELO virus was the first avian adenovirus with a fully characterized viral genome (6). Sequence determination of additional avian adenovirus serotypes is revealing a substantial and unexpected diversity in these viruses (e.g., egg drop syndrome virus [21] and hemorrhagic enteritis virus [44]). The CELO virus genome has substantial homology with mastadenovirus genomes over regions encoding replication functions (E2 region) and capsid proteins (late genes) which constitute the central part of the viral genome (6). No significant homology exists to the mastadenovirus early regions E1, E3, and E4 (6). The leftmost 5 kb and the rightmost 13 kb of the CELO virus genome were identified as being unique to CELO virus (Fig. 1). The similar life cycles of human and avian adenoviruses suggest a conservation of basic viral functions and regulation mechanisms. For example, it was assumed that an adenovirus should possess a gene that impairs the host apoptotic response. Using a screen for such antiapoptotic functions, GAM-1 was identified as a functional homolog of human adenovirus E1B 19K protein (5). The present report describes our efforts to identify an E1A-like, E2F-activating function in the CELO virus genome.
FIG. 1.
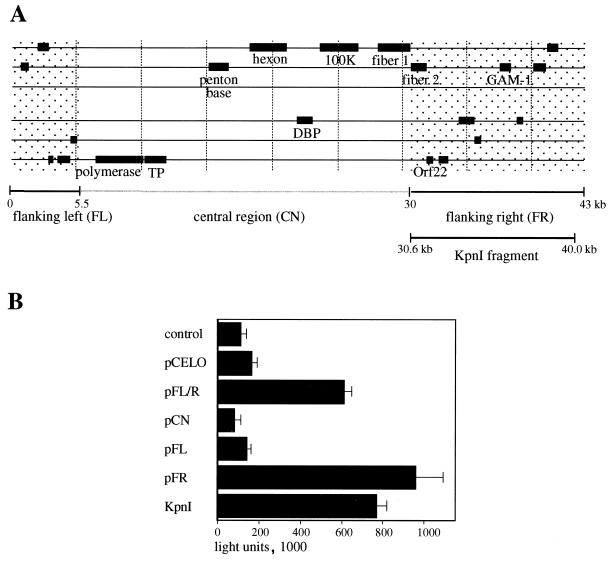
E2F activation assay with different CELO virus constructs. (A) Schematic organization of the CELO virus genome. Some of the conserved E2 and late genes are shown. The shaded regions flanking the central part (CN) on the left and right (FL and FR) indicate sequences that are unique to CELO virus. (B) Reporter construct E2-Luc (0.2 μg) was transfected into CEF cells as described in Materials and Methods (control lane). For cotransfection assays, 0.4 μg of the following plasmids were transfected in addition to E2-Luc: pCELO (full-length CELO virus sequence), pFL/R (kb 0 to 5.5) and 30 to 43 of CELO sequence), pCN (kb 5.5 to 30 of CELO sequence), pFL (kb 0 to 5.5 of CELO sequence), pFR (kb 30 to 43 of CELO sequence), and KpnI (kb 30.6 to 40.0 of CELO sequence); 24 h after transfection, 20 μl of cell lysate was analyzed for luciferase activity.
The E1A gene of human adenovirus is the first gene that is expressed during an adenovirus infection (33, 41). E1A promotes expression of several viral transcription units (E2, E4, and late genes) primarily by recruiting components of the cellular transcription machinery. E1A also influences key targets of the host cell. One of these cellular targets is the retinoblastoma protein (pRb), an important regulator of G1-to-S-phase entry during the cell cycle (10, 14, 15, 51, 52). Binding of pRb by E1A is thought to release the cellular transcription factor E2F and thus activate important S-phase-specific genes. This allows progression of the host cell into S phase and therefore supplies the virus with cellular metabolites that are essential for the efficient replication of the viral genome.
Because of the importance of moderating the inhibitory effects of pRb on E2F function, all small DNA viruses characterized to date have been found to encode proteins that influence this pathway. To identify regions in the CELO virus genome that encode an activity analogous to the E1A region of human adenoviruses, we designed a screen for E2F activation. The adenovirus type 5 (Ad5) E2 promoter strongly depends on E2F for its function (28). We cloned this promoter upstream of a luciferase cDNA and used the resulting construct to search for open reading frames in the CELO virus genome that could upregulate this promoter resulting in increased luciferase expression. We identified two CELO virus gene products that were able to activate this E2F-dependent promoter. The first is a product of open reading frame 22 (Orf22) which thus far has not been characterized. A second CELO protein, GAM-1, was also found to activate the E2F pathway. The two proteins synergize in E2F activation. Consistent with the E2F activation, both proteins were found to bind to pRb, albeit with distinct sites on the cellular protein. However, neither the Orf22 protein nor GAM-1 exhibit any significant sequence homology to human adenovirus E1A proteins, and the two proteins appear to define novel types of pRb-binding partners.
MATERIALS AND METHODS
Cells and transfection.
The chicken embryonic fibroblast (CEF) cell line was grown in Dulbecco’s modified Eagle medium (DMEM; BioWhittaker) containing 8% fetal calf serum and 2% chicken serum. A549 cells and Leghorn male hepatoma (LMH) cells were grown in DMEM containing 10% fetal calf serum. Both media were supplemented with 100 μg each of streptomycin and penicillin per ml.
Transfection complexes were prepared by the polyethyleneimine (PEI) technique (1, 3). Cells were seeded 1 to 2 days before transfection either in 24-well plates at a density of 4 × 104 cells/well or in 6-well plates at a density of 4 × 105 cells/well. Transfection was carried out when cells had reached 80 to 90% confluence. For 24-well plates, 50 μl of transfection complex (containing 0.6 μg of DNA) was added in 250 μl of serum-free medium; for transfection in 6-well plates, 250 μl of complex containing 3 μg of plasmid DNA was used in 1 ml of serum-free medium. Cells were incubated with complexes for 4 h, after which the serum-free medium was replaced by normal growth medium.
For luciferase assays, 4 × 104 cells were lysed in 150 μl of lysis buffer (0.25 M Tris buffer [pH 7.5], 1% Triton X-100), of which 40 μl was measured in a Berthold luminometer as previously described (9).
Generation of antiserum.
For the generation of antibodies against Orf22 protein, rabbits were immunized with a peptide homologous to amino acids (aa) 13 to 30 of the protein (HQQ RRQ QEA ERE EEV GDD C). Antibodies against CELO virus late proteins were raised by infecting rabbits with inactivated CELO virus particles (39).
Plasmids.
pE2-Luc was constructed by cloning the PvuII fragment from Ad5 containing the E2A promoter (nucleotides 26990 to 28837) upstream from a luciferase cDNA-simian virus 40 intron/polyadenylation signal. The pwtE2F-Luc construct contains three synthetic E2F-binding sites upstream from a minimal promoter-luciferase cDNA-simian virus 40 intron/polyadenylation signal, while pmuE2F-Luc carries mutations in the E2F-binding sites. Both constructs were gifts from Wilhelm Krek, Friedrich Miescher Institute, Basel, Switzerland, and are described elsewhere (29). Plasmid pCELO contains the entire CELO virus genome flanked by SpeI sites cloned into a pBR327 derivative as described elsewhere (39). The construct pFL/R carrying the left- and right-end sequences which are unique to CELO virus was generated by deleting central regions of pCELO with an HpaI digest and subsequent religation; pFL/R thus contains only the left HpaI fragment of CELO virus (bp 1 to 5503) and the right fragment (bp 30502 to 43804). The separate constructs of plasmids pFL and pFR resulted from digestion of plasmid pFL/R with HpaI and SpeI and ligation of the resulting fragments into pBluescript. The central region pCN was recovered as a 26,620-bp XbaI fragment (bp 1989 to 28608) of pCELO cloned into pBR327. Deletion constructs named by restriction enzymes refer to fragments that were released by the specified enzymes, which were then ligated into pBluescript (Stratagene). Myc-tagged versions of Orf22 and E1A (pSG9MOrf22 and pSG9ME1A) were generated by PCR amplification of their reading frames followed by cloning into pSG9M (20). Construction of Myc-tagged GAM-1 and E1A 19K plasmids was described previously (5). Constructs of Orf22 with N- and C-terminal deletions were generated by PCR using primers that carried either a new starting site for translation or a premature stop codon. Amplified fragments were cloned into the EcoRV site of pSG9M. Correct orientation was determined by restriction analysis and sequencing. The human pRb (hRb) construct pCMV-hRb, a gift from Meinrad Busslinger and Dirk Eberhard, Institute of Molecular Pathology, Vienna, Austria, contains the full-length hRb sequence under the control of a cytomegalovirus promoter. The glutathione-S-transferase (GST)–pRb fusion plasmids GST-Rb, GST-Rb-Δ21, and GST-Rb-ΔC are described in reference 27.
Phage mutagenesis.
Alteration of the LxCxD coding motif of Orf22 was performed by phage mutagenesis (30, 31). All mutants were analyzed for the presence of the expected mutations by restriction digests and sequencing.
Immunofluorescence.
CEF cells were plated on glass slides at a density of 3 × 105 cells/well and transfected with 3 μg of plasmid DNA. The next day, cells were fixed in 4% formaldehyde (in phosphate-buffered saline) and then incubated in 0.25% Triton X-100. Nonspecific binding was blocked with 5% nonfat milk. Purified anti-Myc antibody 9E10 (16) (Calbiochem) was added at a dilution of 1:100 in blocking solution for 1 h. After repeated washing with blocking solution, an anti-mouse antibody coupled to fluorescein isothiocyanate (DAKO) was added at a dilution of 1:40 for 1 h. Slides were repeatedly washed with phosphate-buffered saline; 4′,6-diamidino-2-phenylindole (DAPI) stain was included in the last washing step to visualize the nuclear DNA. Slides were mounted with Mowiol and analyzed by fluorescence microscopy.
Time course of CELO virus infection.
LMH cells were seeded at a density of 3 × 105 cells/well in six-well plates. Infection with CELO virus was carried out with 1,000 virus particles per cell in DMEM without serum. Where indicated, 1-β-arabinofuranosylcytosine (AraC; Sigma) was added at a final concentration of 20 μg/ml to block adenovirus DNA replication (19). Infected cells were harvested at the indicated time points and analyzed by Western blotting.
Immunoprecipitation.
CEF cells were seeded in six-well plates and transfected with 3 μg of plasmid DNA. Cells were lysed in 600 μl of lysis buffer (150 mM NaCl, 50 mM Tris-buffer [pH 8.0], 5 mM EDTA, 1% Nonidet P-40) 48 h after transfection. Insoluble material was spun down, and the supernatant was preadsorbed with 10 μl of protein A/G-agarose (Calbiochem). The precleared material was incubated with 0.1 to 1 μg of specific antibody for at least 4 h, after which 15 μl of protein A/G-agarose was added and the mixture was incubated for an additional 2 h. The complexed material was pelleted and repeatedly washed with lysis buffer containing 150 or 500 mM NaCl. The resulting pellet was resuspended in 25 μl of sodium dodecyl sulfate (SDS)-sample buffer and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE).
GST-Rb binding assay.
Escherichia coli BL21 (Stratagene) was transformed with GST-Rb constructs and grown as fresh overnight cultures in LB medium containing ampicillin (100 μg/ml). Dilutions (1:20) of this starting material were grown to an optical density at 600 nm of 0.5 to 0.7 and induced with isopropyl-β-d-thiogalactopyranoside at a final concentration of 0.1 mM. After 3 h of induction, cells were harvested and lysed on ice in 1/10 volume of NETN buffer (20 mM Tris buffer [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40) supplemented with a protease inhibitor cocktail (Sigma). The supernatant was recovered after centrifugation at 10,000 × g for 5 min, and aliquots (600 μl) were incubated with 20 μl of equilibrated glutathione-Sepharose (Pharmacia). Samples were gently agitated at 4°C for 30 min and then washed with NETN buffer containing 0.5% milk powder. To screen for binding partners of pRb, CEF cells were transfected with the constructs of interest. Expression levels of the various constructs were analyzed by Western blotting. On the basis of these results, equal amounts of expressed proteins were incubated with recombinant GST-Rb molecules for 1 h at 4°C. After repeated washing with NETN buffer, the pellets were resuspended in sample buffer and analyzed by SDS-PAGE (for more details, see reference 27).
Peptide competition assay.
The peptides were dissolved in 50% dimethyl sulfoxide-HBS (HEPES-buffered saline [150 mM NaCl plus 20 mM HEPES, pH 7.4]) and adjusted to a final concentration of 0.5 mg/ml. GST-Rb proteins were expressed and recovered as described above. Recombinant pRb proteins were saturated with peptides and incubated at 4°C for 1 h. Subsequently the samples were incubated with cell lysates containing the proteins of interest. Addition of cell lysates and recovery of the complexes formed was as described above.
RESULTS
Identification of two E2F-activating regions in the CELO virus genome.
We established an E2F activation assay to screen the CELO virus genome for E1A-like activities. A reporter plasmid (pE2-Luc) carrying the firefly luciferase gene under the control of the E2F-inducible Ad5 E2 promoter was generated and shown to respond to the control E1A signal with an approximate 50-fold induction of luciferase expression (data not shown). Using this assay, we found that a plasmid encoding the full-length CELO virus genome (pCELO) was capable of modestly activating the Ad5 E2a promoter (Fig. 1B). A second construct with all nonconserved DNA sequences of CELO virus, i.e., sequences flanking the central region on the left and right side (FL/R), possessed a more potent activation function than the full-length CELO virus genome (Fig. 1; note that equal mass, rather than equal molecule numbers of the test plasmids, were transfected, which may account for the relatively poor activation observed with the large [46-kb] pCELO plasmid). A construct containing only the conserved central part of the CELO virus genome (pCN), i.e., the region encoding capsid proteins and E2 functions, showed no luciferase induction (Fig. 1B). The E2F-activating function was further localized to the right end of the CELO virus genome (pFR) and not to the left end (pFL) (Fig. 1B). Further deletion studies identified a large KpnI fragment from the right end of the virus genome (bp 30639 to 40060) which exhibited E2F activation comparable to that of the right-end fragment (KpnI and pFR) (Fig. 1B and 2A).
FIG. 2.
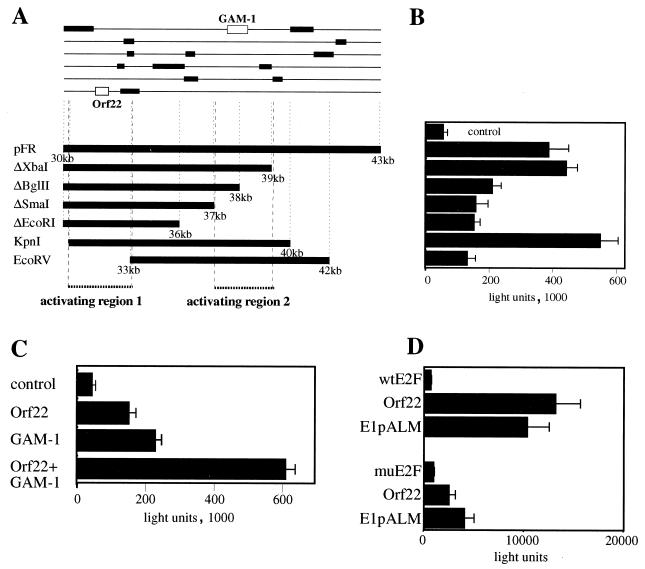
Deletion analysis of pFR. (A) Scheme of pFR deletion constructs indicating open reading frames larger than 25 aa (top) and the deletions introduced by restriction digestion (bottom). (B) A549 cells were transfected with PEI-DNA complexes containing 0.3 μg of E2-Luc reporter construct plus 0.3 μg of the control plasmid pBluescript (control) or 0.3 μg of E2-Luc plus 0.3 μg of the indicated CELO constructs. At 24 h after transfection, luciferase activity was measured as described in Materials and Methods. Each bar shows the average of three transfections with a standard deviation indicated. (C) A549 cells were transfected with PEI-DNA complexes. Control, 0.3 μg of E2-Luc reporter construct plus 0.3 μg of pSG9M; Orf22, 0.3 μg of E2-Luc reporter construct plus 0.15 μg of pSG9MOrf22 plus 0.15 μg of pSG9M; GAM-1, 0.3 μg of E2-Luc reporter construct plus 0.15 μg of pSG9MOrf22 plus 0.15 μg of pSG9M; Orf22+GAM-1, 0.3 μg of E2-Luc reporter construct plus 0.15 μg of pSG9MOrf22 plus 0.15 μg of pSG9MGAM-1. Cells were harvested 2 days after transfection and assayed for luciferase activity. Each bar shows the average of three transfections with a standard deviation indicated. (D) A549 cells were transfected with PEI-DNA complexes. Top: wtE2F, 0.3 μg of pwtE2F-Luc plus 0.3 μg of control plasmid pSG9M; Orf22, 0.3 μg of pwtE2F-Luc plus 0.3 μg of pSG9MOrf22; E1pALM, 0.3 μg of pwtE2F-Luc plus 0.3 μg of the E1 expression plasmid E1pALM. Bottom: muE2F, 0.3 μg of pmuE2F-Luc plus 0.3 μg of control plasmid pSG9M; Orf22, 0.3 μg of pmuE2F-Luc plus 0.3 μg of pSG9MOrf22; E1pALM, 0.3 μg of pmuE2F-Luc plus 0.3 μg of the E1 expression plasmid E1pALM. Cells were harvested 2 days after transfection and assayed for luciferase activity. Each bar shows the average of three transfections with a standard deviation indicated.
The major open reading frames of the right-end fragment are shown in Fig. 2A (see also reference 6). We analyzed the contribution of these reading frames to the observed E2F induction by preparing a series of deletion constructs (Fig. 2A). Progressive deletions from either end of the right-end fragment reduced the luciferase expression to almost 30% of the originally monitored activation (Fig. 2B), suggesting that two separate regions were involved in the observed E2F activation (activating regions [Fig. 2A]). Two reading frames appear to be required for full activity; the first encodes the previously identified antiapoptotic protein GAM-1 (5), and the second (Orf22; bp 31802 to 32430) encodes a previously uncharacterized CELO virus protein product. To facilitate further studies of the Orf22-encoded protein, the reading frame was subcloned and modified to include an amino-terminal Myc epitope similar to a construct previously prepared for GAM-1 (5). Both pSG9MOrf22 and pSG9MGAM-1 were individually capable of E2F activation (Fig. 2C). Notably, coexpression of Orf22 and GAM-1 resulted in an enhanced activation of E2F, comparable to the activity displayed by the full-length right-end fragment pFR (Fig. 2B and C). In contrast, Myc-tagged E1B 19K showed no specific activation of the reporter system (data not shown). Consistent with the idea that the observed E2F activation occurs via release of E2F from pRb, no activation of the reporter construct was obtained with Orf22 and GAM-1 in Saos-2 cells, which carry a truncated and therefore functionally inactive Rb protein (data not shown). Furthermore, the activation by Orf22 is observed with a synthetic wild-type (wt) E2F-dependent promoter but is severely impaired when an identical promoter bearing mutated E2F sites is used (Fig. 2D).
Cellular localization and sequential expression of Orf22 during CELO virus infection.
To determine the cellular localization of Orf22, Myc-tagged Orf22 and GAM-1 expressed in CEF cells were subjected to immunofluorescent detection with anti-Myc antibody. We find that Orf22 localizes to the nucleus of transfected cells similarly to GAM-1 (Fig. 3A) (5).
FIG. 3.
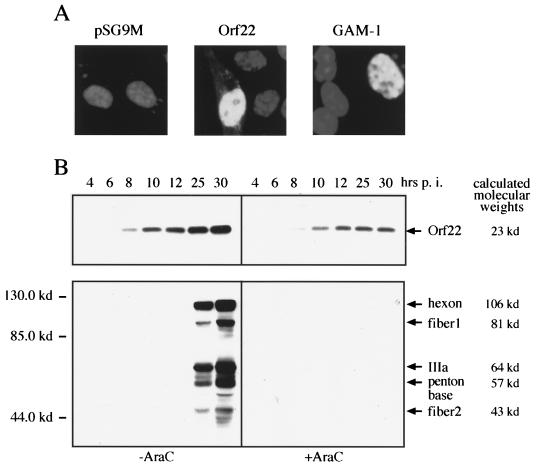
Expression and localization of Myc-tagged Orf22 and GAM-1. (A) CEF cells were transfected with 3 μg of Myc-tagged Orf22, GAM-1, or pSG9M alone. After 3 days, cells were fixed with paraformaldehyde and permeabilized with Triton X-100. Expressed proteins were detected with anti-Myc antibody (Calbiochem) and visualized by fluorescein isothiocyanate-labeled secondary antibody (DAKO). (B) LMH cells were infected with CELO virus at a multiplicity of infection of 1,000 particles per cell. AraC was added at a final concentration of 20 μg/ml. Cells were harvested at the indicated time points and analyzed by Western blotting with anti-Orf22 (top) and anti-CELO rabbit serum (bottom).
To analyze the sequential expression pattern of Orf22 during CELO virus infection, rabbit serum was raised against aa 14 to 30 of the Orf22 protein. LMH cells were infected with CELO virus and incubated for up to 30 h postinfection (p.i.). For the identification of early transcription units, one set of infected cells was incubated with the DNA polymerase inhibitor AraC, which, by preventing the replication of viral genomes, results in a block of adenoviral late gene expression (19). Infected cells were harvested at different time points after infection and assayed for the expression of Orf22 and CELO virus capsid proteins. Staining with anti-Orf22 antiserum revealed that the protein is expressed as early as 6 h p.i. and is still accumulating at 30 h p.i. (Fig. 3B). Orf22 protein was equally detectable both in the presence and absence of AraC, while the expression of CELO virus capsid genes was completely inhibited by AraC (Fig. 3B). These data classify Orf22 as an early transcription product, a result that is supported by studies on CELO virus RNA products where Orf22 transcripts were detectable from 2 h p.i. (42).
Orf22 and GAM-1 interact with pRb.
Since E2F activation by E1A requires its binding to pRb, we analyzed if Orf22 and GAM-1 also interact with pRb. Cells were transfected with Myc-tagged Orf22, GAM-1, or control (E1B 19K) construct. Due to the low expression levels of endogenous pRb, the first set of experiments included a cotransfected hRb expression construct (pCMV-hRB). Orf22 and GAM-1 complexes were harvested from the transfected cells by anti-Myc antibody precipitation. The precipitated complexes were analyzed for the presence of pRb by SDS-PAGE and Western blotting. Under these experimental conditions, we found that precipitates of Orf22 and GAM-1 included pRb (Fig. 4A). As a control, extracts of 293 cells, which constitutively express Ad5 E1A proteins, were precipitated with anti-E1A antibody. Analysis of the precipitated material by anti-pRb Western blotting revealed a band of the same size for pRb, i.e., 105 kDa, as was seen for Orf22 and GAM-1 precipitations. Transfection with the control construct (Myc-tagged E1B 19K) did not show any complex formation with pRb (Fig. 4A).
FIG. 4.
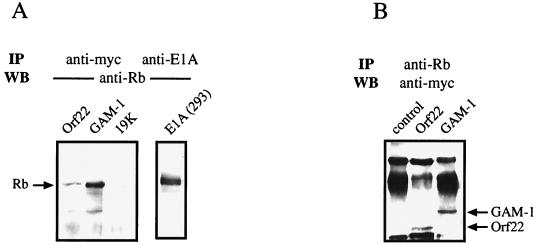
Immunoprecipitation of transfected CEF cell lysates. (A) CEF cells were transfected with 1.5 μg of the indicated Myc-tagged expression constructs together with 1.5 μg of a plasmid coding for hRb (pCMV-hRb). Lysates of transfected CEF cells and 293 cells were immunoprecipitated (IP) either with anti-Myc antibody or anti-E1A antibody as indicated, followed by Western blot (WB) analysis with anti-hRb antibody. (B) CEF cells were transfected with 3 μg of the indicated Myc-tagged constructs. Two days after transfection, cells were lysed and precipitated with anti-hRb antibody. The precipitated material was separated by SDS-PAGE and analyzed by Western blotting with anti-Myc antibody.
We next assessed whether Orf22 and GAM-1 interact with endogenous pRb. In this experiment, cells were solely transfected with Orf22 and GAM-1 expression constructs and precipitated with anti-pRb antibody. Analysis of the immunoprecipitated complexes with anti-Myc antibody revealed the presence of the Myc-tagged Orf22 (30 kDa) and GAM-1 (35 kDa) proteins in endogenous pRb complexes (Fig. 4B). Nontransfected cells showed only background immunoglobulin bands, which were nonspecifically recognized by the Myc antibody. Thus, it appears that Orf22 and GAM-1 can associate with both cellular and overexpressed pRb.
Interaction of Orf22 and GAM-1 with pRb mutants.
The interactions between tumor virus proteins and pRb are well characterized (23, 40, 50). To analyze which regions of pRb interact with Orf22 and GAM-1, we concentrated on the C-terminal part of pRb and the pRb pocket region, since both regions are known to be important protein interaction sites (49). In particular, the pocket region is essential for E1A-pRb interactions (22, 26).
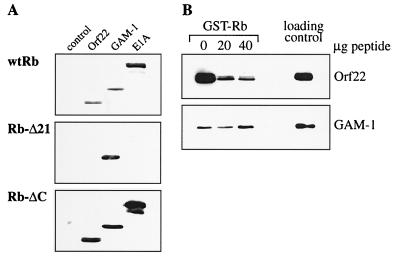
To investigate the importance of both pRb regions for complex formation with Orf22 and GAM-1, we compared the binding properties of both proteins to wt pRb with those of two mutant pRb constructs carrying either a partial deletion of the pocket region (Δ21) or a C-terminal truncation (ΔC). All pRb constructs were fused to GST (7, 27), expressed in bacteria, and purified on glutathione-coupled Sepharose. Constructs encoding Myc-tagged Orf22 and GAM-1 as well as E1A 12S, a protein known to bind to the pocket region of pRb, were transiently transfected into CEF cells. CEF cell lysates were prepared and incubated with purified pRb proteins. Expression levels of the three Myc-tagged proteins were normalized after Western blot analysis (data not shown). Complex formation between pRb and Orf22, GAM-1, or E1A was monitored by Western blotting with anti-Myc antibody. All three proteins bound to GST-wt Rb, while no pRb-binding activity was detectable in lysates of untransfected CEF cells (Fig. 5A, wtRb). Mutant pRb carrying a deletion in the pocket domain was no longer able to bind E1A 12S (Fig. 5A, Rb-Δ21), which is consistent with previous reports (22, 26). The Orf22-pRb interaction was also largely impaired in the absence of a functional pRb pocket region. However, GAM-1–pRb interactions were not affected by this pRb mutation and therefore appear to occur at a different site (Fig. 5A, Rb-Δ21). To test whether Orf22 and GAM-1 interact with the C-terminal region of pRb, the Rb-ΔC mutant was used. In vitro binding assays revealed that all three Myc-tagged proteins (Orf22, GAM-1, and E1A 12S) were able to form a complex with the Rb-ΔC mutant (Fig. 5A, Rb-ΔC). Therefore, the C-terminal part of pRb is not involved in binding reactions with Orf22, GAM-1, or E1A. While Orf22 interacts largely with the pocket region of pRb, the site of interaction for GAM-1 is distinct from the pRb pocket domain and the C terminus.
FIG. 5.
Complex formation of Orf22 and GAM-1 with pRb mutants. (A) CEF cells were transfected with 3 μg of the indicated expression constructs and lysed 24 h after transfection. Equal amounts of the expressed proteins were incubated with recombinant GST-Rb proteins as described in Materials and Methods. Complex formation was monitored by subsequent Western analysis with anti-Myc antibody. (B) Competition assay with T peptide. Myc-tagged constructs of Orf22, GAM-1, and E1A 19K were expressed in CEF cells. Lysates of these cells were incubated with recombinant GST-Rb together with increasing amounts of T peptide. Lane 1, no peptide; lane 2, 20 μg of peptide; lane 3, 40 μg of peptide; lane 4, lysate of transfected CEF cells (40 μl).
To further analyze the interaction of Orf22 with the pocket domain of pRb, we established a competition assay. The pocket binding region of T antigen (T peptide) (10), which is similar to the pocket-binding region of E1A (2, 4), was incubated with purified GST-Rb to bind and occupy the pocket domain. Lysates from CEF cells transfected with Myc-tagged Orf22 were added, and complexes were resolved by SDS-PAGE; the presence of Myc-tagged proteins was verified by anti-Myc antibody. We found that approximately 90% of the Orf22-pRb interaction can be eliminated with the T peptide (Fig. 5B, Orf22). This result is comparable to E1A-pRb interactions, and thus binding of Orf22 to pRb resembles the interaction of pRb with T antigen or E1A. However, the presence of the same quantities of peptide had no effect on GAM-1–pRb interactions demonstrating that GAM-1 interacts with pRb at regions other than the pocket domain (Fig. 5B, GAM-1).
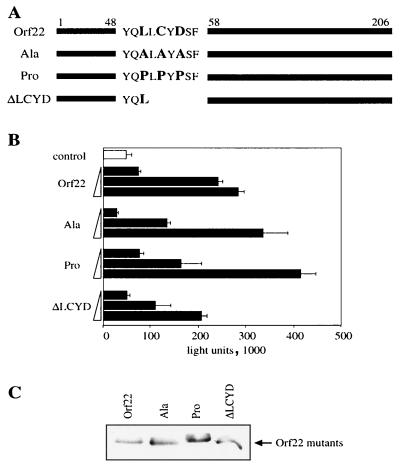
Mutation of an internal LxCxD motif of Orf22.
The pRb-binding region of E1A consists of an LxCxE motif flanked by acidic residues. This binding motif is conserved among the viral oncoproteins of adenovirus E1A, SV40 T antigen, and human papillomavirus E7, all of which interact with the pRb pocket domain (18, 49). In contrast, the protein sequence of Orf22 revealed no significant homology to viral pRb-binding motifs except for an LLCYD sequence at aa 51 to 55. However, no acidic sequences were found adjacent to this motif. As this was the only recognizable motif shared with other viral pRb-binding proteins, we determined its importance for E2F activation and pRb binding by site-directed mutagenesis. The introduced alterations include deletion of two of the conserved amino acids cysteine and aspartic acid as well as alterations of the leucine, cysteine, and aspartic acid to either alanine or proline (Fig. 6A). The modified constructs were expressed at comparable levels (results not shown) and tested for E2F activation as described above. Alterations of the LLCYD motif had no effect on the E2F activity (Fig. 6B). Consistent with the E2F activation, interaction of the mutant Orf22 molecules with pRb, as monitored by immunoprecipitation studies, was unaffected by any of the mutations (Fig. 6C). Thus, it appears that the analogous functions of Orf22 and E1A in binding to pRb and activating E2F are mediated by two different protein motifs.
FIG. 6.
Mutation analysis of the LxCxD motif of Orf22. (A) Schematic diagram showing modifications of the LxCxD motif. (B) CEF cells were transfected with increasing amounts (0.003, 0.03, and 0.3 μg) of the indicated constructs. All complexes contained 0.3 μg of E2F-Luc reporter construct. At 24 h after transfection, cells were lysed and analyzed for luciferase activity. (C) Precipitation of transfected CEF cells with anti-hRb antibody followed by Western analysis with anti-Myc antibody. All cells were cotransfected with 1.5 μg of plasmid pSG9MOrf22 and 1.5 μg of pSG9MGAM-1.
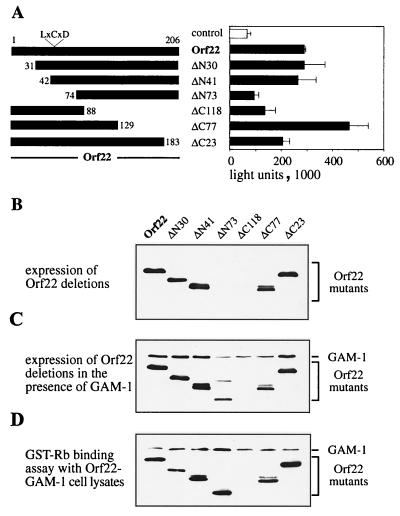
In efforts to identify the pRb-binding region of Orf22, we introduced progressive deletions from either the amino terminus removing 30, 41, or 73 residues (ΔN30, ΔN41, or ΔN73) or from the carboxy terminus removing 23, 77, or 118 residues (ΔC23, ΔC77, or ΔC118) of the Orf22 protein (Fig. 7A). The E2F activation capacity of the deletion clones revealed that deletion of 41 residues from the amino terminus had no effect on protein function. Further removal of the N-terminal 73 residues abolished E2F activation. Of the carboxy-terminal deletions, removal of 23 or 77 residues did not impair E2F activation (instead, the ΔC77 truncation produced a modest but reproducible activation). A more substantial deletion of ΔC118 disrupted the activating function of Orf22 (Fig. 7A). When the protein levels of the truncated proteins were monitored, all but the two largest deletions (ΔN73 and ΔC118) were found to be expressed at levels comparable to wt Orf22. Protein levels of ΔN73 and ΔC118, however, were severely reduced (Fig. 7B). Thus the impaired E2F activation of these two molecules could be due to lower protein levels rather than loss of pRb interaction. It was observed that in the presence of GAM-1, expression of the Orf22 mutant ΔN73 could be rescued to almost wt levels (the nature of this rescue event is not understood). The Orf22 mutant ΔC118, carrying the largest deletion, however, was still poorly detectable (Fig. 7C). Addition of GAM-1 stabilized the synthesis of truncated Orf22 proteins sufficiently to test their pRb-binding capacity. Extracts of CEF cells expressing mutant Orf22 proteins in the presence of GAM-1 were incubated with recombinant GST-Rb (extracts were normalized for constant Orf22 protein levels). Complex formation with pRb was monitored by Western analysis with anti-Myc antibody. Amino-terminal Orf22 mutants ΔN30, ΔN41, and ΔN73 (which removes the LLCYD motif) and carboxy-terminal truncations ΔC23 and ΔC77 bound to pRb to the same extent as wt Orf22 (Fig. 7D). This analysis further supports our finding that the LLCYD motif in Orf22 is neither required for E2F activation nor involved in interactions with pRb. It appears that pRb interaction does not occur in the regions outside aa 88 to 129. However, the instability of the proteins lacking this region made it difficult to assess such interaction directly. An alternate approach was taken.
FIG. 7.
Deletion analysis of the Orf22 construct. (A) Diagram of N- and C-terminal deletions of Orf22. CEF cells were transfected with 0.3 μg of E2F-Luc reporter plasmid and 0.3 μg of Orf22 construct. Two days after transfection, cells were lysed and assayed for luciferase activity. (B to D) GST-Rb was expressed in bacteria and recovered on glutathione-Sepharose. CEF cells were transfected with each 1.5 μg of Orf22 constructs and GAM-1 plasmid. Two days after transfection, cells were assayed for recombinant protein expression by Western blotting (B and C). Equal amounts of the expressed Myc-tagged protein were incubated with recombinant GST-Rb. Resulting complex formation was analyzed by Western blotting with anti-Myc antibody (D).
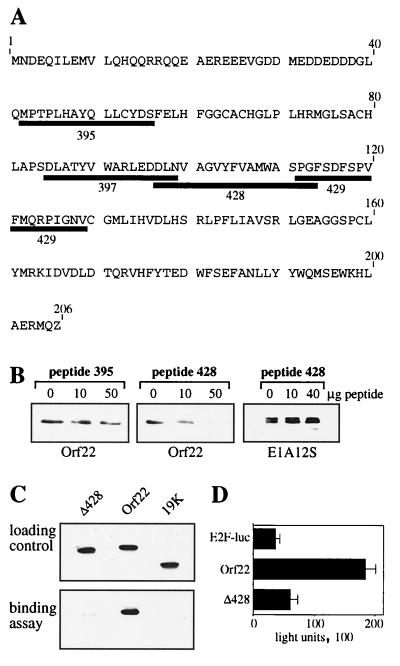
Analysis of aa 88 to 129 in Orf22.
To evaluate the importance of aa 88 to 129 in Orf22 for pRb binding, we designed a series of overlapping peptides of 15 to 18 aa homologous to this region. Figure 8 shows the amino acid sequences and locations relative to the Orf22 sequence for four of the synthesized peptides (395, 397, 428, and 429). Peptide 395 contains the LLCYD region of Orf22 and was included in the experiment (Fig. 8A). Competition assays with various peptides were performed as described for the T-peptide assay. Briefly, GST-wt pRb was incubated with single peptides, Orf22 extracts were added, and complex formation with pRb was monitored by Western analysis. Orf22 and pRb alone formed complexes as shown before. Addition of increasing amounts of peptide 395 had only a slight effect on Orf22-Rb complex formation (Fig. 8B), confirming our previous findings. A similar result was obtained for most of the other peptides that were homologous to the aa 88 to 129 region of Orf22, i.e., peptides 397 and 429 (data not shown). In contrast, peptide 428 efficiently interfered with the binding of Orf22 to pRb. Increasing amounts of the peptide nearly completely abolished complex formation between Orf22 and pRb, while similar levels of the control peptide had no effect on binding (Fig. 8B). This finding supports the involvement of aa 97 to 114 of Orf22 in binding to pRb. The specificity of the competition was analyzed by testing the ability of peptide 428 to compete for E1A-Rb interactions. Under the same conditions that revealed peptide 428-Orf22 competition, there was no effect on the complex formation between Myc-tagged E1A 12S and pRb (Fig. 8B). Thus, the ability of peptide 428 to disrupt Orf22-pRB interactions appears to be specific for Orf22 and does not impair binding of E1A.
FIG. 8.
Analysis of the Orf22 mutant Δ428. (A) Amino acid composition of Orf22 and spatial arrangement of four of the peptides used in competition assays. (B) GST-Rb was expressed in bacteria and recovered on glutathione-Sepharose; 0, 10, or 50 μg of peptide 395 or 428 was incubated with GST-Rb before addition of lysates of pSG9MOrf22-transfected cells (left and middle) or pSG9ME1A12S-transfected cells (right). The resulting complexes were analyzed by Western blotting with anti-Myc antibody. (C) CEF cells were transfected with 3 μg of Myc-tagged Orf22, Δ428, or E1A 19K construct; 30 h after transfection, cells were lysed and incubated with recombinant GST-Rb. Complex formation was monitored by Western analysis with anti-Myc antibody. (D) A549 cells were cotransfected with 0.12 μg of E2F-Luc reporter construct and 0.16 μg of Orf22 and Δ428 plasmids. Cells were assayed for luciferase activity 24 h after transfection.
To examine the Orf22-pRB interaction in an alternate manner, we generated a Myc-tagged Orf22 mutant construct that carried an internal deletion of aa 97 to 114 (pSG9MΔ428) (Fig. 8C). Protein stability was not impaired by this internal deletion (Fig. 8C, top) and allowed us to determine the importance of the aa 97 to 114 region for pRb binding. Binding assays revealed that Orf22 formed a complex with pRb, while a control protein (E1B 19K) showed no association with pRb. Deletion of the internal aa 97 to 114 fragment severely impaired the binding of Orf22 to pRb (Fig. 8C, bottom). We therefore conclude that the pRb-binding region of Orf22 is within the region between aa 97 and 114.
Our starting model was based on the finding that E2F activation is directly linked to the inactivation of pRb, i.e., by viral oncoproteins. After having identified Orf22 as an E2F-activating and pRb-binding protein, we analyzed whether elimination of its pRb-binding activity would result in a reduction or loss of its E2F activation potential. Comparing the E2F-activating functions of Orf22 and the mutant construct Δ428, we found, as shown earlier, that Orf22 clearly activates E2F. Deletion of the internal aa 97 to 114 fragment, however, severely impaired E2F activation (Fig. 8D). This assay demonstrated that binding of Orf22 to pRb and activation of E2F are both dependent on an internal region between aa 97 and 114.
DISCUSSION
We have identified two CELO virus proteins that, like the human adenovirus E1A proteins, are capable of binding pRb and activating E2F-dependent transcription. The E2F-activating and pRb-binding properties of E1A serve as essential initiating steps for viral replication, especially in nondividing cells. As both viruses infect terminally differentiated cells, a similar mechanism can be expected for avian adenoviruses. A screen of the CELO virus genome for E2F activation identified GAM-1 and Orf22 as viral proteins that were capable of inducing an E2-Luc reporter system individually; the combination of both proteins was synergistic for the activation. The molecular basis for this synergy is not clear at the molecular level, complex formation between GAM-1 and Orf22 has not yet been observed.
Orf22 and GAM-1 bind to recombinant pRb in vitro and to both endogenous and overexpressed pRb in transfected cells. While Orf22 interacts with the pocket region of pRb, GAM-1 binds neither to the pocket domain nor to the C-terminal part of pRb. As both proteins apparently interact with different regions of pRb, it is possible that a complex composed of GAM-1, Orf22, and pRb exists. Only the formation of such a complex might ensure the complete inactivation of the repressing functions of pRb. This model would account for the cooperative effect of Orf22 and GAM-1 on E2F activation. Whether such a complex forms during CELO virus infection is not known. Initial studies on the sequential appearance of CELO virus transcripts revealed that Orf22 transcripts are detectable 2 h p.i. and therefore belong to the early transcription units (42), a feature shared with human adenovirus E1A. These RNA data have been confirmed by Western blotting with serum raised against the Orf22 protein. Orf22 protein can be detected as early as 6 h p.i.; protein levels increase until at least 30 hours p.i., and Orf22 protein expression occurs independent of viral DNA replication. For GAM-1, Northern analysis detected first transcripts at late stages of infection (24 h p.i.) (42). The presence of a N-terminal bipartite leader sequence on the GAM-1 transcript suggests that expression is regulated by the major late promoter. Therefore, GAM-1 appears to be a protein which is expressed late in infection. The pRb-E2F interactions required for initiation of virus infection would thus be primarily the function of Orf22, with triple complex formation between pRb, Orf22, and GAM-1 occurring only at late stages of viral infection; the importance of these GAM-1–pRb interactions late in infection are not immediately clear.
Analysis of the pRb-binding region of Orf22 identified a region of 18 aa (aa 97 to 114) that was able to mediate Orf22 binding. Deletion of this region resulted in strongly reduced pRb binding and loss of E2F activation. These 18 aa share no homology with the conserved LxCxE motif of E1A or other viral pRb-binding proteins and thus far have been found to have homology in the database with only CELO virus and fowl adenovirus type 8 sequences. One could speculate that rather than sharing a conserved peptide sequence, the VAGVYFVAM sequence of Orf22 may present important chemical moieties in the appropriate three-dimensional organization to bind the pRb pocket. This structure, in combination with the acidic domain adjacent to this motif, could be sufficient to bind pRb and displace E2F. However, in the absence of crystallographic data on E1A-pRb interactions, it is difficult to examine this hypothesis in greater detail. Although different motifs are used by Orf22 and E1A, their interactions with pRb seem to be similar in some aspects: both proteins bind to the pocket region of pRb, and both compete with the same peptide (T peptide) for binding to the pRb pocket domain. However, peptide 428 was not capable of disrupting E1A-pRB interactions, demonstrating that the interactions do not strictly overlap.
The similar pRb-binding properties of Orf22 and E1A and their activation of the E2F pathway lead to further speculations. E1A has been demonstrated to exhibit transforming activity. Essential for the transforming capacity of E1A are both inactivation of pRb and binding to p300/CBP, a family of proteins with histone acetylase activity. Preliminary studies showed that Orf22 also interacts with p300 (32a). The ability of Orf22 to interact with both pRb and p300 makes it tempting to speculate that Orf22 possesses transforming activity, and experiments to examine this possibility are in progress.
ACKNOWLEDGMENTS
We thank Susanna Chiocca, Wilhem Krek, Martin Scheffner, Meinrad Busslinger, and Dirk Eberhard for sharing plasmids with us. We are grateful to Dirk Eberhard for much advice, and we thank Jola Glotzer and Gerhard Christofori for their comments on the manuscript.
REFERENCES
- 1.Baker A, Saltik M, Lehrmann H, Killisch I, Mautner V, Lamm G, Christofori G, Cotten M. Polyethylenimine (PEI) is a simple, inexpensive and effective reagent for condensing and linking plasmid DNA to adenovirus for gene delivery. Gene Ther. 1997;4:773–782. doi: 10.1038/sj.gt.3300471. [DOI] [PubMed] [Google Scholar]
- 2.Barbosa M S, Edmonds C, Fisher C, Schiller J T, Lowy D R, Vousden K H. The region of the HPV E7 oncoprotein homologous to adenovirus E1a and Sv40 large T antigen contains separate domains for Rb binding and casein kinase II phosphorylation. EMBO J. 1990;9:153–160. doi: 10.1002/j.1460-2075.1990.tb08091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boussif O, Lezoualc’h F, Zanta M A, Mergny M D, Scherman D, Demeneix B, Behr J P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chellappan S, Kraus V B, Kroger B, Munger K, Howley P M, Phelps W C, Nevins J R. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc Natl Acad Sci USA. 1992;89:4549–4553. doi: 10.1073/pnas.89.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiocca S, Baker A, Cotten M. Identification of a novel antiapoptotic protein, GAM-1, encoded by the CELO adenovirus. J Virol. 1997;71:3168–3177. doi: 10.1128/jvi.71.4.3168-3177.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiocca S, Kurzbauer R, Schaffner G, Baker A, Mautner V, Cotten M. The complete DNA sequence and genomic organization of the avian adenovirus CELO. J Virol. 1996;70:2939–2949. doi: 10.1128/jvi.70.5.2939-2949.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chittenden T, Livingston D M, Kaelin W G., Jr The T/E1A-binding domain of the retinoblastoma product can interact selectively with a sequence-specific DNA-binding protein. Cell. 1991;65:1073–1082. doi: 10.1016/0092-8674(91)90559-h. [DOI] [PubMed] [Google Scholar]
- 8.Chomiak T W, Luginbuhl R E, Helmboldt C F. Tissue culture propagation and pathology of CELO virus. Avian Dis. 1961;5:313–320. [Google Scholar]
- 9.Cotten M, Wagner E, Birnstiel M L. Receptor-mediated transport of DNA into eukaryotic cells. Methods Enzymol. 1993;217:618–644. doi: 10.1016/0076-6879(93)17092-j. [DOI] [PubMed] [Google Scholar]
- 10.DeCaprio J A, Ludlow J W, Lynch D, Furukawa Y, Griffin J, Piwnica-Worms H, Huang C M, Livingston D M. The product of the retinoblastoma susceptibility gene has properties of a cell cycle regulatory element. Cell. 1989;58:1085–1095. doi: 10.1016/0092-8674(89)90507-2. [DOI] [PubMed] [Google Scholar]
- 11.Dhillon A S, Jack O K. The oncogenic potential of eleven avian adenovirus strains. Avian Dis. 1997;41:247–251. [PubMed] [Google Scholar]
- 12.Dutta S K, Pomeroy B S. Electron microscopic structure of chicken embryo lethal orphan virus. Proc Soc Exp Biol Med. 1963;114:539–541. doi: 10.3181/00379727-114-28726. [DOI] [PubMed] [Google Scholar]
- 13.Dutta S K, Pomoroy B S. Electron microscopic studies of quail bronchitis virus. Am J Vet Res. 1967;28:296–299. [Google Scholar]
- 14.Dynlacht B D. Regulation of transcription by proteins that control the cell cycle. Nature. 1997;389:149–152. doi: 10.1038/38225. [DOI] [PubMed] [Google Scholar]
- 15.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 16.Evan G I, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fadly A M, Winterfield R W, Olander H J. The oncogenic potential of some avian adenoviruses causing diseases in chickens. Avian Dis. 1976;20:139–145. [PubMed] [Google Scholar]
- 18.Figge J, Breese K, Vajda S, Zhu Q L, Eisele L, Andersen T T, MacColl R, Friedrich T, Smith T F. The binding domain structure of retinoblastoma-binding proteins. Protein Sci. 1993;2:155–164. doi: 10.1002/pro.5560020204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flangan J F, Ginsberg H S. Synthesis of virus specific polymers in adenovirus infected cells: Effects of 5-fluorodeoxyuridine. J Exp Med. 1962;116:141. doi: 10.1084/jem.116.2.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green S, Issemann I, Sheer E. A versatile in vivo and in vitro eukaryotic expression vector for protein engineering. Nucleic Acids Res. 1988;16:369. doi: 10.1093/nar/16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hess M, Blocker H, Brandt P. The complete nucleotide sequence of the egg drop syndrome virus: an intermediate between mastadenoviruses and aviadenoviruses. Virology. 1997;238:145–156. doi: 10.1006/viro.1997.8815. [DOI] [PubMed] [Google Scholar]
- 22.Hu Q J, Dyson N, Harlow E. The regions of the retinoblastoma protein needed for binding to adenovirus E1A or SV40 large T antigen are common sites for mutations. EMBO J. 1990;9:1147–1155. doi: 10.1002/j.1460-2075.1990.tb08221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jansen-Dürr P. How viral oncogenes make the cell cycle. Trends Genet. 1996;12:270–275. doi: 10.1016/0168-9525(96)81455-7. [DOI] [PubMed] [Google Scholar]
- 24.Jasty V, Yates V J, Anderson J, Fry D, Chang P W, Pendola R. Replication of an avian adenovirus (CELO) large-plaque mutant in chick kidney cells. 1. An electron-microscope study. Avian Dis. 1973;17:49–65. [PubMed] [Google Scholar]
- 25.Jones R F, Asch B B, Yohn D S. On the oncogenic properties of chicken embryo lethal orphan virus, an avian adenovirus. Cancer Res. 1970;30:1580–1585. [PubMed] [Google Scholar]
- 26.Kaelin W G, Jr, Ewen M E, Livingston D M. Definition of the minimal simian virus 40 large T antigen- and adenovirus E1A-binding domain in the retinoblastoma gene product. Mol Cell Biol. 1990;10:3761–3769. doi: 10.1128/mcb.10.7.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaelin W G, Jr, Pallas D C, DeCaprio J A, Kaye F J, Livingston D M. Identification of cellular proteins that can interact specifically with the T/E1A-binding region of the retinoblastoma gene product. Cell. 1991;64:521–532. doi: 10.1016/0092-8674(91)90236-r. [DOI] [PubMed] [Google Scholar]
- 28.Kovesdi I, Reichel R, Nevins J R. Identification of a cellular transcription factor involved in E1A transactivation. Cell. 1986;45:219–228. doi: 10.1016/0092-8674(86)90386-7. [DOI] [PubMed] [Google Scholar]
- 29.Krek W, Livingston D M, Shirodkar S. Binding to DNA and the retinoblastoma gene product promoted by complex formation of different E2F family members. Science. 1993;262:1557–1560. doi: 10.1126/science.8248803. [DOI] [PubMed] [Google Scholar]
- 30.Künkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Künkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 32.Laver W G, Younghusband H B, Wrigley N G. Purification and properties of chick embryo lethal orphan virus (an avian adenovirus) Virology. 1971;45:598–614. doi: 10.1016/0042-6822(71)90175-9. [DOI] [PubMed] [Google Scholar]
- 32a.Lehrmann, H., and M. Cotten. Unpublished data.
- 33.Lewis J B, Mathews M B. Control of adenovirus early gene expression: a class of immediate early products. Cell. 1980;21:303–313. doi: 10.1016/0092-8674(80)90138-5. [DOI] [PubMed] [Google Scholar]
- 34.Li P, Bellett A J, Parish C R. Structural organization and polypeptide composition of the avian adenovirus core. J Virol. 1984;52:638–649. doi: 10.1128/jvi.52.2.638-649.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacPherson I A, Wildy P, Stoker M G P, Horne R W. The fine structure of GAL—an avian orphan virus. Virology. 1961;13:146–149. doi: 10.1016/0042-6822(61)90046-0. [DOI] [PubMed] [Google Scholar]
- 36.Maeda M, Okaniwa A, Kawamura H. Morphological studies on intranuclear inclusion bodies in chicken kidney cell culture infected with avian adenovirus. Natl Inst Anim Health Q (Tokyo) 1967;7:164–177. [PubMed] [Google Scholar]
- 37.McFerran J B, Adair B M. Avian adenoviruses—a review. Avian Pathol. 1977;6:189–217. doi: 10.1080/03079457708418228. [DOI] [PubMed] [Google Scholar]
- 38.McFerran J B, Clarke J K, Connor T J. Serological classification of avian adenoviruses. Arch Gesamte Virusforsch. 1972;39:132–139. doi: 10.1007/BF01241536. [DOI] [PubMed] [Google Scholar]
- 39.Michou A I, Lehrmann H, Saltik M, Cotten M. Mutational analysis of the avian adenovirus CELO, which provides a basis for gene delivery vectors. J Virol. 1999;73:1399–1410. doi: 10.1128/jvi.73.2.1399-1410.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nevins J R. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 41.Nevins J R, Ginsberg H S, Blanchard J M, Wilson M C, Darnell J E., Jr Regulation of the primary expression of the early adenovirus transcription units. J Virol. 1979;32:727–733. doi: 10.1128/jvi.32.3.727-733.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Payet V, Arnauld C, Picault J P, Jestin A, Langlois P. Transcriptional organization of the avian adenovirus CELO. J Virol. 1998;72:9278–9285. doi: 10.1128/jvi.72.11.9278-9285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petek M, Felluga B, Zoletto R. Biological properties of CELO virus: stability to various agents, and electron-microscopic study. Avian Dis. 1963;7:38–49. [PubMed] [Google Scholar]
- 44.Pitcovski J, Mualem M, Rei-Koren Z, Krispel S, Shmueli E, Peretz Y, Gutter B, Gallili G E, Michael A, Goldberg D. The complete DNA sequence and genome organization of the avian adenovirus, hemorrhagic enteritis virus. Virology. 1998;249:307–315. doi: 10.1006/viro.1998.9336. [DOI] [PubMed] [Google Scholar]
- 45.Sarma P S, Huebner R J, Lane W T. Induction of tumors in hamsters with an avian adenovirus (CELO) Science. 1965;149:1108. doi: 10.1126/science.149.3688.1108. [DOI] [PubMed] [Google Scholar]
- 46.Sharpless G R, Levine S, Davies M C, Englert M E. GAL virus: its growth in tissue culture and some of its properties. Virology. 1961;13:315–322. [Google Scholar]
- 47.Shenk T. Adenoviridae: the viruses and their replication, B. N. Fields et al. (ed.), Fields Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. [Google Scholar]
- 48.Stenback W A, Anderson J P, McComick K J, Trentin J J. Induction of tumors in the liver of hamsters by an avian adenovirus (CELO) J Natl Cancer Inst. 1973;50:963–970. doi: 10.1093/jnci/50.4.963. [DOI] [PubMed] [Google Scholar]
- 49.Taya Y. RB kinases and RB-binding proteins: new points of view. Trends Biochem Sci. 1997;22:14–17. doi: 10.1016/s0968-0004(96)10070-0. [DOI] [PubMed] [Google Scholar]
- 50.Vousden K H. Regulation of the cell cycle by viral oncoproteins. Semin Cancer Biol. 1995;6:109–116. doi: 10.1006/scbi.1995.0014. [DOI] [PubMed] [Google Scholar]
- 51.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 52.Whyte P, Buchkovich K J, Horowitz J M, Friend S H, Raybuck M, Weinberg R A, Harlow E. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]