Abstract
Productive infection by human immunodeficiency virus type 1 (HIV-1) requires the activation of target cells. Infection of quiescent peripheral CD4 lymphocytes by HIV-1 results in incomplete, labile, reverse transcripts. We have previously identified G1b as the cell cycle stage required for the optimal completion of the reverse transcription process in T lymphocytes. However, the mechanism(s) involved in the blockage of reverse transcription remains undefined. In this study we investigated whether nucleotide levels influence viral reverse transcription in G0 cells. For this purpose the role of the enzyme ribonucleotide reductase was bypassed, by adding exogenous deoxyribonucleosides to highly purified T cells in the G0 or the G1a phase of the cell cycle. Our data showed a significant increase in the efficiency of the reverse transcription process following the addition of the deoxyribonucleosides. To define the stability and functionality of these full reverse transcripts, we used an HIV-1 reporter virus that expresses the murine heat-stable antigen on the surfaces of infected cells. Following activation of infected quiescent cells treated with exogenous nucleosides, no increased rescue of productive infection was seen. Thus, in addition to failure to complete reverse transcription, there was an additional nonreversible blockage of productive infection in quiescent T cells. These experiments have important relevance in the gene therapy arena, in terms of improving the ability of lentivirus vectors to enter metabolically inactive cells, such as hematopoietic stem cells.
Retroviral replication is greatly influenced by the metabolic and activation states of the target cell at the time of infection (1, 19, 20, 24–27). Our previous studies have established that, in contrast to stimulated lymphocytes, quiescent CD4+ T cells allow entry of human immunodeficiency virus (HIV) but fail to allow completion of viral reverse transcription (26, 27). In addition, we have recently studied the extent of the T-cell activation that is required for optimal completion of the HIV-1 reverse transcription process. We found that in highly purified quiescent T cells, activation through both the T-cell receptor (TCR)-CD3 complex and the costimulatory molecule CD28 are needed to allow efficient reverse transcription and productive infection to occur (9). When quiescent T cells, free of contaminating antigen-presenting cells, are stimulated through the TCR alone, they progress only to the G1a phase of the cell cycle and are not permissive for full reverse transcription. Furthermore, although cellular division is not needed for reverse transcription to occur (12, 20, 25), we have shown that progression past the G1a phase of the cell cycle is required for the completion of the reverse transcription process.
The mechanisms involved in the blockage of reverse transcription in quiescent cells and cells arrested at the G1a phase of the cell cycle remain unidentified. Some potential mechanisms for this arrest may be a decrease in the function of the enzyme reverse transcriptase, as well as the presence of cellular inhibitors in quiescent cells or reverse transcription potentiators in activated cells. It has been suggested by others that low levels of deoxynucleoside triphosphates (dNTPs) may contribute to this phenomenon (5, 13, 15, 16). Previous studies involving productive infection of nondividing, cultured mononuclear phagocytes have shown that full-length viral DNA accumulated with a slow kinetics, which could be accelerated by addition of exogenous nucleotide precursors, although not to the rate seen in activated T lymphocytes (8, 16). Most circulating peripheral T lymphocytes and many in lymphoid tissues are in a G0 resting state. Diverse events, including exposure to growth factors and antigen-mediated activation are involved in the stimulation of these cells and may result in different states of cellular activation and cell cycle progression. The intracellular concentration of deoxynucleotides is greatly effected by the differentiation and activation states of the cells (2, 14, 18). The activities of a number of enzymes involved in purine and pyrimidine metabolism, as well as the enzyme ribonucleotide reductase, which reduces ribonucleotides to deoxyribonucleotides, vary with the phase of the cell cycle (3, 22). Thus, there are very low intracellular concentrations of dNTP pools in resting T lymphocytes and a significant increase in these concentrations occurs following activation and progression into the cell cycle (2, 14, 18). Thus, as others have suggested, it seems likely that the low levels of dNTP pools in G0 cells contribute to the premature termination of the reverse transcription process. Indeed, it was demonstrated by Goulaouic et al. (7) that the addition of high concentrations of exogenous nucleosides to nonproliferating murine fibroblasts newly infected with Moloney murine leukemia virus allowed the completion of the reverse transcription process, without initiating cell cycle progression.
In this study we investigated whether intracellular nucleotide levels influence HIV-1 reverse transcription in G0- and G1a-arrested lymphocytes. Cells in these phases of the cell cycle rely mostly on the salvage of exogenous deoxynucleosides rather than on the reduction of de novo-synthesized ribonucleotides (2). In our system the role of the enzyme ribonucleotide reductase was bypassed, by exogenously adding all four deoxyribonucleosides to highly purified T cells in the G0 or the G1a phase of the cell cycle. PCR analysis of DNA extracts showed a significant increase in the efficiency of the reverse transcription process with the addition of deoxyribonucleosides prior to infection with HIV-1. This treatment, however, did not fully restore the levels of complete reverse transcripts to those seen in cells costimulated through the CD3 and CD28 activation pathways. To define the stability and functionality of these full reverse transcripts, we have used HIV-1 reporter viruses that express the murine heat-stable antigen (HSA) on the surfaces of infected cells (9). Following anti-CD3 and anti-CD28 costimulation of infected quiescent cells treated with exogenous nucleosides, no increased rescue of productive infection was seen in comparison to the level of rescue seen in cells not treated with nucleosides. Thus, there is an additional nonreversible blockage of productive infection of newly activated T cells.
MATERIALS AND METHODS
Cells and conditions.
Peripheral blood was obtained from healthy HIV-seronegative blood donors, and peripheral blood lymphocytes (PBLs) were separated over a Ficoll-Hypaque gradient. Nonadherent (NA) cells were obtained after macrophages were depleted by adherence to plastic for 3 h. The highly purified DR− population was enriched by removal of HLA-DR+ cells as previously described (11). In addition, PBLs were enriched for CD4+ cells, to at least 90% purity, by depletion of the CD8+ cells by a panning technique as previously described (11). Purified cells were cultured in RPMI 1640 supplemented with 10% human AB serum, 100 U of penicillin per ml, 100 mg of streptomycin per ml, and 2 mM glutamine. Cells were stimulated with 1 μg of OKT3 (anti-CD3) monoclonal antibody (MAb) (Ortho Diagnostics, Inc., Raritan, N.J.) per ml immobilized on goat anti-mouse antibody (Fisher)-coated plates alone to mimic TCR stimulation or costimulated with the simultaneous addition of soluble anti-CD28 antibody (Pharmingen, San Diego, Calif.) at a concentration of 0.1 μg/ml. Some of the quiescent cells were treated with either a 3.5 mM concentration of the cell cycle inhibitor n-butyrate or with a 5 μM concentration of the ribonucleotide reductase inhibitor hydroxyurea (HU) (Sigma, St. Louis, Mo.) for 2 h prior to their costimulation. To assess the effects of exogenous nucleosides, unstimulated DR− cells were cultured in the presence or absence of 10 to 50 μM concentrations of each of the nucleosides: 2′-deoxycytidine, 2′-deoxyadenosine, 2′-deoxyguanosine, and 2′-deoxythymidine (Sigma) for 2 h prior to infection with HIV-1. To prevent virus spread and achieve a single-cycle infection, some of the cells were treated with a 100 nM concentration of a protease inhibitor (PI) (Indinavir; Merck, Rahway, N.J.) following infection.
Viruses and infections.
Stocks of the HIV-1 molecular clone NL4-3 and the NL4-3-based reporter construct NL-r-HSAS (9) were obtained from 24-h harvests of supernatants from CEM cells electroporated with full-length infectious cloned viral DNA. The reporter virus construct used in this study was obtained by cloning the cell surface murine HSA into the vpr region of HIV-1NL4-3 (9). These supernatants generally contained 2 to 2.5 mg of viral p24 per ml as assessed by enzyme-linked immunosorbent assay. Based on results of limiting-dilution assays, when 1 ml of virus was used to infect 106 cells, the multiplicity of infection was approximately 0.2. To reduce the amount of contamination with viral DNA derived from cells lysed during culture, supernatants were filtered and treated with 2 μg of DNase (Worthington, Lakewood, N.J.) per ml for 30 min at room temperature in the presence of 0.01 M MgCl2. Infection was accomplished by incubating cells for 1 to 2 h with virus in the presence of Polybrene (10 μg/ml). Cells were washed with medium three times to remove residual free virus and recultured under the appropriate conditions. Heat-inactivated virus controls were prepared by incubating the virus for 30 min at 60°C. In some experiments whose results are not shown, a reporter virus containing the murine HSA cDNA inserted into the nef gene was used similarly, to rule out effects due to loss of vpr function.
Flow cytometry for surface markers.
To assess the purity of the cell populations, 5 × 105 cells were costained with MAbs (Becton Dickinson, Mountain View, Calif.) against HLA-DR (major histocompatibility complex class II), CD25 (interleukin 2R), CD19 (B-cell marker), and CD14 (macrophage marker) cell surface markers as previously described (11) (data not shown). To determine viral expression in quiescent and activated cells, cells cultured under different conditions were costained with MAbs to CD4 (Becton Dickinson) and murine HSA (Pharmingen). These antibodies were directly conjugated to phycoerythrin (CD4) and fluorescein isothiocyanate (murine HSA). Under each of the conditions and at each time point, mouse immunoglobulin G1 (fluorescein isothiocyanate- and phycoerythrin-conjugated immunoglobulin G1) were used as antibody isotype controls. Cells were then fixed in 2% paraformaldehyde. Five thousand to 10,000 events were acquired on a FACStarplus flow cytometer (Becton Dickinson). Live cells were gated by using forward–versus–side-scatter dot plots. Data were analyzed by using the CellQuest program (Becton Dickinson).
Cell cycle analysis.
Cells (5 × 105) under each set of conditions were stained for DNA and RNA content with 7-amino-actinomycin D and pyronin Y, respectively, as previously described (23) with some modifications. Briefly, cells were suspended in a buffer containing 0.03% saponin (Sigma). Fifty microliters of 400 μM 7-amino-actinomycin D (Calbiochem, La Jolla, Calif.) was added at a final concentration of 20 μM. Cells were incubated at room temperature for 30 min and cooled on ice for at least 5 min, and 3 μl of 1.7 mM pyronin Y (Polysciences, Warrington, Pa.) was added to achieve a final concentration of 5 μM, after which the cells were incubated for an additional 10 min on ice and analyzed. Data were accumulated on a FACStarplus flow cytometer and analyzed with the CellQuest program.
Quantitative PCR.
Cells to be subjected to quantitative PCR were harvested and washed, and DNAs were extracted as previously described (26). Cells were washed in phosphate-buffered saline, lysed in urea lysis buffer (4.7 M urea, 1.3% [wt/vol] sodium dodecyl sulfate, 0.23 M NaCl, 0.67 mM EDTA [pH 8.0], 6.7 mM Tris-HCl [pH 8.0]), and subjected to phenol-chloroform extraction and ethanol precipitation. Total nucleic acids resulting from this extraction procedure were used for PCR amplification. Quantitative PCR was performed with primers specific for HIV-1 sequences as previously described (26). The primer pairs M667 and AA55 (R and U5) and M667 and M661 (long terminal repeat [LTR] and gag) were used to detect initiation and completion of the HIV-1 reverse transcription process, respectively. Primers specific for the human β-globin gene were used to determine input of cellular DNA. One primer from each pair was end labeled with 32P. Following 25 cycles of PCR, samples were resolved on a 6% polyacrylamide gel and quantitation was performed by value comparison to a standard curve of known amounts of HIV-1 DNA or cellular DNA from uninfected human PBLs with an Ambis (San Diego, Calif.) radioanalytic imager.
Statistical analysis.
Rates of reverse transcription in nucleoside-treated and untreated cells were compared by paired-sample t tests.
RESULTS
Effect of exogenous nucleosides on HIV reverse transcription.
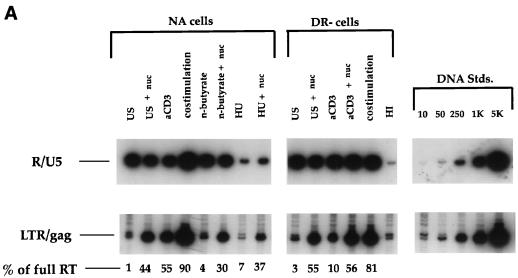
We studied the effect of nucleoside addition on the efficiency of the HIV-1 reverse transcription process in highly purified, CD4+ T cells arrested in the G0 or G1a stage of the cell cycle. Unstimulated T cells are in the G0 state prior to progressing into the cell cycle. In contrast, DR− T cells partially activated with anti-CD3 antibodies alone or NA cells costimulated with anti-CD3 and anti-CD28 antibodies in the presence of the cell cycle inhibitor n-butyrate are arrested at the G1a phase of the cell cycle (11). Since the intracellular dNTP concentration in cells at these phases of the cell cycle is very low and is controlled by the salvaging of extracellular deoxynucleosides, cells were cultured in the presence or absence of a 10 μM concentration of all four deoxynucleosides 2 h prior to, as well as immediately following, infection with the CXCR4-tropic HIV-1NL4-3 viral strain. To assess the efficiency of the reverse transcription process under each set of conditions, we employed quantitative PCR, using the R-U5 and LTR-gag oligonucleotide primer pairs to detect the first region of the viral genome synthesized and a region present only in essentially completed reverse transcripts, respectively. In Fig. 1, NA cell populations were used for experiments in which cells were treated with the drugs n-butyrate and HU, as well as for control experiments for the different activation pathways. Figure 1A shows that following pretreatment with exogenous nucleosides, there was a substantial increase in the amount of full-length reverse transcripts in either the NA or DR− cell populations, arrested in the G0 or G1a phase of the cell cycle. A similar increase occurred in cells treated prior to costimulation with HU, a ribonucleotide reductase inhibitor, indicating that the salvage of deoxynucleosides can override the function of the enzyme ribonucleotide reductase. These results implicate the levels of nucleotide pools in the efficiency of the reverse transcription process.
FIG. 1.
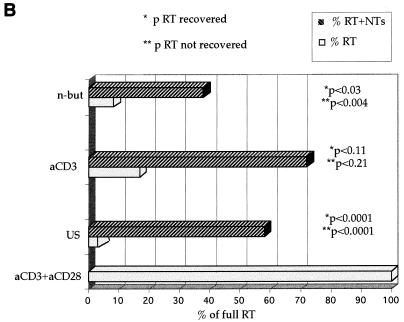
(A) Effect of exogenous nucleosides on HIV reverse transcription. Cells treated as indicated were infected with NL4-3 in the presence or absence of 10 μM of deoxynucleosides (nuc) per ml. US, unstimulated cells; αCD3, cells stimulated with αCD3 alone; costimulation, cells stimulated with αCD3 and αCD28; n-butyrate, costimulated cells treated with n-butyrate prior to costimulation; HU, costimulated cells treated with HU prior to costimulation; HI, cells infected with a heat-inactivated virus, as a negative control for reverse transcription; RT, reverse transcripts. At 17 h postinfection, DNA was harvested and subjected to quantitative PCR with the primer pairs for the R and U5 regions and the LTR and gag regions to detect initiation and completion of the HIV-1 reverse transcription process. Quantitative standards (Stds.) for 10 to 5,000 copies of viral DNA amplified in parallel are shown on the right. (B) Statistical analysis to assess the significance of the nucleoside-induced rescue of the reverse transcription process. The data are compiled from seven experiments to assess the effect of nucleoside addition on amounts of complete reverse transcripts in infected cells arrested in the G0 (US [for unstimulated]) or G1a (αCD3 and n-but [for n-butyrate]) phase of the cell cycle, compared to that on fully activated cells (αCD3+αCD28). *p, P values for the significance of the level of reverse transcripts (RT) recovered compared to the level recovered in cells without nucleoside addition; **p, P values for the significance of the level of reverse transcripts not recovered in nucleotide (NT)-treated cells compared to the level not recovered in the costimulated control cells.
We next assessed the extent of the rescue of the nucleoside-induced reverse transcription process in cells arrested in the G0 or G1a phase of the cell cycle. Data were collected from multiple experiments to analyze the effect of nucleoside addition on amounts of complete reverse transcripts. The reverse transcription in G0- or G1a-arrested cells was compared to that in fully activated cells costimulated with anti-CD3 and anti-CD28 antibodies. Statistical analysis of the results, summarized in Fig. 1B, indicated a significantly increased efficiency of the reverse transcription process in cells treated with nucleosides prior to and immediately following infection. However, the increased levels of reverse transcripts were also significantly lower than levels seen in cells costimulated prior to infection. Similar results were seen following treatment of cells with either 10 or 50 μM nucleosides. Thus, while nucleoside addition increases the efficiency of reverse transcription, it does not fully reconstitute the potential levels seen for reverse transcription in activated cells.
Effect of exogenous nucleosides on the cell cycle.
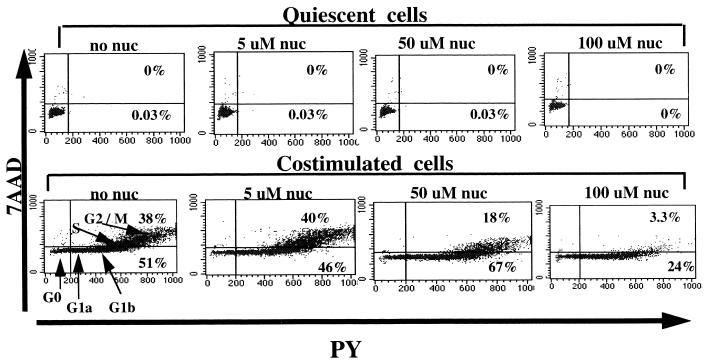
We next determined whether the addition of exogenous nucleosides to the culture medium alters the state of activation and cell cycle progression of quiescent or activated cells. Noninfected, unstimulated cells and cells costimulated with anti CD3 and CD28 antibodies were cultured in the absence or presence of increasing concentrations of exogenous nucleosides for 2 days. Cell cycle progression under all conditions was determined daily by a flow-cytometric technique that allows DNA and RNA quantitation, as described elsewhere (11) and in Materials and Methods. As seen in Fig. 2, there is little change in the state of the cell cycle of the unstimulated (upper panels) or costimulated (lower panels) cells on day 2 following culture with up to 50 μM exogenous nucleosides. However, the addition of 100 μM nucleosides 2 h prior to infection and again immediately thereafter prevented the progression of costimulated cells from the G1b to the S phase of the cell cycle (lower panels). Thus, high concentrations of exogenously added nucleosides can be inhibitory to proliferating cells. As such, subsequent studies were performed with 10 μM nucleosides.
FIG. 2.
Effect of exogenous nucleosides on cell cycle. Cell cycle analysis of unstimulated (upper panels) and costimulated (lower panels) cells cultured in the absence or presence of increasing concentrations of exogenous nucleosides (nuc) for 2 days. The different phases of the cell cycle are shown in the lower left panel. Percentages of cells at the different phases of the cell cycle are indicated inside the respective quadrants for each of the conditions. 7AAD, 7-amino-actinomycin D. PY, pyronin Y.
The stability and functionality of the recovered reverse transcripts in G0 cells.
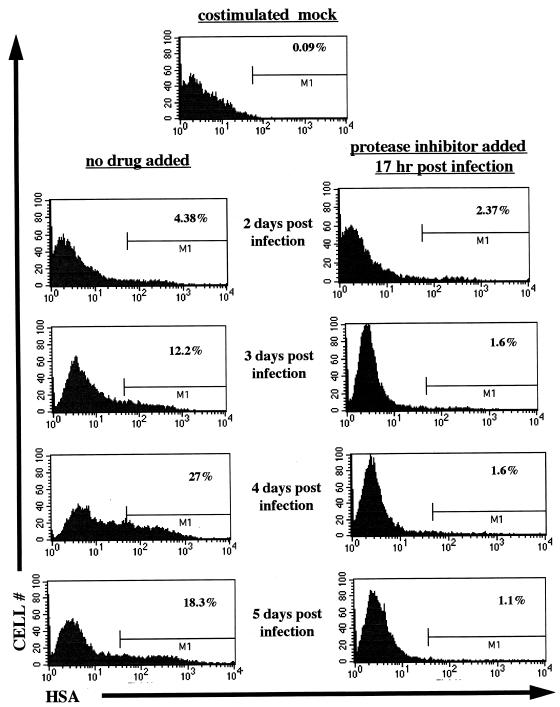
Our previous studies have shown an inefficient viral rescue following mitogenic stimulation of quiescent cells infected with replication-defective HIV-1JRCSF pseudotyped with a murine amphotropic envelope glycoprotein (27). To define the stability and functionality of the recovered reverse transcripts following the addition of exogenous nucleosides to G0 cells, we used NL-r-HSAS, a replication-competent HIV-1 reporter virus, that expresses the murine HSA on the surfaces of infected cells (9). The HSA gene is inserted in place of the HIV-1NL4-3 vpr gene, and expression can be detected on infected cells by flow cytometry, thus identifying productive infection on a per cell basis. Infected cells were treated with the PI Indinavir following exposure to virus, in order to prevent viral replication and obtain a single-cycle infection. To examine the efficiency of the PI treatment, prestimulated cells were infected with NL-r-HSAS and 17 h postinfection 100 nM PI was added. Following infection, cells were stained and analyzed for HSA expression. As shown in Fig. 3, 2 days postinfection, HSA+ cells were detected in about the same amounts in both PI-treated and nontreated cells. However, at the later time points, viral spread was clearly noticeable in cells not treated with PI, as indicated by the increasing numbers of HSA+ cells. In cells treated with PI, the initial number of HSA+ cells remained constant for a period before declining. This loss of HSA+ cells at the later time point is most likely due to death of infected cells. Thus, viral expression is not altered by PI treatment; however, spread to new cells is inhibited. This method provides a single-cycle replication assay to assess levels of productively infected cells.
FIG. 3.
Postinfection addition of PI prevents HIV replication. (Right panels) PI (100 nM) was added 17 h following infection of costimulated cells with the reporter virus NL-r-HSAS. (Left panels) Cells that received no drug. At the indicated time points, cells were collected and stained for the virus-encoded HSA surface marker. Percentages of HSA+ cells are indicated at the M1 gate in the corresponding histograms. Results with mock-infected cells are shown in the top panel.
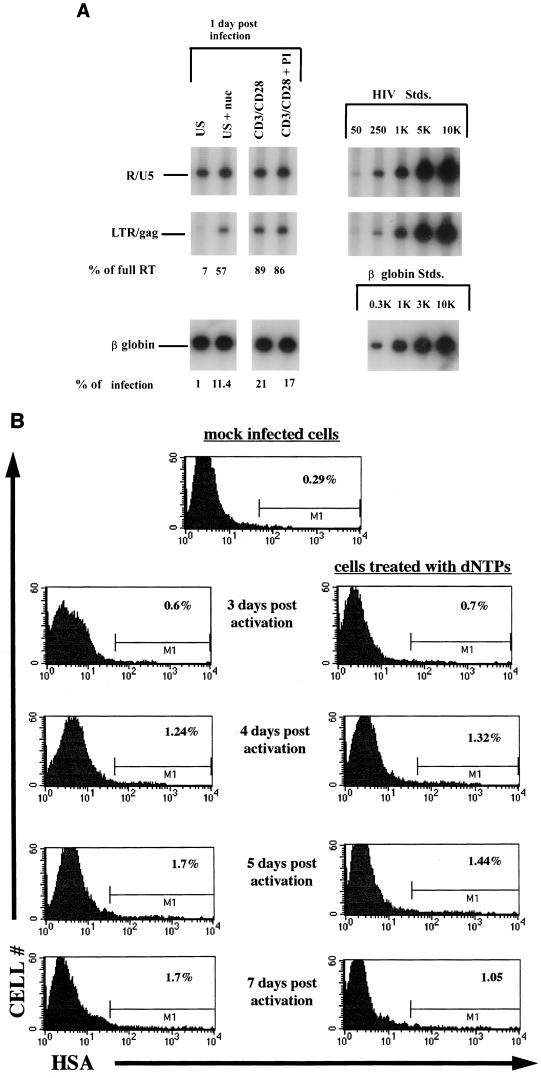
To determine if the increased levels of reverse transcripts seen following nucleoside treatment were functional, highly purified CD4+ T cells in the G0 phase of the cell cycle, as well as previously costimulated cells, were infected with the NL4-3-based reporter virus NL-r-HSAS. Half of the cells were treated with 10 μM deoxyribonucleosides 2 h prior to and an additional 10 μM immediately following the infection. Seventeen hours postinfection, some of the cells were harvested and DNA was extracted for PCR analysis. The remainder of the unstimulated cells were costimulated with anti-CD3 and anti-CD28 antibodies in the presence or absence of 100 nM PI. Figure 4A shows the predicted increase in efficiency of reverse transcription from 7 to 57% following the addition of exogenous nucleosides to G0 cells. In addition, precostimulated cells treated with PI exhibited a similar level of efficiency in reverse transcription, indicating no effect caused by PI on the reverse transcription process itself. To determine the effect of nucleoside addition on virus expression following costimulation, cells were costained for HSA and CD4 surface markers at subsequent time points. As shown in Fig. 4B, the levels of productive infection in quiescent cells not treated with exogenous nucleosides were consistent with the low level of full reverse transcription seen (Fig. 4A). Interestingly, at all time points tested, the numbers of HSA+ cells in cultures treated with exogenous deoxynucleosides were not different from those in cultures that were not treated. Thus, despite an eightfold increase in levels of complete reverse transcripts, no increased rescue of productive infection was seen following activation of nucleoside-treated cells. To rule out the possibility that infection was affected by the usage of a vpr-negative virus, we performed all experiments in parallel with a reporter virus containing a functional vpr gene and HSA sequences inserted into the nef gene. We obtained similar results with this reporter; thus, the lack of rescue of productive infection following activation was not due specifically to the loss of vpr or nef.
FIG. 4.
(A) HIV-1 reverse transcription of cells infected with NL-r-HSAS following addition of exogenous nucleosides. Unstimulated and costimulated cells were infected with 3 μg of p24 of NL-r-HSAS per 106 cells in the presence or absence of 10 μM of deoxynucleosides (nuc) per ml. Seventeen hours later DNA was harvested and subjected to quantitative PCR with the primer pairs for the R-U5 and LTR-gag regions in the viral DNA and with a primer for the β-globin gene of the genomic DNA. Percentages of initiated reverse transcripts that completed the reverse transcription process (% of full RT) as well as percentages of cells in the population that harbor complete reverse transcription as determined by assessing levels of LTR and gag per β-globin signal (% of infection) are indicated for each of the conditions. US, unstimulated; CD3+CD28, costimulated. Quantitative standards (Stds.) are shown on the right for each primer pair and for the β-globin primer. (B) Effect of nucleoside addition on viral rescue. The quiescent or nucleoside-treated quiescent cells shown in panel A were costimulated 17 h postinfection in the presence of 100 nM PI. At the indicated time points cells were collected and stained for the virus-encoded HSA surface marker. Percentages of HSA+ cells are indicated in each of the corresponding histograms. The results are representative of three separate experiments.
DISCUSSION
Although full activation of T cells is not necessary for proviral DNA synthesis, we have previously shown that transition into the G1b phase of the cell cycle was required to efficiently complete the reverse transcription process (11). In the present work we studied the potential role of dNTP concentrations in the HIV-1 reverse transcription block in quiescent cells and cells arrested at the G1a phase of the cell cycle. We have used our previously described system of highly purified DR− CD4+ enriched primary T-cell cultures. In this system, resting lymphocytes are in the G0 phase of the cell cycle, while cells activated with anti-CD3 antibodies alone, or cells treated with the cell cycle inhibitor n-butyrate prior to costimulation with anti-CD3 and anti-CD28 antibodies, are arrested in the G1a phase of the cell cycle (11). Here we show that the addition of all four deoxyribonucleosides to culture media of cells in the G0 and G1a phase of the cell cycle partially alleviated the blockage of complete reverse transcription without having an effect on the state of the cell cycle. While the increased efficiency of the reverse transcription was significant, it was also significantly lower than that seen in fully activated T lymphocytes costimulated with both anti-CD3 and anti-CD28 antibodies. These results suggest that in addition to dNTP concentration, some other factor(s) may be needed to fully restore the levels of complete reverse transcripts in quiescent and partially activated T cells.
In studies designed to investigate the intracellular nucleotide pool, Cohen et al. (2) demonstrated a 10- to 40-fold increase in the dGTP, dCTP, and dTTP concentrations, and a 100-fold increase in dATP levels in resting T lymphocytes following exogenous addition of the corresponding deoxynucleosides. In addition, several studies have demonstrated the ability of resting T lymphocytes to phosphorylate exogenously added dideoxynucleoside analogs, which then compete with intracellular dNTPs and consequently inhibit HIV-1 proviral synthesis (4, 6, 17). Thus, while uptake of these nucleosides is less efficient in quiescent than in activated cells, our experiments show that these increased levels are sufficient to allow complete reverse transcription of the HIV genome. It was also previously shown that the addition of dNTPs to isolated HIV-1 virions could stimulate intravirion reverse transcription activity, leading to higher levels of virions carrying incomplete reverse transcripts (28, 29). In our system CD4+ T cells rather than cell-free virions were cultured in the presence of all four deoxynucleosides prior to and following, but not during, infection. Thus, the increase in the level of full reverse transcripts seen in our study is likely the result of higher levels of intracellular dNTPs rather than higher levels of dNTPs invirions.
We further assessed the stability and functionality of the full reverse transcripts in these cells. For this purpose, we used an HIV-1 reporter construct that expresses the murine HSA on the surfaces of infected cells and treated the cells with PI to prevent spread of infection as a result of viral replication. We showed that following costimulation of cells 17 h postinfection, no increased rescue of productive infection was seen in cells treated with exogenous nucleosides. These results indicate that either the complete reverse transcripts seen in cells treated with exogenous nucleosides may be defective or there may be an additional blockage of productive infection of the newly activated T cells. The latter possibility is consistent with some of our previous data (27) showing inefficient rescue of viral production by stimulation with phytohemagglutinin 15 h postinfection. Furthermore, our results support those of a previously published report by Tang et al. (21) that showed no rescue of viral expression in highly purified quiescent T cells activated postinfection. Thus, the newly synthesized partial or complete reverse transcripts may be highly labile in G0 cells prior to nuclear import and integration. A recent report by Kinoshita (10) looked at potential involvement of some early events in T-cell activation in the blockage of the HIV reverse transcription process. Kinoshita identified the transcription factor NFATc as a host factor that may be involved in controlling productive infection, possibly by improving the reverse transcription process in suboptimaly activated CD4+ cells. Our results are consistent with those conclusions; however, additional work is needed to further elucidate the fate of proviral DNA at the different stages of cellular activation.
ACKNOWLEDGMENTS
We thank Steve Cole for providing the statistical analysis for Fig. 1B.
This work was supported by NIH grants AI33259 and HL55205, the University of California—Los Angeles CFAR, and an AMGEN Fellowship from the UCLA AIDS Institute (to Y.D.K.). J.A.Z. is an Elizabeth Glaser Scientist supported by the Pediatric AIDS Foundation.
REFERENCES
- 1.Chen I S, Temin H M. Establishment of infection by spleen necrosis virus: inhibition in stationary cells and the role of secondary infection. J Virol. 1982;41:183–191. doi: 10.1128/jvi.41.1.183-191.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen A, Barankiewicz J, Lederman H M, Gelfand E W. Purine and pyrimidine metabolism in human T lymphocytes. Regulation of deoxyribonucleotide metabolism. J Biol Chem. 1983;258:12334–12340. [PubMed] [Google Scholar]
- 3.Eriksson S, Thelander L, Akerman M. Allosteric regulation of calf thymus ribonucleoside diphosphate reductase. Biochemistry. 1979;18:2948–2952. doi: 10.1021/bi00581a005. [DOI] [PubMed] [Google Scholar]
- 4.Gao W Y, Agbaria R, Driscoll J S, Mitsuya H. Divergent anti-human immunodeficiency virus activity and anabolic phosphorylation of 2′,3′-dideoxynucleoside analogs in resting and activated human cells. J Biol Chem. 1994;269:12633–12638. [PubMed] [Google Scholar]
- 5.Gao W Y, Cara A, Gallo R C, Lori F. Low levels of deoxynucleotides in peripheral blood lymphocytes: a strategy to inhibit human immunodeficiency virus type 1 replication. Proc Natl Acad Sci USA. 1993;90:8925–8928. doi: 10.1073/pnas.90.19.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao W Y, Shirasaka T, Johns D G, Broder S, Mitsuya H. Differential phosphorylation of azidothymidine, dideoxycytidine, and dideoxyinosine in resting and activated peripheral blood mononuclear cells. J Clin Investig. 1993;91:2326–2333. doi: 10.1172/JCI116463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goulaouic H, Subra F, Mouscadet J F, Carteau S, Auclair C. Exogenous nucleosides promote the completion of MoMLV DNA synthesis in G0-arrested Balb c/3T3 fibroblasts. Virology. 1994;200:87–97. doi: 10.1006/viro.1994.1166. [DOI] [PubMed] [Google Scholar]
- 8.Heinzinger N, Baca-Regen L, Stevenson M, Gendelman H E. Efficient synthesis of viral nucleic acids following monocyte infection by HIV-1. Virology. 1995;206:731–735. doi: 10.1016/s0042-6822(95)80097-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jamieson B D, Zack J A. In vivo pathogenesis of a human immunodeficiency virus type 1 reporter virus. J Virol. 1998;72:6520–6526. doi: 10.1128/jvi.72.8.6520-6526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinoshita S. Host control of HIV-1 parasitism in T cells by the nuclear factor of activated T cells. Cell. 1998;95:595–604. doi: 10.1016/s0092-8674(00)81630-x. [DOI] [PubMed] [Google Scholar]
- 11.Korin Y D, Zack J A. Progression to the G1b phase of the cell cycle is required for completion of human immunodeficiency virus type 1 reverse transcription in T cells. J Virol. 1998;72:3161–3168. doi: 10.1128/jvi.72.4.3161-3168.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li G, Simm M, Potash M J, Volsky D J. Human immunodeficiency virus type 1 DNA synthesis, integration, and efficient viral replication in growth-arrested T cells. J Virol. 1993;67:3969–3977. doi: 10.1128/jvi.67.7.3969-3977.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lori F, Malykh A, Cara A, Sun D, Weinstein J N, Lisziewicz J, Gallo R C. Hydroxyurea as an inhibitor of human immunodeficiency virus-type 1 replication. Science. 1994;266:801–805. doi: 10.1126/science.7973634. [DOI] [PubMed] [Google Scholar]
- 14.Marijnen Y M, de Korte D, Haverkort W A, den Breejen E J, van Gennip A H, Roos D. Studies on the incorporation of precursors into purine and pyrimidine nucleotides via ‘de novo’ and ‘salvage’ pathways in normal lymphocytes and lymphoblastic cell-line cells. Biochim Biophys Acta. 1989;1012:148–155. doi: 10.1016/0167-4889(89)90088-8. [DOI] [PubMed] [Google Scholar]
- 15.Meyerhans A, Vartanian J P, Hultgren C, Plikat U, Karlsson A, Wang L, Eriksson S, Wain-Hobson S. Restriction and enhancement of human immunodeficiency virus type 1 replication by modulation of intracellular deoxynucleoside triphosphate pools. J Virol. 1994;68:535–540. doi: 10.1128/jvi.68.1.535-540.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Brien W A, Namazi A, Kalhor H, Mao S H, Zack J A, Chen I S. Kinetics of human immunodeficiency virus type 1 reverse transcription in blood mononuclear phagocytes are slowed by limitations of nucleotide precursors. J Virol. 1994;68:1258–1263. doi: 10.1128/jvi.68.2.1258-1263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirasaka T, Chokekijchai S, Yamada A, Gosselin G, Imbach J L, Mitsuya H. Comparative analysis of anti-human immunodeficiency virus type 1 activities of dideoxynucleoside analogs in resting and activated peripheral blood mononuclear cells. Antimicrob Agents Chemother. 1995;39:2555–2559. doi: 10.1128/aac.39.11.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spaapen L J, Scharenberg J G, Zegers B J, Rijkers G T, Duran M, Wadman S K. Intracellular purine and pyrimidine nucleotide pools of human T and B lymphocytes. Adv Exp Med Biol. 1986;195A:567–573. doi: 10.1007/978-1-4684-5104-7_95. [DOI] [PubMed] [Google Scholar]
- 19.Stevenson M, Brichacek B, Heinzinger N, Swindells S, Pirruccello S, Janoff E, Emerman M. Molecular basis of cell cycle dependent HIV-1 replication. Implications for control of virus burden. Adv Exp Med Biol. 1995;374:33–45. doi: 10.1007/978-1-4615-1995-9_4. [DOI] [PubMed] [Google Scholar]
- 20.Stevenson M, Stanwick T L, Dempsey M P, Lamonica C A. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 1990;9:1551–1560. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang S, Patterson B, Levy J A. Highly purified quiescent human peripheral blood CD4+ T cells are infectible by human immunodeficiency virus but do not release virus after activation. J Virol. 1995;69:5659–5665. doi: 10.1128/jvi.69.9.5659-5665.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thelander L, Reichard P. Reduction of ribonucleotides. Annu Rev Biochem. 1979;48:133–158. doi: 10.1146/annurev.bi.48.070179.001025. [DOI] [PubMed] [Google Scholar]
- 23.Toba K, Winton E F, Koike T, Shibata A. Simultaneous three-color analysis of the surface phenotype and DNA-RNA quantitation using 7-amino-actinomycin D and pyronin Y. J Immunol Methods. 1995;182:193–207. doi: 10.1016/0022-1759(95)00050-k. [DOI] [PubMed] [Google Scholar]
- 24.Varmus H E, Padgett T, Heasley S, Simon G, Bishop J M. Cellular functions are required for the synthesis and integration of avian sarcoma virus-specific DNA. Cell. 1977;11:307–319. doi: 10.1016/0092-8674(77)90047-2. [DOI] [PubMed] [Google Scholar]
- 25.Zack J A. The role of the cell cycle in HIV-1 infection. Adv Exp Med Biol. 1995;374:27–31. doi: 10.1007/978-1-4615-1995-9_3. [DOI] [PubMed] [Google Scholar]
- 26.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 27.Zack J A, Haislip A M, Krogstad P, Chen I S. Incompletely reverse-transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retroviral life cycle. J Virol. 1992;66:1717–1725. doi: 10.1128/jvi.66.3.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Dornadula G, Pomerantz R J. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenvironments: an important stage for viral infection of nondividing cells. J Virol. 1996;70:2809–2824. doi: 10.1128/jvi.70.5.2809-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Zhang Y, Spicer T P, Abbott L Z, Abbott M, Poiesz B J. Reverse transcription takes place within extracellular HIV-1 virions: potential biological significance. AIDS Res Hum Retroviruses. 1993;9:1287–1296. doi: 10.1089/aid.1993.9.1287. [DOI] [PubMed] [Google Scholar]