Abstract
The genome of hepatitis delta virus (HDV) is a 1,679-nucleotide (nt) single-stranded circular RNA that is predicted to fold into an unbranched rodlike structure. During replication, two complementary RNAs are also detected: an exact complement, referred to as the antigenome, and an 800-nt polyadenylated RNA that could act as the mRNA for the delta antigen. We used a 5′ rapid amplification of cDNA ends procedure, followed by cloning and sequencing, to determine the 5′ ends of the polyadenylated RNAs produced during HDV genome replication following initiation under different experimental conditions. The analyzed RNAs were from the liver of an infected woodchuck and from a liver cell line at 6 days after transfection with either an HDV cDNA or ribonucleoprotein (RNP) complexes assembled in vitro with HDV genomic RNA and purified recombinant small delta protein. In all three situations the 5′ ends mapped specifically to nt 1630. In relationship to what is called the top end of the unbranched rodlike structure predicted for the genomic RNA template, this site is located 10 nt from the top, and in the middle of a 3-nt external bulge. Following transfection with RNP, such specific 5′ ends could be detected as early as 24 h. We next constructed a series of mutants of this predicted bulge region and of an adjacent 6-bp stem and the top 5-nt loop. Some of these mutations decreased the ability of the genome to undergo antigenomic RNA synthesis and accumulation and/or altered the location of the detected 5′ ends. The observed end located at nt 1630, and most of the novel 5′ ends, were consistent with transcription initiation events that preferentially used a purine. The present studies do not prove that the detected 5′ ends correspond to initiation sites and do not establish the hypothesis that there is a promoter element in the vicinity, but they do show that the location of the observed 5′ ends could be controlled by nucleotide sequences at and around nt 1630.
Three major stable RNA species are thought to be involved in the replication of the hepatitis delta virus (HDV) RNA genome: (i) the genomic 1,679-nucleotide (nt) circular single-stranded RNA, (ii) its exact complement, the antigenome, and, least abundant, (iii) an 800-nt polyadenylated RNA that is presumed to be the mRNA for the delta protein (17). Using primer extension on the polyadenylated RNA extracted from cultured cells undergoing HDV genome replication, we reported a 5′ end that mapped to nt 1631 ± 1 (6). For unexplained reasons, this polyadenylated species was not always detectable by Northern analyses and/or primer extension (6). Nevertheless, three subsequent studies report confirmation of a 5′ end in the vicinity of nt 1631 (1, 12, 18) and a recent study has reported that nonpolyadenylated antigenomic RNAs can have a 5′ end at nt 1646 (12).
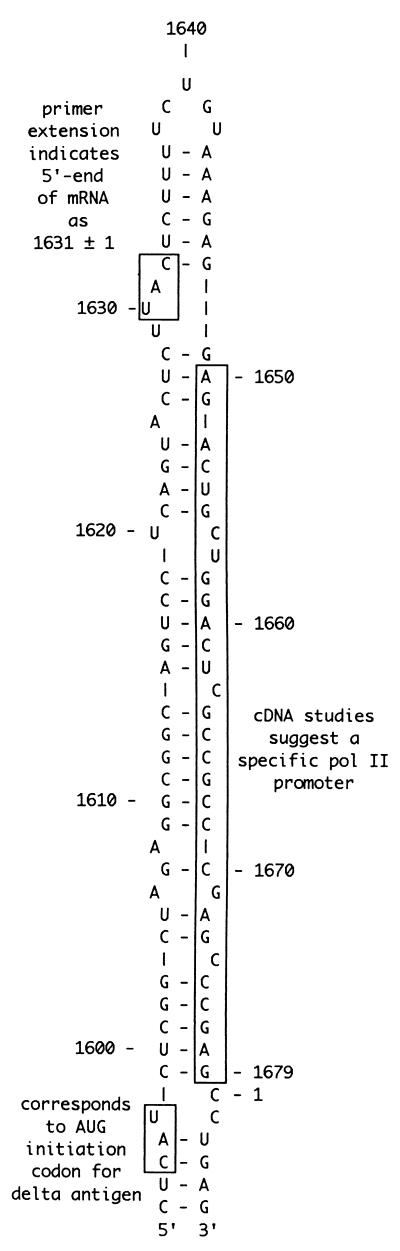
The site at nt 1631, as indicated in Fig. 1, is located 9 nt from what we refer to as the top end of the predicted rodlike genomic RNA. One hypothesis is that the 5′ end corresponds to a site of transcription initiation by RNA polymerase II (17). However, if initiation is at nt 1631, it will begin with UTP, which is puzzling, since transcription by Pol II, at least from the normal DNA templates, usually begins preferentially with ATP or GTP (4). A second hypothesis, which is linked to the first, is that nearby sequence elements define a promoter for Pol II (10). There are data, obtained by using cDNA constructs, which suggest that a sequence as short as 30 nt (indicated in Fig. 1) can, when expressed as a double-stranded DNA, act as a promoter for RNA polymerase II (10). There are also in vitro transcription studies with HDV genomic RNA added to nuclear extracts, which are claimed to support both initiation of transcription near nt 1631 and the concept of a nearby promoter (1).
FIG. 1.
Predicted rodlike folding of nucleotide sequences present at one end of the genome HDV RNA. The nucleotide sequence and numbering are as in Kuo et al. (9). This end is sometimes referred to as the top of the rod (21). The three sequence features indicated by boxes are the site corresponding to the initiation codon for the small delta protein (19), the 5′ end of the polyadenylated mRNA as determined by primer extension (6), and the location of a 30-nt sequence which, when expressed as a double-stranded cDNA, is reported to have promoter activity in transfected cells (10).
To test these two hypotheses further, we and others have previously investigated mutations in this region of the rodlike structure and observed some significant effects on genome replication (1, 18, 21). However, more relevant for understanding initiation are the determination of the 5′ end(s) for the polyadenylated RNAs and an analysis of whether the location of this end(s) is changed by alterations in the local sequence and structure of the rodlike genomic RNA template.
The initial aim of the present study, therefore, was to apply a sensitive and specific 5′ rapid amplification of cDNA ends (RACE) procedure to characterize the 5′ ends of the polyadenylated HDV antigenomic RNAs, synthesized under different conditions of HDV genome replication. Primer extension involves the assumption that the elongated species are 100% identical to the template strand; it would neither detect nontemplated 5′ nucleotides, as occurs naturally for influenza virus (15), nor the artifactual addition of nontemplated nucleotides by reverse transcriptase (14). In contrast, 5′ RACE determines the actual sequence of the transcript. With this approach, we observed specificity for 5′ ends located at nt 1630. We then went on to use this 5′-RACE method to study the 5′ ends obtained during replication of various mutants of HDV, especially those with base changes near the site at nt 1630.
MATERIALS AND METHODS
RNA from the liver of an infected woodchuck.
A woodchuck chronically infected with woodchuck hepatitis virus was superinfected with HDV. After 25 days the animal was sacrificed, and a liver sample (0.5 g) was taken and homogenized in a buffer containing guanidine isothiocyanate. Cesium chloride was then added, and after centrifugation to equilibrium, the RNA band was collected and the RNA was precipitated by the addition of salt and ethanol, all as previously described (3).
Oligo(dT) fractionation of liver RNA.
The total liver RNA in 0.5 M NaCl–0.01 M Tris (pH 7.5)–0.1% sodium dodecyl sulfate, was applied to a column of 0.5 g of oligo(dT)-cellulose (Sigma). The column was washed with the same buffer and then eluted with 0.01 M Tris (pH 7.5)–0.1% sodium dodecyl sulfate. This cycle was repeated a total of six times. From 0.5 g of liver we obtained 10 to 50 μg of poly(A)-containing RNA. The quality of the HDV mRNA was confirmed by Northern analysis.
Genome mutants.
Mutations were constructed in pDL553, a plasmid based on the vector pSVL (Pharmacia) and containing 1.2 cDNA copies of the HDV genome. According to the nucleotide sequence of Kuo et al. (9), the insert is from the unique NheI site (nt 430) to NheI to XbaI (nt 781). Four of the mutants used in this study were described previously (21). Additional mutants were constructed by using the strategy described by Wu et al. (21). All the mutations were made near the top of the rod, i.e., between nucleotide positions 1629 and 1648 on the genome.
In vitro RNA transcription and RNP assembly.
Genomic RNA was transcribed from plasmid pTW101, which contains 1.1 copies of the HDV genome (StyI [nt 621] to StyI to XbaI [nt 781]) inserted between a T7 promoter and terminator (21). Ribonucleoprotein (RNP) was assembled by combining this RNA (500 ng) with purified recombinant small delta protein (200 ng) (provided by Zuccola and Hogle), as previously described (5).
Transfection, RNA extraction, and Northern analysis.
Huh7 cells (13) in a 35-mm-diameter culture dish were grown to approximately 70% confluence before transfection with cDNA, RNA, or RNP with Lipofectamine or Lipofectamine Plus (Life Sciences) according to the manufacturer’s instructions. Total RNA was harvested with Tri reagent (Molecular Research Center, Inc.).
Northern analysis was performed to measure the accumulation of HDV RNAs during the replication of wild-type and mutant genomes, both in infected livers and in cells transfected with cDNA or RNP. RNA (1 to 5 μg) was glyoxylated and submitted to electrophoresis through a gel of 1.5% agarose (21). After electrotransfer to a charged nylon membrane (Zeta-probe; Bio-Rad), genomic or antigenomic RNA was detected with RNA probes transcribed in the presence of [α-32P]UTP (DuPont). Radioactivity was detected and quantitated with a phosphorimager system (Fuji Bio-Imaging).
5′ RACE.
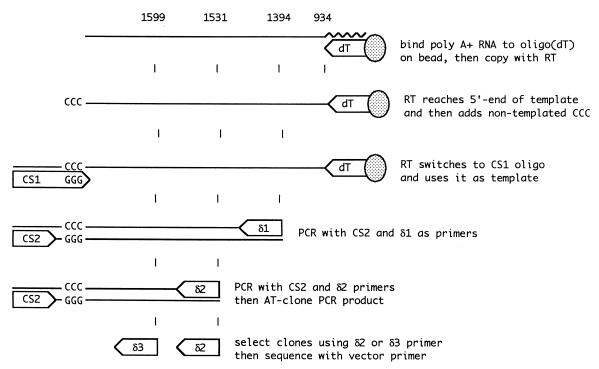
5′ RACE was performed with a Clontech CapFinder kit, which is designed to allow amplification of the 5′ end of messenger RNAs by reverse transcription (RT)-PCR. Our amplification strategy was based on the manufacturer’s recommendations and is represented in Fig. 2. As input RNA for this procedure, we used either polyadenylated RNAs selected from infected liver tissue or total RNAs extracted from transfected cells. These RNAs (10 μg), in water, were heat denatured for 2 min at 95°C and then specifically bound in 1 M NaCl–20 mM Tris (pH 7.5)–2 mM EDTA to avidin-coated superparamagnetic beads (10 μl; Dynal) that had been preincubated in the same buffer with an excess of 5′ biotin-(dT)30. The immobilized RNA was added to an RT reaction mixture (10 μl) containing 50 mM Tris (pH 8.3), 6 mM MgCl2, 75 mM KCl, 2 mM dithiothreitol, 1 mM deoxynucleoside triphosphates, and 100 U of Superscript II reverse transcriptase (Life Sciences) and then incubated at 42°C for 90 min. The reaction mixture also contained 10 pmol of Capswitch oligonucleotide 1 (CS1) (5′-TACGGCTGCGAGAAGACGACAGAAGGG-3′). This oligonucleotide is considered to function as follows. When reverse transcriptase reaches the authentic 5′ end of the mRNA, its terminal transferase activity adds several nontemplated nucleotides to the 3′ end of the cDNA (14). If these extra nucleotides are CCC, the sequence GGG at the 3′ end of CS1 can hybridize and allow the reverse transcriptase to switch templates from the mRNA to CS1. This results in a cDNA that extends past the 5′ end of the mRNA to incorporate a copy of the CS1 sequence. For the subsequent PCRs, the cDNA then has a specific primer-binding site at its 3′ end.
FIG. 2.
The 5′-RACE strategy. As described in Materials and Methods, a CapFinder kit from Clontech was adapted to determine the 5′ ends of polyadenylated antigenomic HDV RNAs. Three of the numbers at the top of the diagram represent the 5′ ends of the three HDV primers, using the genome numbering of Kuo et al. (9). The fourth number, 934, indicates the site of poly(A) addition (6).
To make the 5′ RACE specific for HDV mRNA, a seminested PCR was performed on this cDNA. The first PCR was done with the kit Capswitch primer 2 (CS2) (5′-TACGGCTGCGAGAAGACGACAGAA-3′) and an HDV-specific primer, δ1 (5′-GGGAGCCCCCTCTCCATCCTTATCCTTCTTTCCGA-3′), located at nt 1392 to 1426. (In some cases we used, as an alternative to δ1, the primer 5′-GAGCCCCCTCTCCATCCTTATCC-3′, located at nt 1394 to 1416.) The predicted product size was approximately 240 bp, given the suspected location of the 5′ end of the polyadenylated RNA at nt 1630. The second PCR was done with CS2 and a nested HDV primer, δ2 (5′-CCGGCCACCCACTGCTCGA-3′), located at nt 1531 to 1549, which would give a product of approximately 130 bp. The reaction conditions were as follows: primers, 1 μM each; deoxynucleoside triphosphates, 0.2 mM; Klentaq Advantage DNA polymerase buffer (40 mM Tricine-KOH [pH 9.2], 15 mM potassium acetate, 3.5 mM magnesium acetate, and 75 μg of bovine serum albumin/ml) (Clontech), and 1× Advantage Polymerase Mix (Clontech). Amplifications were performed with an Idaho Technologies 1605 air thermocycler. An initial denaturation at 95°C for 2 min was followed by 35 cycles of 95°C for 1 s, annealing at 58°C for 1 s, and extension at 68°C for 30 s for the first PCR or 10 s for the second PCR.
The products of the seminested PCR were extracted with phenol and precipitated with ethanol. Then they were cloned via the T overhang of the PCR product into the vector pCRII (Invitrogen). In later experiments the PCR products were cloned with a TOPO-2 cloning kit (Invitrogen). Positive colonies were identified by hybridization with an HDV [γ-32P]ATP-labeled oligonucleotide, δ3 (5′-CTCGGCTAGAGGCGGCAGTCCTCAGTA-3′), located at nt 1599 to 1625. Plasmid DNA was isolated by a miniprep procedure (PerfectPrep; 5 Prime 3 Prime), and the sequence of the insert was determined by automated sequencing (Applied Biosystems). In >90% of the clones, the 3′ limit of the HDV genomic sequence was followed by GGGG, and in <10% it was followed by GGG. In this approach, if the 5′ end of the antigenomic template corresponds to C, we can have a corresponding uncertainty in a 5′-RACE determination of the 5′ end.
To test the specificity of this 5′-RACE procedure, we used an RNA with 3′ polyadenylation and a known 5′ end, as synthesized in vitro by T7 RNA polymerase. For such transcription a double-stranded DNA template was generated by expression PCR (7). The primers were 5′-biotin-GGATCCTAATACGACTCACTATAGGGAGGagaaaagagtaagag-3′ and 5′-(T)25gagtggaaacccgct-3′. The first primer encodes a T7 promoter joined to 15 nt of HDV antigenomic sequences (lowercase) beginning at nt 1640. The second primer ends transcription of HDV sequences (lowercase) at nt 935, which corresponds to the natural polyadenylation site of HDV mRNA (6). The RNA product of in vitro transcription was then submitted to the 5′-RACE procedure. The product of the second PCR was homogeneous in size. It was cloned with the TOPO-2 kit. Five clones were sequenced, and each began with the sequence 5′-GGGAGGagaaaag-3′. This 5′ end was as expected for T7 polymerase initiation on the DNA template.
RESULTS
HDV RNA from liver tissue and cultured cells in which the wild-type genome was replicating.
An 800-nt polyadenylated HDV RNA was first observed in the liver of an infected chimpanzee by Northern analysis (2). By primer extension on polyadenylated RNA from cells transfected with HDV cDNA, the 5′ end was mapped to nt 1631 ± 1 (6), using the genome numbering of Kuo et al. (9). As an independent approach to detecting these 5′ ends, we used a 5′-RACE procedure, as summarized in Fig. 2 and described in Materials and Methods, and applied it to the polyadenylated RNA isolated from the liver of a woodchuck at the peak of an HDV infection. All 27 of 27 clones sequenced indicated a 5′ end corresponding to nt 1630. A similar study was made with the RNAs from Huh7 cells at 6 days after transfection with a cDNA construct, pSVL(D3), which is known to initiate HDV genome replication (8). In this experiment 27 of 32 clones indicated 5′ ends at nt 1630. The other five clones indicated ends at nt 1620, 1627, 1655, 1664, and 1667.
Thus, our results with 5′ RACE were within 1 nt of those obtained earlier by primer extension but were still not the same. Thus, as a control for the specificity of the 5′-RACE procedure, we applied it to a polyadenylated RNA with a known 5′ end. As described in Materials and Methods, we found that our observations corresponded to the expected 5′ end. Thus, we trust data obtained by 5′ RACE over that obtained by primer extension. We speculate that interpretation of the primer extension data was deceived by what is now known to be a property of reverse transcriptase, namely, the ability to add one or more nontemplated nucleotides to the 3′ end of a primer extension product (14).
In summary, these 5′-RACE studies provide an answer different from that obtained by primer extension. Furthermore, they show that even for different experimental situations of wild-type HDV genome replication, nt 1630 is the predominant site for the 5′ end of the polyadenylated RNA generated.
Thus, if nt 1630 is an initiation site, then the RNA transcript begins with an adenosine, which is more consistent with initiation by RNA polymerase II (see Discussion). Furthermore, if the predicted rodlike structure of the genomic RNA is relevant to initiation, then this initiation occurs with an unpaired uridine, located in the middle of a 3-nt external bulge, as a template (Fig. 1).
HDV RNA from cells in which mutated HDV genomes were replicating.
The above-mentioned 5′-RACE studies showed that transfection of cells with cDNAs gave 5′ ends largely in agreement with those detected for RNA isolated from the liver of an infected woodchuck. Therefore, we used this cDNA strategy to examine the importance of the nucleotide sequence and predicted secondary structure of the genomic HDV RNA template in determining this initiation site (Fig. 1). As will be explained, we found that the introduction of mutations could cause a decrease in the accumulation of antigenomic RNA and/or changes in the position of the 5′ end. As summarized in Table 1, the locations focused on three areas: the 3-nt external bulge, which includes nt 1630 and also the top 5-nt loop, and the adjacent 6-bp stem.
TABLE 1.
Summary of genome mutations and corresponding sites on genome for 5′ ends deduced from 5′ RACE of polyadenylated RNA
| HDV genome mutagenesis targeta | Predicted changes of top structure elementsb | Antigenomic RNA accumulation (%) | Total no. of clones sequencedc | Occurrence of 5′ endsd
|
|
|---|---|---|---|---|---|
| Predominant site | Additional site(s) | ||||
| 3-nt external bulge | |||||
| U(1630)C | Bulge sequence | 25 | 7 | C(1630)7 | |
| U(1630)G | Bulge sequence | 6 | 11 | U(1629)9 | U(1642)2 |
| U(1630)A | Bulge sequence | 80 | 10 | U(1629)9 | A(1630)1 |
| U(1629)UU | Bulge + 1 nt | 9 | 9 | U(1642)7 | U(1630)1, U(1654)1 |
| G(1648)GU | Bulge − 1 nt | 47 | 7 | U(1630)7 | |
| A(1631)G | Bulge sequence | 100 | 26 | C(1618)9, U(1630)4, U(1629)3, U(1638)3, G(1641)3, U(1642)2, C(1621)1, C(1663)1 | |
| U(1629)A | Bulge sequence | 100 | 49 | C(1628)21, U(1642)13, U(1640)5, A(1672)5, C(1668)3, U(1630)1, U(1654)1 | |
| G(1648)– | Bulge + 1 nt | 44 | 19 | U(1630)11 | U(1642)5, C(1628)1, U(1629)1, C(1656)1 |
| U(1629)–[= U(1630)–] | Bulge − 1 nt | 42 | 18 | U(1630)11 | U(1636)2, C(1674)2, C(1632)1, C(1656)1, C(1663)1 |
| G(1648)GUA | Bulge − 2 nt | <1e | 21 | C(1663)4, U(1642)3, U(1654)3, U(1630)2, C(1618)1, C(1621)1, U(1624)1, C(1632)1, U(1637)1, A(1643)1, U(1649)1, C(1656)1, C(144)1 | |
| G(1648)GUAA | Bulge − 3 nt | <1e | 0 | ||
| 5-nt top loop | |||||
| U(1637)UCU | Top loop + 2 nt | 100 | 21 | U(1630)19 | U(1642)2 |
| U(1637)UCUCU | Top loop + 4 nt | 80 | 17 | U(1630)6, A(1625)3, C(1628)2, U(1635)2, U(1624)1, U(1633)1, U(1636)1, U(1642)1 | |
| 6-bp stem | |||||
| U(1637)UG, U(1642)UC | Stem + 1 bp | 60 | 19 | U(1630)6, C(1621)2, U(1662)2, C(1618)1, U(1637)1, U(1642)1, A(1650)1, C(1656)1, U(1657)1, G(1671)1, U(13)1, C(105)1 | |
| U(1637)UGA, U(1642)UUC | Stem + 2 bp | 40 | 15 | U(1630)4, U(1640)3, U(1624)1, A(1625)1, A(1652)1, A(1660)1, U(1662)1, G(1667)1, G(1671)1, A(1678)1 | |
Each mutant is described in terms of the genomic nucleotides before the change, the site of change (in parentheses), and the nucleotides after the change.
These theoretical predictions were obtained by using the program MFOLD (version 3.0) provided by Michael Zuker. In some cases, alternative, closely related structures were also obtained.
We used the 5′-RACE strategy as applied to polyadenylated RNAs isolated from cells transfected with the indicated HDV mutants. Colonies were isolated, and the total number sequenced is indicated.
Results of the 5′ ends determined, expressed in terms of the nucleotides on the genomic RNA template, are listed as predominant sites if at least 50% were the same; otherwise, they are listed as additional sites. For each of the detected sites, the number of individual clones is indicated as a superscript. A limitation of the 5′-RACE strategy, as indicated in Materials and Methods, is that if the indicated template nucleotide corresponds to C, it is also possible that the actual end corresponds to the 3′-adjacent genomic nucleotide that is not C.
The ability of these two constructs to accumulate antigenomes could not be rescued by the provision of small delta protein in trans (reference 17 and data not shown).
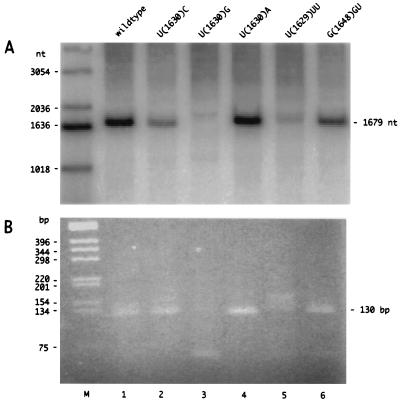
Consider first the results obtained for the following five mutations in the 3-nt external bulge: U(1630)C, U(1630)G, U(1630)A, U(1629)UU, and G(1648)GU. At 6 days after cDNA transfection, total cellular RNA was extracted. As shown in Fig. 3A, Northern analysis was then used to quantitate levels of antigenomic RNA. Relative to the unmodified wild-type genome as 100%, all five mutants gave measurable levels of antigenome accumulation, with U(1630)G, at 6%, being the lowest. Next, we tested each of the RNAs in the 5′-RACE assay. The results, as shown in Fig. 3B, were that four of the mutants gave a predominant species of PCR product similar in size and intensity to that obtained with the wild type, although additional, larger bands were sometimes present. Only a fainter band of 130 bp was detected with the fifth mutant, U(1630)G.
FIG. 3.
Antigenomic RNA accumulation and 5′ RACE for HDV RNAs produced by cells transfected with cDNAs mutated at and around a 3-nt external bulge predicted for the genomic RNA. Huh7 cells were transfected with cDNA corresponding to either the wild type or five specific mutants (as described in the text). After 6 days, the total RNA was extracted and examined by glyoxylation, agarose gel electrophoresis, and Northern analysis to detect antigenomic RNA (A) or by nested RT-PCR and nondenaturing agarose gel electrophoresis and ethidium staining of the 5′-RACE product (B). In panel A, lane M contains end-labeled 1-kb ladder (Life Sciences) and at the right is indicated the position of 1,679-nt unit-length antigenomic RNA. In panel B, lane M is unlabeled 1-kb ladder and at the right is indicated the position of the 130-bp PCR product. In lane 3, the ∼60-bp product corresponds to a primer-dimer artifact.
The PCR products shown in Fig. 3B were cloned and sequenced to determine the 5′ ends. The results for each mutant are summarized in Table 1. For U(1630)C, all the clones had the same 5′-end location as the wild type, although the first nucleotide synthesized would now be G rather than A. For mutants U(1630)G and U(1630)A, most of the 5′ ends were moved to nt 1629. This shift would maintain the first nucleotide incorporated as an A, just as for the wild type. Mutant U(1629)UU increased the predicted bulge by only 1 nt, but it moved most of the 5′ ends to nt 1642. In contrast, mutant G(1648)GU decreased the bulge by 1 nt and yet the ends were unchanged. We note that for these five mutants there was not a clear correlation between the level of antigenomic RNA accumulation, as detected by Northern analysis (Fig. 3A) and the maintenance of the 5′ end of the polyadenylated RNA at nt 1630 (Table 1).
Table 1 summarizes the results for these five mutants. We next constructed six more mutants in order to better evaluate the importance of the sequence and structure of the 3-nt-bulge region.
U(1629)A and A(1631)G each changed the nucleotide immediately adjacent to nt 1630. These changes had no effect on the accumulation of HDV RNA and yet they moved most of the 5′ ends away from nt 1630 and also created a significant heterogeneity for the other detected sites.
The other four mutants either increased or decreased the size of the predicted bulge. G(1648)–, like U(1629)UU, increased the bulge by 1 nt; however, in this case the majority of 5′ ends remained at nt 1630. U(1629)–, like G(1648)GU, reduced the size of the bulge by 1 nt. The predominant 5′ end was still U(1630), but now several minor sites were also observed. With G(1648)GUA we reduced the bulge by 2 nt; this greatly reduced genome accumulation (<1%), and there was no longer a predominant 5′ end. Finally, with G(1648)GUAA, which eliminated the predicted bulge, not only was the genome accumulation undetectable by Northern analysis (<1%), but so were the 5′ ends.
To summarize these results so far, it seems that the sequence and/or the size of the predicted 3-nt bulge can control whether the 5′ ends are located at nt 1630. We note that changing the nucleotides adjacent to position 1630 had a more profound effect than changes at 1630 itself. Also, when the size of the bulge was reduced from 3 to 1 or 0 nt, there were profound effects on both RNA accumulation and the ability to detect 5′ ends at nt 1630.
In addition to the 3-nt external bulge, we targeted two other sites which have been previously studied and shown to be essential for HDV genome accumulation (21). Between the external bulge (which includes nt 1630) and what we refer to as the top of the rod are two other secondary-structure elements: a 6-bp stem and a terminal 5-nt loop (Fig. 1). Our next aim was to determine the importance of these elements in the specificity of 5′ ends. Previously, we have reported studies of HDV genome replication for a series of 22 mutants made in these regions (21). Studies with some of these mutants were carried out as before. As summarized in Table 1, we tested increases in the size of the top loop of 2 and 4 nt. Neither increase had a significant affect on RNA accumulation. The 5′ ends were not changed by the 2-nt increase, but with the 4-nt increase, there were many sites in addition to nt 1630. A similar result was obtained when the length of the stem was increased by 1 or 2 bp. However, for each of these changes in either the top loop or the adjacent stem, the most frequent 5′ end was still at nt 1630.
In summary, the above mutagenesis studies allow certain generalizations as to the role of genomic RNA sequence and structure on the accumulation of antigenomic RNA and, in parallel, the detection of specific 5′ ends. The examined mutants in the top loop and adjacent stem were able to reduce the specificity of the 5′ end. In contrast, some of the mutations at and around the predicted 3-nt external bulge had more profound effects in that they created specific sites other than nt 1630. Overall, these findings support the hypothesis that the sequence and structure near the top of the rodlike structure, and especially the predicted 3-nt bulge, are important for the specificity of the 5′ ends of the polyadenylated mRNA species.
HDV RNA from cultured cells at early times after transfection with RNP.
In each of the above-mentioned studies, using RNAs from either an infected animal or transfected cells, we examined the 5′ ends of those polyadenylated RNAs present at times when genome replication was well under way. The sensitivity of the 5′-RACE procedure allowed us to look at earlier times and test the possibility that during replication there was somehow a time-dependent selection for mRNAs with such specific 5′ ends.
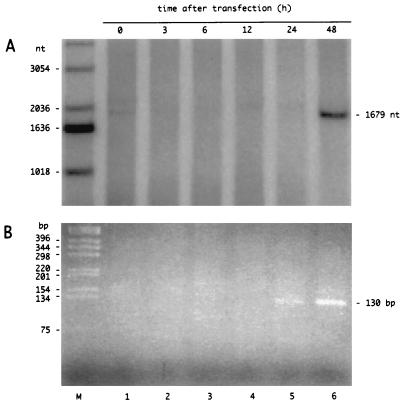
To do this, we made use of our recent findings that linear HDV genomic RNA synthesized in vitro can be complexed with recombinant small delta protein to make RNPs, which following transfection into cells, will lead to HDV genome replication (5). Cultures of Huh7 cells were transfected with such RNPs, and total RNA was isolated at 0, 3, 6, 12, 24, and 48 h. We first used Northern analyses to detect unit-length antigenomic RNA. As shown in Fig. 4A, we could readily detect such RNA at 48 h. Next, we examined the polyadenylated RNA by the more sensitive 5′-RACE procedure and detected a major band of RT-PCR product not only from the RNA at 48 h but also at 24 h (Fig. 4B). This band was the same size, 130 bp, as that produced from the RNA isolated from the liver of an infected woodchuck (data not shown). To obtain more precise data, we cloned the RT-PCR products and sequenced multiple clones (data not shown). For RNA isolated at 24 h, we found that in 11 of 12 clones the 5′ end was at nt 1630 (the exception was at nt 1668). Similarly, at 48 h, nine of nine were at nt 1630. Even with the increased sensitivity of the 5′-RACE procedure, analysis of the RT-PCR products corresponding to RNAs harvested at 3, 6, and 12 h did not yield valid 5′ ends. (Clones detected by hybridization were rare and contained sequence discontinuities considered to be artifacts of PCR amplification.)
FIG. 4.
Time course for RNP transfection. Linear HDV genomic RNA was combined in vitro with purified recombinant small delta protein and then used to transfect cultures of Huh7 cells, as previously described (5). At the indicated times after transfection, total RNA was extracted and analyzed, as for Fig. 3, by Northern analysis to detect 1,679-nt unit-length antigenomic RNA (A) and by 5′ RACE to detect RT-PCR products (B), with the ethidium bromide-stained band at 130 bp being a preliminary indication of a polyadenylated antigenomic RNA species with a 5′ end near nt 1630.
In summary, these results show not only that we detected the mRNA species as early as 24 h after initiation of replication but also that the specificity for 5′ ends at nt 1630 was already the same as that detected at much later times, when replication was well under way.
DISCUSSION
This application of a 5′-RACE procedure (Fig. 2) to the polyadenylated antigenomic RNAs of HDV has provided several findings which increase our understanding of HDV genome replication. (i) The 5′ site located at nt 1630 was reproducible in our studies. For example, 27 of 27 sequences from liver tissue were at nt 1630. Moreover, we obtained the same answer for RNAs from infected liver tissue and for cultured cells transfected with either a cDNA construct or an RNP assembled in vitro. (ii) The new result was different from previous primer extension studies that indicated a 5′ end at nt 1631. (iii) Exploiting the sensitivity of the 5′-RACE procedure, we were able to detect 5′ ends for RNAs isolated from cells as early as 24 h after transfection with RNP (Fig. 4). We thus found that these 5′ ends were at nt 1630, just like those detected at much later times after the initiation of replication. (iv) Finally, when 5′ RACE was applied to RNAs from cells transfected with mutated HDV genomes, we were able to detect situations in which the 5′ ends were relocated, sometimes without a major inhibition of genome replication as assayed by accumulation of antigenomic RNA (Fig. 3 and Table 1). In most cases the novel sites, like nt 1630, were consistent with initiation by a purine, usually an adenosine (Table 2). However, it must be pointed out that not only Pol II but also Pol I and Pol III preferentially initiate with a purine (4, 11, 16, 20).
TABLE 2.
Deduced 5′ ends other than those at U(1630)a
| Nature of 5′ end expressed in terms of:
|
No. of occurrences | Frequency (%) | |
|---|---|---|---|
| Template | Product | ||
| U | A | 93 | 51 |
| C | G | 65 | 36 |
| G | C | 9 | 5 |
| A | U | 15 | 8 |
For each determined 5′ site other than those at U(1630), we deduced the 5′ nucleotide of the template and product RNAs, as indicated. The values were extracted from the data in Table 1.
An obvious interpretation of the 5′ ends that we have detected by 5′ RACE is that they are sites for the initiation of RNA-directed RNA synthesis. Objectively, further studies are needed to establish this. Furthermore, other interpretations need to be excluded. For example, it might be that 3′-end processing by polyadenylation of antigenomic precursor RNAs is coupled with some form of 5′ processing to make an mRNA that is both translatable and stable; polyadenylated RNAs with more sequence at the 5′ end would be able to fold into the rodlike structure, which could make them less likely to be translated and less stable or perhaps make them substrates for RNA editing.
While our data support the interpretation that the sequence and predicted structure of the genomic RNA around nt 1630 are important, we nevertheless consider that there is not yet sufficient evidence to conclude that any RNA structure in the vicinity actually defines a promoter. Some of the mutations we have characterized here were able to change the location of the detected 5′ ends, and in some cases, especially when we reduced the size of the predicted 3-nt external bulge region, they were able to reduce RNA accumulation to undetectable levels, and yet all the sites of mutagenesis considered were only adjacent to the region that has been claimed to contain a promoter element (as indicated in Fig. 1). As mentioned in the introduction, it has been claimed that a double-stranded cDNA version of the genomic sequence will act in transfected cells as a promoter for RNA polymerase II (10). However, in such studies mapping of the 5′ end by primer extension either failed or yielded multiple 5′ ends (10). Furthermore, in our own studies, we consider that all of the 5′ ends we obtained were derived from RNA transcripts that were RNA directed rather than DNA directed. And, as an additional test to exclude the latter possibility, we studied polyadenylated RNAs expressed in cells transfected with a cDNA construct that contained a 2-nt deletion at nt 1425, about 200 nt away from most 5′ ends. As previously characterized, this genome, with a mutation in the open reading frame of the delta protein, is only able to undergo genome replication when delta protein is provided from a separate plasmid (8). When this mutant alone was transfected into cells and the polyadenylated RNAs were tested by the 5′-RACE procedure, we were unable to detect any clones whose sequences indicated 5′ ends at nt 1630 or anywhere in the vicinity (data not shown). Thus, we can infer, and thereby further support the interpretation, that the 5′ ends detected for wild-type genomes and for the replication-competent mutants described in Table 1, were all obtained for polyadenylated RNA species that were RNA directed rather than DNA directed.
ACKNOWLEDGMENTS
S.G. and K.D. contributed equally to this study.
J.T. was supported by grants AI-26522 and CA-06927 from the N.I.H. and by an appropriation from the Commonwealth of Pennsylvania.
Purified recombinant small delta protein was provided by Harmon Zuccola and James Hogle. Vadim Bichko and Huan Zhou gave valuable advice. More than 600 constructs were sequenced by Anita Cywinski and the DNA Sequencing Facility. Tony Yeung and the DNA Synthesis Facility provided essential oligonucleotides. Finally, Glenn Rall, Richard Katz, Hans Jurgen Netter, and William Mason gave constructive comments on the manuscript.
REFERENCES
- 1.Beard M R, Macnaughton T B, Gowans E J. Identification and characterization of a hepatitis delta virus RNA transcriptional promoter. J Virol. 1996;70:4986–4995. doi: 10.1128/jvi.70.8.4986-4995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen P-J, Kalpana G, Goldberg J, Mason W, Werner B, Gerin J, Taylor J. Structure and replication of the genome of hepatitis δ virus. Proc Natl Acad Sci USA. 1986;83:8774–8778. doi: 10.1073/pnas.83.22.8774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chirgwin J, Przybyla A, MacDonald R, Rutter W. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 4.Darnell J, Lodish H, Baltimore D. Molecular cell biology. New York, N.Y: Scientific American Books; 1986. p. 346. [Google Scholar]
- 5.Dingle K, Bichko V, Zuccola H, Hogle J, Taylor J. Initiation of hepatitis delta virus genome replication. J Virol. 1998;72:4783–4788. doi: 10.1128/jvi.72.6.4783-4788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsieh S-Y, Chao M, Coates L, Taylor J. Hepatitis delta virus genome replication: a polyadenylated mRNA for delta antigen. J Virol. 1990;64:3192–3198. doi: 10.1128/jvi.64.7.3192-3198.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kain K, Orlans P, Lanar D. Universal promoter for gene expression without cloning: expression-PCR. BioTechniques. 1991;10:366–374. [PubMed] [Google Scholar]
- 8.Kuo M Y-P, Chao M, Taylor J. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J Virol. 1989;63:1945–1950. doi: 10.1128/jvi.63.5.1945-1950.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuo M Y-P, Goldberg J, Coates L, Mason W, Gerin J, Taylor J. Molecular cloning of hepatitis delta virus RNA from an infected woodchuck liver: sequence, structure, and applications. J Virol. 1988;62:1855–1861. doi: 10.1128/jvi.62.6.1855-1861.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macnaughton T B, Beard M R, Chao M, Gowans E J, Lai M M C. Endogenous promoters can direct the transcription of hepatitis delta virus RNA from a recircularized cDNA template. Virology. 1993;196:629–636. doi: 10.1006/viro.1993.1519. [DOI] [PubMed] [Google Scholar]
- 11.McBryant S J, Kassavetis G A, Gottesfeld J M. Repression of vertebrate RNA polymerase III transcription by DNA binding proteins located upstream from the transcription start site. J Mol Biol. 1995;250:315–326. doi: 10.1006/jmbi.1995.0379. [DOI] [PubMed] [Google Scholar]
- 12.Modahl L E, Lai M M C. Transcription of hepatitis delta antigen mRNA continues throughout hepatitis delta virus (HDV) replication: a new model of HDV RNA transcription and regulation. J Virol. 1998;72:5449–5456. doi: 10.1128/jvi.72.7.5449-5456.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 14.Patel P H, Preston B D. Marked infidelity of human immunodeficiency virus type 1 reverse transcriptase at RNA and DNA template ends. Proc Natl Acad Sci USA. 1994;91:549–553. doi: 10.1073/pnas.91.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plotch S J, Bouloy M, Ulmanen I, Krug R M. A unique cap(m7GpppXm)-dependent influenza virion dependent endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 16.Riggs D L, Nomura M. Specific transcription of Saccharomyces cerevisiae 35S rDNA by RNA polymerase I in vitro. J Biol Chem. 1990;265:7596–7603. [PubMed] [Google Scholar]
- 17.Taylor J. Hepatitis delta virus: cis and trans functions needed for replication. Cell. 1990;61:371–373. doi: 10.1016/0092-8674(90)90516-h. [DOI] [PubMed] [Google Scholar]
- 18.Wang H-W, Wu H-L, Chen D-S, Chen P-J. Identification of the functional regions required for hepatitis D virus replication and transcription by linker-scanning mutagenesis of viral genome. Virology. 1997;239:119–131. doi: 10.1006/viro.1997.8818. [DOI] [PubMed] [Google Scholar]
- 19.Weiner A J, Choo Q-L, Wang K-S, Govindarajan S, Redeker A G, Gerin J L, Houghton M. A single antigenomic open reading frame of the hepatitis delta virus encodes the epitope(s) of both hepatitis delta antigen polypeptides p24 and p27. J Virol. 1988;62:594–599. doi: 10.1128/jvi.62.2.594-599.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkinson J A K, Miller K G, Sollner-Webb B. Dinucleotide primers facilitate convenient identification of the mouse ribosomal DNA transcription initiation site. J Biol Chem. 1983;258:13919–13928. [PubMed] [Google Scholar]
- 21.Wu T-T, Netter H J, Lazinski D W, Taylor J M. Effects of nucleotide changes on the ability of hepatitis delta virus to transcribe, process and accumulate unit-length, circular RNA. J Virol. 1997;71:5408–5414. doi: 10.1128/jvi.71.7.5408-5414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]