Abstract
Herpes simplex virus types 1 and 2 (HSV1 and HSV2) enter and reactivate from latency in sensory neurons, although the events governing these processes are little understood. During latency, only the latency-associated transcripts (LATs) are produced. However, although the LAT RNAs were described ≈10 years ago, their function remains ambiguous. Mutations affecting the LATs have minimal effects other than a small reduction in establishment of and reactivation from latency in some cases. Mutations in putative LAT-contained open reading frames (ORFs) have so far shown no effect. The LATs consist of a large species from which smaller (≈2 kb), nuclear, nonlinear LATs which are abundant during latency are spliced. Thus, translation of ORFs in these smaller LATs would not usually be expected to be possible, and if expressed at all, their expression might be tightly regulated. Here we show that deregulated expression of the largest HSV1 2-kb LAT-contained ORF in various cells of neuronal and nonneuronal origin greatly enhances virus growth in a manner specific to HSV1—the HSV1 LAT ORF has no effect on the growth of HSV2. Similar results of enhanced growth were found when the HSV1 LAT ORF was constitutively expressed from within the HSV1 genome. The mechanism of LAT ORF action was strongly suggested to be by substituting for deficiencies in immediate-early (IE) gene expression (particularly ICP0), because deregulated LAT ORF expression, as well as enhancing wild-type virus growth, was also found to allow efficient growth of viruses with mutations in ICP0 or VMW65. Such viruses otherwise exhibit considerable growth defects. IE gene expression deficiencies are often the block to productive infection in nonpermissive cells and are also evident during latency. These results, which we show to be protein- rather than RNA-mediated effects, strongly suggest a function of the tightly regulated expression of a LAT ORF-encoded protein in the reactivation from HSV latency.
Herpes simplex virus types 1 and 2 (HSV1 and -2) enter latency in sensory neurons, after an initial peripheral infection, from which they reactivate at intervals causing the symptoms of cold sores and genital herpes, respectively. During latency, the virus genome is quiescent, except for the transcription of the latency-associated transcripts (LATs) from the long repeat regions of the genome (30) (see Fig. 1). Here a large, ≈8-kb RNA is transcribed from a TATA-containing promoter which is spliced to release smaller (≈2-kb) RNAs which are the predominant LATs present in latently infected neurons (14). All strains of HSV1 and HSV2 so far sequenced contain potential open reading frames (ORFs) of significant size within the 2-kb LATs (11, 34). The largest of these ORFs in HSV1 (≈274 amino acids [aa], here designated the LAT ORF) is conserved in amino acid sequence between HSV1 strains, including the region that is not overlapped by the gene encoding ICP0. The ORF in the HSV2 2-kb LAT (419 aa) (9) is not, however, similar in sequence to the ORFs in HSV1. Differences in sequence between the HSV1 and HSV2 LAT regions have previously been speculatively attributed to the different anatomical sites at which the viruses enter latency (20)—the sequences of the HSV1 and HSV2 genomes are otherwise very similar—which could explain the difference between the sequences of the LAT ORFs. However, the possibility of a LAT-encoded protein function has largely been discounted (described below), not least due to the lack of a detectable phenotype when LAT ORFs are mutated (14).
FIG. 1.
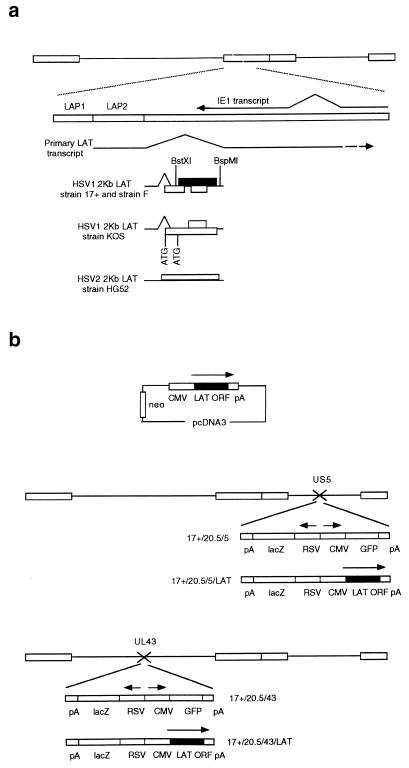
The HSV DNA genome, the LAT region, and constructs used in this study. (a) The LATs are transcribed from the long terminal repeat regions of the genome (larger rectangles), resulting in production of a large primary transcript, antisense to the IE1 transcript, which is spliced to release smaller LATs that are abundant during latency. Potential 2-kb LAT-contained ORFs in HSV1 and HSV2 are shown, as are regions with promoter activity (LAP1 and LAP2). LAP1 is predominantly responsible for 2-kb LAT production in latency (7). The HSV1 LAT ORF studied here is shaded. (b) Constructs and viruses used.
While the function of the LATs is not well understood, it seems likely that they have a function in latency. Data suggest that the LATs may reduce the expression of ICP0 by an antisense effect (24, 32), which could aid the establishment and maintenance of latency, and, with various LAT deletion mutants, that a portion of the LAT region (in a region 5′ to the 2-kb LAT and the LAT ORFs under discussion here) is important for the wild-type reactivation phenotype (3, 4, 26, 27, 33). No LAT-encoded proteins have reliably been detected (10, 22). However, we previously found that the leader sequence from the 2-kb LAT, which precedes the LAT ORF in HSV1 (≈950 bases and containing an intron selectively spliced in neurons), allows efficient translation of a downstream protein if present in a capped and polyadenylated RNA, even though such expression from within the 2-kb LAT is not possible (9). Thus, if the 2-kb LAT were ever produced as a nonnuclear species, rather than being retained in the nucleus in a lariat structure as is usually the case (28, 35), translation of LAT ORFs might be possible. This could occur, for example, by activation of the LAP2 promoter, a promoter not usually responsible for 2-kb LAT production during latency (7, 16) (see Fig. 1). We thus set out to identify any potential function of the larger HSV1 2-kb LAT-contained ORF by artificially deregulating its expression both in cell lines and from within the virus genome. This artificial deregulation of LAT ORF expression allowed such analysis to be performed without first identifying the circumstances which might usually make LAT ORF translation possible during the virus life cycle and which would have previously made identification of a LAT ORF function difficult. These experiments showed that deregulated expression of the LAT ORF greatly enhanced the growth of HSV1 and could overcome deficiencies in immediate-early (IE) gene expression and that the protein was thus likely to have a role in the reactivation from virus latency.
MATERIALS AND METHODS
Viruses, cell lines, and constructs used.
The LAT ORF was subcloned from a 3.5-kb NotI fragment from the LAT region by using BstXI and BspMI and inserted into pcDNA3 (Invitrogen), giving pcDNA3LAT, before stable transfection under neomycin selection into BHK and ND7 cells (Fig. 1). Viruses were produced by insertion of the LAT ORF in place of the GFP gene in the pR20.5/5 and pR20.5/43 shuttle vectors, giving pR20.5/5/LAT and pR20.5/43/LAT. The pR20.5 cassette contains lacZ and green fluorescent protein (GFP) genes under Rous sarcoma virus (RSV) and cytomegalovirus (CMV) promoter control in a back-to-back orientation, and in pR20.5/5 and pR20.5/43 it is flanked by HSV1 US5 sequences (inserted at nucleotide 137,945 [SacI]) and UL43 sequences (inserted at the unique NsiI site), respectively. pR20.5/5, pR20.5/5/LAT, pR20.5/43, and pR20.5/43/LAT were then recombined into the HSV1 strain 17+ genome by standard techniques, and lacZ-expressing plaques were further purified, giving viruses 17+/pR20.5/5, 17+/pR20.5/5/LAT, 17+/pR20.5/43, and pR20.5/43/LAT. The DNA structures of the viruses were confirmed by Southern blot analysis. Revertant viruses in which the LAT ORF in 17+/pR20.5/5/LAT was replaced by GFP by recombination with pR20.5/5 were also produced, and these gave results identical to those for 17+/pR20.5/5 in the experiments described (not shown). Mutant LAT ORFs were generated by digestion with SalI (which cuts once in the LAT ORF after aa 163), treatment with T4 DNA polymerase, and religation prior to insertion into pcDNA3 (for cells) or pR20.5/5 (for viruses).
Cell culture and growth curves.
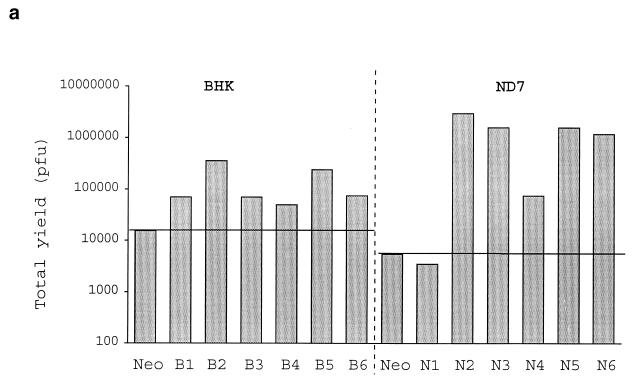
BHK and ND7 cells were cultured in Dulbecco’s modified Eagle’s medium–10% fetal calf serum (FCS) and Leibovitz L15–10% FCS–3.5% glucose, respectively, including 800 μg of neomycin per ml for selection where necessary. Cell lines were produced following standard calcium phosphate transfection of ND7 and BHK cells with pcDNA3LAT and antibiotic selection. Clones shown in Fig. 2a represent pairs of neomycin-resistant colonies picked from separate transfections in each case. Growth curves were obtained by infecting 80% confluent BHK or ND7 cell monolayers in six-well plates at the relevant multiplicity of infection (MOI [duplicate monolayers/time point]) in serum-free medium for 1 h at 37°C. Serum-free medium was then replaced with full-growth medium. At each time point, monolayers were harvested, virus was released by freeze-thawing, and virus yields were determined by the titers on BHK cells.
FIG. 2.
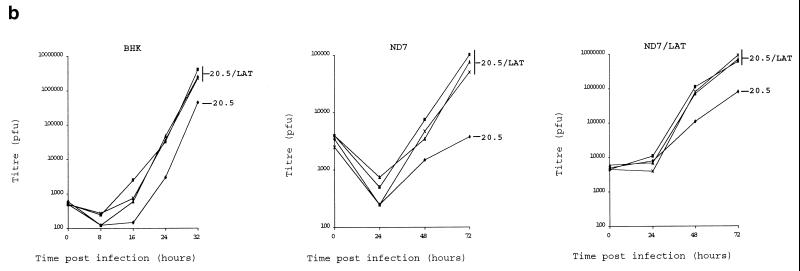
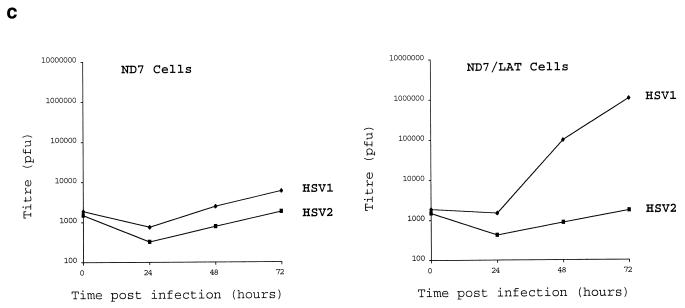
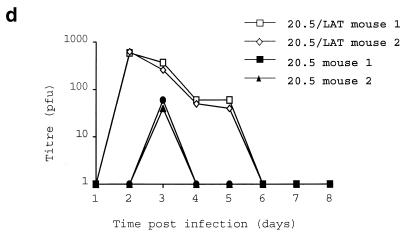
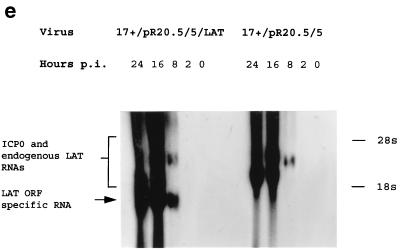
Deregulated expression of the HSV1 LAT ORF enhances virus growth. (a) HSV1 growth curves on six randomly picked ND7 and BHK clones stably transfected with pcDNA3LAT compared to that of pcDNA3-transfected control cells. (b) Growth curves of three separately plaque-purified 17+/pR20.5/5/LAT virus stocks compared to that of a 17+/pR20.5/5 control virus on unmanipulated BHK or ND7 cells or ND7 cells stably transfected with pcDNA3LAT (Lat N5 in panel a). MOI = 0.1 and 0.01 for ND7 and BHK cells, respectively. (c) Growth curves of HSV1 and HSV2 (strain HG52) on unmanipulated ND7 cells and ND7 cells stably transfected with pcDNA3/LAT (Lat N5). MOI = 0.01. (d) Growth of HSV1 expressing the LAT ORF in vivo. Twenty 3-week-old female BALB/c mice were footpad inoculated with 105 PFU of either 17+/pR20.5/5 or 17+/pR20.5/5/LAT. Pairs of mice from each group were killed on days 1 to 10 after inoculation, DRG L1 to L6 were harvested and manually homogenized, and titers of samples were determined for virus content on BHK cells. The points represent results from individual mice. (e) Northern blot showing LAT ORF encoding RNA expressed from the CMV promoter in 17+/pR20.5/5/LAT at the indicated times after infection of BHK cells (MOI = 1) and that of 17+/pR20.5 control virus (see Materials and Methods). p.i., postinfection.
Northern blots.
Total RNA was prepared from BHK or ND7 cells by standard methods (8). For detection of the CMV promoter-driven expression of LAT ORF-encoding RNA from recombinant viruses, 10 μg of total RNA was separated on a 1.5% agarose–morpholinepropanesulfonic acid (MOPS)–formaldehyde gel (29). RNA was transferred to a Hybond N membrane (Amersham) according to the manufacturer’s instructions and probed with a LAT ORF-specific 32P-labelled DNA probe (excised from pSP72/LAT, with the BstXI-BspMI DNA fragment used to construct the viruses and cell lines above subcloned into pSP72 [Promega]). For detection of LAT ORF and mutant LAT ORF-specific RNA in stably transfected cell lines, 10 μg of total RNA was separated on a 1.5-mm-thick 6% polyacrylamide gel containing 7 M urea in 1× TBE (Tris-borate-EDTA). RNA was then electroblotted in 1× TAE onto a Hybond N+ membrane (Amersham). This was hybridized at 72°C in a mixture of 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 5× Denhardt’s solution, 0.5% sodium dodecyl sulfate, and 10 μg of nonspecific DNA per ml, with a LAT-specific antisense RNA probe generated by standard techniques with T7 RNA polymerase and [α-32P]CTP (29) following linearization of the pSP72/LAT template. Other methods resulted in cross-hybridization of the probe with rRNA sequences.
RESULTS
Cell lines constitutively expressing the LAT ORF show enhanced permissiveness for HSV1.
Cell lines were produced in which the LAT ORF, precisely excised from an HSV1 genomic clone, was expressed under CMV promoter control followed by a BGH pA sequence. Both ND7 and BHK cells were used. ND7 cells are a neuronal cell line derived from a dorsal root ganglion (DRG) neuron-neuroblastoma fusion and are of low permissiveness for HSV due to the presence of transcription factors repressing HSV IE gene expression (19), and BHK cells are a fibroblast cell line of high permissiveness for HSV. Thus, here in each case, the LAT ORF is present in an RNA which would be expected to be processed as a normal cellular RNA and from which protein expression, i.e., translation of the LAT ORF, would be expected to be possible.
Growth curves showed that randomly chosen clones of ND7 and BHK cells stably transfected with the LAT ORF (Fig. 2a) gave considerably enhanced permissiveness for HSV growth compared to control cells transfected with Neo only. Expression of the LAT ORF encoding RNA in these cells is shown by Northern blotting in Fig. 4. Neo-transfected cells behaved as did untransfected cells. The growth enhancement shown in LAT ORF-containing cells was considerably more marked in ND7 than in BHK cells (up to ≈1,000-fold growth enhancement). ND7 cells are usually far less permissive than BHK cells for HSV growth (see above). In all cases, the growth enhancement shown by LAT ORF-containing cells was most evident at a relatively low MOI.
FIG. 4.
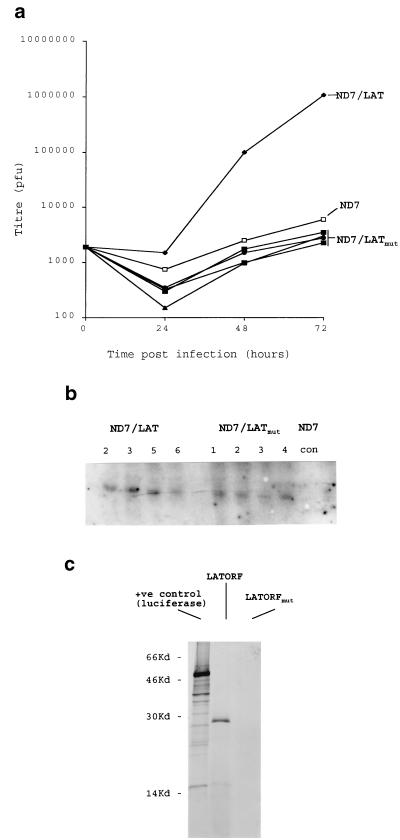
The HSV1 LAT ORF functions as a protein. (a) Viruses and cell lines were produced as described in Fig. 1, except where a frameshift mutation was inserted into the LAT ORF. Growth curves for wild-type HSV1 are shown for four representative ND7 cell lines (ND7LATmut) compared to that of a representative unaltered LAT ORF-expressing line (Lat N5 in Fig. 2a). A Northern blot (b) showing LAT ORF- and mutant LAT ORF-specific RNA expression in these cells is also shown (LAT ORF cells N2, N3, N5, and N6 in Fig. 2, and four representative mutant LAT ORF cell lines [see Materials and Methods]). con, control pcDNA3 transfected cells. (c) LAT ORF protein expression in vitro. [35S]Met-labelled in vitro transcription/translation reactions (TnT; Promega) were performed with pcDNA3LAT, pcDNA3LATmut, and a control luciferase-encoding plasmid. Reactions were run on a 15% polyacrylamide gel before autoradiographic detection of protein products.
Viruses with deregulated LAT ORF expression show enhanced replication.
Viruses were also constructed in which the LAT ORF under CMV promoter control and terminated with a BGH pA was inserted into nonessential sites in the HSV1 genome (US5 and UL43 [Fig. 1b]). LAT ORF expression was confirmed by Northern blotting (Fig. 2e). It has previously been shown that inactivation of US5 or UL43 does not otherwise affect virus growth in vitro or virulence or the kinetics of latency in vivo (2, 23). Viruses expressing the LAT ORF were tested by comparison of their growth curves with those of control viruses, and these, like the cell lines above, showed enhanced virus growth in (unmanipulated) ND7 and BHK cells (Fig. 2b). Insertion in US5 and UL43 gave viruses with identical growth kinetics in vitro, and thus only results for viruses in which insertions have been made in one of these sites (US5) are shown. This enhancement of growth was even greater when LAT ORF-expressing viruses were used to infect cells which already expressed the LAT ORF. Thus, deregulated expression of the LAT ORF either in cells or from within the virus genome can greatly enhance the growth of HSV1.
Deregulated expression of the HSV1 LAT ORF has no effect on HSV2.
The effect of HSV1 LAT ORF expression in BHK and ND7 cells was also tested with regard to the growth of HSV2. (Figure 2c shows the growth of both HSV1 and HSV2 on a representative ND7 cell line expressing the LAT ORF in comparison with growth on a LAT ORF-non-expressing cell line.) The results showed that the growth enhancement provided by deregulated HSV1 LAT ORF expression was specific for HSV1 in both BHK and ND7 cells, with no effect on HSV2. Thus, LAT ORF expression does not generally affect the permissiveness of cells for herpesvirus growth but acts in a manner which is specific for HSV1. Therefore, it would appear that the LAT ORF must interact either directly or through cellular factors with either HSV1-encoded protein(s), DNA, or RNA to enhance virus growth, such interactions not being possible for HSV2. The reasons for this lack of effect on the growth of HSV2 may be of help in identifying the precise mechanism of action and function in vivo of the LAT ORF and must relate in some way to sequence differences between HSV1 and HSV2.
LAT ORF-expressing viruses show enhanced replication in vivo.
Growth in vivo of LAT ORF-expressing viruses was assessed by quantifying virus content in DRG at days 1 to 10 after footpad inoculation of mice and comparing it to that of control LAT ORF-non-expressing viruses. This resulted, as in cell culture, in enhanced virus growth (Fig. 2d).
Constitutive LAT ORF expression can overcome deficiencies in IE gene expression.
The results presented above show that deregulated expression of the LAT ORF significantly enhances the growth of HSV both in vitro and in vivo, particularly in otherwise relatively nonpermissive cells, and that, therefore, the LAT ORF is likely to have a function in the HSV life cycle. However, as yet, the results have given no indication as to the mechanism by which the LAT ORF might function, other than that the LAT ORF appears to function in a manner specific to HSV1.
We have previously found that the LAT ORF does not have a direct activating or repressing effect on HSV1 IE promoters in cotransfection experiments (9). It is thus unlikely that the effects on virus growth of deregulated LAT ORF expression are mediated by a direct effect on stimulating or repressing IE gene expression, which could have provided the mechanism of action of the LAT ORF (9). To explore the level at which the LAT ORF might function, we tested the effect of the LAT ORF in the cell lines described above on a number of mutant viruses to see if LAT ORF expression could compensate for deficiencies in IE gene expression. Such deficiencies in IE gene expression are a characteristic of the latent state (if IE genes were expressed during latency, the virus would be expected to reactivate) and are the likely cause of the limited replication of HSV in nonpermissive cells such as ND7 cells (19). Thus if important in latency or in enhancing cellular permissivity, the LAT ORF might function by substituting for IE genes with expression defects. Of the IE genes tested by using the mutant viruses, ICP0 is important for efficient reactivation from latency (21), although not essential for lytic growth (31), ICP27 is an essential IE gene required for growth in all cells (and thus also for reactivation) (21), and VMW65 is a virion protein responsible for transactivating IE genes after infection, although it is not itself necessary for reactivation (12). ICP0 and VMW65 mutants usually show greatly reduced growth in vitro compared to wild-type virus, particularly at a low MOI (1, 31).
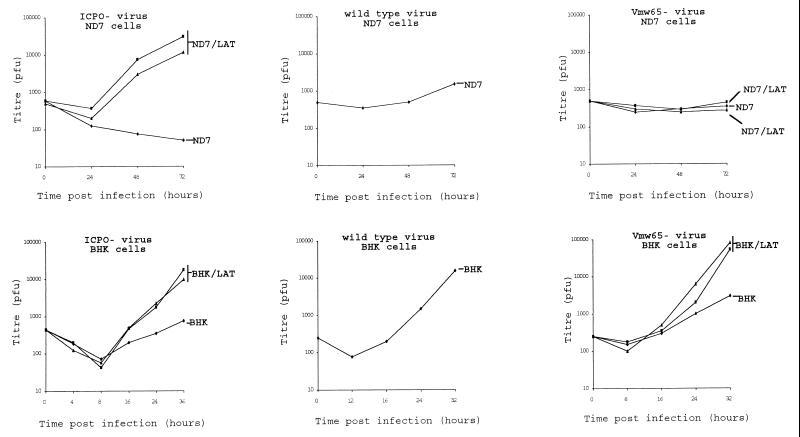
These experiments showed that expression of the LAT ORF has no effect on viruses with mutations in ICP27 (not shown), such viruses being incapable of supporting a productive infection in both LAT ORF-containing and control cells. However, LAT ORF expression can completely compensate for deficiencies in ICP0 in both BHK and ND7 cells and for VMW65 (in1814 mutant) (1) in BHK but not ND7 cells (Fig. 3). Indeed, on LAT-expressing ND7 cells, ICP0 mutants grew considerably better than does wild-type virus on unmanipulated ND7 cells. Thus, deficiencies in IE gene expression, importantly of ICP0, can be overcome in vitro by deregulated expression of the LAT ORF. The reason for the lack of compensation by the LAT ORF for a VMW65 deficiency in ND7 cells remains to be investigated, but it probably relates in some way to the manner in which the LAT ORF functions and may relate to cellular targets with which either VMW65 or the LAT ORF interacts.
FIG. 3.
Effect of deregulated LAT ORF expression on HSV1 ICP0 mutants. Growth curves of an HSV1 strain 17+ ICP0 deletion mutant (dl1403) (28) and VMW65 insertional mutant (in1814) (1) on ND7 (Lat N3, N5, and Neo in Fig. 2a) and BHK (Lat B2, B5, and Neo in Fig. 2a) cells stably transfected with pcDNA3/LAT or pcDNA3 and to those of wild-type virus (strain 17+) on pcDNA3-transfected BHK and ND7 cells are shown. MOI = 0.01 in each case.
The HSV1 LAT ORF functions as a protein.
While the experiments described above show that the HSV1 LAT ORF has a profound effect on virus growth under circumstances in which translation of the ORF is expected, unlike from within the 2-kb LAT RNA (9), the experiments do not conclusively show a protein (rather than RNA or DNA)-mediated effect. To confirm a protein-mediated effect, a number of experiments were performed. First mutant LAT ORFs in which a frameshift mutation had been inserted in the center of the ORF were expressed in both cell lines and viruses. Northern blots showed that mutant and nonmutant LAT ORF-expressing cells contained similar levels of LAT ORF-specific RNA (Fig. 4a). These cell lines showed none of the LAT ORF-mediated effects, and there was no enhancement of growth of either wild-type virus or ICP0 mutants—indeed, growth was slightly reduced for wild-type viruses. Viruses containing the mutant LAT ORFs grew very poorly, considerably worse than the wild type (not shown). This and the slight reduction in virus growth on the mutant LAT ORF-expressing cell lines were concluded to be due to an ICP0 antisense effect, because RNA antisense to ICP0 is here being transcribed in high abundance from the CMV promoter but is not being counteracted by the nonmutant LAT ORF’s capability of compensating for deficiencies in ICP0. These data strongly suggest a protein-mediated effect involving an intact LAT ORF. Finally, in vitro transcription/translation reactions performed with plasmids containing unaltered and mutant LAT ORFs (Fig. 4c) showed a protein close to the expected size of 30 kDa to be efficiently and stably produced with the unaltered ORF, but it was not present when a point frameshift mutation was inserted.
DISCUSSION
The data presented above show that the HSV1 LAT ORF encodes a protein which can profoundly effect (enhance) wild-type virus growth in a manner which is specific to HSV1. This enhanced growth is particularly the case in cells which are otherwise relatively nonpermissive for HSV. Indeed, as well as in ND7 cells, we have shown greatly enhanced growth of HSV1 in embryonic stem cells by deregulated LAT ORF expression (data not shown), again a cell type which is otherwise relatively nonpermissive for HSV. The data show that the mechanism by which this enhanced growth occurs is probably a compensation for deficiencies in IE gene expression—IE mutants show greatly enhanced permissiveness on LAT ORF-expressing cells. Such deficiencies can either be generalized, as caused by mutation to VMW65, or be specific to ICP0. Thus, the LAT ORF may encode a protein with a function and/or mechanism of action similar to that of ICP0. ICP0 has previously been strongly implicated as being necessary early in the reactivation process, and if expression of the LAT ORF can substitute for ICP0 activity, this strongly suggests that the LAT ORF has a role in the reactivation from latency. Indeed it has previously been suggested that a cellular factor might substitute for a lack of ICP0 expression early in reactivation (5), an activity which could in reality be provided by the LAT ORF.
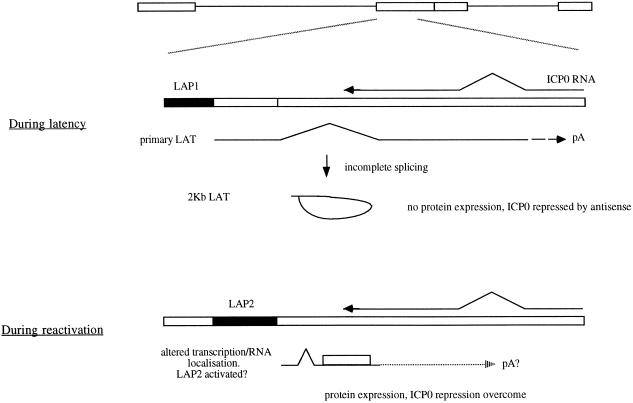
Thus, while the LAT ORF-encoded protein could function to aid the initial replication and thus the establishment of latency, or at other times in the virus life cycle, the most likely function is in reactivation, where tightly regulated expression would be required. Previous work has shown that a LAT-encoded function can reduce productive-cycle gene expression both in vitro and in vivo (6, 14, 15, 24), this possibly being caused by an antisense effect on ICP0 expression. Thus while this previous work might appear contradictory to the results reported here, both activating and repressing functions of the LAT region can be accommodated in a common model (Fig. 5). This would propose two functions for LAT expression, first providing an antisense effect to ICP0 in damping down gene expression during the establishment and maintenance of latency and in agreement with previous results (for example, see references 6, 15, and 24). Here no LAT ORF-encoded protein product would be produced due to the nuclear, nonlinear nature of the LAT ORF-encoding RNA, which has previously been described (28, 35). However, during reactivation, the expression/localization of the LAT ORF-encoding RNA would change in response to as yet unidentified stimuli such that translation of the LAT ORF becomes possible (described below). Because LAT ORF expression compensates for deficiencies in ICP0 expression, this would be expected to relieve the usual ICP0 antisense effect of the LATs during latency and thus aid the reactivation process. This model requires tight regulation of expression of the LAT ORF, which may explain the lack of detection of the protein previously, the lack of similarity of animal models to the situation in a latently infected patient, and possibly the lack of a previously identified phenotype when LAT ORFs have been mutated.
FIG. 5.
Model of LAT function during HSV latency and reactivation.
Experiments aimed at confirming LAT ORF-mediated effects on the reactivation phenotype of HSV were performed by using LAT ORF-expressing viruses as part of this study, and while more virus can be reactivated by explant from latently infected ganglia than with controls (not shown), this could be due to the enhanced initial replication we have observed in vivo rather than a direct effect on reactivation itself. Thus, no further conclusion is drawn from the experiments presented here, further work being necessary. To effectively study the effects of the LAT ORF on reactivation, it is necessary to either (i) produce viruses or transgenic animals in which LAT ORF expression can be switched on or off or (ii) identify the signals which might usually allow LAT ORF expression in vivo.
The mechanism of regulation of expression of the LAT ORF is currently under investigation by using constructs and viruses in which GFP has either replaced or been fused to the LAT ORF, and generation of an antiserum is under way. These reagents will also further facilitate the detection and study of the LAT ORF-encoded protein in vivo. However, it seems possible that in the LAT region regulatory elements which have previously been identified but for which as yet no function has been definitively ascribed may have a role in the regulatory process. For example, the LAP2 promoter, which has previously been identified upstream of the 2-kb LAT (16), while initially potentially thought to be important for 2-kb LAT production during latency, has since been found to be inactive during this time (7). LAP2 promoter function could thus conceivably be activated in response to reactivation signals, allowing production of a 2-kb LAT-like RNA that has not been produced by splicing and from which LAT ORF expression might be possible.
The necessity for tightly regulated LAT ORF expression could also explain the complexity of the HSV LAT region compared to the LAT regions of other alphaherpesviruses. In bovine herpes virus 1, for example, in which a LAT-encoded protein is present throughout latency (possibly preventing the death of latently infected cells by inhibition of cyclin-dependent kinase 2) (18), regulation of expression would not need to be so tightly regulated because the protein is constantly required. Thus, here, a linear, polyadenylated RNA from which the LAT protein can be directly translated is produced in latently infected neurons (17).
Thus, overall, the work reported has shown that while expression of the LAT ORF-encoded protein is unlikely to be necessary for reactivation, as has been evidenced in various animal models, its profound effects on HSV growth and ability to compensate for deficiencies in IE gene expression show that it is likely to be necessary for the full wild-type characteristics of latency and reactivation in an infected individual. Indeed the considerable differences in LAT ORF sequence between HSV1 and HSV2 may in part explain the different anatomical sites from which the viruses can reactivate (the HSV1 LAT ORF has no effect on HSV2), because otherwise the protein-coding regions of these two viruses are very similar. Experiments swapping LAT ORFs between viruses will be important in further studies to see if this is the case.
ACKNOWLEDGMENTS
This work was funded by a Medical Research Council Infrastructure Award and Glaxo Wellcome.
We thank Nigel Stow, Chris Preston, and Alasdair Maclean of the MRC Virology Unit, Glasgow, United Kingdom, for virus strains dl1403, in1814, and HG52 and Kate Bankhead for assistance with Northern blotting.
REFERENCES
- 1.Ace C I, McKee T A, Ryan M, Cameron J M, Preston C M. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J Virol. 1989;63:2260–2269. doi: 10.1128/jvi.63.5.2260-2269.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balan P, Davispoynter N, Bell S, Atkinson H, Browne H, Minson T. An analysis of the in vitro and in vivo phenotypes of mutants of herpes-simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J Gen Virol. 1994;75:1245–1258. doi: 10.1099/0022-1317-75-6-1245. [DOI] [PubMed] [Google Scholar]
- 3.Block T M, Deshmane S, Masonis J, Maggioncalda J, Valyi N T, Fraser N W. An HSV LAT null mutant reactivates slowly from latent infection and makes small plaques on CV-1 monolayers. Virology. 1993;192:618–630. doi: 10.1006/viro.1993.1078. [DOI] [PubMed] [Google Scholar]
- 4.Bloom D C, Hill J M, Devi-Rao G B, Wagner E K, Feldman L T, Stevens J G. A 348-base-pair region in the latency-associated transcript facilitates herpes simplex virus type 1 reactivation. J Virol. 1996;70:2449–2459. doi: 10.1128/jvi.70.4.2449-2459.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai W, Schaffer P A. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J Virol. 1989;63:4579–4589. doi: 10.1128/jvi.63.11.4579-4589.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen S-H, Kramer M F, Schaffer P S, Coen D M. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J Virol. 1997;71:5878–5884. doi: 10.1128/jvi.71.8.5878-5884.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Schmidt M C, Goins W F, Glorioso J C. Two herpes simplex virus type 1 latency-active promoters differ in their contributions to latency-associated transcript expression during lytic and latent infections. J Virol. 1995;69:7899–7908. doi: 10.1128/jvi.69.12.7899-7908.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Coffin R S, Thomas S K, Thomas D P, Latchman D S. The herpes simplex virus 2Kb latency associated transcript (LAT) leader sequence allows efficient expression of downstream proteins which is enhanced in neuronal cells: possible functions of LAT ORFs. J Gen Virol. 1998;79:3019–3026. doi: 10.1099/0022-1317-79-12-3019. [DOI] [PubMed] [Google Scholar]
- 10.Doerig C, Pizer L I, Wilcox C I. An antigen encoded by the latency-associated transcript in neuronal cell cultures latently infected with herpes simplex virus type 1. J Virol. 1997;65:2724–2727. doi: 10.1128/jvi.65.5.2724-2727.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolan A, Jamieson F E, Cunningham C, Barnett B C, McGeoch D J. The genome sequence of herpes simplex virus type 2. J Virol. 1998;72:2010–2021. doi: 10.1128/jvi.72.3.2010-2021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ecob-Prince M S, Rixon F J, Preston C M, Hassan K, Kennedy P G E. Reactivation in vivo and in vitro of herpes simplex virus from mouse dorsal root ganglia which contain different levels of latency associated transcripts. J Gen Virol. 1993;74:995–1002. doi: 10.1099/0022-1317-74-6-995. [DOI] [PubMed] [Google Scholar]
- 13.Fareed M U, Spivack J G. Two open reading frames (ORF1 and ORF2) within the 2.0-kilobase latency-associated transcript of herpes simplex virus type 1 are not essential for reactivation from latency. J Virol. 1994;68:8071–8081. doi: 10.1128/jvi.68.12.8071-8081.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrel M J, Dobson A T, Feldman L T. Herpes simplex virus latency-associated transcript is a stable intron. Proc Natl Acad Sci USA. 1991;88:790–794. doi: 10.1073/pnas.88.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garber D A, Schaffer P A, Knipe D M. A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. J Virol. 1997;71:5885–5893. doi: 10.1128/jvi.71.8.5885-5893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goins W F, Sternberg L R, Croen K D, Krause P R, Hendricks R L, Fink D J, Straus S E, Levine M, Glorioso J C. A novel latency-active promoter is contained within the herpes simplex virus type 1 UL flanking repeats. J Virol. 1994;68:2239–2252. doi: 10.1128/jvi.68.4.2239-2252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hossain A, Schang L M, Jones C. Identification of gene products encoded by the latency-related gene of bovine herpesvirus 1. J Virol. 1995;69:5345–5352. doi: 10.1128/jvi.69.9.5345-5352.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Y, Hossain A, Winkler M T, Holt T, Doster A, Jones C. A protein encoded by the latency-related gene of bovine herpesvirus 1 is expressed in trigeminal ganglionic neurons of latently infected cattle and interacts with cyclin-dependent kinase 2 during productive infection. J Virol. 1998;72:8133–8142. doi: 10.1128/jvi.72.10.8133-8142.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kemp L M, Latchman D S. Regulated expression of herpes simplex virus immediate early genes in neuroblastoma cells. Virology. 1989;171:607–610. doi: 10.1016/0042-6822(89)90631-4. [DOI] [PubMed] [Google Scholar]
- 20.Krause P R, Ostrove J M, Straus S E. The nucleotide sequence, 5′ end, promoter domain, and kinetics of expression of the gene encoding the herpes simplex virus type 2 latency-associated transcript. J Virol. 1991;65:5619–5623. doi: 10.1128/jvi.65.10.5619-5623.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leib D A, Coen D M, Bogard C L, Hicks K A, Yager D R, Knipe D M, Tyler K L, Schaffer P A. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol. 1989;63:759–768. doi: 10.1128/jvi.63.2.759-768.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luganoff M, Roizman B. Expression of a herpes simplex virus 1 open reading frame antisense to the γ134.5 gene and transcribed by an RNA 3′ coterminal with the unspliced latency-associated transcript. J Virol. 1994;68:6021–6028. doi: 10.1128/jvi.68.9.6021-6028.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacLean C A, Efstathiou S, Elliott M L, Jamieson F E, McGeoch D J. Investigation of herpes simplex virus type 1 genes encoding multiply inserted membrane proteins. J Gen Virol. 1991;72:897–906. doi: 10.1099/0022-1317-72-4-897. [DOI] [PubMed] [Google Scholar]
- 24.Mador N, Goldenberg D, Cohen O, Panet A, Steiner I. Herpes simplex virus type 1 latency-associated transcripts suppress viral replication and reduce immediate-early gene mRNA levels in a neuronal cell line. J Virol. 1998;72:5067–5075. doi: 10.1128/jvi.72.6.5067-5075.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perng G-C, Dunkel E C, Geary P A, Slanina S M, Ghiasi H, Kaiwar R, Nesburn A B, Wechsler S L. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J Virol. 1994;68:8045–8055. doi: 10.1128/jvi.68.12.8045-8055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perng G-C, Ghiasi H, Slanina S M, Nesburn A B, Wechsler S L. The spontaneous reactivation function of the herpes simplex virus type 1 LAT gene resides completely within the first 1.5 kilobases of the 8.3-kilobase primary transcript. J Virol. 1996;70:976–984. doi: 10.1128/jvi.70.2.976-984.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perng G-C, Slanina S M, Ghiasi H, Nesburn A B, Wechsler S L. A 371-nucleotide region between the herpes simplex virus type 1 (HSV-1) LAT promoter and the 2-kilobase LAT is not essential for efficient spontaneous reactivation of latent HSV-1. J Virol. 1996;70:2014–2018. doi: 10.1128/jvi.70.3.2014-2018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rødahl E, Haarr L. Analysis of the 2-kilobase latency-associated transcript expressed in PC12 cells productively infected with herpes simplex virus type 1: evidence for a stable, nonlinear structure. J Virol. 1997;71:1703–1707. doi: 10.1128/jvi.71.2.1703-1707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Stevens J G, Wagner E K, Devi-Rao G B, Cook M L, Feldman L T. RNA complementary to a herpesvirus α gene mRNA is prominent in latently infected neurons. Science. 1987;235:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 31.Stow N D, Stow E C. Isolation and characterisation of a herpes simplex type 1 mutant containing a deletion in the gene encoding the immediate early polypeptide Vmw110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 32.Thompson R L, Sawtell N M. The herpes simplex virus type 1 latency-associated transcript gene regulates the establishment of latency. J Virol. 1997;71:5432–5440. doi: 10.1128/jvi.71.7.5432-5440.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trousdale M D, Steiner I, Spivack J G, Deshmane S L, Brown S M, MacLean A R, Subak-Sharpe J H, Fraser N W. In vivo and in vitro reactivation impairment of a herpes simplex virus type 1 latency-associated transcript variant in a rabbit eye model. J Virol. 1991;65:6989–6993. doi: 10.1128/jvi.65.12.6989-6993.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wechsler S L, Nesburn A B, Zwaagstra J, Ghiasi H. Sequence of the latency related gene of herpes simplex virus type 1. Virology. 1989;168:168–172. doi: 10.1016/0042-6822(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 35.Wu T-T, Su Y-H, Block T M, Taylor J M. Evidence that two latency-associated transcripts of herpes simplex virus type 1 are nonlinear. J Virol. 1996;70:5962–5967. doi: 10.1128/jvi.70.9.5962-5967.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]