Abstract
Expression of the region of the feline calicivirus (FCV) ORF1 encoded by nucleotides 3233 to 4054 in an in vitro rabbit reticulocyte system resulted in synthesis of an active proteinase that specifically processes the viral nonstructural polyprotein. Site-directed mutagenesis of the cysteine (Cys1193) residue in the putative active site of the proteinase abolished autocatalytic cleavage as well as cleavage of the viral capsid precursor, suggesting that this “3C-like” proteinase plays an important role in proteolytic processing during viral replication. Expression of the region encoding the C-terminal portion of the FCV ORF1 (amino acids 942 to 1761) in bacteria allowed direct N-terminal sequence analysis of the virus-specific polypeptides produced in this system. The results of these analyses indicate that the proteinase cleaves at amino acid residues E960-A961, E1071-S1072, E1345-T1346, and E1419-G1420; however, the cleavage efficiency is varied. The E1071-S1072 cleavage site defined the N terminus of a 692-amino-acid protein that contains sequences with similarity to the picornavirus 3C proteinase and 3D polymerase domains. Immunoprecipitation of radiolabeled proteins from FCV-infected feline kidney cells with serum raised against the FCV ORF1 C-terminal region showed that this “3CD-like” proteinase-polymerase precursor protein is apparently stable and accumulates in cells during infection.
Feline calicivirus (FCV), a major agent of respiratory illness in cats, is classified in the genus Vesivirus of the family Caliciviridae. The positive-sense single-strand RNA genomes of viruses in the family Caliciviridae are organized into two or three open reading frames (ORFs), a feature that distinguishes this family from its closest relative, the Picornaviridae. The nonstructural proteins of the caliciviruses are encoded by a large ORF (ORF1) beginning near the 5′ end of the genome. Similar to maturation of the picornavirus nonstructural proteins, the maturation of such proteins of the caliciviruses involves the proteolytic processing of a large polyprotein. The proteinase responsible for this activity has been mapped in the rabbit hemorrhagic disease virus (RHDV) (5, 42) and the human Southampton virus (SHV) (26) calicivirus genomes.
The 7.6-kb FCV RNA genome (URB strain) is organized into three major ORFs: ORF1 (nucleotides [nt] 20 to 5308), ORF2 (nt 5314 to 7317), and ORF3 (nt 7317 to 7634). ORF1 contains regions that have sequence similarity with the 2C helicase, 3C cysteine proteinase, and 3D RNA-dependent RNA polymerase regions of the picornaviruses (31). ORF2 encodes the capsid protein precursor (73 kDa) that is cleaved by the FCV ORF1-encoded proteinase into the mature capsid protein (60 kDa) (8, 39). ORF3 encodes a small protein of unknown function found in infected cells in a truncated form (19). Previous studies of the proteins synthesized in feline kidney cells following infection with FCV demonstrated the presence of nonstructural proteins with sizes of approximately 96, 75, 39, 36, and 27 kDa (7). It was proposed that these proteins were cleavage products (intermediate-sized and mature forms) derived from a larger polyprotein. The presence in infected cells of “intermediate” viral proteins suggested the possibility of regulatory mechanisms involved in processing of this polyprotein and, correspondingly, in viral replication.
Our present study was undertaken to examine proteolytic processing of the nonstructural polyprotein of FCV. The boundaries of the mature nonstructural proteins had not yet been mapped for FCV, and little was known concerning the role of proteolytic processing in the replication of this virus. Our previous work demonstrated that the viral proteinase was responsible for processing of the capsid precursor protein during viral infection. Our present study demonstrates that the same proteinase mediates at least four additional cleavages in the viral nonstructural polyprotein when analyzed in vitro. In addition, one of these cleavages generates a precursor protein (designated Pro-Pol) containing both proteinase and polymerase motifs that is stable in vitro and in infected cells.
MATERIALS AND METHODS
Cells and virus.
Crandell-Rees feline kidney cells (CRFK), were maintained in Eagle’s minimum essential medium containing amphotericin B (2.5 μg/ml), chlortetracycline (25 μg/ml), penicillin (250 U/ml), and streptomycin (250 μg/ml) and supplemented with 10% heat-inactivated fetal bovine serum. The source and molecular characterization of the Urbana (URB) strain of FCV have been described (GenBank accession no. L40021) (38).
Radiolabeling of virus-specific proteins.
CRFK monolayers (106 cells) were mock infected or infected with FCV at a multiplicity of infection of 4 and incubated at 37°C. For radiolabeling of proteins, the cells were washed at 1.5, 2.5, and 4.5 h postinfection with methionine-free growth medium and incubated in the same medium for 30 min. [35S]methionine (>1,000 Ci/mmol; Amersham) was added to cells at a concentration of 100 μCi/ml, and the cells were incubated for an additional hour. The monolayer was washed with phosphate-buffered saline before lysis in 500 μl of radioimmunoprecipitation assay (RIPA) buffer (36).
Plasmid construction.
Standard recombinant DNA methods were used for plasmid constructions (36).
Plasmid pETF-1 was constructed by subcloning the 5,282-bp AspI-NspV fragment of plasmid pQ14 (38) into NcoI-digested pET-29c vector (Novagen) after filling in the protruding restriction ends of the plasmid and the fragment. The resulting plasmid contained the entire ORF1 of the FCV genome (with the exception of the two C-terminal codons) downstream from the T7 RNA polymerase promoter. Plasmids pETFΔXm, pETFΔB, pETFΔXh, and pETFΔR were created by the removal of the XmaIII, BamHI, XhoI, and EcoRI fragments, respectively, from plasmid pETF-1, followed by recircularization of the plasmid. The resulting plasmids contained FCV ORF1 sequence starting at the first AUG and truncated at nt 4566, 3481, 2651, and 1186, respectively (Fig. 1A).
FIG. 1.
Mapping the FCV proteinase gene responsible for cleavage of the FCV ORF1 polyprotein. (A) ORF1 map and localization of clones analyzed in the present study. Plasmids were engineered as described in Materials and Methods. The nucleotide positions of restriction sites used for plasmid construction are shown in parentheses. (B) SDS-PAGE analysis of the products obtained in coupled TNT reactions from plasmids that contained ORF1 sequences. Radiolabeled TNT products derived from ORF1 clones were loaded onto a 10 to 20% Tris-glycine gel (Novex) as indicated.
Plasmid pNB was constructed by subcloning the 2,755-bp ClaI-BamHI fragment of plasmid pQ14 into NspV-BamHI-digested pET-29c vector. Plasmid pNBΔXh was generated by excision of the 870-bp XhoI fragment from plasmid pNB, followed by recircularization of the plasmid. Plasmid pNBΔN was constructed by insertion of the 2,707-bp XhoI fragment excised from pETF-1 into the XhoI site of pNBΔXh. Restriction of pNBΔN with XmaIII, followed by removal of a 784-bp fragment and further religation of the plasmid, resulted in construction of plasmid pNBΔXm. Plasmids pNBΔN, pNBΔXm, pNB, and pNBΔXh contained the FCV ORF1 sequence starting at nt 726 (ClaI site) and truncated at nt 5301, 4566, 3481, and 2651, respectively.
Plasmid pBSX was constructed as follows: plasmid pETFΔXm was cleaved with BsaI and Acc65.1, followed by sequential incubation with Klenow, first in the presence of dGTP, then in the presence of dATP, and then in the presence of dTTP. The plasmid, with partially filled in protruding ends, was religated.
To truncate the sequence of the proteinase-polymerase precursor protein (Pro-Pol) downstream from the E1345-T1346 cleavage site, a DNA fragment was amplified from plasmid pBSX by PCR with a sense primer, A1, corresponding to nt 3406 to 3426 and an antisense primer (5′ AGATAGCTCGAGttaTTCTGAGGAAATGTTCAC 3′) corresponding to nt 4037 to 4054 of the FCV genome. The antisense primer contained a terminator codon (lowercase) and an XhoI site (underlined). The purified fragment was treated with XhoI and NcoI and ligated into XhoI-NcoI-digested pBSX. The resulting plasmid, designated pPro, was verified by sequencing analysis.
Plasmids pfΔ20, pTMF-1, and pVPP were constructed previously (39) and contained virus-specific sequences as follows: pfΔ20 contained the entire 3′-terminal part of the FCV genome starting at nt 5302 through the poly(A) tract, pTMF-1 contained the entire ORF1 of the FCV genome with the exception of the two C-terminal codons, and pVPP contained nt 2843 to 5303 of the URB genome encoding the 820 amino acid residues of the C-terminal part of the ORF1 polyprotein (Fig. 1A).
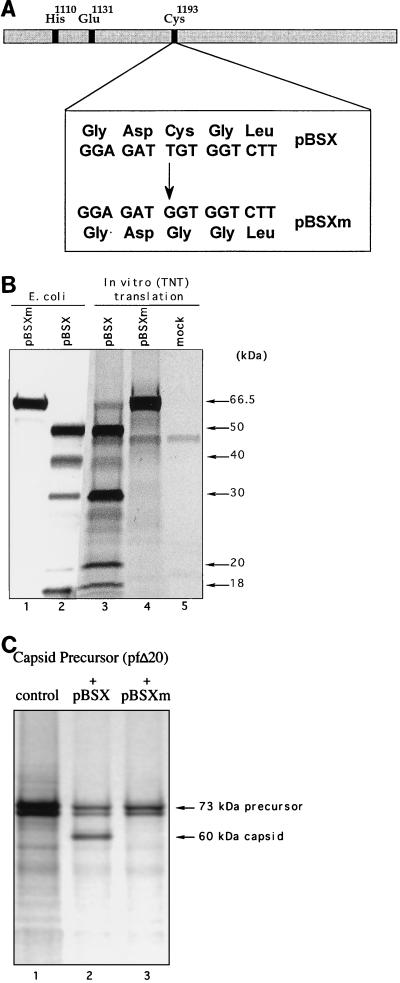
Site-directed mutagenesis of the cysteine 1193.
To introduce a Cys1193→Gly1193 substitution into the sequence of the pBSX-encoded protein, PCR-mutagenesis was employed. Two DNA fragments were amplified from plasmid pBSX by PCR, the first with the sense primer A1 (described above) and an antisense primer, A2 (5′ GTAGGGAAGACCACCATCTCCTGGGTGAGTTTC 3′), corresponding to nt 3578 to 3610 of the genome and the second with a sense primer, B1 (5′ CACCCAGGAGATGGTGGTCTTCCCTACATTG 3′), corresponding to nt 3584 to 3614, and an antisense primer, B2, corresponding to the T7 terminator primer from Novagen. Primers A2 and B1 contained the nucleotide change converting the TGT codon of cysteine into a GGT codon of glycine (underlined above). Purified PCR-generated DNA fragments were digested with BbsI, ligated, and treated with NcoI and XhoI. The resulting fragment was used to replace the authentic NcoI-XhoI fragment in plasmid pBSX, and the selected plasmid was designated pBSXm. pBSXm was sequenced in order to confirm the presence of the desired mutation.
Bacterial expression of the Pro-Pol protein, purification, and production of specific antiserum.
The expression of pVPP in Escherichia coli BL21(DE3) cells was performed as previously described (39), and the 78-kDa protein was fused to a C-terminal His6 tag and purified from the insoluble fraction of the bacterial lysate. Briefly, the insoluble fraction was solubilized in lysis buffer containing 8 M urea, 0.1 M NaH2PO4, and 0.01 M Tris-Cl (pH 8.0), and the solution was clarified from the remaining insoluble material by centrifugation. Ni-nitrilotriacetic acid (NTA) agarose (Qiagen) was added to the supernatant, and the His6 tag-containing proteins were purified according to the protocol supplied by the manufacturer. The fraction containing the expressed protein was dialyzed against phosphate-buffered saline, and aliquots of the solution (100 μg) were used to immunize a guinea pig. Inoculations were administered by the intramuscular route in three doses 2 weeks apart. The first dose was given with Freund’s complete adjuvant, and the second and third doses were administered with Freund’s incomplete adjuvant. Serum (designated αPro-Pol) was collected 1 week following the third immunization.
Protein sequencing.
To conduct direct N-terminal sequence analysis, proteins expressed in bacteria were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with tricine running buffer (Novex), transferred to a ProBlott membrane (Applied Biosystems), and visualized by staining with 0.1% Coomassie blue R-250 in 40% methanol and 1% acetic acid, followed by destaining in 50% methanol. The band of interest was excised and subjected to N-terminal sequence analysis with a model 477A protein sequencer coupled to a model 120A phenylthiohydantoin (PTH) analyzer (Applied Biosystems) according to the manufacturer’s program NORMAL-1.
In vitro coupled transcription and translation experiments, immunoprecipitation, and Western blot analysis.
One to 5 μg of plasmid DNA was used as the template in a coupled transcription and translation reaction (TNT T7 Coupled Reticulocyte Lysate System; Promega). For radiolabeling of synthesized protein, [35S]methionine (>1,000 Ci/mmol) from ICN or Amersham was used at a concentration of 1.5 mCi/ml.
Immunoprecipitation of viral proteins from infected cell lysates and Western blot analysis were performed as described previously (39).
For immunoprecipitation analysis of FCV proteins synthesized in the TNT time-course experiment, 15-μl aliquots of the TNT reaction were taken at each time point, diluted with 80 μl of RIPA buffer, and heated for 10 min at 60°C. The mixtures were then incubated for 1 h at room temperature with 5 μl of guinea pig pre- or postimmunization serum raised against the 78-kDa FCV Pro-Pol protein expressed in E. coli, and the immune complexes were precipitated with protein A beads (Sigma Chemical Co.). The binding and washing conditions were the same as those described previously (39).
RESULTS
Mapping of the proteinase gene responsible for cleavage of the FCV ORF1 polyprotein.
We previously reported that expression of the entire FCV ORF1 by plasmid pTMF-1 in an in vitro translation system led to the appearance of several intermediate-sized proteins that were consistent with autocatalytic cleavage of the polyprotein by an encoded proteinase (39). In order to map the proteinase sequences involved in this processing, a panel of plasmids containing the FCV genome truncated at varying lengths from the 3′ end of ORF1 was constructed (Fig. 1A). Selected viral sequences were then subcloned into the pET-29c vector for expression in a bacterial system.
Comparative analysis of in vitro-translation products synthesized from pETF-1 (a plasmid containing the entire FCV ORF1 fused to a N-terminal S tag sequence and placed downstream from the T7 promoter and a bacterial translation initiation signal in pET-29c) and from pTMF-1 (a plasmid containing the FCV ORF1 engineered downstream from the T7 promoter and the encephalomyocarditis virus IRES element in pTM-1) (39) demonstrated the presence of similar patterns for autocatalytic cleavage of the encoded ORF1 polyprotein (Fig. 1B, lanes 5 and 6). Instead of the predicted polyproteins with estimated sizes of 201 and 195 kDa, respectively, we observed several smaller protein bands with sizes ranging from approximately 30 to 80 kDa. Protein bands with molecular sizes ranging from 90 to 130 kDa, presumably corresponding to higher-molecular-weight precursors, were also detected in the translation products. As previously reported for pTMF-1, translation products from pETF-1 were specifically recognized by antibodies obtained from a cat infected with FCV and corresponded in size to bands detected in FCV-infected CRFK cells in a Western blot reacted with the same serum (data not shown).
Analysis of the translation products derived from the pETFΔ plasmids with truncated 3′-end ORF1 sequences showed that proteolytic processing of the ORF1 polyprotein did not occur until the region marked by an XmaIII site (nt 4,566) was translated (Fig. 1B, lane 4). This region overlapped the area of ORF1 previously found to have sequence motifs related to the picornavirus 3C proteinase sequences. A similar result was obtained in the analysis of the pNB series of plasmids (pNBΔN, pNBΔXm, pNB, and pNBΔXh) that contained ORF1 sequences truncated at the 5′ end from the ClaI site (nt 726) and at the 3′ end, similar to the pETFΔ plasmids (Fig. 1B, lanes 7 to 10).
Analysis of the products generated in the TNT reactions containing the pNBΔN and pETF-1 plasmids with sequences extending to the 3′ end of ORF1 demonstrated the presence of a 78-kDa protein analogous in size to the “3CD-like” proteinase-polymerase protein we had described previously in TNT studies of plasmids pTMF-1 and pVPP (39). Comparison of the TNT products derived from the pETFΔXm and pNBΔXm clones that were truncated at the same site (737 nt from the 3′ end of ORF1) resulted in loss of the 78-kDa protein and appearance of a 50-kDa protein (Fig. 1B, lanes 4 and 8). Of note, a protein that appeared similar in size to the 78-kDa protein was observed in the pETFΔXm TNT products. Further analysis showed that this protein was not recognized by the αPro-Pol serum described below (data not shown). Thus, it is likely that this protein represented a polypeptide derived from the N terminus of FCV ORF1.
Examination of the translation products derived from the pNBΔN or pNBΔXm clones, which were each engineered to begin at amino acid residue 237 of ORF1, showed the accumulation of two proteins with sizes of approximately 43 and 46 kDa (Fig. 1B, lanes 7 and 8). Comparison of these translation products with those from two analogous clones that contained coding sequence beginning from the first methionine of the N terminus, pETF1 and pETFΔXm, showed the disappearance of the 43- to 46-kDa proteins and the accumulation of smaller proteins. This observation suggests that the presence of the N terminus allowed the recognition and processing of cleavage sites that were not recognized by the proteinase in the pNBΔN and pNBΔXm truncated 43- to 46-kDa translation products. Thus, it is possible that the conformation of the nascent polyprotein plays a role in presenting an authentic cleavage site to the proteinase.
Amino-terminal sequencing analysis.
The pVPP plasmid encodes an active proteinase that mediates trans cleavage of the capsid precursor (39). Furthermore, we showed previously that the pVPP-encoded protein undergoes autocatalytic cleavage when expressed in bacteria, producing proteins of approximately 78, 18, and 14 kDa in the insoluble fraction that correspond to those observed in in vitro-translation reactions (Fig. 2A, lane 2). In addition, virus-specific proteins of approximately 30 and 40 kDa could be detected in the insoluble fractions of pVPP-transformed bacterial cells; however, the intensities of these bands varied among experiments.
FIG. 2.
Protein sequence analysis of the products derived by autocatalytic processing of the C-terminal part of the FCV ORF1 polyprotein expressed in bacteria. (A) Profiles of proteins encoded in plasmids pVPP, pBSX, and pPro. Bacteria carrying either the pVPP plasmid or the pET-29c vector plasmid were induced with IPTG (isopropyl-β-d-thiogalactopyranoside), and fractions of the insoluble bacterial products were subjected to SDS-PAGE and visualized with Coomassie blue stain. The arrows point to the pBSX and pVPP proteins used for direct N-terminal sequencing. (B) Schematic representation of proteinase cleavage sites mapped by direct sequencing of proteins produced in bacterial cells. A map of the observed sizes of the corresponding cleavage products used in the sequence analysis is shown above the diagram.
In our present study, direct sequence analysis was performed on the 78-, 18-, and 14-kDa proteins and two cleavage sites were mapped. The 18-kDa protein was found to have the sequence MKETAAAKF at its N terminus, suggesting that it represented the N-terminal part of the protein encoded by pVPP. Analysis of the 14- and 78-kDa proteins identified the sequences AKGKTKSKV and SGPGTKFHK respectively, at their N termini, consistent with the presence of cleavage sites at positions E960-A961 and E1071-S1072. Sites E960-A961 and E1071-S1072 defined the boundaries of a protein with a calculated size of 12.7 kDa (Fig. 2B) that represents the putative viral VPg. Site E1071-S1072 defined the N-terminal border of the 3CD-like 78-kDa Pro-Pol protein that we had mapped previously to the C terminus of the pVPP-encoded protein sequence, and thus, to the 3′ end of ORF1 (39). We previously proposed that this 78-kDa protein could be the active form of the proteinase in FCV-infected cells, similar to the 3CD of the picornaviruses. However, the picornavirus 3CD protein is subsequently cleaved to release an active polymerase. In order to investigate whether the FCV 78-kDa Pro-Pol protein undergoes further cleavage and whether the 30- and 40-kDa proteins observed in bacterial cells could represent its possible cleavage products, we analyzed the amino termini of these proteins by direct sequence analysis. The amino-terminal sequences of these two proteins were identical to that of the 78-kDa protein. The corresponding cleavage counterparts from the remainder of the 78-kDa protein for these proteins were difficult to visualize in the insoluble fraction of bacterial cells with Coomassie blue staining. However, blot analysis of the same fraction with (Ni-NTA)-alkaline phosphatase conjugate demonstrated the presence of two bands corresponding to proteins of about 40 and 50 kDa (data not shown). These 40- and 50-kDa proteins were purified with (Ni-NTA)-agarose; however, sequence analysis of the N termini failed.
The truncation of the putative polymerase gene in plasmid pBSX led to an increase in the observed efficiency of the internal cleavage between the polymerase and proteinase regions. In the insoluble fraction of bacterial cells harboring pBSX, we could detect both 20- and 11-kDa proteins, possibly representing two forms of the truncated polymerase (Fig. 2A, lane 3). These proteins were present in lower concentration than the other cleavage products; however, such amounts were sufficient to carry out direct amino-terminal sequencing. The sequencing data revealed that the 20- and 11-kDa proteins were the C-terminal products of processing of the pBSX-encoded protein at the E1345-T1346 and E1419-G1420 cleavage sites (Fig. 2B). Expression of pBSX also led to the production of five proteins of approximately 50, 40, 30, 18, and 14 kDa (Fig. 2A, lane 3). Proteins with sizes of about 40, 30, 18, and 14 kDa had counterparts in the expression products of pVPP and were shown by direct sequencing to have the same amino termini. The N-terminal sequence of the 50-kDa protein was SGPGTKFHK, consistent with cleavage at E1071-S1072 that would result in a truncated version of the Pro-Pol protein from this construct (Fig. 2B). This protein comigrated with analogous proteins from pETFΔXm and pNBΔXm that were truncated at the same position (data not shown).
Mutagenesis in proteinase active site.
The sites E1071-S1072 and E1345-T1346 defined the boundaries of a protein with a calculated size of 29.6 kDa that contained the conserved His1110, Glu1131, and Cys1193 residues characteristic of the active site of the picornavirus-like cysteine 3C proteinase (1, 12, 16, 31). In order to verify the identity of this protein as the FCV cysteine “3C-like” proteinase, we substituted Gly for Cys1193 in the pBSX sequence (Fig. 3A). This mutation abolished autocatalytic processing of the pBSX-encoded portion of the polyprotein. In addition, a 66.5-kDa protein corresponding to the full-size translation product was found in an in vitro-translation mixture and in induced bacterial cells (Fig. 3B, lanes 1 and 4). Of interest, comparison of protein profiles of pBSX-driven expression showed significant differences in efficiency of cleavage at the E1419-G1420 site in polypeptides expressed either in in vitro-translation experiments or in bacterial cells (Fig. 3B, lanes 2 and 3).
FIG. 3.
Analysis of the effects of amino acid substitutions in the predicted catalytic site of the FCV 3C-like proteinase. (A) Schematic representation of Cys→Gly mutagenesis in the GDC domain of the FCV proteinase. (B) Elimination of specific autocatalytic processing in the ORF1 fragment encoded by pBSX. The expression of pBSX and pBSXm plasmids was analyzed in vitro and in E. coli. Lanes 1 and 2, insoluble fractions of induced bacterial cells transformed with pBSXm and pBSX, respectively (Coomassie blue staining). Lanes 3, and 4, in vitro TNT products derived from analysis of pBSX and pBSXm, respectively (autoradiography). Lane 5, TNT mixture without plasmid DNA. The arrows indicate pBSX-derived proteins identified by N-terminal sequence analysis. (C) Effect of the mutation introduced into the catalytic site of the proteinase on the cleavage of the viral capsid precursor protein encoded in pfΔ20. The radiolabeled capsid precursor protein derived from pfΔ20 was incubated with nonradiolabeled translation products derived from plasmid pBSX or pBSXm (lanes 2 and 3, respectively). The first lane contains the pfΔ20 translation products without treatment.
It should be noted that pBSX-driven expression led to synthesis of a proteinase active in its ability to mediate cleavage of the capsid precursor protein in trans. We found that the Cys→Gly mutation abrogated this activity as well (Fig. 3C).
Uncleaved Pro-Pol is a stable protein both in vitro and in FCV-infected cells.
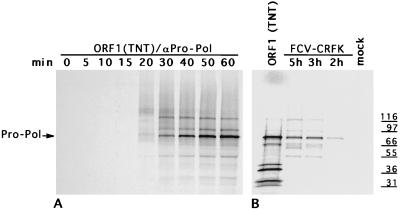
The identification of cleavage sites between the FCV proteinase and polymerase sequences prompted us to examine the efficiency of this cleavage event in infected cells. The cleavage between these proteins appeared inefficient in many of our expression experiments, and the mapping of the cleavage site required overexpression of this region in E. coli. For picornaviruses, it has been suggested that cleavage between 3C and 3D and release of the polymerase is an essential requirement for replication (3, 15, 18).
In order to compare the processing of FCV Pro-Pol in vitro and in infected cells, a specific antiserum was developed against the Pro-Pol region. The 78-kDa Pro-Pol protein expressed in E. coli by the pVPP plasmid was purified with a C-terminal His tag, and antiserum was raised in a guinea pig. Immunoprecipitation of radiolabeled proteins during a time-course experiment of virus infection or translation products of the ORF1 clone showed that an uncleaved 78-kDa Pro-Pol protein accumulated in both lysates (Fig. 4). In both cases, we observed faint bands that were immunoprecipitated with the αPro-Pol serum corresponding to proteins of 90 and 118 kDa that appeared at approximately the same time as the 78-kDa protein but remained at a constant level. These proteins could represent the existence of intermediates formed during processing of the C-terminal part of the polyprotein that resemble those of the corresponding P3 regions of the polyproteins of some picornaviruses, such as the foot-and-mouth disease virus 3ABCD (P100) and 3BCD (P81) (14). An interesting observation in our time-course analysis of FCV-infected cell lysates was the appearance of a triplet of protein bands in the 60-kDa range that was immunoprecipitated with αPro-Pol serum. Although the identities of these proteins were not confirmed, their sizes were consistent with either further cleavage of the Pro-Pol protein or other processing events, such as the cleavage of 3ABCD to yield 3ABC, as described for encephalomyocarditis virus (21). Proteins of 50 kDa or less that could potentially represent products of internal processing of the Pro-Pol protein were detected as faint bands in the in vitro-translation analysis and in infected cells. The relatively small amounts of these proteins suggested that cleavage at the proteinase-polymerase junction might be inefficient both in vitro and in infected cells for FCV.
FIG. 4.
Accumulation of the Pro-Pol protein in FCV-infected cells and in TNT products derived from the ORF1 clone, pTMF-1. Radiolabeled proteins were immunoprecipitated with αPro-Pol serum from time-course points of the in vitro TNT analysis of pTMF-1 (A) and from lysates of FCV-infected CRFK cells (B).
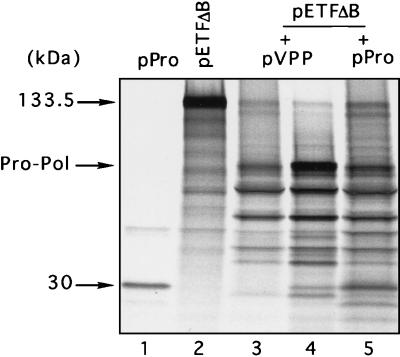
To examine whether the proteinase sequence localized upstream of the E1345-T1346 cleavage site is functional in the absence of polymerase sequence, we removed all ORF1 sequences downstream of the E1345-T1346 cleavage site in the polyprotein encoded by pVPP. Expression of the resulting plasmid, pPro, in bacteria provided evidence for autocatalytic processing of the encoded protein (predicted size, 47 kDa) into three proteins of approximately 18, 14, and 30 kDa (Fig. 2A, lane 4). The last protein was consistent with the predicted size of the mature FCV proteinase (assuming that its C terminus was defined by cleavage at the E1345-T1346 site) and the observed mobilities of the 30-kDa products of autocatalytic cleavage of the pBSX- and pVPP-encoded proteins. To examine whether the pPro-encoded proteinase was capable of cleaving the upstream part of the polyprotein, we coincubated nonradiolabeled TNT products synthesized from pPro with the 133.5-kDa product derived from pETFΔB. We observed trans cleavage of pETFΔB-encoded protein into several smaller proteins (Fig. 5, lane 5). The patterns of trans processing of this protein by pVPP- and pPro-encoded proteinases were similar (Fig. 5, lanes 3 and 5). In addition, an identical cleavage pattern was observed in a cotranslation experiment that included both pVPP and pETFΔB (Fig. 5, lane 4). The proteinase encoded by pPro was also found to cleave the capsid precursor protein efficiently in trans (data not shown).
FIG. 5.
Trans-cleavage analysis of the proteolytic activity of the proteinase encoded by FCV ORF1. Posttranslation or cotranslation incubation of radiolabeled TNT products derived from plasmid pETFΔB with those derived from plasmid pVPP or pPro was performed. Labeled in vitro-translation products derived from plasmids pPro (lane 1) and pETFΔB (lane 2) are shown. The radiolabeled pETFΔB-protein was incubated prior to analysis by SDS-PAGE with nonradiolabeled translation products synthesized from pVPP (lane 3) or from pPro (lane 5). Cotranslation analysis of plasmids pETFΔB and pVPP (lane 4) was also done.
DISCUSSION
Chymotrypsin-like cysteine proteinases (designated 3C) are involved in the proteolytic maturation of structural and nonstructural proteins of viruses in the picornavirus superfamily (17, 33). Varying in size (20 to 30 kDa) and levels of sequence similarity, these proteinases share several features, including the arrangement of the catalytic active center, the presence of cysteine as the nucleophile group, and a genomic location that is immediately upstream of the RNA polymerase gene (1, 12, 25, 28, 33, 40). The proteinases of viruses in the family Caliciviridae have some features similar to those of the 3C-like cysteine proteinases of the picornavirus superfamily, including discrete regions of sequence similarity, a genomic location upstream of the polymerase, and the participation of a cysteine residue in the catalysis of structural and nonstructural protein cleavage sites with E or Q residues in the P1 position (5, 22, 26, 29, 31, 39, 42).
In the present study, we demonstrate that the 3C-like proteinase encoded by the FCV ORF1 polyprotein that mediates processing of the capsid protein precursor is also responsible for the processing of the nonstructural polyprotein. Time-course analysis of the in vitro-translation products derived from a full-length ORF1 cDNA clone showed the early appearance of a diffuse band containing high-molecular-mass precursors ranging from 120 to 140 kDa that were immunoprecipitated with antisera specific for the Pro-Pol protein (Fig. 4A). The sizes of these proteins were consistent with the size of the N-terminal part of the polyprotein that extended into the predicted proteinase sequence in the 3′ end of ORF1 (assuming initiation of translation at the first AUG of ORF1). Further elongation of the polyprotein chain into the region bordered by nt 3233 to 4054 resulted in efficient cotranslational processing of the upstream polyprotein, as the nascent proteinase was apparently expressed. Rapid processing of calicivirus nonstructural proteins during the translation of templates containing the 3C-like proteinase has also been described in the analysis of RHDV and SHV cDNA clones (26, 27). Mutagenesis confirmed that cysteine 1193, located within the putative active site, was essential for the activity of the FCV proteinase. Protein products derived from coupled transcription and translation reactions containing plasmids encoding the N-terminal part of the FCV ORF1 polyprotein corresponded to the predicted sizes in polyacrylamide gels. This observation was consistent with the absence of additional proteinase activity encoded in the N-terminal part of the polyprotein and suggests that the 3C-like proteinase encoded in the region defined by nt 3233 to 4054 of FCV may be the only virus-encoded proteolytic enzyme in ORF1.
Translation extension of the polyprotein chain beyond the proteinase sequence resulted in the synthesis of a stable protein containing both proteinase and polymerase motifs. We previously reported that a virus-specific protein of approximately 78 kDa was observed in FCV-infected cells and in bacterial or reticulocyte lysates expressing recombinant proteins derived from cDNA clones of the FCV ORF1. We proposed that the 78-kDa protein could represent a proteolytically active proteinase-polymerase protein analogous to the picornavirus 3CD (39). Our present study provided additional evidence for the identity of this protein as a potential proteinase-polymerase precursor. This was indicated by immunoprecipitation analysis with region-specific antisera (αPro-Pol) and by its direct N-terminal sequence analysis, which localized an E-S cleavage site at residues 1071 to 1072 of the FCV ORF1 polyprotein. Cleavage of the native FCV polyprotein encoded by ORF1 at this site would lead to the release of a 75.7-kDa protein containing 692 amino acid residues. Time-course experiments confirmed the presence of this protein in infected cells and revealed that it was stable and accumulated with time.
It was only during expression of the C-terminal region of the FCV ORF1 in bacterial cells from plasmids pBSX and pVPP that we could detect cleavage in this region that was efficient enough to allow N-terminal protein sequence analysis of additional cleavage products from the Pro-Pol region. The N-terminal sequence of 20- and 11-kDa proteins in the bacterial lysates from pBSX allowed the identification of cleavage sites at E1345-T1346 and E1419-G1420, respectively. Of interest, both of these sites could be identified in the FCV ORF1 polyprotein as potential cleavage sites for the picornavirus proteinase by computer analysis (4). Cleavage of the Pro-Pol protein at the E1345-T1346 site should lead to the appearance of proteins in infected cells with sizes of 29.6 and 46.1 kDa, with sequences overlapping the proteinase and polymerase motifs, respectively. However, cleavage at E1419-G1420 would result in a putative polymerase lacking the conserved KDELR sequence characteristic of a number of picornavirus polymerases (23). We could not detect the efficient production of the predicted cleavage products from either cleavage site in FCV-infected cells. Thus, it is not yet clear whether these two cleavage sites represent valid processing events or nonspecific cleavages in the expression systems. The predominance of the Pro-Pol protein in infected cells suggests that the inefficient cleavage between 3C and 3D observed in vitro may reflect a mechanism for regulation of the amounts of fully processed proteinase and polymerase during viral replication. Alternatively, the stability of the Pro-Pol protein suggests that it could be a mature protein exhibiting both protease and polymerase activities.
The size of the potential mature FCV proteinase (29.6 kDa) as defined by the E1071-S1072 and E1345-T1346 cleavage sites mapped in the present study was larger than that reported for RHDV (15 kDa). However, comparative analysis of RHDV and FCV sequences showed that the distances between the predicted catalytic residues of the FCV proteinase were similar to those of the RHDV enzyme (5). The observed length difference between the predicted mature proteinases of FCV and RHDV was due to an extended C-terminal region of the FCV proteinase that contained sequences similar to those reported to be part of the RHDV polymerase (42). A 30-kDa protein corresponding to the putative FCV proteinase was observed during analysis of several different ORF1 clones in various expression systems. However, the predominance of the stable proteinase-polymerase protein in infected cells precluded identification of the putative mature forms of both the proteinase and polymerase. Analysis of clone pPro containing the pVPP coding sequence truncated at the E1345-T1346 cleavage site showed that the proteinase could mediate efficient cis and trans cleavage in the absence of downstream polymerase region sequence. In addition, the pPro clone showed no signs of further processing of the 30-kDa protein. Clarification of whether the sequence of the putative 30-kDa FCV proteinase contains an alternative cleavage site that would more closely resemble the border between the RHDV proteinase and polymerase will require additional study.
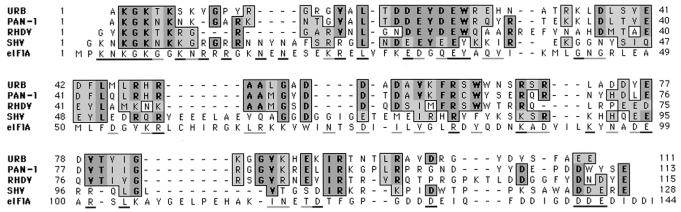
Our present study has mapped a small 12.7-kDa protein (14 kDa, according to its mobility in SDS-PAGE) localized immediately upstream of the proteinase gene. Following picornavirus nomenclature, this protein corresponds to 3B (VPg). In picornaviruses, the VPg is covalently linked through a tyrosine residue to the 5′ end of the viral genomic RNA (2, 35). Calicivirus genomes also have a small VPg protein linked to the genomic and subgenomic RNAs (6, 20, 30, 37), and the VPg has been mapped just upstream of the proteinase for RHDV and primate calicivirus (PAN-1) (11, 43). Comparison of the amino acid sequence of the putative FCV VPg (12.7-kDa protein) with those of RHDV, PAN, and SHV shows similarities of 58, 69, and 36%, respectively, in this region of the genome (Fig. 6). Of interest, these sequences share a common structural feature, with the presence of two hydrophilic amino acid clusters at the N termini that can be detected by analysis of amino acid sequences by the method of Kyte and Doolittle (24) (not shown). The first cluster consists of basic residues, and the second contains acidic residues. The deduced amino acid sequence of the FCV 12.7-kDa protein does not have significant structural similarity to picornavirus VPg sequences. In addition, the picornavirus VPg is smaller and apparently functions in RNA replication and packaging (32, 34). The genomic RNA of picornavirus is infectious and can be efficiently translated in the presence or absence of VPg protein at its 5′ end. However, removal of the calicivirus VPg protein by proteinase treatment leads to loss of RNA infectivity and a significant decrease in the efficiency of translation of the RNA template (20). The latter observation and the infectivity of capped, synthetic RNA derived from an infectious FCV cDNA clone suggests a cap-like function for the calicivirus VPg and its involvement in translation of viral RNAs (20, 38). The mechanism of such an involvement is not yet understood. However, the efficiency of translation of mRNA isolated from FCV-infected cells is not affected in the presence of cap analogue 7mGTP, which suggests that the translation mechanism is independent of cap-binding proteins (20). One possible mechanism may be related to a direct interaction of the calicivirus VPg with ribosomal proteins. In regard to this possibility, the existence of sequence similarity between the calicivirus VPg and one of the eukaryotic initiation factors (eIF) of translation, eIF1A (formerly eIF-4C) (10), is of interest. The similarity reaches the level of 35% between eIF1A and SHV sequences and 26% between eIF1A and FCV sequences. The eIF1A molecule belongs to a group of small protein factors that promote initiation of translation in eukaryotic cells. This protein is essential for transfer of the initiator Met-tRNAf-eIF2-GTP ternary complex to 40S ribosomal subunits to form the 40S preinitiation complex (9). This factor is also thought to stimulate mRNA binding to 40S subunits and to be an accessory for other factors in dissociating 80S ribosomes (13, 41). It will be of interest to examine whether the FCV VPg may play such a role in the initiation of translation.
FIG. 6.
Comparison of the putative FCV VPg with other caliciviruses and human eukaryotic translation initiation factor eIF1A. A ClustalW alignment of the deduced amino acid sequence of the 12.7-kDa protein with the corresponding regions of RHDV (GenBank accession no. M67473), PAN-1 (GenBank accession no. AF091736), and SHV (GenBank accession no. L07418) was created with the following parameters: open gap penalty, 10.0; extend gap penalty, 0.1; gap distance, 8; and similarity matrix, blosum. Afterwards, the alignment was edited manually to include the eIF1A sequence (Protein Information Resource no. C53045). For the FCV sequence (URB), the number 1 position corresponds to position 961 of the ORF1 polyprotein; for PAN-1, it corresponds to position 1072; for RHDV, it corresponds to position 994; and for SHV, it corresponds to position 962. Identical and similar amino acids of calicivirus sequences are shaded in dark and light tones, respectively. Identical residues are also shown in boldface. Identical and similar amino acids conserved between the eIF1A and SHV sequences are underlined in black and gray, respectively.
ACKNOWLEDGMENTS
We thank John Coligan and Mark Garfield of LMS, NIAID, NIH, for assistance with the protein sequence analysis. We thank Jose Valdesuso for his dedicated technical support. We extend our appreciation to Albert Z. Kapikian and Robert M. Chanock, LID, NIAID, NIH, for continuing support.
REFERENCES
- 1.Allaire M, Chernaia M M, Malcolm B A, James M N G. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases. Nature. 1994;369:72–76. doi: 10.1038/369072a0. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V, Baltimore D. Protein is linked to the 5′ end of poliovirus RNA by a phosphodiester linkage to tyrosine. J Biol Chem. 1978;253:5263–5266. [PubMed] [Google Scholar]
- 3.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blom N, Hansen J, Blaas D, Brunak S. Cleavage site analysis in picornaviral polyproteins: discovering cellular targets by neural networks. Protein Sci. 1996;5:2203–2216. doi: 10.1002/pro.5560051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boniotti B M, Wirlblich C, Sibilia M, Meyers G, Thiel H-J, Rossi C. Identification and characterization of 3C-like protease from rabbit hemorrhagic disease virus, a calicivirus. J Virol. 1994;68:6487–6495. doi: 10.1128/jvi.68.10.6487-6495.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burroughs J N, Brown F. Presence of a covalently linked protein on calicivirus RNA. J Gen Virol. 1978;41:443–446. doi: 10.1099/0022-1317-41-2-443. [DOI] [PubMed] [Google Scholar]
- 7.Carter M J. Feline calicivirus protein synthesis investigated by Western blotting. Arch Virol. 1989;108:69–79. doi: 10.1007/BF01313744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter M J, Milton I D, Turner P C, Meanger J, Bennett M, Gaskell R M. Identification and sequence determination of the capsid protein gene of feline calicivirus. Arch Virol. 1992;122:233–235. doi: 10.1007/BF01317185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhuri J, Si K, Maitra U. Function of eukaryotic translation initiation factor 1A (eIF1A) (formerly called eIF-4C) in initiation of protein synthesis. J Biol Chem. 1997;272:7883–7891. doi: 10.1074/jbc.272.12.7883. [DOI] [PubMed] [Google Scholar]
- 10.Dever T E, Wei C L, Benkowski L A, Browning K, Merrick W C, Hershey J W. Determination of the amino acid sequence of rabbit, human, and wheat germ protein synthesis factor eIF-4C by cloning and chemical sequencing. J Biol Chem. 1994;269:3212–3218. [PubMed] [Google Scholar]
- 11.Dunham D M, Jiang X, Berke T, Smith A W, Matson D O. Genomic mapping of a calicivirus VPg. Arch Virol. 1998;143:2421–2430. doi: 10.1007/s007050050471. [DOI] [PubMed] [Google Scholar]
- 12.Gorbalenya A E, Donchenko A P, Blinov V M, Koonin E V. Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases. A distinct protein superfamily with a common structural fold. FEBS Lett. 1989;243:103–114. doi: 10.1016/0014-5793(89)80109-7. [DOI] [PubMed] [Google Scholar]
- 13.Goumans H, Thomas A, Verhoeven A, Voorma H O, Benne R. The role of eIF-4C in protein synthesis initiation complex formation. Biochim Biophys Acta. 1980;608:39–46. doi: 10.1016/0005-2787(80)90131-8. [DOI] [PubMed] [Google Scholar]
- 14.Grubman M J, Robertson B H, Morgan D O, Moore D M, Dowbenko D. Biochemical map of polypeptides specified by foot-and-mouth disease virus. J Virol. 1984;50:579–586. doi: 10.1128/jvi.50.2.579-586.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall D J, Palmenberg A C. Cleavage site mutations in the encephalomyocarditis virus P3 region lethally abrogate the normal processing cascade. J Virol. 1996;70:5954–5961. doi: 10.1128/jvi.70.9.5954-5961.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammerle T, Hellen C U T, Wimmer E. Site-directed mutagenesis of the putative catalytic triad of poliovirus 3C proteinase. J Biol Chem. 1991;266:5412–5416. [PubMed] [Google Scholar]
- 17.Harris K S, Hellen C U T, Wimmer E. Proteolytic processing in the replication of picornaviruses. Semin Virol. 1990;1:323–333. [Google Scholar]
- 18.Harris K S, Reddigari S R, Nicklin M J, Hammerle T, Wimmer E. Purification and characterization of poliovirus polypeptide 3CD, a proteinase and a precursor for RNA polymerase. J Virol. 1992;66:7481–7489. doi: 10.1128/jvi.66.12.7481-7489.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbert T P, Brierley I, Brown T D K. Detection of the ORF3 polypeptide of feline calicivirus in infected cells and evidence for its expression from single, functionally bicistronic, subgenomic mRNA. J Gen Virol. 1996;77:123–127. doi: 10.1099/0022-1317-77-1-123. [DOI] [PubMed] [Google Scholar]
- 20.Herbert T P, Brierly I, Brown T D K. Identification of a protein linked to the genomic and subgenomic mRNAs of feline calicivirus and its role in translation. J Gen Virol. 1997;78:1033–1040. doi: 10.1099/0022-1317-78-5-1033. [DOI] [PubMed] [Google Scholar]
- 21.Jackson R J. A detailed kinetic analysis of the in vitro synthesis and processing of encephalomyocarditis virus products. Virology. 1986;149:114–127. doi: 10.1016/0042-6822(86)90092-9. [DOI] [PubMed] [Google Scholar]
- 22.Jiang X, Wang M, Wang K, Estes M K. Sequence and genomic organization of Norwalk virus. Virology. 1993;195:51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- 23.Koonin E V. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 24.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 25.Lawson M A, Semler B L. Picornavirus protein processing—enzymes, substrates, and genetic regulation. Curr Top Microbiol Immunol. 1990;161:49–88. [PubMed] [Google Scholar]
- 26.Liu B, Clarke I N, Lambden P R. Polyprotein processing in Southampton virus: identification of 3C-like protease cleavage sites by in vitro mutagenesis. J Virol. 1996;70:2605–2610. doi: 10.1128/jvi.70.4.2605-2610.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin Alonso J M, Casais R, Boga J A, Parra F. Processing of rabbit hemorrhagic disease virus polyprotein. J Virol. 1996;70:1261–1265. doi: 10.1128/jvi.70.2.1261-1265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matthews D A, Smith W W, Ferre R A, Condon B, Budahazi G, Sisson W, Villafranca J E, Janson C A, McElroy H E, Gribskov C L, Worland S. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell. 1994;77:761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 29.Meyers G, Wirblich C, Thiel H-J. Rabbit hemorrhagic disease virus—molecular cloning and nucleotide sequencing of a calicivirus genome. Virology. 1991;184:664–676. doi: 10.1016/0042-6822(91)90436-f. [DOI] [PubMed] [Google Scholar]
- 30.Meyers G, Wirblich C, Thiel H-J. Genomic and subgenomic RNAs of rabbit hemorrhagic disease virus are both protein-linked and packaged into particles. Virology. 1991;184:677–686. doi: 10.1016/0042-6822(91)90437-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neill J D. Nucleotide sequence of a region of the feline calicivirus genome which encodes picornavirus-like RNA-dependent RNA-polymerase, cysteine protease and 2C polypeptides. Nucleic Acids Res. 1990;17:145–160. doi: 10.1016/0168-1702(90)90061-f. [DOI] [PubMed] [Google Scholar]
- 32.Nomoto A, Kitamura N, Golini F, Wimmer E. The 5′-terminal structures of poliovirion RNA and poliovirus mRNA differ only in the genome-linked protein VPg. Proc Natl Acad Sci USA. 1977;74:5345–5349. doi: 10.1073/pnas.74.12.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmenberg A C. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- 34.Paul A V, van Boom J H, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 35.Rothberg P G, Harris T J, Nomoto A, Wimmer E. O4-(5′-uridylyl)tyrosine is the bond between the genome-linked protein and the RNA of poliovirus. Proc Natl Acad Sci USA. 1978;75:4868–4872. doi: 10.1073/pnas.75.10.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J E, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Schaffer F L, Ehresmann D W, Fretz M K, Soergel M I. A protein, VPg, covalently linked to 36S calicivirus RNA. J Gen Virol. 1980;47:215–220. doi: 10.1099/0022-1317-47-1-215. [DOI] [PubMed] [Google Scholar]
- 38.Sosnovtsev S V, Green K Y. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VPg for infectivity. Virology. 1995;210:383–390. doi: 10.1006/viro.1995.1354. [DOI] [PubMed] [Google Scholar]
- 39.Sosnovtsev S V, Sosnovtseva S A, Green K Y. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J Virol. 1998;72:3051–3059. doi: 10.1128/jvi.72.4.3051-3059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spall V E, Shanks M, Lomonossoff G P. Polyprotein processing as a strategy for gene expression in RNA viruses. Semin Virol. 1997;8:15–23. [Google Scholar]
- 41.Thomas A, Goumans H, Voorma H O, Benne R. The mechanism of action of eukaryotic initiation factor 4C in protein synthesis. Eur J Biochem. 1980;107:39–45. doi: 10.1111/j.1432-1033.1980.tb04621.x. [DOI] [PubMed] [Google Scholar]
- 42.Wirblich C, Sibilia M, Boniotti M B, Rossi C, Thiel H-J, Meyers G. 3C-like protease of rabbit hemorrhagic disease virus: identification of cleavage sites in the ORF1 polyprotein and analysis of cleavage specificity. J Virol. 1995;69:7159–7168. doi: 10.1128/jvi.69.11.7159-7168.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wirblich C, Thiel H-J, Meyers G. Genetic map of the calicivirus rabbit hemorrhagic disease virus as deduced from in vitro translation studies. J Virol. 1996;70:7974–7983. doi: 10.1128/jvi.70.11.7974-7983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]