Abstract
Mouse mammary tumor virus (MMTV) encodes a superantigen (Sag) that is expressed at the surface of antigen-presenting cells in conjunction with major histocompatibility complex (MHC) type II molecules. The Sag-MHC complex is recognized by entire subsets of T cells, leading to cytokine release and amplification of infected B and T cells that carry milk-borne MMTV to the mammary gland. Expression of Sag proteins from endogenous MMTV proviruses carried in the mouse germ line usually results in the deletion of self-reactive T cells during negative selection in the thymus and the elimination of T cells required for infection by specific milk-borne MMTVs. However, other endogenous MMTVs are unable to eliminate Sag-reactive T cells in newborn mice and cause partial loss of reactive T cells in adults. To investigate the kinetics of Sag-reactive T-cell deletion, backcross mice that contain single or multiple MMTVs were screened by a novel PCR assay designed to distinguish among highly related MMTV strains. Mice that contained Mtv-17 alone showed slow kinetics of reactive T-cell loss that involved the CD4+, but not the CD8+, subset. Deletion of CD4+ or CD8+ T cells reactive with Mtv-17 Sag was not detected in thymocytes. Slow kinetics of peripheral T-cell deletion by Mtv-17 Sag also was accompanied by failure to detect Mtv-17 sag-specific mRNA in the thymus, despite detectable expression in other tissues, such as spleen. Together, these data suggest that Mtv-17 Sag causes peripheral, rather than intrathymic, deletion of T cells. Interestingly, the Mtv-8 provirus caused partial deletion of CD4+Vβ12+ cells in the thymus, but other T-cell subsets appeared to be deleted only in the periphery. Our data have important implications for the level of antigen expression required for elimination of self-reactive T cells. Moreover, these experiments suggest that mice expressing endogenous MMTVs that lead to slow kinetics of T-cell deletion will be susceptible to infection by milk-borne MMTVs with the same Sag specificity.
The mouse mammary tumor virus (MMTV) superantigen (Sag) protein is necessary for the efficient transmission of virus from infected mother’s milk to the mammary glands of offspring (23, 54). MMTV transmission requires infection of both B and T lymphocytes to facilitate transport of virus from the gut of newborns to mammary cells, and lymphocytes appear to be a reservoir of virus in the postnatal period prior to mammary gland development (7, 23, 26). Viral infection of B cells in the gut-associated lymphoid tissue leads to the expression of Sag at the B-cell surface as a type II transmembrane glycoprotein (32, 33). The C-terminal portion of Sag in conjunction with major histocompatibility complex (MHC) class II protein is recognized by entire classes of T cells that have specific β chains as part of the T-cell receptor (TCR) (4, 54, 59). Experiments that switch the N-terminal and C-terminal regions of different Sags as well as site-directed Sag mutations suggest that the C-terminal 30 to 40 amino acids are critical for TCR interaction (38, 54, 59). Interaction of Sag with the TCR leads to a T-cell signaling pathway that results in the production of cytokines and/or T-cell proliferation (3, 45, 50). The production of cytokines leads to further B- and T-cell proliferation and amplification of MMTV-infected cells (3, 29).
Another consequence of T-cell stimulation by MMTV Sag is the ultimate loss of Sag-reactive T cells from the immune repertoire of the infected mouse (36). For example, milk-borne infection with C3H MMTV leads to stimulation and, in subsequent months, deletion of Vβ14+ T cells reactive with C3H Sag (5, 10). Similarly, endogenous MMTVs express Sag proteins at the surface of antigen-presenting cells, and Sag expression results in the deletion of specific T-cell subsets (52). Previous data have shown that expression of the milk-borne C3H MMTV Sag from a transgene results in complete deletion of Vβ14+ T cells that are required for C3H virus transmission to the mammary gland (23). Such sag transgenic mice are resistant to infection by C3H MMTV through nursing on infected mothers. These experiments suggested that T-cell deletion induced by endogenous MMTV Sags would provide protection against exogenous MMTV infection with the same Sag specificity for T cells (23, 28, 36).
Deletion of T-cell classes due to endogenous MMTV proviruses may be incomplete (43). For example, both the Mtv-8 and Mtv-9 proviruses delete Vβ5+, Vβ11+, and Vβ12+ T cells. The Mtv-8 provirus causes incomplete deletion of Vβ5+ and Vβ11+ cells, whereas the Mtv-9 provirus causes complete deletion of these T-cell subsets (17, 43, 52), despite the fact that the Mtv-8 and Mtv-9 long terminal repeats (LTRs) are nearly identical (8). Because complete deletion of a T-cell subset by an endogenous MMTV Sag appears to require effective antigen presentation in the thymus to eliminate self-reactive T cells during development of the immune repertoire (3, 44), incomplete deletion of Sag-reactive cells may reflect a failure to express certain endogenous MMTVs in thymic antigen-presenting cells, perhaps dendritic cells (39). Incomplete deletion also may reflect the expression and presentation of these viral Sags on other cell types that result in the deletion of self-reactive cells in the periphery (31, 49). Alternatively, some endogenous MMTVs may be expressed in thymic antigen-presenting cells at a reduced level, resulting in an avidity that borders the threshold for T-cell signaling necessary to eliminate self-reactive cells by apoptosis (50). Because little is known about the level of endogenous MMTV sag-specific expression (56), we investigated whether there was a correlation between thymic expression of spliced sag mRNA and the kinetics of Sag-specific T-cell deletion. Using mice bred to contain single endogenous MMTV proviruses, we showed that partial deletion of Sag-reactive T cells was correlated with characteristics typical for the establishment of T-cell tolerance in the peripheral immune system. Slow kinetics of Sag-reactive T-cell deletion also correlated with poor expression of sag-specific mRNA in the thymus.
MATERIALS AND METHODS
Mice.
C58/J, BALB/cJ, and PERA/Ei (Peru-Atteck) mice were obtained from the Jackson Laboratories (Bar Harbor, Maine); these animals, F1 hybrids, and backcross animals were bred in the Animal Resources Center at the University of Texas at Austin. Sentinel animals tested negative for the presence of mouse hepatitis virus and other common murine pathogens (except for those tested for the experiments reported in Table 1).
TABLE 1.
Deletion of specific T-cell classes in (C58 × PERA)F1 and (C58 × PERA) × PERA N1 backcross mice
| Mice | Provirus(es) | na | % of CD4+ lymph node cellsb
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Vβ3 | Vβ6 | Vβ8 | Vβ9 | Vβ11 | Vβ12 | Vβ14 | |||
| C58 | Mtv-3, -7, -17 | 3 | 0.4 ± 0.6 | 0.2 ± 0.4 | 10.2 ± 1.2 | 0.1 ± 0.2 | 10.0 ± 0.3 | 6.7 ± 2.3 | 7.3 ± 0.8 |
| PERA | None | 3 | 4.2 ± 0.7 | 6.7 ± 0.6 | 12.2 ± 1.3 | 1.0 ± 0.1 | 7.2 ± 0.4 | 9.5 ± 0.5 | 4.8 ± 0.1 |
| (C58 × PERA)F1 | Mtv-3, -7, -17 | 4 | 0.1 ± 0.1 | 0.5 ± 0.3 | 14.9 ± 0.7 | 0.2 ± 0.1 | 7.5 ± 1.8 | 3.1 ± 1.6 | 8.6 ± 0.8 |
| Backcross N1 | None | 2 | 3.2 | 6.6 | 11.6 | 0.9 | 6.8 | 7.4 | 4.4 |
| Backcross N1 | Mtv-3 | 4 | 0.2 ± 0.2 | 8.6 ± 1.8c | 15.2 ± 1.2 | 0.9 ± 0.2 | 8.0 ± 0.8 | 10.1 ± 0.4 | 5.2 ± 0.9 |
| Backcross N1 | Mtv-7 | 1 | 4.3 | 0.4 | 10.6 | 0.1 | 8.4 | 9.5 | 6.8 |
| Backcross N1 | Mtv-17 | 3 | 4.4 ± 0.9 | 9.4 ± 3.1c | 15.4 ± 0.6 | 1.0 ± 0.2 | 6.9 ± 3.9 | 1.7 ± 1.3 | 5.7 |
| Backcross N1 | Mtv-3, -7 | 2 | 0.2 | 1.0 | 11.1 | 0.1 | 11.9 | 12.7 | 6.6 |
| Backcross N1 | Mtv-7, -17 | 2 | 0.3 | 9.6 | 16.6 | 1.2 | 6.8 | 3.5 | 6.2 |
| Backcross N1 | Mtv-7, -17 | 1 | 5.3 | 1.2 | 9.0 | 0.1 | 6.0 | NDd | 6.0 |
| Backcross N1 | Mtv-3, -7, -17 | 2 | 0.2 | 0.5 | 12.3 | 0.1 | 9.1 | 4.3 | 8.4 |
Number of mice analyzed. The age of mice tested was ≥5 months.
Average ± standard deviation.
The high standard deviation observed in these samples was due to the use of limiting antibody in early experiments and was not observed in later experiments.
ND, not done.
Antibodies and FACS analysis.
Lymph nodes (in most cases, only inguinal nodes) were removed, and cells were released into fluorescence-activated cell sorter (FACS) wash buffer (phosphate-buffered saline containing 0.1% NaN3 and 2.5% bovine serum) by being crushed with the blunt end of a 3-ml syringe. Clumps were removed on ice by settling for 5 min, and single cells were washed several times in FACS buffer before staining. Peripheral blood lymphocytes were obtained and purified as described previously (53, 54). Cells (approximately 106) were incubated with antibody specific for Vβ3, -5, -6, -7, -8, -9, -11, -12, or -14 labeled with fluorescein and CD4- or CD8-specific antibody labeled with phycoerythrin (PharMingen, San Diego, Calif.). Thymocytes were prepared similarly except that the staining was performed with fluorescein-labeled Vβ antibodies, phycoerythrin-labeled CD4, and Cy-chrome-labeled CD8 (also from PharMingen). Cells were incubated for 45 min on ice prior to being washed with FACS buffer and fixed in 1% paraformaldehyde in FACS buffer. Cells were analyzed by using the CELLQuest program and a FACSCalibur cytometer (Becton Dickinson, San Jose, Calif.). Statistical analysis was performed with a two-tailed Student t test.
RNA extractions and RNase protection assays.
The guanidine isothiocyanate method was employed for RNA extractions as described previously (55). DNA and low-molecular-weight RNAs were removed by precipitation with sodium acetate (41). RNase protection assays were performed essentially as described by Yang and Dudley (58) except that hybridizations were performed at 56°C. The riboprobe was derived from the Sau3A fragment (−455 to −116 relative to the +1 start site for transcription at the U3/R junction of the LTR of Mtv-17 (24). This probe should detect all known MMTV mRNAs.
DNA extractions and Southern blotting.
High-molecular-weight DNA was obtained from tails or livers of backcross mice as described by Choi et al. (11) or Dudley and Risser (13). In later experiments, a simplified procedure was used for isolating tail DNA. A tail section (ca. 1 in.) was added to 1 ml of 20 mM Tris-HCl (pH 7.4)–25 mM EDTA–0.5% sodium dodecyl sulfate–75 mM NaCl–100 μg of proteinase K per ml and digested for at least 3 h at 53°C. The protease was inactivated by being boiled for 5 min, and the solution was cooled to 4°C prior to centrifugation at the same temperature for 10 min at 1,700 × g. The supernatant (500 μl) was clarified further by centrifugation at 10,000 × g for 5 min at 4°C to remove sodium dodecyl sulfate prior to ethanol precipitation. Pellets were washed with 70% ethanol and resuspended in 500 μl of 10 mM Tris-HCl (pH 7.4)–0.1 mM EDTA. Southern blotting of high-molecular-weight DNA or PCR products was performed as described by Dudley and Risser (13), and blots were hybridized to a C3H LTR probe (13) followed by autoradiography.
PCR and reverse transcription-PCR (RT-PCR).
In the majority of experiments, PCR was used for determining the number and identity of endogenous MMTVs in individual mice of the BALB/cJ × PERA/Ei and C58/J × PERA/Ei crosses. In BALB/c crosses, mice were typed for Mtv-6, -8, and -9, whereas in C58/J crosses, mice were typed for Mtv-3, -7, and -17. Although C58/J mice also appear to contain the Mtv-30 provirus, expression of this provirus has not been detected (43) and may represent a solo LTR; therefore, Mtv-30 was not considered in this analysis. Since all endogenous MMTVs are highly related (8), primers were designed by using variable portions of the LTR and/or the DNA sequence flanking the provirus. Flanking sequences adjacent to the 3′ ends of the Mtv-8, -9, and -17 proviruses were obtained by sequencing of plasmid subclones of DNA extracted from the lambda phages AACl14, AACl7, and AACl6, respectively (14, 15).
PCR mixtures for provirus typing contained 1 μl of each primer (diluted to 50 ng/μl), 3 μl (ca. 400 ng) of tail DNA, and 45 μl of PCR SuperMix (Gibco BRL, Gaithersburg, Md.). The following primer pairs were used to detect different MMTV proviruses: Mtv-3 and Mtv-6, LTR926+ (5′ AGGCATTGCCCTTAGCTTTC 3′) and LTR 1191− (5′ GTGAATGTTAGGACTGTTGCA 3′) (product size, 265 bp); Mtv-7, LTR970+ (5′ ATACAATCAGGTCTACTTGC 3′) and LTR 1191− (product size, 252 bp); Mtv-17, LTR958+ (5′ AACCTTTATGAGCCCAACCTTG 3′) and AC6A− (flanking DNA) (5′ GTTCCCCATTCAAGAAAGCCCT 3′) (product size, 434 bp); Mtv-8, LTR 958+ and Mtv-F2− (flanking DNA) (5′ GGAATAGAGGAGAATGAAGATTCC 3′) (product size, 488 bp); and Mtv-9, LTR958+ and pCl7F− (5′ GTATAAGAGTCCCCCAAGAGGCT 3′) (product size, 537 bp). PCR mixtures for Mtv-3, -7, and -17 were incubated at 94°C for 5 min and for 44 cycles with denaturing at 94°C for 1 min, annealing at 50°C for 1 min, and polymerization at 72°C for 1 min followed by incubation at 72°C for 5 min. PCRs for Mtv-6, -8, and -9 were performed similarly except that the annealing temperature was 46°C.
RT-PCRs were performed essentially as described previously (55, 56) with the following modifications. After RNA extractions, each sample was digested for 20 min in a 500-μl reaction mixture containing 50 mM KCl, 20 mM Tris-HCl (pH 7.4), 2 mM MgCl2, 50 U of RNase-free DNase I (Boehringer Mannheim, Indianapolis, Ind.), and 20 U of RNasin (Promega, Madison, Wis.). Control RT-PCRs with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers were performed as described previously (55). After addition of EDTA to 100 mM, reaction mixtures were extracted with phenol-chloroform-isoamyl alcohol (25:24:1). RNA was precipitated with ethanol, and the concentration was determined by absorbance at 260 nm. For cDNA synthesis, 4 μg of total RNA was used in a 25-μl reaction mixture containing 240 U of murine leukemia virus reverse transcriptase (Gibco BRL). One-tenth of each reaction mixture was added directly to PCR mixtures as described previously (56).
RESULTS
Partial deletion of cognate T cells by Mtv-17.
Previously, it has been reported that Vβ5+, Vβ11+, and Vβ12+ T cells are not deleted in C58/J mice (1, 2, 20, 46), although C58 mice harbor the endogenous proviruses Mtv-3, -7, -17, and -30 (43). Mtv-8, -9, -11 (also called Mlsf [1, 20]), and -17 Sags are highly related and have been shown to cause complete or partial deletion of Vβ11+ and Vβ12+ T-cell subsets in inbred strains resulting from a CBA/CaJ × C58/J cross (43). Furthermore, C58/J mice have the I-Ek haplotype that should allow efficient presentation of Sag on antigen-presenting cells (43, 46). To confirm this result, we tested lymph nodes from C58/J mice for the percentage of CD4+ T cells bearing different TCR β chains (Table 1). Compared to PERA/Ei mice that lack endogenous MMTV proviruses (58a), there was a 30% deletion of Vβ12+ cells, but this deletion was variable (6.7% ± 2.3% in C58/J mice compared to PERA mice [9.5% ± 0.5%]), and no deletion of Vβ11+ cells was observed. Therefore, deletion of cognate T cells due to the Mtv-17 provirus appears to be minimal and somewhat variable in C58/J mice, as previously described (43, 46). In contrast, there was virtually complete deletion of Vβ3+ T cells reactive with Mtv-3 Sag and Vβ6+ and Vβ9+ T cells reactive with Mtv-7 Sag as noted previously (3, 44) (Table 1).
We also analyzed the deletion of specific classes of CD4+ T cells from the lymph nodes of F1 hybrids between C58/J and PERA/Ei mice (Table 1). In these animals, the deletion of CD4+Vβ12+ T cells was approximately 70% but variable, and some animals showed minimal deletion of Vβ11+ cells. Thus, as previously observed (46), F1 animals that were haploid for each MMTV provirus, including Mtv-17, showed increased deletion of Mtv-17 cognate T cells compared to animals that were diploid for the provirus. This may be due to competition for limiting MHC class II molecules (35).
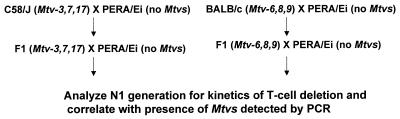
To further analyze deletion of specific T cells by individual MMTVs, we tested mice from the (C58/J × PERA) × PERA backcross (Fig. 1). Although a relatively small number of animals were tested, results from such backcross mice again indicated that Mtv-3 deleted CD4+Vβ3+ T cells, Mtv-7 deleted CD4+Vβ6+ and -9+ T cells, and Mtv-17 variably deleted CD4+Vβ12+ T cells (Table 1).
FIG. 1.
Scheme for generation of Mtv single-positive mice. Each animal was analyzed in a series of three PCRs containing primers that were specific for the MMTV proviruses in C58/J or BALB/cJ parents. Animals negative for all proviruses were checked for the integrity of DNA samples by PCRs with GAPDH primers.
Analysis of (C58/J × PERA) × PERA backcross mice.
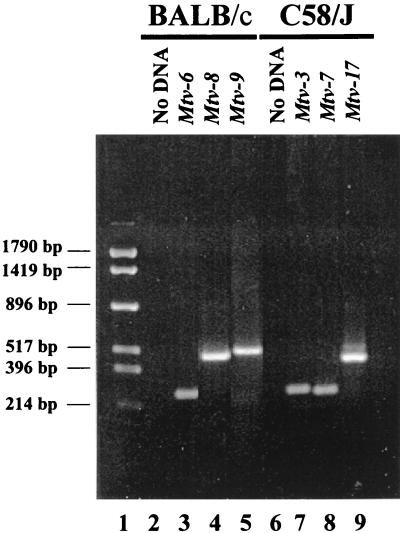
Because we observed considerable variability of T-cell loss due to the Mtv-17 provirus, we analyzed a larger number of animals from the (C58/J × PERA) × PERA cross for the kinetics of T-cell deletion. The initial backcross mice were tested for the presence of specific MMTV proviruses by Southern blotting analysis since the pattern of proviral integration will distinguish highly related MMTV proviruses in different chromosomal positions (12). However, the testing of a larger number of animals was expedited by the development of PCR assays that could distinguish among highly related MMTV proviruses expressed in C58/J mice (Fig. 2).
FIG. 2.
PCR assay for specific endogenous Mtv proviruses. BALB/cJ (lanes 3 to 5) or C58/J (lanes 7 to 9) DNA was used in reactions containing primers specific for the indicated MMTV proviruses. The primers used to detect Mtv-3 and Mtv-6 were the same. Reaction mixtures were analyzed on a 2% agarose gel and stained with ethidium bromide. Lanes 2 and 6 show PCR mixtures lacking template DNA. Molecular size markers are shown in lane 1.
Analysis for the presence of the Mtv-3 and Mtv-7 proviruses was based on a 5′ primer with sequence polymorphisms in the MMTV LTR hypervariable region that encodes the C-terminal portion of Sag (8) and a 3′ conserved primer within the LTR. Testing for the Mtv-17 provirus used a 5′ primer from the LTR polymorphic region combined with a 3′ primer specific for the cellular DNA flanking this provirus. With appropriate conditions, fragments of 265, 252, and 434 bp were obtained with primer sets specific for Mtv-3, -7, and -17, respectively (Fig. 2, lanes 7 to 9). These bands were not obtained in the absence of DNA template (lane 6) and could be used to distinguish among the three MMTV proviruses in DNAs from backcross mice previously tested by Southern blotting (data not shown).
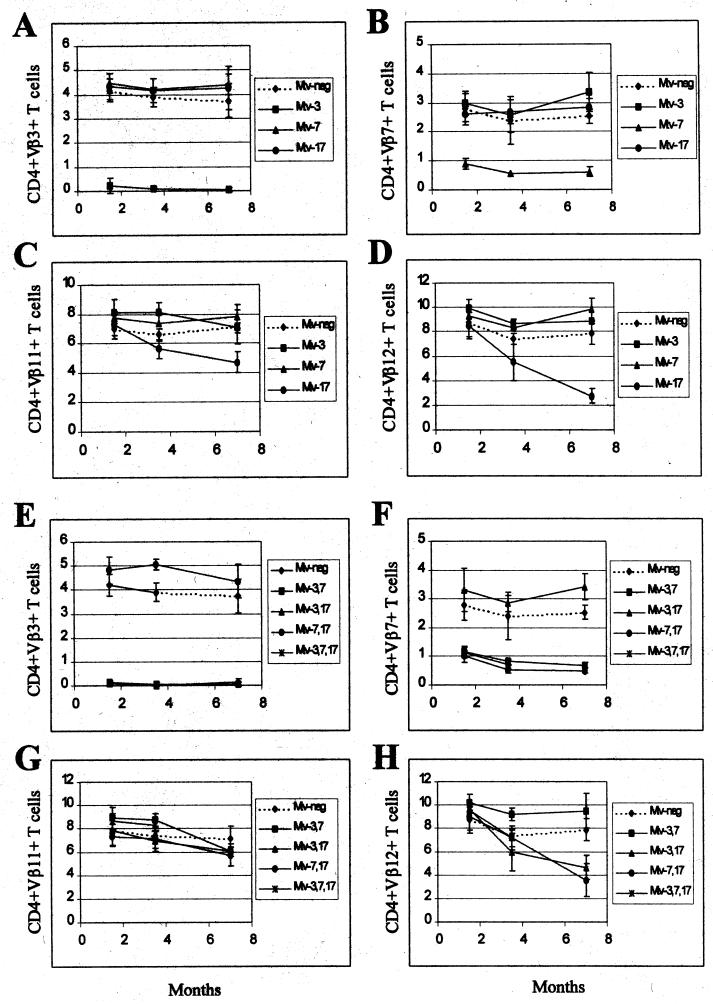
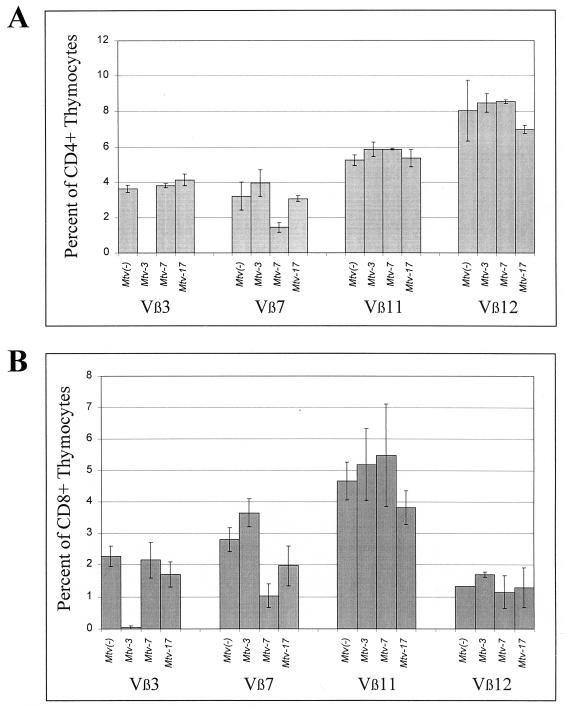
Backcross mice containing single MMTV proviruses were tested over a period of 7 months for deletion of CD4+ T cells expressing Vβ3, Vβ7, Vβ11, and Vβ12 (Fig. 3A to D). As expected, Mtv-3-only animals showed complete deletion of Vβ3-specific T cells but not T cells expressing Vβ7, -11, or -12, and this deletion was complete when the mice were 6 weeks of age. Animals containing only Mtv-7 had 68% deletion of Vβ7-specific T cells by 6 weeks after birth, whereas deletion of T cells expressing Vβ3, -11, and -12 was unaffected. Interestingly, Mtv-17-only mice showed no deletion of the T-cell subsets when tested at 6 weeks of age; however, deletion of Vβ11- and Vβ12-specific T cells was apparent after 3 to 7 months. At 7 months, deletion of Vβ11-specific cells was only 34% and deletion of Vβ12-specific cells was 65%. Scherer et al. reported 11% deletion of CD4+Vβ11+ and 35% of CD4+Vβ12+ cells in Mtv-17-only mice, but the age of the mice tested was not reported (43). These results suggested that deletion of T cells specific for Mtv-7 and Mtv-3 Sag occurred intrathymically. In contrast, Mtv-17 Sag-specific cells were deleted in the peripheral immune system.
FIG. 3.
Kinetics of T-cell deletion in N1 mice from the C58/J × PERA cross. Percentages of CD4+Vβ3+ (A and E), CD4+Vβ7+ (B and F), CD4+Vβ11+ (C and G), and CD4+Vβ12+ (D and H) T cells were determined. All values were compared to the percentage of T cells in Mtv-negative mice from the same cross. Each point represents the average of values from 3 to 12 mice, except that two animals with Mtv-7 alone were tested at 3.5 months. No N1 animals with Mtv-3, -7, and -17 were tested at 7 months. Standard deviations are represented by vertical bars spanning each point.
To confirm and extend these observations, we also tested Sag-specific deletion of CD4+ T cells in backcross animals that expressed multiple endogenous MMTVs (Fig. 3E to H). As anticipated, all mice that had Mtv-3 or Mtv-7 also had deletion of Vβ3- or Vβ7-specific T cells, respectively (Fig. 3E and F). In addition, all animals with the Mtv-17 provirus and either Mtv-3 or Mtv-7 showed slow kinetics of Vβ12-specific deletion of CD4+ cells (Fig. 3H). Because deletion of T cells bearing Vβ12 was not detectable in very young mice, these data support the idea that peripheral, but not intrathymic, deletion is mediated by Mtv-17 Sag. In contrast to results observed with mice containing single MMTV proviruses, deletion of Vβ11-specific cells was not detected in mice with Mtv-17 in combination with other endogenous MMTVs (Fig. 3G); this may be due to compensatory changes in the percentage of T-cell deletion caused by sag expression from other proviruses (43).
We also attempted to analyze deletion of Vβ12+ T cells in Mtv-17-only mice by testing whether deletion of these cells was detectable in the peripheral CD8+ subset. If deletion of Vβ12+ cells is initiated intrathymically when the majority of cells are CD4+ CD8+, then deletion of Vβ12-specific cells should be detectable in both CD4 and CD8 single-positive subsets released from the thymus (23). Although there was a small difference between the levels of CD8+Vβ12+ T cells in Mtv-17-positive mice at 4 to 6 months of age and those in mice that lack MMTV proviruses, this was not statistically significant (P = 0.12) (Table 2); this result could be due to the low number of animals tested. As expected, Mtv-7-only mice deleted Vβ7+ T cells (80% compared to Mtv-negative animals) in the CD8+ subset (Table 2). These data and results from Scherer et al. (43) are consistent with the idea that Mtv-17 causes peripheral deletion of CD4+Vβ12+ T cells.
TABLE 2.
Deletion of CD8+ T cells in mouse strains with single MMTV proviruses
| Mouse straina | % of CD8+ lymph node cellsb
|
|||
|---|---|---|---|---|
| Vβ3+ | Vβ7+ | Vβ11+ | Vβ12+ | |
| Mtv (−) | 2.5 ± 0.2 | 3.5 ± 0.3 | 7.5 ± 0.8 | 1.9 ± 0.4 |
| Mtv-7 | 3.3 ± 0.3 | 0.7 ± 0.2 | 8.7 ± 1.1 | 2.7 ± 0.9 |
| Mtv-17 | 2.4 ± 0.3 | 2.9 ± 0.4 | 7.5 ± 1.4 | 1.3 ± 0.4 |
Four animals were tested from each strain.
Average percentage ± standard deviation in animals tested at 4 to 6 months of age. Animals could not be tested at older ages because many of them developed leukemia.
Analysis of (BALB/cJ × PERA) × PERA backcross mice.
Other endogenous MMTVs, such as Mtv-8, also have been reported to cause partial deletion of specific T-cell subsets (17, 43); however, the kinetics of this deletion have not been reported. Therefore, we analyzed N1 progeny from a backcross between the BALB/c strain (containing Mtv-6, -8, and -9) and Mtv-negative PERA/Ei mice (Fig. 1). To determine the proviral content of these mice, we again used PCR assays that could discriminate among the three endogenous MMTVs of BALB/c mice. The primer pair used to detect Mtv-6 proviruses was the same as that used for Mtv-3 in the C58/J cross, whereas the virtually identical Mtv-8 and Mtv-9 proviruses were distinguished with a 5′ primer within the sag hypervariable region and a 3′ primer derived from the cellular flanking region. Using optimal conditions, PCR products of 265, 488, and 537 bp were obtained for Mtv-6, -8, and -9, respectively, with BALB/c DNA (Fig. 2, lanes 3 to 5).
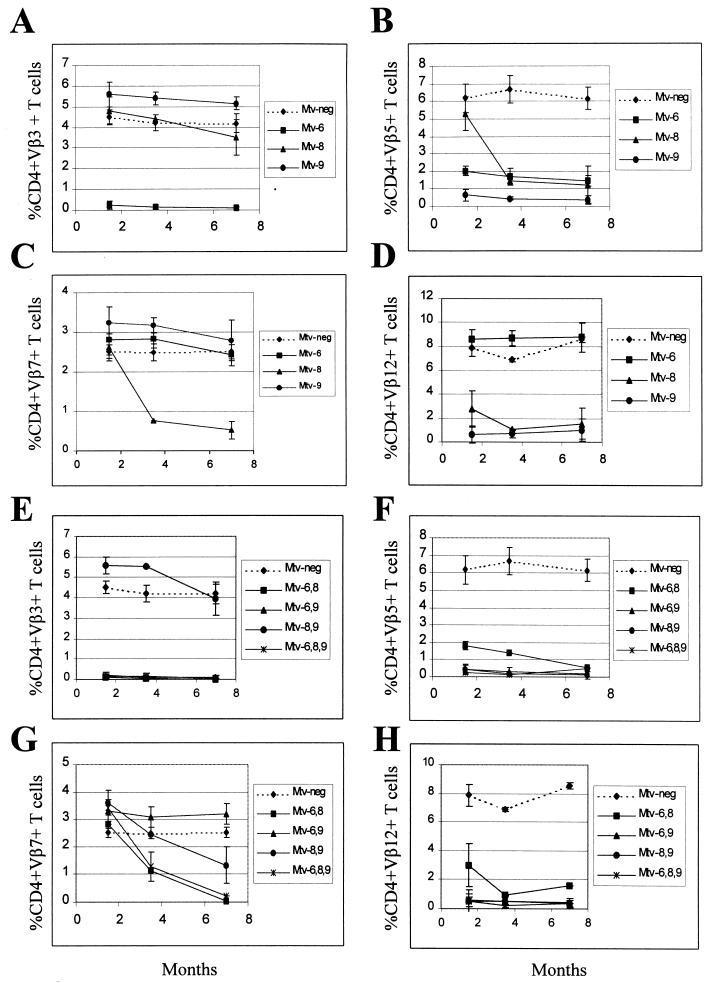
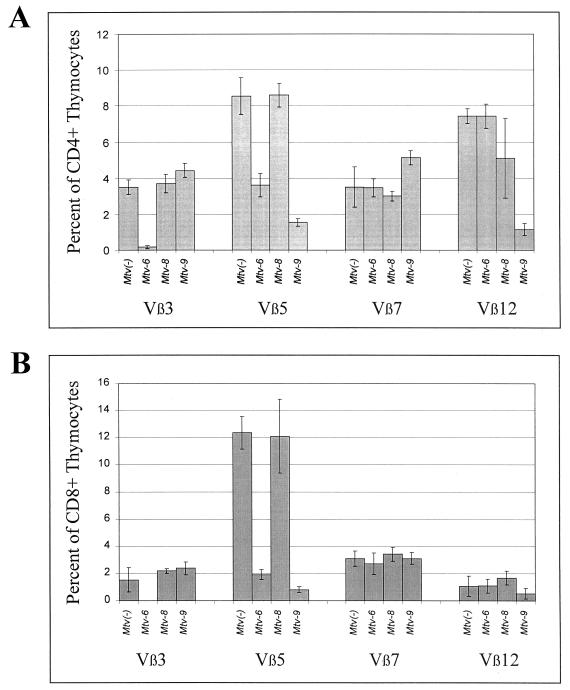
Backcross mice containing single MMTVs were examined for deletion of CD4+ T cells expressing Vβ3, -5, -7, and -12 (Fig. 4A to D). As expected from earlier experiments (48), mice containing only Mtv-6 showed deletion of Vβ3-specific and Vβ5-specific T cells, respectively, at the earliest time point tested. Mice containing only Mtv-9 deleted 90% of Vβ5-specific T cells by 6 weeks of age (Fig. 4B); these T-cell levels were lower than those achieved by the Mtv-6 or Mtv-8 proviruses (68 and 79% deletions, respectively, by 7 months). Mtv-9 also caused very efficient deletion of Vβ12+ cells in 6-week-old mice (Fig. 4D). Interestingly, Mtv-8 showed different kinetics of deletion for different T-cell subsets. Deletion of Vβ5- and Vβ7-specific T cells was not detectable in 6-week-old mice, but deletion of 79% of Vβ5+ cells and 70% of Vβ7+ cells was observed by 3.5 months. However, an average of 65% of Vβ12+ cells were deleted in 6-week-old mice with Mtv-8 only, but this was variable, and these levels declined slightly in the subsequent months (Fig. 4D). Therefore, the data indicate that Mtv-8 causes relatively rapid deletion of certain T-cell subsets (Vβ12+ cells), yet the same provirus causes much slower deletion of Vβ5+ and Vβ7+ cells in the peripheral lymphoid system.
FIG. 4.
Kinetics of T-cell deletion in N1 mice from the BALB/cJ × PERA cross. Percentages of CD4+Vβ3+ (A and E), CD4+Vβ5+ (B and F), CD4+Vβ7+ (C and G), and CD4+Vβ12+ (D and H) T cells were determined. Each point represents the average of values from 3 to 11 mice, except that two animals with Mtv-8 alone were tested at 3.5 months; two animals with both Mtv-6 and Mtv-8 were tested at 3.5 and 7 months; two animals with Mtv-6, -8, and -9 were tested at 7 months; and one animal with Mtv-8 and -9 was tested at 3.5 months. All values were compared to the percentage of T cells in Mtv-negative mice from the same cross. Standard deviations are given by vertical bars spanning each point.
BALB/c backcross mice with multiple MMTV proviruses also were analyzed (Fig. 4E to H). Again, all mice with the Mtv-6 provirus deleted CD4+Vβ3+ T cells, whereas mice with any of the three endogenous MMTVs of BALB/c mice deleted Vβ5+ T cells. Mice containing both Mtv-6 and Mtv-8 appeared to delete CD4+Vβ5+ cells with slightly slower kinetics than mice having other proviral combinations (Fig. 4F). As expected, mice with Mtv-8 in combination with other endogenous MMTVs had slow deletion of Vβ7+ T cells; however, the presence of Mtv-9, in the absence of Mtv-6, appeared to slow deletion even further (Fig. 4G). This result may be due to compensatory changes in the percentage of Vβ7+ T cells caused by deletion of other T-cell subsets by the Mtv-9 provirus (43). Rapid kinetics of Vβ12+ T-cell deletion was observed again with Mtv-8+ mice, but the deletion appeared to be more complete and less variable in the presence of the Mtv-9 provirus, which also deletes Vβ12+ T cells (Fig. 4H).
Intrathymic deletion of Sag-reactive thymocytes in mice containing single MMTV proviruses.
To test directly whether individual MMTV proviruses caused intrathymic deletion, we analyzed thymocytes from Mtv single-positive mice. In Mtv-3 single-positive mice from the C58/J cross, we observed complete deletion of Vβ3+ T cells in both the CD4 and the CD8 single-positive populations (Fig. 5). However, in Mtv-7-only mice, there was 55% deletion of CD4+Vβ7+ thymocytes and 63% deletion of CD8+Vβ7+ thymocytes. This is in agreement with the observation that there is partial deletion of CD4+ T cells in the periphery of Mtv-7 single-positive mice, and this deletion changes little with time (Fig. 3B). Mtv-17-only mice showed no significant deletion of thymocytes in either the CD4+ or CD8+ subset. Thus, the Sag proteins encoded by the Mtv-3 and Mtv-7 proviruses appear to cause intrathymic deletion of specific T-cell subsets as reported previously (16, 48), whereas Mtv-17 Sag does not (Fig. 5).
FIG. 5.
Sag-specific T-cell deletion in CD4+ and CD8+ thymocytes from mice containing Mtv-3, Mtv-7, and Mtv-17 only. Percentages of CD4+ (A) and CD8+ (B) thymocytes in mice containing single MMTV proviruses were compared to those in Mtv-negative mice derived from the same cross. Most animals tested (three to six mice from each strain) were 4 weeks old.
We also analyzed Sag-specific deletion of thymocytes in mice derived from the BALB/c backcross (Fig. 6). Mtv-6-only mice showed 95% or greater deletion of Vβ3+ thymocytes in both the CD4+ and CD8+ populations; these mice also deleted 58% of CD4+Vβ5+ and 85% of CD8+Vβ5+ thymocytes. Mtv-9-only mice deleted 82% of CD4+Vβ5+ and 94% of CD8+Vβ5+ thymocytes as well as 84 and 51% of CD4+Vβ12+ and CD8+Vβ12+ thymocytes, respectively. In Mtv-8-only mice, we observed approximately 31% deletion of Vβ12+CD4+ T cells; this partial deletion was not statistically significant (P = 0.13), largely because it was variable among different animals. The partial deletion of Vβ12+ cells was not observed in the CD8+ thymocytes. Therefore, as concluded from kinetic analysis of peripheral lymphocyte populations, it appears that Mtv-6 and Mtv-9 cause intrathymic deletion of T-cell subsets, whereas Mtv-8 causes variable and partial deletion of CD4+Vβ12+ cells intrathymically.
FIG. 6.
Sag-specific T-cell deletion in CD4+ and CD8+ thymocytes from mice containing Mtv-6, Mtv-8, and Mtv-9 only. Percentages of CD4+ (A) and CD8+ (B) thymocytes in mice containing single MMTV proviruses were compared to those in Mtv-negative mice derived from the same cross. Animals tested (three mice from each strain) were 4 to 8 weeks old.
Although we also analyzed deletion of specific T-cell subsets in CD4+ CD8+ thymocytes from Mtv single-positive mice, the percentages were too low for us to reliably assess their significance (data not shown).
Expression of Mtv-17.
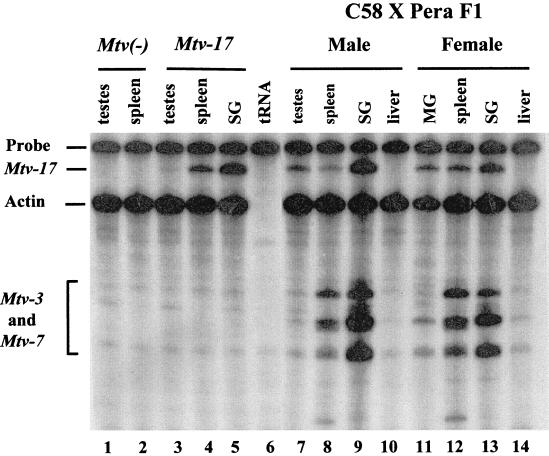
Sags from two endogenous MMTV proviruses, Mtv-8 and Mtv-17, appear to cause peripheral deletion of their cognate T cells. At least one report has suggested that the Mtv-17 provirus is expressed poorly in lymphoid tissues, particularly thymus (43). In addition, sequencing of the Mtv-17 3′ LTR indicated that there is a mutation in the binding site for the transcription factor NF-1, and this mutation may greatly reduce transcriptional activity of the provirus (34). To examine the tissue distribution of Mtv-17 expression, we used RNase protection assays in conjunction with a riboprobe spanning the sag hypervariable region and the promoter of the Mtv-17 provirus. As expected, hybridization of the Mtv-17 riboprobe to RNA extracted from the testes or spleen of an Mtv-negative backcross animal showed no specific protection of the probe from RNase digestion (Fig. 7, lanes 1 and 2). However, RNA obtained from the testes, spleen, and salivary glands of a male backcross mouse harboring only the Mtv-17 provirus protected 340 nucleotides from digestion, consistent with Mtv-17 expression in all of these tissues (lanes 3 to 5). Each of the samples showed similar expression of the control actin gene. Therefore, the Mtv-17 promoter does not appear to be generally defective for transcription.
FIG. 7.
RNase protection assays for the expression of endogenous MMTVs. Total RNA (40 μg) was hybridized to a riboprobe containing the Sau3A fragment of the Mtv-17 LTR (24), including the polymorphic region of the sag gene. An actin probe (Ambion, Austin, Tex.) was used as a control for the quality of the RNA. Hybridizations contained RNA from the following sources: tissues from an MMTV-negative backcross mouse (lanes 1 and 2), tissues from a backcross animal containing Mtv-17 only (lanes 3 to 5), yeast RNA (lane 6), tissues from a male (C58/J × PERA)F1 animal (lanes 7 to 10), and tissues from a female (C58/J × PERA)F1 animal (lanes 11 to 14). Abbreviations: SG, salivary gland; MG, mammary gland. The faster-migrating bands in these lanes are due to partial protection of the riboprobe by Mtv-3- and Mtv-7-specific RNA transcripts. The Mtv-3 and Mtv-7 transcripts cannot be distinguished in this assay.
To determine whether Mtv-17 RNA expression was affected significantly by the presence of other proviruses, we also examined RNA extracted from male and female tissues of (C58/J × PERA)F1 mice (Fig. 7). Again, Mtv-17 expression was highest in the salivary gland of a male animal (lane 9), and this expression was comparable to that observed for the Mtv-3 and/or Mtv-7 proviruses (bands migrating faster than actin). A virgin female animal showed significant expression of Mtv-17 in salivary gland, spleen, and mammary gland (lanes 11 to 13), although the expression of the Mtv-3 and -7 proviruses was diminished considerably in virgin mammary gland (lane 11). Expression of Mtv-17 RNA was enhanced in lactating compared to virgin mammary gland (data not shown), suggesting that proviral transcription is hormone inducible. Thus, Mtv-17 expression was easily detectable and similar to other endogenous MMTV proviruses in certain tissues, notably salivary gland.
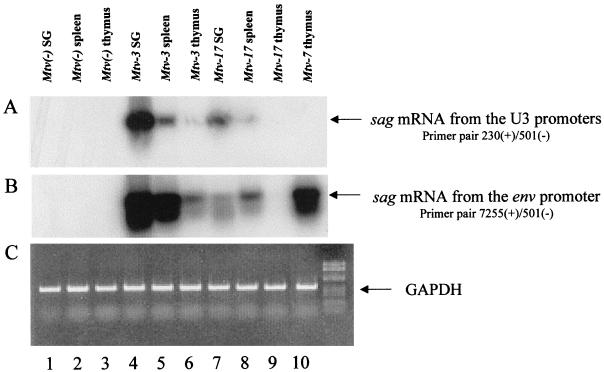
Four promoters have been described for MMTV sag gene expression (6, 18, 25, 47, 51), and at least two of these promoters are used by the Mtv-17 provirus for transcription of spliced sag mRNAs (56). Because RNase protection assays are not sufficiently sensitive to detect sag gene expression and to discriminate among different sag promoters, we used RT-PCR assays to detect spliced Mtv-17 sag mRNA in the thymus (Fig. 8). RT-PCR was performed with RNA from the salivary glands, spleens, and thymi of Mtv-negative, Mtv-3-only, or Mtv-17-only mice. To increase the sensitivity for detection of sag mRNAs, PCR products then were subjected to Southern blotting followed by hybridization with an MMTV LTR probe. Strikingly, the spliced sag mRNAs initiated from the LTR were much more abundant than those initiated from the intragenic envelope promoter originally described by Elliott et al. (18); transcripts from the LTR promoters could be detected after 20 PCR cycles (Fig. 8A), whereas the envelope promoter transcripts could not (data not shown). Expression of both Mtv-3 and -7 sag RNAs was detected from the LTR promoters as well as the envelope promoter (Fig. 8A and B; data not shown). Levels of Mtv-3 expression appeared to be highest in the salivary gland, intermediate in spleen, and lowest in the thymus for both sag mRNAs. Mtv-3 had higher levels of LTR-directed transcripts in the thymus than did the Mtv-7 provirus. As previously reported, Mtv-17 sag mRNAs were detected from both LTR and envelope promoters (56). However, by comparison to Mtv-3, Mtv-17 appeared to have lower levels of sag mRNA in salivary gland and spleen, and no expression was observed in the thymus (Fig. 8A and B, lanes 7 to 9). Additional PCR assays indicated that the Mtv-3 sag-specific RNAs from the envelope promoter were at least 100- to 500-fold more abundant in thymus than were transcripts from the Mtv-17 provirus (data not shown). Levels of sag transcripts from LTR promoters were approximately 10- to 50-fold higher in thymi from Mtv-7-only mice than in thymi from Mtv-17-only mice (data not shown).
FIG. 8.
Detection of sag-specific spliced transcripts from the LTR and envelope promoters. RT-PCRs were performed with RNA extracted from the salivary gland (SG) (lanes 1, 4, and 7), spleen (lanes 2, 5, and 8), and thymus (lanes 3, 6, 9, and 10). RNA samples were derived from Mtv-negative animals (lanes 1 to 3), Mtv-3-only animals (lanes 4 to 6), Mtv-17-only animals (lanes 7 to 9), or Mtv-7-only animals (lane 10). RNAs were derived from pools of organs from 6-week-old mice (three to five animals). Each cDNA was used in three separate PCRs containing primers for the LTR promoters (A), the envelope promoter (B), and GAPDH (C). The PCRs in panel A were performed for 20 cycles, and the PCRs in panel B were performed for 35 cycles. RT-PCR mixtures (one-third of the reaction mixture) were analyzed on 2% NuSieve agarose gels. After electrophoresis, DNA was transferred to nylon membranes, hybridized to an MMTV LTR probe (A and B), and subjected overnight to autoradiography. Longer exposures of the autoradiogram in panel A show Mtv-7 expression in the thymus. Mtv-17 expression in the thymus was not detected after 35 PCR cycles with primers for the LTR promoters or long exposures of the autoradiograms. GAPDH reaction mixtures were stained with ethidium bromide after electrophoresis as a control for the integrity of cDNAs.
DISCUSSION
We have investigated the kinetics of T-cell deletion resulting from the expression of Sags from six different endogenous MMTV proviruses, Mtv-3, -6, -7, -8, -9, and -17, in mice. Previous data have described the T-cell subsets deleted due to Sag expression from each of these proviruses (3, 43, 44). However, the kinetics of the deletion induced has not been addressed for many MMTV proviruses.
Expression from four of these proviruses, Mtv-3, -6, -7, and -9, gave 50% or more deletion of the reactive T-cell subsets tested, and this deletion was apparent intrathymically and in peripheral T cells of animals that were 6 weeks old. This is consistent with previous data that show intrathymic deletion of cognate T cells for the Mtv-6 and Mtv-7 proviruses (40, 48). In contrast, deletion of reactive T cells by the Sag proteins encoded by the Mtv-8 and Mtv-17 proviruses largely was undetectable in thymocytes or in peripheral T cells by 6 weeks but could be detected in peripheral T cells over a period of 7 months. Our data confirmed previous experiments indicating that Sags from Mtv-3, -6, -7, and -9 caused T-cell deletion during negative selection in the thymus of MHC class II I-E+ mice (19, 21, 22, 27, 44, 52).
Deletion of T cells reactive with Mtv-17 Sag.
Expression of Sags from Mtv-17 resulted in slow kinetics of T-cell deletion that was detectable in the peripheral lymphoid system. Deletion of thymocytes reactive with Mtv-17 Sag was not detected. In agreement with this and data published by Scherer et al. (43), deletion of Vβ12+ T cells due to Mtv-17 Sag was detected only in peripheral CD4+ T cells, not in the CD8+ subset. Thus, presentation of Mtv-17 Sag by MHC class II on antigen-presenting cells in the periphery will stimulate and delete mature CD4 single-positive cells.
What are the factors that influence the kinetics of Sag-mediated T-cell deletion? One of the key factors influencing early deletion of T-cell subsets is the ability of MMTV proviruses to be transcribed in the thymus. RT-PCR analysis showed that Mtv-17-only mice had no detectable expression in the thymus as previously reported (55), whereas Mtv-3 and Mtv-7 both produced detectable expression of spliced sag mRNA from the LTR and envelope promoters (Fig. 8 and data not shown). The results of Scherer et al. suggested that the Mtv-17 provirus was expressed at low levels in the thymus; however, their assay did not distinguish between total (spliced and unspliced) MMTV RNA and sag-specific transcripts (43). Greater sensitivity for detection of sag mRNAs in our assays was achieved by Southern blotting of RT-PCR products, but the blotting did not allow detection of Mtv-17 sag transcripts in the thymus. Thus, it appears likely that negative selection of Mtv-17 Sag-reactive T cells cannot take place in the thymus because the appropriate transcripts or a threshold level of these transcripts is not synthesized (40). In support of this, Morishima et al. showed that low expression of total Mtv-1 RNA in the thymus led to peripheral deletion of Vβ3+ T cells, whereas eightfold-higher expression of the related Mtv-6 provirus in the thymus gave intrathymic deletion of Vβ3+ T cells (40). Waanders et al. also suggested that the levels of sag mRNA in the thymus correlated with intrathymic deletion of Sag-reactive T cells (48).
Deletion of T cells reactive with Mtv-8 Sag.
CD4+Vβ12+ T cells in Mtv-8-only mice showed partial and variable deletion intrathymically (Fig. 6A). Deletion of CD8+Vβ12+ T cells was not detected in thymocytes (Fig. 6B). Preliminary results from a 7-month-old mouse with the Mtv-8 provirus indicate that deletion of Vβ12+ cells, but not of other T-cell subsets, is detectable in peripheral CD8 single-positive cells (data not shown) as well as in the CD4+ subset (Fig. 4D). These data are consistent with a previous report that Mtv-8 expression deleted Vβ12+ T cells in the CD4+ and CD8+ subsets (43). Although the ages of mice tested were not reported, the Mtv-8-only mice deleted Vβ5+, Vβ7+, and Vβ11+ T cells only in the CD4+ subset (43).
The results of Mtv-8 expression on T-cell deletion are more complex than those observed for Mtv-17. We have not been able to test this directly because of breeding problems with the Mtv-8-only mice. However, we expect that Mtv-8 will be expressed in the thymus because this provirus caused deletion of peripheral CD4+Vβ12+ T cells at the earliest time point tested and variable deletion of CD4+Vβ12+ thymocytes. However, other T-cell subsets (Vβ5+ and Vβ7+ cells) were not deleted in the thymus. Such cells were deleted with slow kinetics in the periphery, consistent with extrathymic deletion of reactive T cells. Therefore, it is likely that Mtv-8 is expressed at very low levels in the thymus and that this level is close to the threshold required for elimination of cells expressing self-reactive antigens. The Mtv-8 provirus is located in the Vκ locus on mouse chromosome 6 (57) and appears to be expressed in B cells following certain light chain rearrangements that allow close proximity of the Mtv-8 provirus and the Vκ enhancers (58). Thus, expression of Mtv-8 in certain mature B cells likely leads to the peripheral deletion observed for the Vβ5+ and Vβ7+ T-cell subsets. In contrast, reaction of a limited amount of Mtv-8 Sag on the surface of thymic antigen-presenting cells may allow negative selection of thymocytes if there is high affinity of this Sag for a given TCR (e.g., Vβ12). Similarly, experiments by Chervonsky et al. indicated that expression of sag-specific mRNA in the thymus was sufficient to get peripheral, but not intrathymic, deletion of Vβ14+ T cells in mice expressing low amounts of the C3H MMTV transgene (9). Therefore, complete intrathymic deletion of self-reactive T cells will be determined both by the level of Sag expression and by the affinity for TCR (19). It will be interesting to determine the level of Mtv-8 sag-specific expression in the thymus since it may help to define the minimum level of expression needed for intrathymic deletion. Based on results with Mtv-3 sag mRNA with a combination of RT-PCR and Southern blotting (Fig. 8) and complete intrathymic deletion of cells reactive with Mtv-3 Sag, negative selection of thymocytes appears to be an exquisitely sensitive process.
Both Mtv-8 and Mtv-17 cause peripheral deletion of T cells. What is the antigen-presenting cell used for peripheral elimination of Sag-reactive T cells? Although we have presented no definitive data here, previous experiments indicate that B cells express Mtv-8 (30, 58). Moreover, our RT-PCR experiments suggest that expression of Mtv-17 sag mRNA is detectable in spleen, a tissue rich in mature B cells (42), but not thymus. Previous experiments also have detected Mtv-17 transcripts in gut-associated lymphocytes (55). Since MMTV expression in B cells is a requirement for exogenous MMTV transmission (7) and B cells are expanded during exposure of adult mice to Mtv-7-expressing spleen cells (49), B cells are likely candidates for presentation of Mtv-8 and -17 Sag in the peripheral immune system. However, CD8+ T cells also may cause Sag-specific T-cell deletion in the periphery (49). As expected, B cells are not required for deletion of cognate T cells by endogenous Mtv-7 and Mtv-9, two proviruses which cause intrathymic T-cell deletion (7).
Selection for endogenous MMTVs with Sag function.
Previous experiments have shown that expression of milk-borne MMTV Sag in transgenic animals leads to the intrathymic deletion of cognate T cells. Such transgenic animals are resistant to infection by milk-borne MMTVs that express the same Sag proteins (23). Thus, it has been proposed that endogenous MMTVs that express Sag have been retained by most mouse strains because these MMTVs protect against milk-borne virus infections (23, 28, 36). The existence of endogenous MMTVs that largely cause slow kinetics of T-cell deletion appears to contradict this proposal. Our data would suggest that mice that have Mtv-8 and/or Mtv-17 would be susceptible to exogenous MMTVs with similar Sag proteins since T cells reactive with Mtv-8 and -17 Sags would be present during the neonatal period when milk-borne infection occurs. Moreover, these mice would be susceptible to tumorigenesis by certain recombinants between different endogenous MMTVs (e.g., Mtv-17 recombinants with Mtv-2 are integrated in GR tumors) (24). Alternatively, endogenous viruses such as Mtv-8 and -17 may provide limited protection against milk-borne MMTV infection by anergizing Sag-reactive T cells. However, these proviruses may provide better resistance to bacterial or other viral infections that may occur later in life and that require a response from particular T-cell subsets (29, 37).
ACKNOWLEDGMENTS
We thank Susan Ross for useful discussions and for comments on the manuscript.
This work was supported by grants CA34780 and CA52646 from the National Institutes of Health. F.M. is a recipient of an NRSA award from the National Institutes of Health.
REFERENCES
- 1.Abe R, Foo-Phillips M, Granger L G, Kanagawa O. Characterization of the Mlsf system. I. A novel “polymorphism” of endogenous superantigens. J Immunol. 1992;149:3429–3439. [PubMed] [Google Scholar]
- 2.Abe R, Kanagawa O, Sheard M A, Malissen B, Foo-Phillips M. Characterization of a new minor lymphocyte stimulatory system. I. Cluster of self antigens recognized by “I-E-reactive” Vβs, Vβ5, Vβ11, and Vβ12 T cell receptors for antigen. J Immunol. 1991;147:739–749. [PubMed] [Google Scholar]
- 3.Acha-Orbea H, Held W, Waanders G A, Shakhov A N, Scarpellino L, Lees R K, MacDonald H R. Exogenous and endogenous mouse mammary tumor virus superantigens. Immunol Rev. 1993;131:5–25. doi: 10.1111/j.1600-065x.1993.tb01527.x. [DOI] [PubMed] [Google Scholar]
- 4.Acha-Orbea H, MacDonald H R. Superantigens of mouse mammary tumor virus. Annu Rev Immunol. 1995;13:459–486. doi: 10.1146/annurev.iy.13.040195.002331. [DOI] [PubMed] [Google Scholar]
- 5.Acha-Orbea H, Shakhov A N, Scarpellino L, Kolb E, Muller V, Vessaz-Shaw A, Fuchs R, Blochlinger K, Rollini P, Billotte J, et al. Clonal deletion of Vβ14-bearing T cells in mice transgenic for mammary tumour virus. Nature. 1991;350:207–211. doi: 10.1038/350207a0. [DOI] [PubMed] [Google Scholar]
- 6.Arroyo J, Winchester E, McLellan B S, Huber B T. Shared promoter elements between a viral superantigen and the major histocompatibility complex class II-associated invariant chain. J Virol. 1997;71:1237–1245. doi: 10.1128/jvi.71.2.1237-1245.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beutner U, Kraus E, Kitamura D, Rajewsky K, Huber B T. B cells are essential for murine mammary tumor virus transmission, but not for presentation of endogenous superantigens. J Exp Med. 1994;179:1457–1466. doi: 10.1084/jem.179.5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandt-Carlson C, Butel J S, Wheeler D. Phylogenetic and structural analyses of MMTV LTR ORF sequences of exogenous and endogenous origins. Virology. 1993;193:171–185. doi: 10.1006/viro.1993.1113. [DOI] [PubMed] [Google Scholar]
- 9.Chervonsky A V, Golovkina T V, Ross S R, Janeway C A., Jr Differences in the avidity of TCR interactions with a superantigenic ligand affect negative selection but do not allow positive selection. J Immunol. 1995;155:5115–5123. [PubMed] [Google Scholar]
- 10.Choi Y, Kappler J W, Marrack P. A superantigen encoded in the open reading frame of the 3′ long terminal repeat of mouse mammary tumour virus. Nature. 1991;350:203–207. doi: 10.1038/350203a0. [DOI] [PubMed] [Google Scholar]
- 11.Choi Y W, Henrard D, Lee I, Ross S R. The mouse mammary tumor virus long terminal repeat directs expression in epithelial and lymphoid cells of different tissues in transgenic mice. J Virol. 1987;61:3013–3019. doi: 10.1128/jvi.61.10.3013-3019.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen J C, Varmus H E. Endogenous mammary tumour virus DNA varies among wild mice and segregates during inbreeding. Nature. 1979;278:418–423. doi: 10.1038/278418a0. [DOI] [PubMed] [Google Scholar]
- 13.Dudley J, Risser R. Amplification and novel locations of endogenous mouse mammary tumor virus genomes in mouse T-cell lymphomas. J Virol. 1984;49:92–101. doi: 10.1128/jvi.49.1.92-101.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dudley J P. Mouse mammary tumor proviruses from a T-cell lymphoma are associated with the retroposon L1Md. J Virol. 1988;62:472–478. doi: 10.1128/jvi.62.2.472-478.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dudley J P, Arfsten A, Hsu C L, Kozak C, Risser R. Molecular cloning and characterization of mouse mammary tumor proviruses from a T-cell lymphoma. J Virol. 1986;57:385–388. doi: 10.1128/jvi.57.1.385-388.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dyson P J, Elliott J, Antoniou A N, Corley K T. Efficient presentation of endogenous superantigen by H-2Aq. Eur J Immunol. 1998;28:1034–1039. doi: 10.1002/(SICI)1521-4141(199803)28:03<1034::AID-IMMU1034>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 17.Dyson P J, Knight A M, Fairchild S, Simpson E, Tomonari K. Genes encoding ligands for deletion of Vβ11 T cells cosegregate with mammary tumour virus genomes. Nature. 1991;349:531–532. doi: 10.1038/349531a0. [DOI] [PubMed] [Google Scholar]
- 18.Elliott J F, Pohajdak B, Talbot D J, Shaw J, Paetkau V. Phorbol diester-inducible, cyclosporine-suppressible transcription from a novel promoter within the mouse mammary tumor virus env gene. J Virol. 1988;62:1373–1380. doi: 10.1128/jvi.62.4.1373-1380.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fink P J, Fang C A, Turk G L. The induction of peripheral tolerance by the chronic activation and deletion of CD4+Vβ5+ cells. J Immunol. 1994;152:4270–4281. [PubMed] [Google Scholar]
- 20.Foo-Phillips M, Kozak C A, Principato M A, Abe R. Characterization of the Mlsf system. II. Identification of mouse mammary tumor virus proviruses involved in the clonal deletion of self-Mlsf-reactive T cells. J Immunol. 1992;149:3440–3447. [PubMed] [Google Scholar]
- 21.Frankel W N, Rudy C, Coffin J M, Huber B T. Linkage of Mls genes to endogenous mammary tumour viruses of inbred mice. Nature. 1991;349:526–528. doi: 10.1038/349526a0. [DOI] [PubMed] [Google Scholar]
- 22.Gollob K J, Palmer E. Divergent viral superantigens delete Vβ5+ T lymphocytes. Proc Natl Acad Sci USA. 1992;89:5138–5141. doi: 10.1073/pnas.89.11.5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golovkina T V, Chervonsky A, Dudley J P, Ross S R. Transgenic mouse mammary tumor virus superantigen expression prevents viral infection. Cell. 1992;69:637–645. doi: 10.1016/0092-8674(92)90227-4. [DOI] [PubMed] [Google Scholar]
- 24.Golovkina T V, Prakash O, Ross S R. Endogenous mouse mammary tumor virus Mtv-17 is involved in Mtv-2-induced tumorigenesis in GR mice. Virology. 1996;218:14–22. doi: 10.1006/viro.1996.0161. [DOI] [PubMed] [Google Scholar]
- 25.Gunzburg W H, Heinemann F, Wintersperger S, Miethke T, Wagner H, Erfle V, Salmons B. Endogenous superantigen expression controlled by a novel promoter in the MMTV long terminal repeat. Nature. 1993;364:154–158. doi: 10.1038/364154a0. [DOI] [PubMed] [Google Scholar]
- 26.Held W, Shakhov A N, Izui S, Waanders G A, Scarpellino L, MacDonald H R, Acha-Orbea H. Superantigen-reactive CD4+ T cells are required to stimulate B cells after infection with mouse mammary tumor virus. J Exp Med. 1993;177:359–366. doi: 10.1084/jem.177.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Held W, Shakhov A N, Waanders G, Scarpellino L, Luethy R, Kraehenbuhl J P, MacDonald H R, Acha-Orbea H. An exogenous mouse mammary tumor virus with properties of Mls-1a (Mtv-7) J Exp Med. 1992;175:1623–1633. doi: 10.1084/jem.175.6.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Held W, Waanders G A, Shakhov A N, Scarpellino L, Acha-Orbea H, MacDonald H R. Superantigen-induced immune stimulation amplifies mouse mammary tumor virus infection and allows virus transmission. Cell. 1993;74:529–540. doi: 10.1016/0092-8674(93)80054-i. [DOI] [PubMed] [Google Scholar]
- 29.Huber B T, Hsu P N, Sutkowski N. Virus-encoded superantigens. Microbiol Rev. 1996;60:473–482. doi: 10.1128/mr.60.3.473-482.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jarvis C D, Germain R N, Hager G L, Damschroder M, Matis L A. Tissue-specific expression of messenger RNAs encoding endogenous viral superantigens. J Immunol. 1994;152:1032–1038. [PubMed] [Google Scholar]
- 31.Kang J, Ido E, Pawling J, Beutner U, Huber B T, Hozumi N. Expression of Mtv-7 sag gene in vivo using a retroviral vector results in selective inactivation of superantigen reactive T cells. J Immunol. 1994;152:1039–1046. [PubMed] [Google Scholar]
- 32.Karapetian O, Shakhov A N, Kraehenbuhl J P, Acha-Orbea H. Retroviral infection of neonatal Peyer’s patch lymphocytes: the mouse mammary tumor virus model. J Exp Med. 1994;180:1511–1516. doi: 10.1084/jem.180.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korman A J, Bourgarel P, Meo T, Rieckhof G E. The mouse mammary tumour virus long terminal repeat encodes a type II transmembrane glycoprotein. EMBO J. 1992;11:1901–1905. doi: 10.1002/j.1460-2075.1992.tb05242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuo W L, Vilander L R, Huang M, Peterson D O. A transcriptionally defective long terminal repeat within an endogenous copy of mouse mammary tumor virus proviral DNA. J Virol. 1988;62:2394–2402. doi: 10.1128/jvi.62.7.2394-2402.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lund F E, Randall T D, Woodland D L, Corley R B. MHC class II limits the functional expression of endogenous superantigens in B cells. J Immunol. 1993;150:78–86. [PubMed] [Google Scholar]
- 36.Marrack P, Kushnir E, Kappler J. A maternally inherited superantigen encoded by a mammary tumour virus. Nature. 1991;349:524–526. doi: 10.1038/349524a0. [DOI] [PubMed] [Google Scholar]
- 37.Marrack P, Winslow G M, Choi Y, Scherer M, Pullen A, White J, Kappler J W. The bacterial and mouse mammary tumor virus superantigens; two different families of proteins with the same functions. Immunol Rev. 1993;131:79–92. doi: 10.1111/j.1600-065x.1993.tb01531.x. [DOI] [PubMed] [Google Scholar]
- 38.McMahon C W, Traxler B, Grigg M E, Pullen A M. Transposon-mediated random insertions and site-directed mutagenesis prevent the trafficking of a mouse mammary tumor virus superantigen. Virology. 1998;243:354–365. doi: 10.1006/viro.1998.9071. [DOI] [PubMed] [Google Scholar]
- 39.Moore N C, Anderson G, McLoughlin D E, Owen J J, Jenkinson E J. Differential expression of Mtv loci in MHC class II-positive thymic stromal cells. J Immunol. 1994;152:4826–4831. [PubMed] [Google Scholar]
- 40.Morishima C, Norby-Slycord C, McConnell K R, Finch R J, Nelson A J, Farr A G, Pullen A M. Expression of two structurally identical viral superantigens results in thymic elimination at distinct developmental stages. J Immunol. 1994;153:5091–5103. [PubMed] [Google Scholar]
- 41.Palmiter R D. Magnesium precipitation of ribonucleoprotein complexes. Expedient techniques for the isolation of undergraded polysomes and messenger ribonucleic acid. Biochemistry. 1974;13:3606–3615. doi: 10.1021/bi00714a032. [DOI] [PubMed] [Google Scholar]
- 42.Picker L J, Siegelman M H. Lymphoid tissues and organs. In: Paul W E, editor. Fundamental immunology. New York, N.Y: Raven Press; 1993. pp. 145–197. [Google Scholar]
- 43.Scherer M T, Ignatowicz L, Pullen A, Kappler J, Marrack P. The use of mammary tumor virus (Mtv)-negative and single-Mtv mice to evaluate the effects of endogenous viral superantigens on the T cell repertoire. J Exp Med. 1995;182:1493–1504. doi: 10.1084/jem.182.5.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simpson E, Dyson P J, Knight A M, Robinson P J, Elliott J I, Altmann D M. T-cell receptor repertoire selection by mouse mammary tumor viruses and MHC molecules. Immunol Rev. 1993;131:93–115. doi: 10.1111/j.1600-065x.1993.tb01532.x. [DOI] [PubMed] [Google Scholar]
- 45.Subramanyam M, Mohan N, Mottershead D, Beutner U, McLellan B, Kraus E, Huber B T. Mls-1 superantigen: molecular characterization and functional analysis. Immunol Rev. 1993;131:117–130. doi: 10.1111/j.1600-065x.1993.tb01533.x. [DOI] [PubMed] [Google Scholar]
- 46.Vacchio M S, Ryan J J, Hodes R J. Characterization of the ligand(s) responsible for negative selection of Vβ11- and Vβ12-expressing T cells: effects of a new Mls determinant. J Exp Med. 1990;172:807–813. doi: 10.1084/jem.172.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Ooyen A J, Michalides R J, Nusse R. Structural analysis of a 1.7-kilobase mouse mammary tumor virus-specific RNA. J Virol. 1983;46:362–370. doi: 10.1128/jvi.46.2.362-370.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waanders G A, Lees R K, Held W, MacDonald H R. Quantitation of endogenous mouse mammary tumor virus superantigen expression by lymphocyte subsets. Eur J Immunol. 1995;25:2632–2637. doi: 10.1002/eji.1830250934. [DOI] [PubMed] [Google Scholar]
- 49.Waanders G A, Shakhov A N, Held W, Karapetian O, Acha-Orbea H, MacDonald H R. Peripheral T cell activation and deletion induced by transfer of lymphocyte subsets expressing endogenous or exogenous mouse mammary tumor virus. J Exp Med. 1993;177:1359–1366. doi: 10.1084/jem.177.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weber G F, Abromson-Leeman S, Cantor H. A signaling pathway coupled to T cell receptor ligation by MMTV superantigen leading to transient activation and programmed cell death. Immunity. 1995;2:363–372. doi: 10.1016/1074-7613(95)90144-2. [DOI] [PubMed] [Google Scholar]
- 51.Wheeler D A, Butel J S, Medina D, Cardiff R D, Hager G L. Transcription of mouse mammary tumor virus: identification of a candidate mRNA for the long terminal repeat gene product. J Virol. 1983;46:42–49. doi: 10.1128/jvi.46.1.42-49.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woodland D L, Happ M P, Gollob K J, Palmer E. An endogenous retrovirus mediating deletion of αβ T cells? Nature. 1991;349:529–530. doi: 10.1038/349529a0. [DOI] [PubMed] [Google Scholar]
- 53.Wrona T, Dudley J P. Major histocompatibility complex class II I-E-independent transmission of C3H mouse mammary tumor virus. J Virol. 1996;70:1246–1249. doi: 10.1128/jvi.70.2.1246-1249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wrona T J, Lozano M, Binhazim A A, Dudley J P. Mutational and functional analysis of the C-terminal region of the C3H mouse mammary tumor virus superantigen. J Virol. 1998;72:4746–4755. doi: 10.1128/jvi.72.6.4746-4755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu L, Wrona T J, Dudley J P. Exogenous mouse mammary tumor virus (MMTV) infection induces endogenous MMTV sag expression. Virology. 1996;215:113–123. doi: 10.1006/viro.1996.0014. [DOI] [PubMed] [Google Scholar]
- 56.Xu L, Wrona T J, Dudley J P. Strain-specific expression of spliced MMTV RNAs containing the superantigen gene. Virology. 1997;236:54–65. doi: 10.1006/viro.1997.8717. [DOI] [PubMed] [Google Scholar]
- 57.Yang J N, Boyd R T, Gottlieb P D, Dudley J P. The endogenous retrovirus Mtv-8 on mouse chromosome 6 maps near several kappa light chain markers. Immunogenetics. 1987;25:222–227. doi: 10.1007/BF00404691. [DOI] [PubMed] [Google Scholar]
- 58.Yang J N, Dudley J. Endogenous Mtv-8 or a closely linked sequence stimulates rearrangement of the downstream Vκ9 gene. J Immunol. 1992;149:1242–1251. [PubMed] [Google Scholar]
- 58a.Yang, J.-N., and J. Dudley. Unpublished data.
- 59.Yazdanbakhsh K, Park C G, Winslow G M, Choi Y. Direct evidence for the role of COOH terminus of mouse mammary tumor virus superantigen in determining T cell receptor Vβ specificity. J Exp Med. 1993;178:737–741. doi: 10.1084/jem.178.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]