Abstract
Human immunodeficiency virus type 1 (HIV-1) infection is highly compartmentalized, with distinct viral genotypes being found in the lungs, brain, and other organs compared with blood. CCR5 and CXCR4 are the principal HIV-1 coreceptors, and a number of other molecules support entry in vitro but their roles in vivo are uncertain. To address the relationship between tissue compartmentalization and the selective use of entry coreceptors, we generated functional env clones from primary isolates derived from the lungs and blood of three infected individuals and analyzed their use of the principal, secondary, orphan, and virus-encoded coreceptors for fusion. All Env proteins from lung viruses used CCR5 but not CXCR4, while those from blood viruses used CCR5 or CXCR4 or both. The orphan receptor APJ was widely used for fusion by Env proteins from both blood and lung viruses, but none used the cytomegalovirus-encoded receptor US28. Fusion mediated by the secondary coreceptors CCR2b, CCR3, CCR8, and CX3CR1 and orphan receptors GPR1, GPR15, and STRL33 was variable and heterogeneous, with relatively broad utilization by env clones from isolates of one subject but limited use by env clones from the other two subjects. However, there was no clear distinction between blood and lung viruses in secondary or orphan coreceptor fusion patterns. In contrast to fusion, none of the secondary or orphan receptors enabled efficient productive infection. These results confirm, at the level of cofactor utilization, previous observations that HIV-1 populations in the lungs and blood are biologically distinct and demonstrate diversity within lung-derived as well as blood-derived quasispecies. However, the heterogeneity in coreceptor utilization among clones from each isolate and the lack of clear distinction between lung- and blood-derived Env proteins argue against selective coreceptor utilization as a major determinant of compartmentalization.
Initial events in the entry of human immunodeficiency virus type 1 (HIV-1) require interactions between the viral envelope glycoprotein (Env) and two cellular receptors, CD4 and a chemokine receptor (reviewed in references 6 and 39). The chemokine receptors that function as coreceptors for HIV-1 entry are members of the family of seven-transmembrane (7TM) G-protein-coupled receptors, important for the recruitment of leukocytes to sites of inflammation. While CD4 is the primary high-affinity receptor for all HIV-1 Env glycoproteins, the fusogenic activity of each chemokine receptor is limited to a subset of Env variants. The differential utilization of chemokine receptors by HIV-1 variants explains in large part the molecular basis of HIV-1 target cell tropism. Macrophage (M)-tropic strains infect primary macrophages and lymphocytes but not transformed cell lines, do not induce syncytia in infected lymphoid cells (non-syncytium inducing [NSI]), and use mainly CCR5 (R5 variants [7]). T-cell-line (T)-tropic strains infect lymphocytes and CD4-positive cell lines but not primary macrophages, form syncytia in infected targets (syncytium inducing [SI]), and use mainly CXCR4 (X4 variants). Dualtropic variants infect all three cell types and can use either coreceptor (R5X4 variants). In addition to the principal coreceptors CCR5 and CXCR4, other chemokine receptors including CCR3, CCR2b, CCR8, and CX3CR1 can support fusion and entry by more restricted groups of strains (11, 23, 46) and a growing number of orphan 7TM receptors including STRL33, GPR1, GPR15, and APJ mediate entry by various HIV-1, HIV-2, or simian immunodeficiency virus (SIV) isolates (10, 21, 25, 26, 36, 46). US28, a chemokine receptor encoded by cytomegalovirus (CMV), also supports fusion by several strains (42), which may be particularly relevant because HIV-infected people are frequently coinfected with CMV (38, 43, 48).
The fusion cofactor selectivity of HIV-1 variants is important in pathogenesis, at least for CCR5 and CXCR4. In vivo, new infections are generally associated with M-tropic NSI R5 variants that depend on CCR5 for entry, and individuals who are homozygous for a defective allele of CCR5 are largely resistant to HIV-1 infection (37, 50). During the course of HIV-1 infection, SI T-tropic X4 or dually tropic R5X4 variants that use CXCR4 instead of or in addition to CCR5 frequently emerge, and their appearance is associated with accelerated progression to AIDS (19, 52). These variants often also show broadened use of other coreceptors in addition to CXCR4 (19), but the contribution of these other molecules to pathogenesis remains to be elucidated.
While lymphoid tissues are the major sites of viral replication and the source of infected cells and virions in the circulation, HIV-1 also infects other organs, where it contributes to organ-specific dysfunction. Microglia are productively infected in the brain, and both CCR3 and CCR5 may mediate microglial infection, suggesting a role for CCR3 in the pathogenesis of AIDS dementia (31). In the lungs, HIV-1 infects mainly macrophages, and lung infection may play a role in local immune system dysfunction and inflammatory processes and as a reservoir for viral persistence and dissemination to T cells (1, 9, 12, 55). Consistent with the cellular reservoir in the lungs, HIV-1 isolates from the lungs are typically M-tropic and NSI (29, 52). Sequence analysis has shown that viral species in the lungs are genetically distinct from those in the blood (3, 34), which suggests the possibility of HIV-1 variants specifically adapted to lung cells either by independent evolution within the pulmonary compartment or by selective recruitment from circulating virions or infected cells. Since multiple coreceptors can facilitate HIV-1 entry in vitro, selective utilization of specific coreceptors in vivo may be involved in viral selection and tissue compartmentalization. In this study we analyzed the use of the principal coreceptors, secondary chemokine receptor coreceptors, and orphan and virus-encoded coreceptors for fusion by molecularly cloned HIV-1 Env variants derived from the lungs and blood.
MATERIALS AND METHODS
Lung and blood cell isolation.
HIV-1 isolation was carried out on lung and blood cells obtained from sequential HIV-1-seropositive patients undergoing diagnostic bronchoscopy for suspected lung disease. To obtain bronchoalveolar lavage (BAL) fluid from the lungs, 100 to 200 ml of saline was instilled through a fiberoptic bronchoscope wedged into a subsegmental bronchus, followed by gentle aspiration. Approximately 35 ml of the aspirated BAL fluid was filtered through sterile gauze to remove mucus and debris. The cells were then pelleted, washed twice in Hanks buffered salt solution, and plated in 24-well plates at 5 × 105 cells per well in macrophage medium (Dulbecco’s minimal essential medium containing 10% fetal bovine serum, 1% penicillin-streptomycin, 1 mM l-glutamine, 50 U of macrophage colony-stimulating factor [Genetics Institute, Cambridge, Mass.], and 1% nonessential amino acids). After overnight incubation, the nonadherent cells were removed and the adherent macrophage cultures were washed vigorously and then incubated with fresh macrophage medium. To obtain macrophage-depleted lung lymphocytes, the nonadherent cells were subjected to another round of overnight adherence, and if sufficient numbers of nonadherent cells remained, they were plated at 5 × 105 cells per well in round-bottom 96-well plates in 200 μl of lymphocyte medium (RPMI containing 20% fetal bovine serum 1% penicillin-streptomycin, 1 mM l-glutamine, and 1% nonessential amino acids). Any bronchoscopy specimen that was visibly contaminated with blood was not used. Peripheral blood lymphocytes (PBL) from the same subject undergoing bronchoscopy were obtained from peripheral blood mononuclear cells (PBMC) that were serially depleted of macrophages as previously described (15) and were suspended in lymphocyte medium at 2 × 106 per well in 24 well plates.
Virus isolation.
Virus was isolated by using PBMC obtained from heparinized blood of healthy donors identified as homozygous for the wild-type CCR5 allele as previously described (44), suspended at 2 × 106 cells/ml in PBL medium, and stimulated with phytohemagglutinin (PHA; 5 μg/ml) for 3 to 5 days. For virus isolation from lung macrophages and blood lymphocytes, 1.5 × 106 donor PBMC were used per well for coculture in 24-well plates or 5 × 105 PBMC were added to lung lymphocyte cultures in 96-well plates. The cultures were then supplemented with interleukin-2 (IL-2; 20 U/ml [Boehringer Mannheim Biochemicals, Indianapolis, Ind.]) sheep anti-alpha-interferon neutralizing antibody (20 U/ml [ICN, Costa Mesa, Calif.]), and Polybrene (2 μg/ml). Twice weekly, ∼50% of the supernatant was removed from the lung macrophage and blood lymphocyte cultures and replaced with fresh medium. Lung lymphocyte cultures were expanded twice weekly with fresh medium until ∼1 ml was reached, after which they were split as above. Supernatant was tested twice weekly for viral p24gag antigen by enzyme-linked immunosorbent assay (ELISA; Dupont, Wilmington, Del.), and when the p24 level reached ≥1 ng/ml, the cultures were expanded with fresh PHA- and IL-2-stimulated donor PBMC. When the p24 antigen reached ≥25 ng/ml, the supernatant was filtered (0.45-μm-pore-size filters) and stored (−80°C) for use as virus stocks, and the cellular DNA was used for PCR amplification of viral env genes.
Full-length functional env gene cloning.
env genes were cloned from cellular DNA of PBMC in which viruses were isolated. Cells were lysed (100 mM KCl, 20 mM Tris, 0.1% Nonidet P-40, 500 μg of proteinase K per ml) and subjected to PCR amplification with nested env primers modified from the method of Gao et al. (28). rTth-XL polymerase (Perkin-Elmer, Foster City, Calif.) was used for amplification to obtain enhanced fidelity. The first reaction was done with the upstream primer 5′-GGCTTAGGCATCTCCTATGGCAGGAAGAA-3′ and the downstream primer 5′-CTGCCAATCAGGGAAGTAGCCTTGTGT-3′; the mixture was heated to 95°C for 1 min and then subjected to 35 cycles of 94°C for 1 min, 54°C for 5 min, and 72°C for 7 min, followed by a final extension at 72°C for 10 min. For the second round of PCR, the products of the first reaction were amplified under similar conditions with the upstream primer 5′-AGAAAGAGCAGAAGACAGTGGCAATGA-3′ and the downstream primer 5′-TAGCCCTTCCAGTCCCCCCTTTTCTTTTA-3′. The amplification products were extracted with phenol-chloroform, precipitated, and treated for 30 min with Pfu polymerase (Stratagene, La Jolla, Calif.) to generate blunt ends, under the conditions recommended by the manufacturer. The products were then purified and ligated downstream of the T7 promoter into the plasmid vector pCR-Blunt (Invitrogen, Carlsbad, Calif.). Clones carrying properly sized and oriented 2.7-kb env inserts were identified by restriction analysis and then screened for function by a luciferase reporter gene assay for cell-cell fusion.
Analysis of Env-mediated fusion.
To analyze fusion mediated by Env and CD4-coreceptors, 293T effector cells were infected with the recombinant vaccinia virus vP11T7gene1 (vTF1.1), which expresses the T7 polymerase (2), and transfected with the T7-driven env clones. These effector cells, expressing Env glycoprotein and T7 polymerase, were then mixed with quail QT6 target cells that had been cotransfected with a plasmid containing the luciferase gene under control of the T7 promoter and plasmids expressing CD4 along with the various chemokine receptor plasmids or control vector. Cell-cell fusion results in content mixing and transactivation of the target cell reporter construct by effector cell T7 polymerase, and fusion was determined by measuring the luciferase activity in cell lysates 16 h later. Details of this assay have been reported previously (23, 46). CD4, CCR5, CXCR4, CCR2b, CCR3, CCR8, CX3CR1, STRL33, GPR1, GPR15, APJ, and US28 plasmids were kindly provided by R. Doms (University of Pennsylvania) and have been described previously (23, 25, 46). Controls used in each experiment included effector cells without env and target cells with CD4 but no chemokine receptor. Fusion assays were carried out a minimum of three times with each combination of env and coreceptor, and the data shown are means of replicate experiments. env clones were not subjected to further analysis if they did not show at least fivefold greater fusion with at least one chemokine receptor than with target cells expressing CD4 only and at least fivefold greater luciferase activity than that of control effector cells lacking env.
Infection studies.
For primary cell infections, 25 ng of p24 antigen of each virus was used to infect 1-week-old cultures of monocyte-derived macrophages (MDM) or PBL that had been stimulated for 3 to 4 days with PHA and maintained with IL-2. Supernatant was then sampled periodically for p24 antigen by ELISA. The isolation of MDM from PBMC by serial adherence, macrophage depletion of PBMC to yield PBL, and methods used for infections have been described previously (15). For coreceptor-specific infection studies, we used U87-CD4 and HOS-CD4 cells stably expressing CCR2, CCR3, CCR5, and CXCR4 (8, 20) and U87-CD4 and HOS-CD4 cells transiently transfected by the calcium-phosphate method with coreceptor plasmids. One day after plating and/or transfection, the cells were infected overnight with 20 ng of p24 antigen of each virus, washed, and then replated into new wells to eliminate any residual inoculum. Supernatant was sampled on day 4 or 5 and day 7 or 9 for p24 antigen measurement by ELISA.
Sequence analysis.
Each functional clone was subjected to automated sequencing of a 500-bp region that included the V3 through V5 hypervariable domains. Sequences were compared by using the University of Wisconsin GCG sequence analysis package.
RESULTS
Isolation and biological characterization of matched lung and blood viruses.
To address the biological features and entry cofactor utilization patterns of HIV-1 quasispecies in tissue compartments, we isolated viral variants simultaneously from the lungs and blood of HIV-1-infected individuals with AIDS and pulmonary disease. We focused on this group because individuals with advanced disease have a higher lung virus burden and more frequently have virus detectable in the lungs (13, 55). In addition, late disease is more likely to reflect multiple rounds of replication and/or selection within the compartment and thus is more likely to reflect compartment-specific adaptation. Seven consecutive patients with active lung disease were studied, and we obtained both lung and blood isolates from three. The characteristics of these subjects are shown in Table 1. All had depressed CD4 counts and opportunistic lung infections. Two subjects (patients 6 and 7) were on antiretroviral monotherapy. In all three patients we isolated virus from alveolar macrophages and blood PBMC. Virus was isolated from nonadherent lung lymphocytes in only one subject (patient 7), and no isolates were obtained from stringently purified blood MDM in any individuals from whom lung isolates were also obtained.
TABLE 1.
Primary HIV-1 isolates used for this study
| Patient | pulmonary diagnosis | CD4 count (cells/mm3) | virus source | Isolate | Syncytium phenotypea | No. of functional env clones |
|---|---|---|---|---|---|---|
| 1 | Histoplasmosis | 45 | Blood lymphocytes | PBL-1 | NSI | 8 |
| Alveolar macrophages | AM-1 | NSI | 6 | |||
| 6 | M. avium infection | 50 | Blood lymphocytes | PBL-6 | SI | 5 |
| Alveolar macrophages | AM-6 | NSI | 6 | |||
| 7 | P. carinii infection | 100 | Blood lymphocytes | PBL-7 | NSI | 7 |
| Alveolar macrophages | AM-7 | NSI | 7 | |||
| Lung lymphocytes | LL-7 | NSI | 6 |
The syncytium-inducing phenotype was determined in MT-2 cells and is indicated as SI (syncytium inducing) or NSI (non-syncytium inducing).
All of the primary isolates replicated with similar kinetics in PBL (Fig. 1). Interestingly, the blood isolate from subject 1 replicated to high levels in MDM but the lung isolate replicated to only very low levels. Neither strain from subject 6 replicated to high level in MDM, while all three from subject 7 did. Only one isolate, PBL-6, was an SI strain in MT-2 cells (Table 1). This is consistent with previous reports showing that HIV-1 variants isolated from lungs are nearly uniformly NSI even while variants isolated simultaneously from blood late in disease may be either SI or NSI (29, 52). On the other hand, the limited replication by two lung-derived viruses in MDM was unexpected since lung-derived isolates are also typically M-tropic and these isolates were derived from lung macrophages.
FIG. 1.
Replication of primary isolates in PBL and MDM. Four-day-old PHA- and IL-2-stimulated PBL cultures and 1-week-old MDM cultures were infected overnight with each isolate by using 25 ng of p24gag antigen, washed, and sampled periodically for p24 antigen levels in supernatants. Data are representative of duplicate infections with cells from different donors.
Cloning of full-length env genes.
To define the entry cofactor utilization patterns of quasispecies in lung versus blood cells, functional envelope gene clones were generated by PCR from each isolate. Efforts to amplify functional, full-length env genes (≥2.7 kb) directly from uncultured patient material were not successful. Therefore, DNA was extracted from PBMC in which viruses were isolated after limited culture in vitro (approximately 2 weeks). env genes were cloned under control of the T7 promoter and tested for the ability to mediate cell-cell fusion with cells expressing CD4 along with each of 11 different coreceptors. Those that did not support fusion with at least one cofactor were not analyzed further. About 12 to 15 clones per isolate with full-length properly oriented env inserts were tested to generate a panel of five to eight functional clones for each (Table 1).
Utilization of the principal coreceptors.
Since there is a strong correlation between CCR5 use by M-tropic NSI variants and CXCR4 use by T-tropic SI variants, we first addressed the ability of these cloned Env proteins to mediate fusion with cells expressing CD4 in conjunction with these molecules (Fig. 2). For all fusion studies, cofactor utilization was considered significant if luciferase expression was reproducibly at least 10-fold greater than that seen with control cells expressing CD4 alone (Table 2). Levels that were 5- to 10-fold higher in the presence of the cofactor than with CD4 alone were considered to indicate marginal fusion of indeterminate significance. While there was considerable variability between experiments in the absolute level of fusion resulting from each env-coreceptor combination, the relative level of fusion was consistent, reflected as high (≥20-fold over control [Table 2]), moderate (10- to 20-fold), marginal (5- to 10-fold), or negative.
FIG. 2.
Fusion mediated by the primary-isolate env clones and the principal coreceptors CCR5 and CXCR4. Effector 293T cells were infected with the recombinant vaccinia virus vP11T7gene1, which expresses the T7 polymerase, and transfected with T7-driven env clones. These were then mixed with QT6 cells that were cotransfected with CD4, the indicated coreceptor or control vector, and a plasmid encoding luciferase under control of the T7 promoter. Cell-cell fusion was measured 16 h later on the basis of luciferase expression and is expressed as the fold increase in the luciferase level for the indicated cofactor in conjunction with CD4 compared with CD4 alone. Data represent the means of at least three independent experiments for each env-coreceptor combination.
TABLE 2.
Summary of fusion mediated by lung- and blood-derived Env glycoproteins
| Clone | Fusion with coreceptora
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Principal
|
Secondary
|
Orphan
|
||||||||
| CCR5 | CXCR4 | CCR2b | CCR3 | CCR8 | CX3CR1 | GPR1 | GPR15 | STRL33 | APJ | |
| PBL-1.1 | + | ± | ± | ± | ± | + | ± | + | ||
| PBL-1.3 | + | ± | ± | ± | ± | + | ||||
| PBL-1.4 | ++ | ± | ± | + | ||||||
| PBL-1.5 | + | ± | ± | ± | + | |||||
| PBL-1.14 | ++ | ± | + | ± | + | |||||
| PBL-1.16 | ++ | ± | ± | + | ||||||
| PBL-1.18 | + | ± | ± | ± | + | |||||
| PBL-1.28 | ++ | ± | ± | ± | + | |||||
| AM-1.12 | ++ | ± | ± | ± | + | ++ | ||||
| AM-1.20 | ++ | ± | ± | + | ++ | |||||
| AM-1.22 | ++ | ± | ± | ++ | ||||||
| AM-1.40 | + | ± | ± | ± | ||||||
| AM-1.63 | ++ | ± | ± | |||||||
| AM-1.65 | + | ± | ± | ± | ± | |||||
| PBL-6.3 | + | + | + | + | ++ | |||||
| PBL-6.9 | ± | ± | ± | |||||||
| PBL-6.15 | + | ++ | ± | ± | ± | + | ||||
| PBL-6.18 | ± | ± | + | |||||||
| PBL-6.21 | + | + | ± | ± | ± | |||||
| AM-6.1 | + | + | ± | ± | ++ | |||||
| AM-6.2 | + | ± | ± | ± | + | |||||
| AM-6.3 | + | ± | ± | ± | ± | + | ± | ++ | ||
| AM-6.4 | ++ | ± | ± | ++ | ||||||
| AM-6.7 | ++ | ± | ± | ± | + | |||||
| AM-6.56 | + | ± | + | |||||||
| PBL-7.8 | + | ± | ± | ± | ||||||
| PBL-7.18 | ± | ± | ± | ± | ± | |||||
| PBL-7.28 | + | ± | + | + | ± | ++ | ||||
| PBL-7.66 | ± | ± | ± | ± | ± | ++ | ||||
| PBL-7.68 | + | ++ | ± | ± | + | ++ | ||||
| PBL-7.69 | + | + | ± | + | + | ± | + | ++ | ± | ++ |
| PBL-7.76 | + | + | ± | + | ± | ++ | ++ | |||
| AM-7.12 | + | ± | ± | + | ++ | ++ | ||||
| AM-7.54 | + | + | ++ | ++ | ± | + | ± | ++ | ||
| AM-7.57 | ++ | + | ++ | + | + | ++ | + | ± | ++ | |
| AM-7.64 | + | + | ± | + | ± | ± | ++ | |||
| AM-7.66 | + | + | + | + | + | + | ++ | |||
| AM-7.69 | + | ± | ± | ± | ± | |||||
| AM-7.83 | ± | + | ++ | |||||||
| LL-7.2 | + | ± | ± | ++ | ++ | + | ± | |||
| LL-7.4 | ++ | ± | ± | + | + | + | ± | |||
| LL-7.5 | ++ | ± | ± | ± | ++ | ± | ++ | + | ± | |
| LL-7.10 | ++ | ± | ± | ± | ||||||
| LL-7.11 | ++ | ± | + | + | ± | ++ | + | |||
| LL-7.12 | ± | ± | ± | ++ | ± | ± | ± | + | + | |
Fusion was determined by measurement of luciferase expression as described in Methods and is indicated as follows: ++, >20-fold-greater luciferase expression in the presence of the indicated receptor than for CD4 alone; +, 10- to 20-fold greater expression; +/−, 5- to 10-fold greater. Less than a fivefold increase in fusion compared with CD4 alone was considered negative and is left blank. Boldface type indicates fusion greater than 10-fold over CD4 alone.
We found efficient fusion with CCR5 by the majority of functional lung- and blood-derived Env proteins from all three symptomatic HIV-1-seropositive individuals (Fig. 2 and Table 2). A few lung clones showed marginal use of CXCR4 (5- to 10-fold enhancement of fusion compared with CD4 alone), but none used it efficiently (≥10-fold). In contrast, at least one cloned Env protein from each blood isolate fused efficiently with CXCR4. Most CXCR4-using Env proteins used it in addition to rather than instead of CCR5. Thus, these results are consistent with recent data that dualtropic variants are more common among primary HIV-1 isolates than are CXCR4-restricted variants (56) and are also consistent with the restricted biological features of lung viruses (29, 52). Interestingly, all three sets of env clones from blood contained CXCR4-using variants, even though only one of the isolates (PBL-6) was biologically SI (Table 1). It is not clear why two isolates did not replicate or induce syncytia in MT-2 cells even though they contained CXCR4-using variants. Nevertheless, the data suggest that the evolution to CXCR4 utilization with disease progression may develop at the level of individual env species even prior to the emergence of detectable SI blood variants.
Utilization of secondary coreceptors.
CCR2b, CCR3, CCR8, and CX3CR1 support fusion and entry in vitro by more restricted groups of HIV-1 isolates than do the principal cofactors. To determine if utilization of these chemokine receptors might be related to HIV-1 infection in the lungs, we compared our panel of lung- and blood-derived env genes for fusion with cells expressing CD4 in conjunction with CCR2b, CCR3, CCR8, and CX3CR1 (Fig. 3 and Table 2).
FIG. 3.
Fusion mediated by the primary-isolate env clones and the secondary chemokine receptor coreceptors CCR2b, CCR3, CCR8, and CX3CR1. Fusion assays were performed as described in the legend to Fig. 2. Data represent the means of at least three independent experiments for each env-coreceptor combination.
Neither lung- nor blood-derived Env proteins from subject 1 used any of these chemokine receptors efficiently, and only single clones from lung or blood isolates from subject 6 used CCR2b, CCR3, or CCR8. In contrast, multiple clones from subject 7 used them. For blood-derived Env proteins, this was limited to modest levels of fusion with CCR3 by two of seven clones and with CCR8 by one of seven clones. For lung-derived clones, however, CCR2b and CCR8 supported efficient fusion by more than half of the lung macrophage-derived Env proteins and CCR3 was used by several lung macrophage- and lung lymphocyte-derived Env proteins. CX3CR1 was used by two Env proteins cloned from lung macrophage-derived virus and three cloned from lung lymphocyte-derived virus. Thus, in primary isolates from this individual, there was considerably broader use of the secondary chemokine receptors by lung-derived than by blood-derived env clones. Some Env glycoproteins fused with multiple secondary coreceptors, but others used only one or two of the secondary coreceptors, and there did not appear to be a strong association between the use of each particular coreceptor by multiple clones from each isolate (Table 2).
Utilization of orphan coreceptors.
Several putative 7TM G-protein-coupled receptor proteins that lack known ligands, termed orphan receptors, that support HIV or SIV fusion and/or entry in heterologous systems have been identified. GPR1, GPR15, and STRL33 are used widely by many SIV strains and by smaller numbers of HIV-1 strains (4, 21, 26, 36, 46). APJ is used broadly by both SIV and HIV-1 (10, 25). Importantly, GPR1 and GPR15 are expressed in lung cells (26). Therefore, we tested our panel of lung and blood primary isolate env clones for fusion with these molecules (Fig. 4 and Table 2).
FIG. 4.
Fusion mediated by the primary-isolate env clones and the orphan receptors GPR1, GPR15, STRL33, and APJ. Fusion assays were performed as described in the legend to Fig. 2. Data represent the means of at least three independent experiments for each env-coreceptor combination.
We found remarkably widespread fusion with the orphan receptor APJ by Env proteins from both lung- and blood-derived viruses from all three individuals. For some clones the levels of fusion mediated by APJ were modest compared with those supported by CCR5 or CXCR4, but for many of them APJ facilitated levels of fusion that were equivalent to or greater than that seen with the principal coreceptors. Fusion mediated by the other orphan receptors was more restricted (Fig. 4). GPR1, which has limited function as an HIV-1 coreceptor, was used by a small minority of clones, mainly from subject 7. In most cases the levels were marginal, but 10- to 20-fold enhancement of fusion was seen for a few lung macrophage- and blood lymphocyte-derived Env proteins. GPR15 and STRL33 supported fusion with a few Env proteins from each of the panels as well, although these levels were also low except for those with several clones from subject 7. As was the case for the secondary coreceptors, the clones from subject 7 displayed broader and more efficient utilization of the orphan receptors than did those from the other two subjects. However, within the set of Env proteins derived from a particular isolate, there was marked heterogeneity in the utilization of each orphan receptor. In addition, while some glycoproteins used multiple orphan receptors, no pattern in the coutilization of these molecules was evident. Importantly, we could see no link between clones derived from lung viruses and the utilization of any particular orphan receptors, including two reportedly expressed in the lungs, GPR1 and GPR15.
Utilization of virus-encoded chemokine receptors.
A chemokine receptor encoded by CMV, US28, is distantly related to CCR5 and CXCR4 and can serve as a coreceptor for several strains of HIV-1 and HIV-2 (42). Since the lungs are common sites of coinfection by HIV-1 and CMV, we tested whether these env clones utilized US28. None of the primary isolate clones used US28 as a coreceptor, however, whether they were from lung- or blood-derived viruses (data not shown).
Genetic analysis of lung- and blood-derived clones.
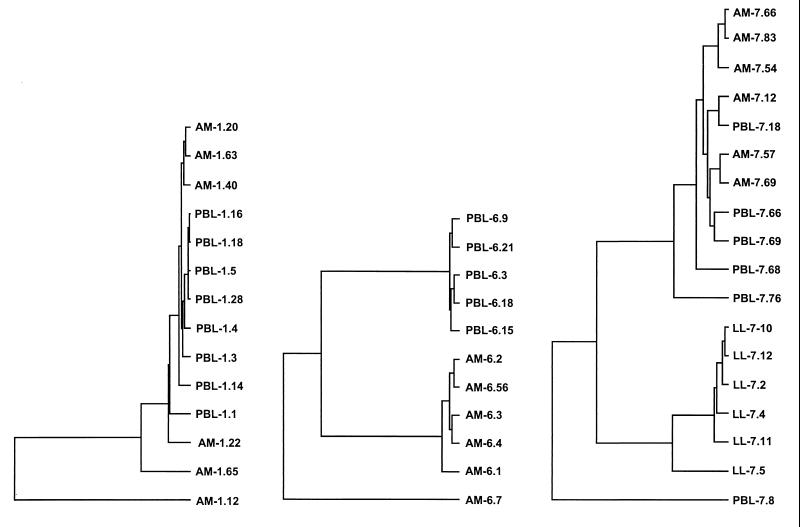
To determine the genetic relatedness between these clones, a region of each env gene was sequenced that included the V3 to V5 hypervariable domains to maximize differences identified. While clones in several of the panels were very closely related, none of them were identical. This confirmed that each clone represented an independent variant within the quasispecies, rather than multiple amplification products of a single virus. The relationships between the clones within this region sequenced are shown in Fig. 5. For subject 1, all clones from the blood isolate and several from the lung isolate were very closely related, while several additional clones from the lung isolate were less closely related. For subject 6, two distinct groups were evident and correlated with the site of origin, except for one less closely related lung-derived variant. For subject 7, for whom the greatest functional differences were seen between lung- and blood-derived env genes, the lung lymphocyte-derived clones clustered separately from the lung macrophage and blood isolate sequences, and one blood isolate clone was less closely related. These results show that the env genes captured by cloning after virus isolation continue, in general, to reflect organ-specific sequence relationships. When the env sequence clustering (Fig. 5) was compared to the overall entry cofactor utilization patterns (Table 2), there was no evidence that env genes within each set which had more closely related V3 to V5 sequences had similar entry cofactor utilization patterns.
FIG. 5.
env clone sequence relationship. Sequencing of each clone was carried out on a 500-bp region that included the hypervariable V3 to V5 domains. The sequences were used to confirm the independence of env clones and to determine relationships between clones from each patient by using the University of Wisconsin GCG program.
Infection supported by entry coreceptors.
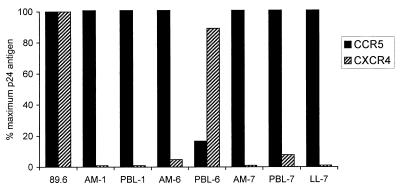
To address infection supported by these coreceptors, we tested the primary isolates for replication in several target cells expressing CD4 in conjunction with specific coreceptors. Both major coreceptors supported productive infection in stably or transiently transfected HOS-CD4 and U87-CD4 cells. Six of the seven strains used CCR5 only for efficient replication, while PBL-6 used CXCR4 efficiently and used CCR5 at very low levels (Fig. 6). This is consistent with the SI and NSI phenotypes, since PBL-6 was the only isolate to produce p24 antigen or form syncytia in MT-2 cells (Table 1). In contrast, none of the secondary coreceptors supported efficient replication, whether expressed in stable lines or following transfection (data not shown). Similarly, none of the orphan coreceptors supported efficient p24 antigen production in transiently transfected cells (data not shown), even APJ, which was used for fusion by multiple clones within each env quasispecies. Although productive replication was not seen with any secondary or orphan coreceptor, PBL-7 and LL-7 produced low levels of p24 antigen in U87-CD4 cells even in the absence of any exogenous coreceptor (∼200 pg of p24 antigen per ml [data not shown]). This probably reflects low-level infection mediated by endogenous coreceptors, and U87 cells have been shown to express GPR1 and STRL33 (21, 24). Thus, while efficient infection did not occur through any pathway other than CCR5 and CXCR4, this is consistent with the observation that isolates from subject 7 used the orphan coreceptors for fusion more efficiently than the other isolates did (Table 2).
FIG. 6.
Coreceptor-mediated productive infection. U87-CD4 cells expressing CCR5 or CXCR4 were infected with 20 ng of p24gag antigen as described in Materials and Methods, washed, and sampled on day 4. Bars represent levels of p24 antigen in supernatant relative to the dualtropic prototype 89.6, which uses both CCR5 and CXCR4 for infection.
DISCUSSION
We addressed the use of 7TM entry coreceptors by paired lung- and blood-derived isolates from individuals with AIDS and lung disease. We found that CCR5 was used by all lung- and most blood-derived Env clones (R5 variants), while a minority of the blood-derived clones used CXCR4 either alone or in addition to CCR5 (X4 or R5X4). These results extend to the level of cofactor usage previous observations that primary isolates from lungs are uniformly M-tropic and NSI, even late in disease, when T-tropic SI variants emerge in blood (29, 52). The data also support the notion that CCR5 is probably the major pathway for infection of pulmonary macrophages (14), the principal infected cell in the lungs. CCR5 is also the major coreceptor used by the prototype strain HIV-1Bal, which was isolated from lungs and extensively passaged in MDM following isolation (29), and by one recently analyzed clone that was derived directly from lung tissue without culture in vitro (22). In contrast, they differ from a report that HIV-1 species predictive of SI variants emerge in the lungs during active tuberculosis infection (40). Interestingly, we found no Env variants that used CXCR4 efficiently in the one isolate obtained from lung lymphocytes (LL-7), even though CXCR4 was used by several blood lymphocyte clones from that subject. The presence of CXCR4-using variants in blood but not lung lymphocyte quasispecies may indicate that macrophages are a reservoir or obligate intermediate for HIV-1 transmission to lung lymphocytes. Further studies are required to confirm this, however, since only one lung lymphocyte isolate was obtained and genetic analysis in this set of env clones (Fig. 5) did not suggest that the clones from lung lymphocytes were more closely related to the lung macrophage-derived clones than to those from blood.
Our data also confirm the persistence of R5 species in blood even after the emergence of R5X4 or X4 variants. Unexpectedly, we found Env proteins that used CXCR4 in blood quasispecies of all 3 subjects even though only one of the primary isolates was SI. This suggests that variants with the capacity to use CXCR-4 may emerge in blood even before SI viruses are evident. It is not certain why two isolates were NSI yet contained CXCR4-using env genes. It is possible that some Env proteins use CXCR4 for fusion in heterologous systems but do not use it efficiently for infection or in all cell types such as MT-2 cells. Of note, not all viruses that used CCR5 established productive replication in macrophages, which is similar to previous observations (22) and is consistent with the findings that primary isolates may be restricted in macrophages at various levels subsequent to entry (27) and by several different genetic determinants (61, 64).
The chemokine receptors other than CCR5 and CXCR4 that support HIV-1 entry in heterologous systems appear to play little role in infection of blood-derived cells (5, 18, 30, 44), but whether they are used selectively for infection in specific organs is an important question. Many HIV-1 isolates use CCR3, found initially in eosinophils but also present in microglia and certain T-cell subsets (16, 31, 49). Microglial infection can be inhibited by blocking CCR3, suggesting that it may play a role in the development of AIDS dementia (31), but a link between CCR3 utilization and brain-derived isolates has not been found by all observers (53). Fewer strains use CCR2b, which is expressed mainly in monocytes and NK cells (17, 54); CCR8, which is expressed mainly in activated monocytes, lymphocytes, and neutrophils (32, 51, 60); and CX3CR1, which is expressed in neural and lymphoid tissues (32, 33, 45). Although late-stage disease is associated with broader cofactor utilization in general (19), no pattern has emerged to link any of these molecules with particular aspects of pathogenesis or with variants from particular organs, except for the possible association between CCR3 and neurological infection. We found relatively distinct although overlapping patterns of secondary coreceptor fusion by clones from lung lymphocytes, lung macrophages, and blood lymphocytes from one of the subjects (patient 7), which suggests the presence of biologically distinct strains in these cellular compartments. Since the preferential use of secondary coreceptors by lung variants was seen only in one of three individuals, however, it is uncertain whether this reflects coreceptor-mediated adaptation to lung cells or some other feature specific to isolates from this subject.
Among the orphan receptors that support virus entry, GPR1 and GPR15 are of particular interest because they are expressed in alveolar macrophages (26, 32). GPR1 and/or GPR15 supported fusion by several clones from subject 7 and fewer from the other subjects, but neither were used preferentially by lung-derived variants. The orphan receptor STRL33 is also used widely by SIV and occasionally by HIV-1 (21, 36). In this panel, STRL33 was also used mainly by clones from subject 7 and more frequently by lung lymphocyte-derived clones. Since we did not have lung lymphocyte isolates from the other two subjects, we cannot determine whether this pattern indicates preferential fusion with STRL33 by lung lymphocyte-derived Env protein or, more likely, reflects generally broader coreceptor use by variants from this subject.
The orphan receptor APJ is expressed in lymphoid and neural cells and supports the entry of several T-tropic and dually tropic HIV-1 strains (10, 25). We found wide use of APJ for fusion, and, unlike the other secondary and orphan coreceptors, APJ mediated relatively high levels of fusion, comparable to those of CCR5 and CXCR4. While our results do not suggest APJ involvement in lung compartmentalization, its broad utilization by these primary isolates does indicate that it might play a role in vivo. On the other hand, the isolates from which these env genes were derived did not use APJ efficiently for infection. Unlike APJ, none used the CMV-encoded β-chemokine receptor US28, which is distantly related to CCR5 and CXCR4 (42). This is important because individuals with AIDS are often also infected with CMV, and CMV coinfection may be associated with accelerated disease progression (43, 48, 57). The lung is one of the most common sites of CMV infection in people with AIDS (38, 62), often is asymptomatic, and is a likely location for HIV-1–CMV interactions since alveolar macrophages, along with several other lung cells, are infected by CMV in vivo (58, 59). However, our results argue against a role for US28 in vivo resulting from CMV coinfection and broadened HIV-1 target cell tropism.
None of our Env proteins used secondary or orphan coreceptors in the absence of any CCR5- or CXCR4-mediated fusion, although a few fused only inefficiently with CCR5 or CXCR4 (PBL-6.18, PBL-7.66, AM-7.83, and LL-7.12). It is possible that the low levels of fusion seen with the principal cofactors are sufficient to support infection and virus replication, and we consider it unlikely that these env genes reflect variants that replicated in culture through such alternative coreceptors (18, 44). Alternatively, these env genes may represent replication-incompetent forms, possibly carried along as pseudotypes with variants that use the principal coreceptors. Studies to analyze the replication capacity and tropism of variants containing these env genes are under way. Of note, we selected as significant a level of 10-fold enhancement of fusion by a coreceptor and considered 5- to 10-fold enhancement marginal, but it is not known what level of fusion, determined in a cell-cell assay, is necessary for infection of cells that naturally express these molecules. Moreover, since the ability to utilize a coreceptor is highly dependent on levels of both coreceptor and CD4 expressed (35, 41, 46), it is likely that the capacity of these Env proteins to support fusion and infection with each specific coreceptor will differ depending on the target cell in which it is expressed. Along these lines, we found that the primary isolates from which these env genes were derived were unable to establish productive infection through coreceptors other than CCR5 and CXCR4 in stable or transiently transfected cell lines. The discrepancy between fusion and infection is an important distinction and probably reflects differences between cell-cell and virus-cell fusion, possibly including differences in env density on effector cells and virions.
One consideration in this analysis is that these clones were generated from primary isolates that were subjected to culture in vitro. Unlike passage in transformed cells, brief passage in primary PBMC results in more limited selection (47, 63). Nevertheless, we cannot exclude the possibility that in vitro isolation influences the biological characteristics of these clones or even excludes env species that support the infection of lung cells but not PBMC. To circumvent this, we attempted to generate full-length functional env clones directly from lung tissue without culture but were not successful. Another consideration is that the blood-derived viruses came from PBL, which may reflect archival species compared with plasma virions, although they would still be valid representatives of extrapulmonary variants. Also, in addition to distinct body compartments, the viruses differ in their cells of origin. Since macrophages are the principal cell type infected in the lungs, it is expected that most lung isolates would be from the macrophages, while lymphocytes but not monocytes are mainly infected in blood. Thus, differences between the env genes from different sites could be linked either to tissue compartmentalization itself or to the cell type within the compartment.
In summary, this is the first study to systematically analyze entry coreceptor utilization by HIV-1 variants from the lungs and compare them with variants from blood. We found CCR5 but not CXCR4 use by lung-derived viruses, which is consistent with the principal cellular reservoir in the lungs. In contrast, we found CCR5 and/or CXCR4 utilization by blood-derived variants, including CXCR4-using variants from the blood of individuals with advanced disease even in the absence of SI isolates. There was marked heterogeneity in the use of most secondary and orphan coreceptors by individual clones within a single isolate, with no evidence to suggest selective use of any specific coreceptor by lung-derived variants. The lack of concordance in the use of secondary coreceptors among clones from a particular isolate may indicate that their utilization is a result of random sequence variations within env quasispecies rather than of biological selection for use of that cofactor. Exceptions were APJ, which was broadly used for fusion, and US28, which was not used at all. In contrast to utilization for fusion by cloned env genes, however, productive infection by the primary isolates was limited to CCR5 and CXCR4. Additional studies are required to discover if any coreceptors other than CCR5 support infection of lung cells, and it remains to be determined what role is played by the secondary and orphan receptors in pathogenesis and in HIV-1 compartmentalization.
ACKNOWLEDGMENTS
We thank S. Isaacs, B. Lee, R. Doms, and M. Rossman for valuable advice and assistance, R. Doms for cofactor plasmids, N. Landau and D. Littman for cell lines obtained through the NIH AIDS Research and Reference Reagent Program, and S. Isaacs, D. Kolson, and D. Weissman for critically reviewing the manuscript.
This work was supported by grant HL 58004 from the National Institutes of Health.
REFERENCES
- 1.Agostini C, Trentin L, Zambello R, Semenzato G. HIV-1 and the lung. Infectivity, pathogenic mechanisms, and cellular immune responses taking place in the lower respiratory tract. Am Rev Respir Dis. 1993;147:1038–1049. doi: 10.1164/ajrccm/147.4.1038. [DOI] [PubMed] [Google Scholar]
- 2.Alexander W A, Moss B, Fuerst T R. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alimohammadi A, Coker R, Miller R, Mitchell D, Williamson J, Clarke J. Genotypic variants of HIV-1 from peripheral blood and lungs of AIDS patients. AIDS. 1997;11:831–832. [PubMed] [Google Scholar]
- 4.Alkhatib G, Liao F, Berger E A, Farber J M, Peden K W C. A new SIV co-receptor, STRL33. Nature. 1997;388:238. doi: 10.1038/40789. [DOI] [PubMed] [Google Scholar]
- 5.Ayehunie S, Garcia-Zepeda E A, Hoxie J A, Horuk R, Kupper T S, Luster A D, Ruprecht R M. Human immunodeficiency virus-1 entry into purified blood dendritic cells through CC and CXC chemokine coreceptors. Blood. 1997;90:1379–1386. [PubMed] [Google Scholar]
- 6.Berger E A. HIV entry and tropism: the chemokine receptor connection. AIDS. 1997;11(Suppl. A):S3–S16. [PubMed] [Google Scholar]
- 7.Berger E A, Doms R W, Fenyö E M, Korber B T M, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 8.Björndal Å, Deng H K, Jansson M, Fiore J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyö E M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chayt K J, Harper M E, Marselle L M, Lewin E B, Rose R M, Oleske J M, Epstein L G, Wong-Staal F, Gallo R C. Detection of HTLV-III RNA in lungs of patients with AIDS and pulmonary involvement. JAMA. 1986;256:2356–2359. [PubMed] [Google Scholar]
- 10.Choe H, Farzan M, Konkel M, Martin K, Sun Y, Marcon L, Cayabyab M, Berman M, Dorf M E, Gerard N, Gerard C, Sodroski J. The orphan seven-transmembrane receptor apj supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. J Virol. 1998;72:6113–6118. doi: 10.1128/jvi.72.7.6113-6118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choe H, Farzan M, Sun Y, Sullivan N, Rollins R, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 12.Clarke J R, Robinson D S, Coker R J, Miller R F, Mitchell D M. AIDS and the lung: update 1995. 4. Role of the human immunodeficiency virus within the lung. Thorax. 1995;50:567–576. doi: 10.1136/thx.50.5.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke J R, Taylor I K, Fleming J, Nukuna A, Williamson J D, Mitchell D M. The epidemiology of HIV-1 infection of the lung in AIDS patients. AIDS. 1993;7:555–560. doi: 10.1097/00002030-199304000-00015. [DOI] [PubMed] [Google Scholar]
- 14.Coffey M J, Woffendin C, Phare S M, Strieter R M, Markovitz D M. RANTES inhibits HIV-1 replication in human peripheral blood monocytes and alveolar macrophages. Am J Physiol Lung Cell Mol Physiol. 1997;272:L1025–L1029. doi: 10.1152/ajplung.1997.272.5.L1025. [DOI] [PubMed] [Google Scholar]
- 15.Collman R, Hassan N F, Walker R, Godfrey B, Cutilli J, Hastings J C, Friedman H, Douglas S D, Nathanson N. Infection of monocyte-derived macrophages with human immunodeficiency virus type 1 (HIV-1). Monocyte-tropic and lymphocyte-tropic strains of HIV-1 show distinctive patterns of replication in a panel of cell types. J Exp Med. 1989;170:1149–1163. doi: 10.1084/jem.170.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Combadiere C, Ahuja S K, Murphy P M. Cloning and functional expression of a human eosinophil CC chemokine receptor. J Biol Chem. 1995;270:16491–16494. doi: 10.1074/jbc.270.28.16491. [DOI] [PubMed] [Google Scholar]
- 17.Combadiere C, Ahuja S K, Van Damme J, Tiffany H L, Gao J L, Murphy P M. Monocyte chemoattractant protein-3 is a functional ligand for CC chemokine receptors 1 and 2B. J Biol Chem. 1995;270:29671–29675. doi: 10.1074/jbc.270.50.29671. [DOI] [PubMed] [Google Scholar]
- 18.Connor R I, Paxton W A, Sheridan K E, Koup R A. Macrophages and CD4+ T lymphocytes from two multiply exposed, uninfected individuals resist infection with primary non-syncytium-inducing isolates of human immunodeficiency virus type 1. J Virol. 1996;70:8758–8764. doi: 10.1128/jvi.70.12.8758-8764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng H, Liu R, Ellmeir W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 21.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 22.Dittmar M T, Simmons G, Donaldson Y, Simmonds P, Clapham P R, Schulz T F, Weiss R A. Biological characterization of human immunodeficiency virus type 1 clones derived from different organs of an AIDS patient by long-range PCR. J Virol. 1997;71:5140–5147. doi: 10.1128/jvi.71.7.5140-5147.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3 and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 24.Edinger A L, Hoffman T L, Sharron M, Lee B, O’Dowd B, Doms R W. Use of GPR1, GPR15, and STRL33 as coreceptors by diverse human immunodeficiency virus type 1 and simian immunodeficiency virus envelope proteins. Virology. 1998;249:367–378. doi: 10.1006/viro.1998.9306. [DOI] [PubMed] [Google Scholar]
- 25.Edinger A L, Hoffman T L, Sharron M, Lee B, Yi Y, Choe W, Kolson D L, Mitrovic B, Zhou Y, Faulds D, Collman R G, Hesselgesser J, Horuk R, Doms R W. An orphan seven-transmembrane domain receptor expressed widely in the brain functions as a coreceptor for human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol. 1998;72:7934–7940. doi: 10.1128/jvi.72.10.7934-7940.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farzan M, Choe H, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, Gerard N, Gerard C, Sodroski J. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J Exp Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fouchier R A M, Brouwer M, Kootstra N A, Huisman H G, Schuitemaker H. HIV-1 macrophage tropism is determined at multiple levels of the viral replication cycle. J Clin Investig. 1994;94:1806–1814. doi: 10.1172/JCI117529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao F, Morrison S G, Robertson D L, Thornton C L, Craig S, Karlsson G, Morgado M, Galvao-Castro B, von Briesen H, et al. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. The WHO and NIAID Networks for HIV Isolation and Characterization. J Virol. 1996;70:1651–1667. doi: 10.1128/jvi.70.3.1651-1667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gartner S, Markovits P, Markovitz D M, Kaplan M H, Gallo R C, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 30.Granelli-Piperno A, Moser B, Pope M, Chen D, Wei Y, Isdell F, O’Doherty U, Paxton W, Koup R, Mojsov S, Bhardwaj N, Clark-Lewis I, Baggiolini M, Steinman R M. Efficient interaction of HIV-1 with purified dendritic cells via multiple chemokine coreceptors. J Exp Med. 1996;184:2433–2438. doi: 10.1084/jem.184.6.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He J L, Chen Y Z, Farzan M, Choe H Y, Ohagen A, Gartner S, Busciglio J, Yang X Y, Hofmann W, Newman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 32.Horuk R, Hesselgesser J, Zhou Y, Faulds D, Halks-Miller M, Harvey S, Taub D, Samson M, Parmentier M, Rucker J, Doranz B J, Doms R W. The CC chemokine I-309 inhibits CCR8-dependent infection by diverse HIV-1 strains. J Biol Chem. 1998;273:386–391. doi: 10.1074/jbc.273.1.386. [DOI] [PubMed] [Google Scholar]
- 33.Imai T, Hieshima K, Haskell C, Baba M, Nagira M, Nishimura M, Kakizaki M, Takagi S, Nomiyama H, Schall T J, Yoshie O. Identification and molecular characterization of fractalkine receptor CX3CR1, which mediates both leukocyte migration and adhesion. Cell. 1997;91:521–530. doi: 10.1016/s0092-8674(00)80438-9. [DOI] [PubMed] [Google Scholar]
- 34.Itescu S, Simonelli P F, Winchester R J, Ginsberg H S. Human immunodeficiency virus type 1 strains in the lungs of infected individuals evolve independently from those in peripheral blood and are highly conserved in the C-terminal region of the envelope V3 loop. Proc Natl Acad Sci USA. 1994;91:11378–11382. doi: 10.1073/pnas.91.24.11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozak S L, Platt E J, Madani N, Ferro F E, Jr, Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao F, Alkhatib G, Peden K W C, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 38.Mann M, Shelhamer J H, Masur H, Gill V J, Travis W, Solomon D, Manischewitz J, Stock F, Lane H C, Ognibene F P. Lack of clinical utility of bronchoalveolar lavage cultures for cytomegalovirus in HIV infection. Am J Respir Crit Care Med. 1997;155:1723–1728. doi: 10.1164/ajrccm.155.5.9154883. [DOI] [PubMed] [Google Scholar]
- 39.Moore J P, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 40.Nakata K, Rom W N, Honda Y, Condos R, Kanegasaki S, Cao Y, Weiden M. Mycobacterium tuberculosis enhances human immunodeficiency virus-1 replication in the lung. Am J Respir Crit Care Med. 1997;155:996–1003. doi: 10.1164/ajrccm.155.3.9117038. [DOI] [PubMed] [Google Scholar]
- 41.Platt E J, Wehrly K, Kuhmann S E, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophage-tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pleskoff O, Tréboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276:1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- 43.Rabkin C S, Hatzakis A, Griffiths P D, Pillay D, Ragni M V, Hilgartner M W, Goedert J J. Cytomegalovirus infection and risk of AIDS in human immunodeficiency virus-infected hemophilia patients. National Cancer Institute Multicenter Hemophilia Cohort Study Group. J Infect Dis. 1993;168:1260–1263. doi: 10.1093/infdis/168.5.1260. [DOI] [PubMed] [Google Scholar]
- 44.Rana S, Besson G, Cook D G, Rucker J, Smyth R J, Yi Y, Turner J D, Guo H-H, Du J-G, Peiper S C, Lavi E, Samson M, Libert F, Liesnard C, Vassart G, Doms R W, Parmentier M, Collman R G. Role of CCR5 in infection of primary macrophages and lymphocytes by M-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from the Δccr5 mutation. J Virol. 1997;71:3219–3227. doi: 10.1128/jvi.71.4.3219-3227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raport C J, Schweickart V L, Eddy R L, Shows T B, Gray P W. The orphan G-protein-coupled receptor-encoding gene V28 is closely related to genes for chemokine receptors and is expressed in lymphoid and neural tissues. Gene. 1995;163:295–299. doi: 10.1016/0378-1119(95)00336-5. [DOI] [PubMed] [Google Scholar]
- 46.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sabino E, Pan L-Z, Cheng-Mayer C, Mayer A. Comparison of in vivo plasma and peripheral blood mononuclear cell HIV-1 quasispecies to short-term tissue culture isolates: an analysis of tat and C2-V3 env regions. AIDS. 1994;8:901–909. [PubMed] [Google Scholar]
- 48.Saillour F, Bernard N, Dequae-Merchadou L, Marimoutou C, Journot V, Dabis F. Predictive factors of occurrence of cytomegalovirus disease and impact on survival in the Aquitaine Cohort in France, 1985 to 1994. J Acquired Immune Defic Syndr Hum Retrovirol. 1998;17:171–178. doi: 10.1097/00042560-199802010-00012. [DOI] [PubMed] [Google Scholar]
- 49.Sallusto F, Mackay C R, Lanzavecchia A. Selective expression of the eotaxin receptor CCR3 by human T helper 2 cells. Science. 1997;277:2005–2007. doi: 10.1126/science.277.5334.2005. [DOI] [PubMed] [Google Scholar]
- 50.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cogniaux J, Forceille C, Muyldermans G, Verhofstede C, Bortonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection of Caucasian individuals bearing mutant alleles of the CCR5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 51.Samson M, Stordeur P, Labbe O, Soularue P, Vassart G, Parmentier M. Molecular cloning and chromosomal mapping of a novel human gene, ChemR1, expressed in T lymphocytes and polymorphonuclear cells and encoding a putative chemokine receptor. Eur J Immunol. 1996;26:3021–3028. doi: 10.1002/eji.1830261230. [DOI] [PubMed] [Google Scholar]
- 52.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, de Goede R E Y, van Steenwijk R P, Lange J M A, Eeftink Schattenkerk J K M, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shieh J T, Albright A V, Sharron M, Gartner S, Strizki J, Doms R W, Gonzalez-Scarano F. Chemokine receptor utilization by human immunodeficiency virus type 1 isolates that replicate in microglia. J Virol. 1998;72:4243–4249. doi: 10.1128/jvi.72.5.4243-4249.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sica A, Saccani A, Borsatti A, Power C A, Wells T N, Luini W, Sozzani S, Mantovani A. Bacterial lipopolysaccharide rapidly inhibits expression of C-C chemokine receptors in human monocytes. J Exp Med. 1997;185:969–974. doi: 10.1084/jem.185.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sierra-Madero J G, Toossi Z, Hom D L, Finegan C K, Hoenig E, Rich E A. Relationship between load of virus in alveolar macrophages from human immunodeficiency virus type 1-infected persons, production of cytokines, and clinical status. J Infect Dis. 1994;169:18–27. doi: 10.1093/infdis/169.1.18. [DOI] [PubMed] [Google Scholar]
- 56.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sinicco A, Raiteri R, Sciandra M, Dassio G, Bechis G, Maiello A. The influence of cytomegalovirus on the natural history of HIV infection: evidence of rapid course of HIV infection in HIV-positive patients infected with cytomegalovirus. Scand J Infect Dis. 1997;29:543–549. doi: 10.3109/00365549709035891. [DOI] [PubMed] [Google Scholar]
- 58.Sinzger C, Grefte A, Plachter B, Gouw A S, The T H, Jahn G. Fibroblasts, epithelial cells, endothelial cells and smooth muscle cells are major targets of human cytomegalovirus infection in lung and gastrointestinal tissues. J Gen Virol. 1995;76:741–750. doi: 10.1099/0022-1317-76-4-741. [DOI] [PubMed] [Google Scholar]
- 59.Sinzger C, Plachter B, Grefte A, The T H, Jahn G. Tissue macrophages are infected by human cytomegalovirus in vivo. J Infect Dis. 1996;173:240–245. doi: 10.1093/infdis/173.1.240. [DOI] [PubMed] [Google Scholar]
- 60.Tiffany H L, Lautens L L, Gao J L, Pease J, Locati M, Combadiere C, Modi W, Bonner T I, Murphy P M. Identification of CCR8: a human monocyte and thymus receptor for the CC chemokine I-309. J Exp Med. 1997;186:165–170. doi: 10.1084/jem.186.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toohey K, Wehrly K, Nishio J, Perryman S, Chesebro B. Human immunodeficiency virus envelope V1 and V2 regions influence replication efficiency in macrophages by affecting virus spread. Virology. 1995;213:70–79. doi: 10.1006/viro.1995.1547. [DOI] [PubMed] [Google Scholar]
- 62.Uberti-Foppa C, Lillo F, Terreni M R, Puglisi A, Guffanti M, Gianotti N, Lazzarin A. Cytomegalovirus pneumonia in AIDS patients: value of cytomegalovirus culture from BAL fluid and correlation with lung disease. Chest. 1998;113:919–923. doi: 10.1378/chest.113.4.919. [DOI] [PubMed] [Google Scholar]
- 63.Wain-Hobson S. Human immunodeficiency virus type 1 quasispecies in vivo and ex vivo. Curr Top Microbiol Immunol. 1992;176:181–193. doi: 10.1007/978-3-642-77011-1_12. [DOI] [PubMed] [Google Scholar]
- 64.Westervelt P, Henkel T, Trowbridge D B, Orenstein J, Heuser J, Gendelman H E, Ratner L. Dual regulation of silent and productive infection in monocytes by distinct human immunodeficiency virus type 1 determinants. J Virol. 1992;66:3925–3931. doi: 10.1128/jvi.66.6.3925-3931.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]