Abstract
Cytoplasmic dynein is a dimeric motor that drives minus-end directed transport on microtubules (MTs). To couple ATP hydrolysis to a mechanical step, a dynein monomer must be released from the MT before undergoing a conformational change that generates a bias towards the minus end. However, the dynamics of dynein stepping have been poorly characterized by tracking flexible regions of the motor with limited resolution. Here, we developed a cysteine-light mutant of yeast dynein and site-specifically labeled its MT-binding domain in vitro. MINFLUX tracking at sub-millisecond resolution revealed that dynein hydrolyzes one ATP per step and takes multitudes of 8 nm steps at physiological ATP. Steps are preceded by the transient movement towards the plus end. We propose that these backward “dips” correspond to MT release and subsequent diffusion of the stepping monomer around its MT-bound partner before taking a minus-end-directed conformational change of its linker. Our results reveal the order of sub-millisecond events that result in a productive step of dynein.
Cytoplasmic dynein (dynein hereafter) conducts minus-end-directed transport of organelles, vesicles, signaling complexes, and mRNA, as well as driving nuclear migration and retrograde transport in neurons1. Dynein also plays central roles in mitosis, such as focusing MTs into spindle poles and pulling on astral MTs to maintain tension on the spindle. Complete knockouts of dynein stop the MT transport machinery and inhibit mitosis. Mutations that alter dynein or dynein-associated factors lead to the pathogenesis of developmental and neurological diseases2.
Dynein is built around a pair of heavy chains that comprise the tail and motor domains2 (Fig. 1a). The motor domain is a ring of six nonidentical AAA sites (AAA1–6)3 that connects to an MT binding domain (MTBD) via an antiparallel coiled-coil stalk. The two rings are connected to a common tail via the linker domains. Dynein motility is primarily driven by coupling the ATPase activity at AAA14 to conformational changes of its stalk/MTBD and linker. ATP binding triggers the release of the motor from the MT by altering the registry of stalk coiled-coils5, 6 and the bending of the linker at the surface of the AAA+ ring (priming stroke)7. After ATP hydrolysis, dynein rebinds the MT, straightens its linker (power stroke), and pulls the cargo forward8. According to this scheme, dynein must be released from the MT before the linker undergoes the priming stroke to generate a productive step2. However, the order of events during stepping could not be directly determined from the structures of dynein captured in distinct nucleotide states8–10.
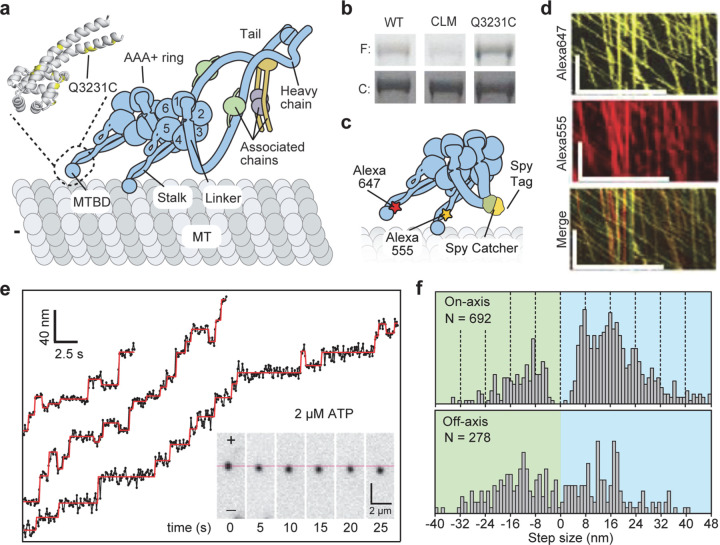
Figure 1.|. Site-specific labeling and tracking of the MTBD of dynein.
| a, The schematic of yeast dynein dimer bound to the MT. (Insert) The atomic model of the MTBD shows the positions of unique cysteines (yellow) introduced after removing surface-exposed cysteines in DynCLM. b, The denaturing gel images show that DynCLM-Q3231C is labeled more efficiently than DynCLM with all surface-exposed cysteines removed (F: fluorescence, C: Coomassie-stained). c, DynCLM-Q3231C monomers were labeled with different dyes at their MTBD and heterodimerized via SpyCatcher-SpyTag at the tail. d, Kymographs show robust processive motility of dual-labeled dynein heterodimer, similar to WT dynein. Scale bars x: 10 µm, y: 100 s. e, Trajectories show FIONA tracking of dynein heterodimers labeled with a single LD655 dye at 2 µM ATP with 100 ms temporal resolution. The red lines show a fit for the step-finding algorithm. f, The histogram of steps taken along the MT on- and off-axis. Dashed lines show increments of 8 nm steps.
Stepping of dynein monomers has been observed by labeling the AAA+ ring with a fluorophore and tracking its position using fluorescence imaging with one-nanometer accuracy (FIONA)11, 12. Dynein moves by uncoordinated stepping of its motor domains, and it frequently takes steps in sideways and backward directions on the MT11, 12. Due to the flexibility of dynein’s structure, these measurements were affected by the thermal fluctuations and different orientations of the AAA+ ring within the dimer13–16. The movement of dynein from one tubulin binding site to the next could only be observed at limited spatiotemporal resolution16 because FIONA requires ~104 photons to localize the center of the diffraction-limited image of a fluorophore with nanometer precision17. In comparison, MINFLUX requires ~100 times fewer photons for the localization by using a toroidal excitation beam18 and tracks the stepping of motors labeled with small (~1 nm in size) organic dyes at sub-millisecond temporal resolution19, 20.
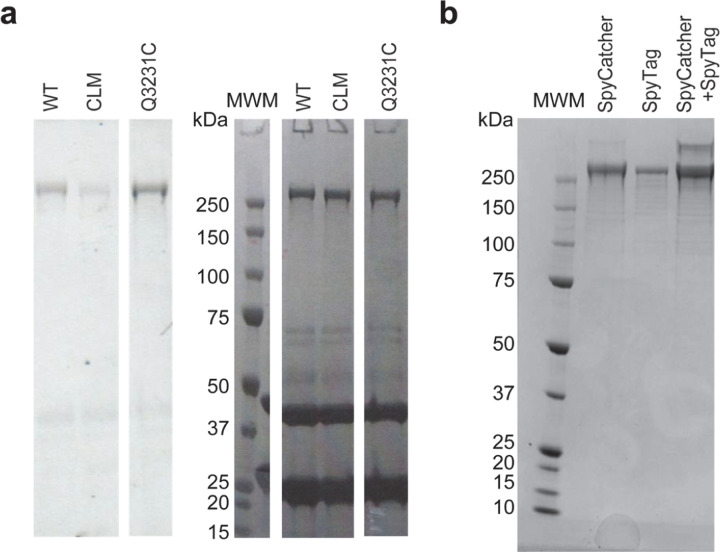
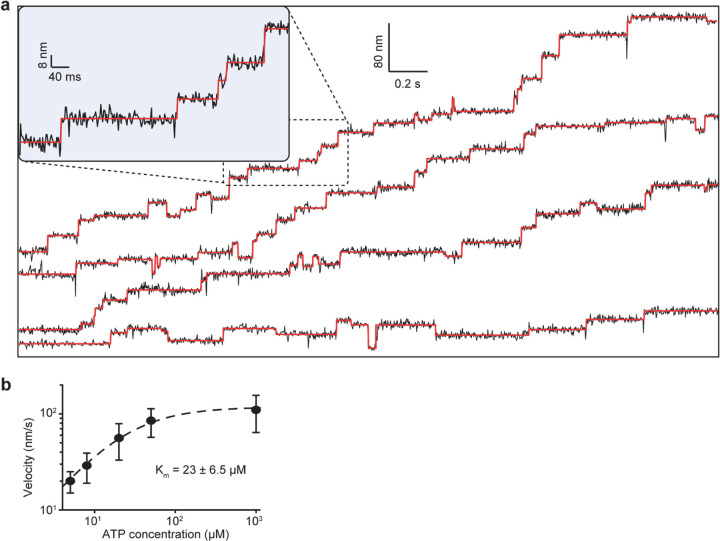
To determine how dynein steps along the MT lattice, we first site-specifically labeled the MTBD of yeast dynein. The artificial dimer of yeast dynein motor domain is an ideal model system to study the intrinsic stepping of dynein since it walks processively in the absence of dynein accessory proteins and has similar stepping properties to the mammalian dynein21, 22. Based on the available structures23, 24, we identified 5 out of 39 cysteines that are more than 5Å exposed to the solvent (Extended Data Table 1) and mutated them to serine (DynCLM). We next introduced a single solvent-accessible cysteine at multiple candidate sites at or near the MTBD for fluorescence labeling (Fig. 1a, Extended Data Fig. 1). We identified a mutant (Q3231C) that was efficiently labeled compared to DynCLM in vitro (Fig. 1b, Extended Data Fig. 2a, Extended Data Table 2). We constructed a heterodimer of DynCLM via SpyCatcher-SpyTag (Fig. 1c, Extended Data Fig. 2b) and confirmed that this motor moves with similar velocity and stepping properties to wild-type (WT) dynein25 (Fig. 1d-f, Extended Data Table 2).
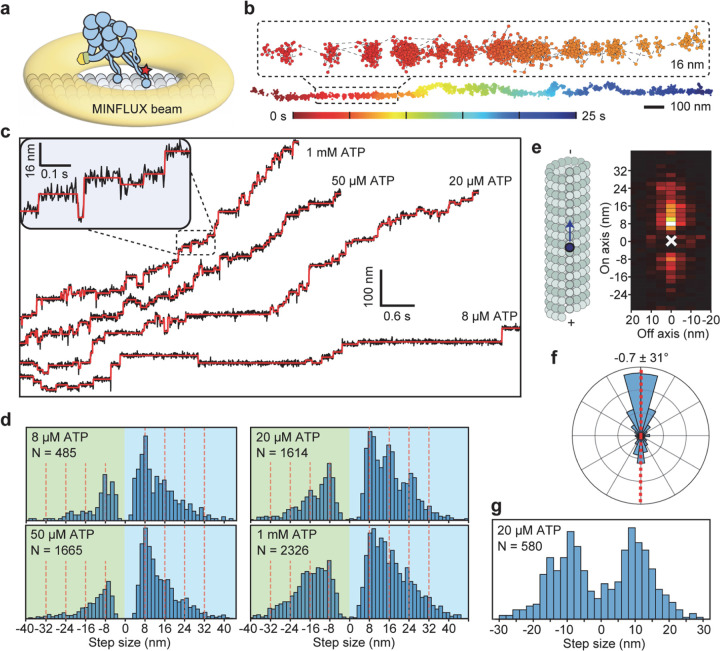
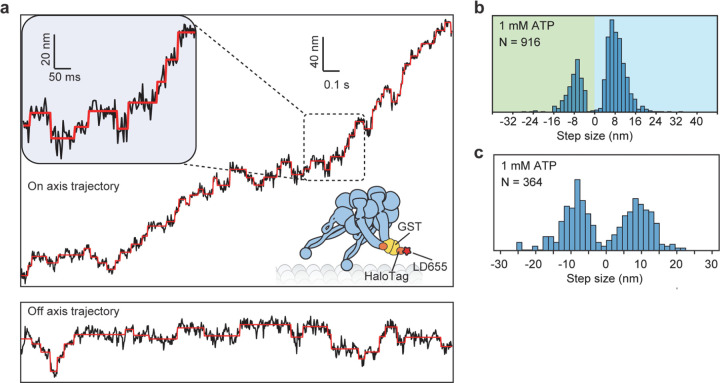
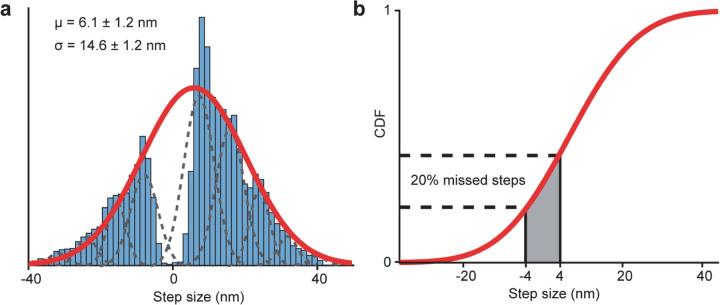
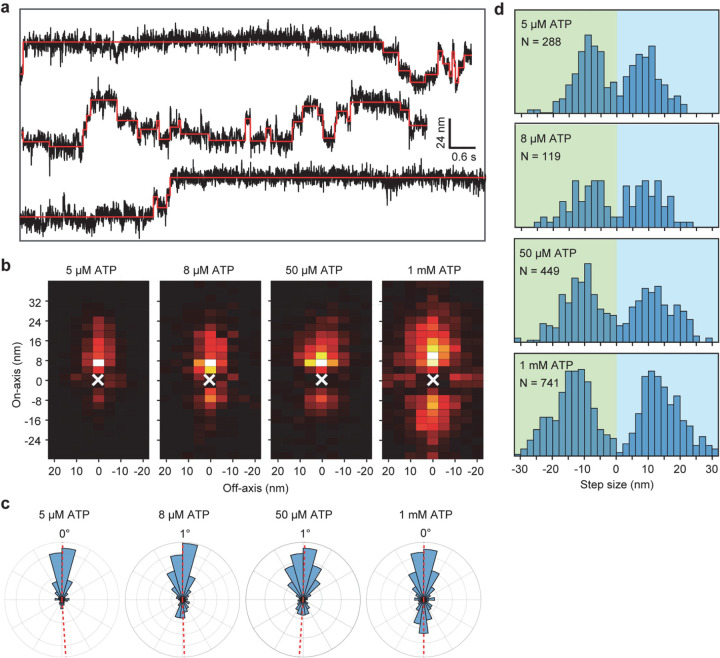
We next labeled motors with a single ultra-stable LD655 dye26 and tracked their motility at 3.0 nm spatial and 2.5 ms temporal resolution of MINFLUX19, 20 to determine dynein stepping behavior at physiological ATP. We confirmed that MINFLUX tracking of tail-labeled dynein exhibits stepping properties comparable to FIONA and optical trapping measurements (Extended Data Fig. 3)11, 25, 27. Trajectories exhibited large variations as the motor dwells between steps (Extended Data Fig. 3), presumably due to thermal fluctuations and conformational heterogeneity of the motor. We next tracked MTBD-labeled dynein at both limiting and saturating ATP concentrations using MINFLUX (Fig. 2a-b). We observed that dynein takes increments of 8 nm steps with ~28% of the steps taken backward (Fig. 2c-d). The periodicity of the dynein step size matches the distance between adjacent tubulin binding sites (8.2 – 8.4 nm)16. The direction and size distribution of dynein steps were unaffected by ATP concentration (Fig. 2d). Fitting the step size histogram to a normal distribution suggests that dynein has a 6 nm net bias to step towards the minus-end with a ±14 nm (s.d.) diffusional component to search for a new tubulin binding site (Extended Data Fig. 4a). Although we cannot detect a step if the monomer lifts off and rebinds to the same tubulin, the step size distribution suggests that 20% of steps are “0 nm” in size (Extended Data Fig. 4b). Dynein also frequently (46%) landed on adjacent protofilaments with nearly equal probability to step left or rightward28, 29 and these off-axis displacements were restricted to the 25 nm diameter of MTs (Fig. 2e-g, Extended Data Fig. 5).
Figure 2 |. MINFLUX tracking of dynein stepping at both limited and physiological ATP.
a, A schematic of MTBD-labeled dynein motility tracked with a toroidal MINFLUX beam (not to scale). b, An example two-dimensional trajectory of dynein at 20 µM ATP. The inset is color-coded by time. c, Example MT on-axis trajectories of dynein stepping under limiting and saturating ATP concentrations with 3.0 nm and 2.5 ms resolution. Red lines show fit to steps. d, On-axis step size histograms under all ATP concentrations tested. Red dashed lines show increments of 8 nm steps ranging from −24 nm to 40 nm (red dashed lines). e, A heat map shows a high probability of dynein accessing neighboring protofilaments while stepping along the MT. f, A histogram of dynein’s stepping angle reveals no directional bias to dynein’s off-axis steps (mean ± s.d.). g, The off-axis step-size histogram of dynein. ATP concentration was 20 µM in e-g.
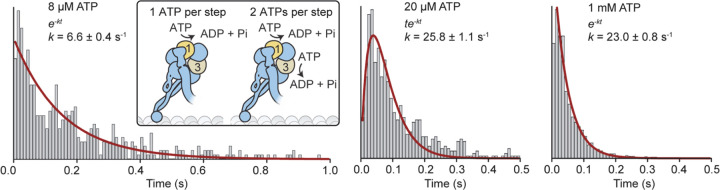
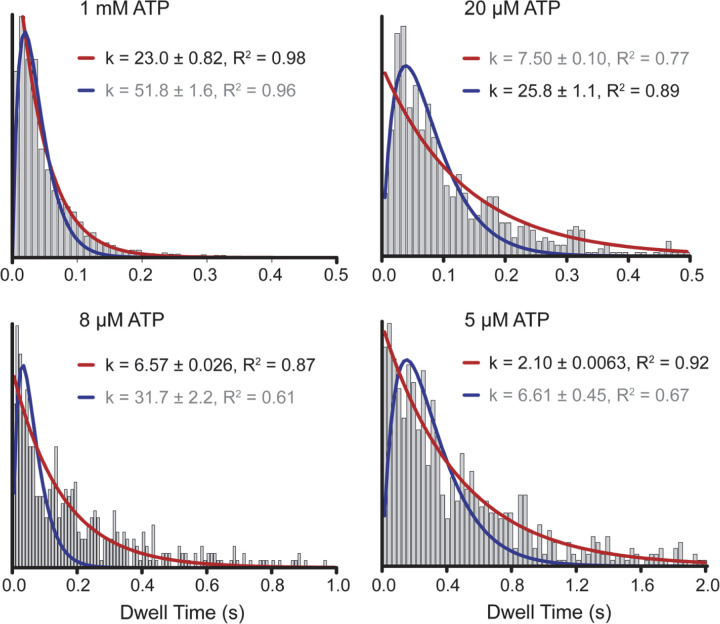
We analyzed the kinetics of dwell times between successive steps to gain insight into the number of ATPs hydrolyzed per step. In addition to AAA1, AAA3 hydrolyzes ATP, and mutations to this site substantially slow dynein motility30. It remains controversial whether only AAA1 hydrolyzes an ATP or AAA3 hydrolyzes ATP in coordination with AAA1 to generate each step31, 32. Solution kinetics studies showed that the ATPase cycle of dynein is rate-limited by ATP binding at low ATP and ADP release at saturating ATP concentrations33. Therefore, if dynein hydrolyzes one ATP per step, stepping would be limited by a single rate constant at both limiting (ATP binding) and saturating (ADP release) ATP and by two equal rate constants at the Michaelis-Menten constant (KM) of dynein for ATP (20 µM)34 because ATP binding and ADP release rates become equal under this condition. In comparison, if AAA1 and AAA3 sequentially hydrolyze ATP to generate each step, the stepping kinetics would be limited by two rate constants even at limiting and saturating ATP. We found that the dwell time distribution of dynein fits best to a model with a single rate-limiting constant under both saturating (1 mM) and limiting (5–8 µM) ATP and with the convolution of two equal rate constants at 20 µM ATP. (Fig. 3, Extended Data Figs. 6 and 7). These results demonstrate that dynein hydrolyzes one ATP per step, consistent with MINFLUX tracking of dynein stepping in neurons22.
Figure 3 |. Dynein hydrolyzes one ATP per stepping cycle.
(Insert) Kinetic schemes of dynein hydrolyzing one or two ATPs per step. The histogram of dwell times between successive on-axis steps of dynein under limiting (8 µM), KM (20 µM), and saturating (1 mM) ATP concentrations. Red curves represent a fit to a single exponential decay (8 µM and 1 mM) or Gamma distribution (20 µM) to determine the stepping rate (k, ± s.e.).
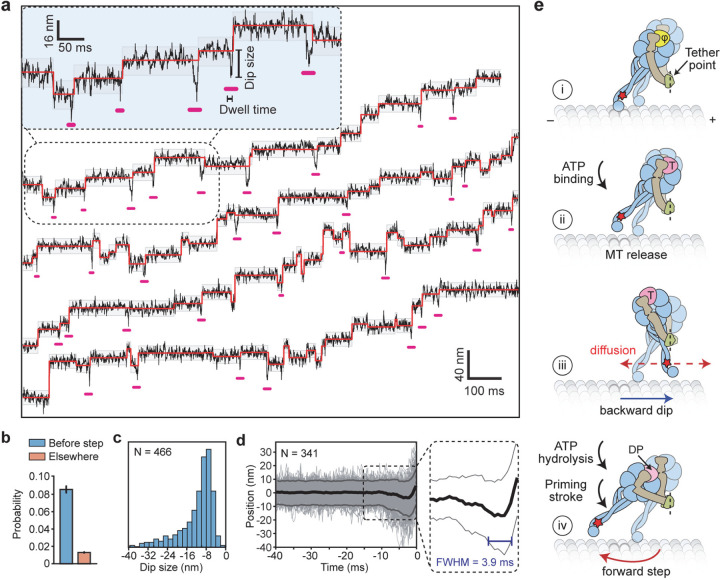
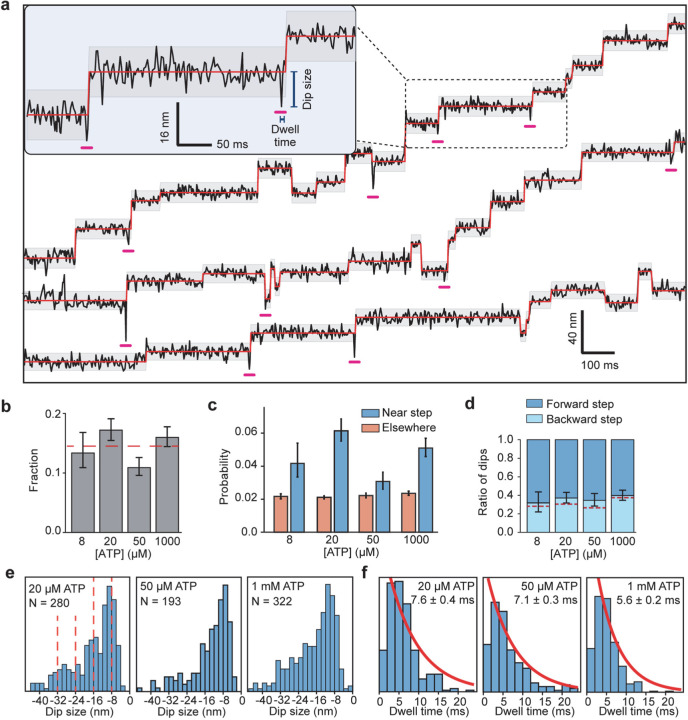
We noticed that the stepping monomer has a characteristic backward movement (referred to as dips) immediately before taking a step (Fig. 2a, Extended Data Fig 8a). However, only 14% of steps were preceded by a dip that typically lasts for a single frame, suggesting that these dips represent transient intermediates of dynein stepping (Extended Data Fig. 8). To detect these dips more reliably, we increased the temporal resolution of MINFLUX to 0.3 ms (Fig. 4a, Extended Data Table 4). We observed a higher fraction (35%) of steps are preceded by dips at this resolution (Fig. 4b). Dip sizes varied from 8 to 24 nm (Fig. 4c) and their lifetime was independent of ATP concentration (Fig. 4d, Extended Data Fig. 8).
Figure 4 |. MINFLUX observes the dynamics of dynein between successive steps.
a, Example trajectories of dynein in 1 mM ATP at 0.28 ms temporal resolution show brief dips (underlined in pink) before a step. Shaded regions represent ±2σ of the dwelling position between successive steps. b, The probability of −2σ deviation from the dwelling position within 7 ms before a step or elsewhere in trajectories. c, Dips occur in the backward direction and range from 8–40 nm in size. d, The cumulative overlay of dynein before a step (t = 0 s) demonstrates the presence of a backward dip. Black and grey curves represent mean ± 2σ, respectively. The average duration of the dips was measured from the full-width half minimum (FWHM) of the −2σ deviation before steps (blue line in the zoomed region). e, Interpretation of the dips observed in MINFLUX trajectories. (i) Dynein is strongly bound to the MT in the nucleotide-free state (φ). (ii) Upon ATP binding (T), a dynein monomer releases from the MT and (iii) moves backward toward the tethering point at the dimerization domain before ATP is hydrolyzed (dashed red line). (iv) The linker undergoes the priming stroke after ATP hydrolysis (DP) and moves the MTBD towards the minus end before dynein rebinds the MT.
We propose that the dips represent the movement of the stepping monomer towards its partner monomer after it binds ATP and releases from the MT (Fig. 4e, Extended Data Fig. 9a). Because the dynein motor domain is tilted relative to the MTBD15, the tether point connecting the two dimers is positioned towards the plus-end, resulting in backward displacement. The size of the dips may depend on the relative position of the dynein monomers (Extended Data Fig. 9b)11. The stepping monomer diffuses around the MT-bound monomer on the order of a millisecond. The transient nature and variable size of the dips provide an explanation for why we observed a third of the steps preceded by a clearly distinguishable dip. Upon ATP hydrolysis, the linker is free to transition between straight, semi-bent, and bent conformations across the ring or it partially detaches from the ring7, 9, 35. The combination of bending of the linker and the requirement of the dynein motor to bind with its stalk pointing forward creates a net bias towards the minus end15. Diffusion of the tethered monomer and different conformations of its linker may contribute to the high variability of the dynein step size (Fig. 4e, Extended Data Fig. 9c). After MT binding, the linker returns to the straight conformation, pulling the cargo forward. A recent cryo-electron microscopy study10 proposed an alternative model, where the linker straightens before MT binding, and a net bias in the step size is generated by further swinging of the linker towards AAA5 after the motor binds the MT. These possibilities can be tested by tracking the stepping of both monomers36 and observing the conformational dynamics of the linker as dynein steps along the MT.
Collectively, we used MINFLUX to obtain key insight into the stepping mechanism of dynein and detect transient conformational changes of the mechanochemical cycle of a motor on millisecond timescales, which cannot be captured by cryo-EM. The ability to site-specifically label DynCLM also enables the real-time visualization of how the AAA+ ring, stalk, and linker contribute to dynein stepping in future studies.
Materials and Methods:
Cloning and molecular biology
S. cerevisiae cytoplasmic dynein heavy chain gene (DYN1) was truncated at the N terminus (encoding amino acids 1219–4093, referred to as Dyn314kDa) as a template for mutagenesis25. An inducible galactose promoter, a tandem protein-A (ZZ) tag, a dual Tev protease site, and a glutathione-S-transferase (GST) tag were added to the N-terminus of Dyn314kDa gene by homologous recombination. The GST tag was replaced with an N-terminal SpyTag or SpyCatch for heterodimerization. The list of constructs used in this study is given in Extended Data Table 3.
The crystal structure of the S. cerevisiae dynein motor domain (PDB ID 4AKG)23 was used to analyze the accessibility of cysteine residues by PyMOL. Cysteine residues with more than 5 Å, between 2.5 – 5 Å, and less than 2.5 Å surface accessibility were classified as surface exposed, partially exposed, and buried, respectively (Extended Data Table 1). The five surface exposed residues on the dynein motor domain were mutated to serine (DynCLM). Partially exposed cysteines of dynein could not be removed as their mutagenesis substantially lowered protein expression (not shown). Surface-exposed cysteines of the GST-tag were also mutated to serine.
Protein expression, purification, and labeling
Dynein proteins were expressed in yeast. A single fresh yeast colony was used to inoculate 10 ml YP media containing 2% glucose. Cultures were grown overnight at 30°C with shaking at 200 rpm. The 100 ml YP media containing 1% raffinose was inoculated with overnight culture for ~10 h at 30°C at 200 rpm agitation until OD600 reached 0.2. The 100 ml culture was used to inoculate 1 L of YP media containing 2% galactose and 100 mg/ml adenine (Sigma) for 24–48 h at 30°C with 200 rpm agitation. Yeast was harvested by centrifugation at 5000 g for 7 min. Pellets were re-suspended in phosphate buffered saline to form a thick paste and frozen by dropwise addition in liquid nitrogen. Frozen yeast was stored at −80°C.
Frozen yeast pellets were ground and thawed in a lysis buffer (150 mM HEPES pH 7.4, 250 mM K-acetate, 10 mM Mg-acetate, 1 mM EGTA, 0.5 mM ATP-Mg2+, 5 mM DTT, 10 mM PMSF). The lysate was mixed with IgG Sepharose affinity beads and then cleaved by TEV protease as described previously25. The resulting protein was concentrated and stored in TEV storage buffer (50 mM Tris HCl pH 7.4, 100 mM KAc, 2 mM MgAc2, 1 mM EGTA, 10% glycerol, 0.2 mM TCEP, 0.1 mM ATP). The proteins were run in a denaturing gel and their concentrations were determined from OD280. 20 pmol SpyCatch dynein was then incubated with 5-fold excess maleimide reactive dyes in TEV storage buffer at room temperature for 1 h. The reaction was quenched with 1 mM DTT. After removing excess dye with a Zeba desalting column, SpyCatch was incubated with SpyTag dynein at 5:1 ratio in room temperature for 10 mins to form heterodimers. Dimerization was confirmed using a native gel and size exclusion chromatography. Fluorescence labeling was detected using a Typhoon gel image scanner. The labeling efficiency (~80% efficiency) was determined from 280 nm and 633 nm absorbance in a spectrophotometer. Purified protein was aliquoted, flash-frozen in liquid nitrogen, and stored at −80°C.
Single-molecule motility assays
Motility assays were performed in custom-made flow chambers comprised of polyethylene glycol (PEG)/PEG-biotin coated coverslips adhered to glass slides by double-sided tape37. The chamber was incubated with 10 µL 0.5 mg/mL streptavidin and washed with 40 µL DLBT buffer (30 mM HEPES pH 7.2, 2 mM MgCl2, 1 mM EGTA, 10% glycerol with added 1 mg/mL casein, 0.5% pluronic acid, and 1 µM Taxol). The chamber was then incubated with Cy3- and biotin-labeled MTs for 3 min before washing with 60 µL DLBCPT. Dynein was then diluted in DLBCPT to desired concentrations and flown into the chamber. After 3 min of incubation, the unbound motor was washed with 10 µL stepping buffer containing DLBCT, 0.8% dextrose, 0.1 mg/mL glucose oxidase, 0.2 mg/mL catalase, and desired ATP concentration. The sample was sealed and imaged for 1 h.
Microscope and imaging
Single-molecule motility assays were performed on a custom-built objective-type total internal reflection fluorescence (TIRF) microscope, equipped with an inverted Nikon Ti-E microscope body (Nikon Ti-Eclipse), perfect focusing system, and 1.45 NA 60× microscope objective. The sample was illuminated with 488, 561, and 632 nm laser beams (Coherent) to excite GFP, Cy3/Alexa555/LD555 and Cy5/Alexa647/LD655 fluorophores, respectively. Movies of LD655-labeled dynein were recorded with a 0.1 – 0.3 s exposure time under 2 mW 632 nm excitation. The fluorescence signal was detected with an electron multiplied CCD camera (Andor, iXon).
Single-molecule tracking measurements were also made on a commercial MINFLUX instrument (Abberior). The microscope is equipped with a 100× 1.4 NA oil immersion objective lens (Olympus), 642 nm excitation laser, two avalanche photodiodes (Excelitas) with a detection range of 650 – 685 nm, and a pinhole size corresponding to 0.78 airy units. The microscope was controlled by Abberior Imspector. An iterative localization approach with a hexagonal array of points was used to localize the fluorophores, as reported previously20, 38. Once the fluorophore was detected, the radius of the donut-shaped focused excitation beam (L) was decreased (284 nm, 302 nm, 151 nm, and 75 nm) and the laser power was increased (47, 47, 94, and 118 μW) in three increments. At this final step, ~100 photons were collected before localization. This final iteration was repeated continuously until the detection signal was lost due to permanent photobleaching of the dye. This procedure achieves ~2.5 ms for tracking of the LD655 dye. The parameters of the tracking algorithm were adjusted to reduce or increase the temporal resolution (Extended Data Table 4).
Data processing and analysis
Recorded TIRF movies were analyzed by a two-dimensional Gaussian fitting algorithm on a custom MATLAB software, YFIESTA available at GitHub (https://github.com/Yildiz-Lab/YFIESTA) to localize the fluorescent spots in the xy plane. All MINFLUX data were also processed, analyzed, and rendered on YFIESTA. Trajectories longer than 100 nm were detected and sorted according to a custom algorithm that relies on time autocorrelation along a linear path to determine fitness. The long axis of the trajectories was defined as the on-axis of the MT tracks. Trajectories that had acquisition frequency greater than 1 kHz suggesting multiple photon emissions, high off-axis noise due to poor MT immobilization and exhibited diffusion or poor motility were excluded from data analysis. Trajectories that passed initial screening were then fit to steps by a Schwartz Information Criterion-based step-fitting algorithm in both the on- and off-axis directions, as described previously39. Fitted trajectories were manually screened for overfitting or missed steps. Steps that were less than 3 frames were removed. If the mean positions of 8 data points before and after a step differ by less than 7.5 nm, these steps are removed by a custom algorithm to eliminate the positional drift being detected as a step. These manual corrections correspond to less than 3% of all on-axis steps. On and off-axis steps that occur within 5 frames (~12 ms) were combined into a “diagonal” step.
Analysis of dips was performed on on-axis trajectories. A custom algorithm detects the deviation of a data point more than two standard deviations (2σ) of the mean dwelling position before a detectable step. Dips were defined as 2σ deviations within 5 (~12 ms) or 20 data points (~7 ms) before a detectable step for trajectories recorded at ~2.5 or 0.28 ms temporal resolution, respectively. The dwell time of a dip was defined as the time between the beginning of a dip and the movement of the stepping monomer to the next position. The statistical significance of dips was calculated from comparing the probability of observing a −2σ deviation within ~7.2 ms (3 and 20 data points at ~2.5 and ~0.28 ms temporal resolution, respectively) around a step versus anywhere in the trajectory. The Beta distribution confirmed that −2σ deviations are not randomly distributed, and instead more likely to occur before steps with >95% confidence.
Extended Data
Extended Data Figure 1 |. Expression of DynCLM mutants in S. cerevisiae.
Denaturing gel pictures of 12 DynCLM constructs listed in Extended Data Table 2.
Extended Data Figure 2 |. Labeling and heterodimerization of the DynCLM mutant.
a, (Left) Fluorescence imaging and (Right) denaturing gel picture of dynein constructs labeled with Alexa647-maleimide under the same experimental conditions (MWM: molecular weight marker; kDa: kilodalton). b, A native gel picture shows the mixing of SpyCatcher-DynCLM and SpyTag-DynCLM at a 5:1 ratio to result in the heterodimerization of these constructs.
Extended Data Figure 3 |. MINFLUX analysis of tail-labeled dynein.
a, An example trajectory of GST-Dyn314kDa labeled at its tail domain at 1 mM ATP. On- and off-axis displacement of the same motor are shown separately. Red horizontal lines represent a fit to the step finder algorithm. Despite high precision in localization (σ = 3.2 nm), the insert shows large fluctuations in the dye position while the motor dwells on the MT between steps. b, The histogram of the size steps taken by dynein in the on- and off-axis direction. c, The dwell time distribution of tail-labeled dynein fits to a single exponential decay (dashed curve, ±s.e.).
Extended Data Figure 4 |. On-axis step size distribution suggests missing 0 nm steps.
a, The combined histogram of dynein step size measured in 5 µM – 1 mM ATP reveals a large gap from −4 nm to 4 nm. The red curve represents a fit to a Gaussian distribution to calculate the mean (µ) and standard deviation (σ; ±s.e.). Dashed curves represent subpopulations located at increments of 8 nm. The fit suggests that a dynein monomer has a 6 nm bias to move towards the minus end while searching for the new tubulin binding site via diffusion. b, The cumulative distribution function (CDF) of the step size histogram in a. CDF values from −4 nm to 4 nm suggest that 20% of the steps are 0 nm in size due to the rebinding of dynein to the same tubulin site.
Extended Data Figure 5 |. Two-dimensional step analysis of MINFLUX trajectories.
a, Additional example trajectories of dynein stepping along the MT off-axis at 20 µM ATP. b, Heat maps of two-dimensional stepping of dynein at different ATP conditions. c, Polar histograms of dynein’s stepping direction reveal the mean angular deviation of dynein from the MT minus-end. d, The histogram of steps taken by dynein in the off-axis direction under different ATP concentrations.
Extended Data Figure 6 |. Velocity of dynein motility under different ATP concentrations.
a, Additional example trajectories of dynein stepping along the MT on-axis at 20 µM ATP. b, The mean velocity of analyzed trajectories under different ATP concentrations. The dashed curve shows that dynein velocity vs. [ATP] follows Michaelis-Menten kinetics with KM near 20 µM (±s.e.).
Extended Data Figure 7 |. Dynein rates at different ATP concentrations.
The dwell time distribution of dynein stepping fits better to a single exponential decay (red curves) at limited (5 and 8 µM) and saturating (1 mM) ATP concentrations. In comparison, the dwell time distribution at KM = 20 µM ATP fits better to a Gamma distribution (blue curves). R2 represents the goodness of fit of a model. The fitting parameter of a better fit is highlighted in black.
Extended Data Figure 8 |. Analysis of dips in trajectories recorded at 2.5 ms temporal resolution.
a, Exemplary on-axis stepping trajectories at 20 µM ATP with dips underscored in pink. b, The probability of a step preceded by a dip is independent of ATP concentration. c, The probability of a dip occurring within 3 frames before a step or elsewhere during the dwell. d, The ratio of dips that occur before a forward or a backward step. The centerline and whiskers represent the mean and s.e. Dashed red line represents the probability of backward steps at given ATP concentrations. e, The histogram of the size of dips that occur within 3 frames before a step at different ATP concentrations. Red dashed lines represent increments of 8 nm. f, The histogram of the dwell time of dips at different ATP concentrations. The average lifetime of dips (±s.e.) is calculated by a fit to a single exponential decay (red curve). The dip lifetime at this lower temporal resolution is higher than the FWHM of the dips observed at 0.28 ms temporal resolution (Fig. 4d), probably due to the limited ability to detect dips that last less than a few ms.
Extended Data Figure 9 |. Models for the productive ATPase cycle and transient dips of dynein stepping.
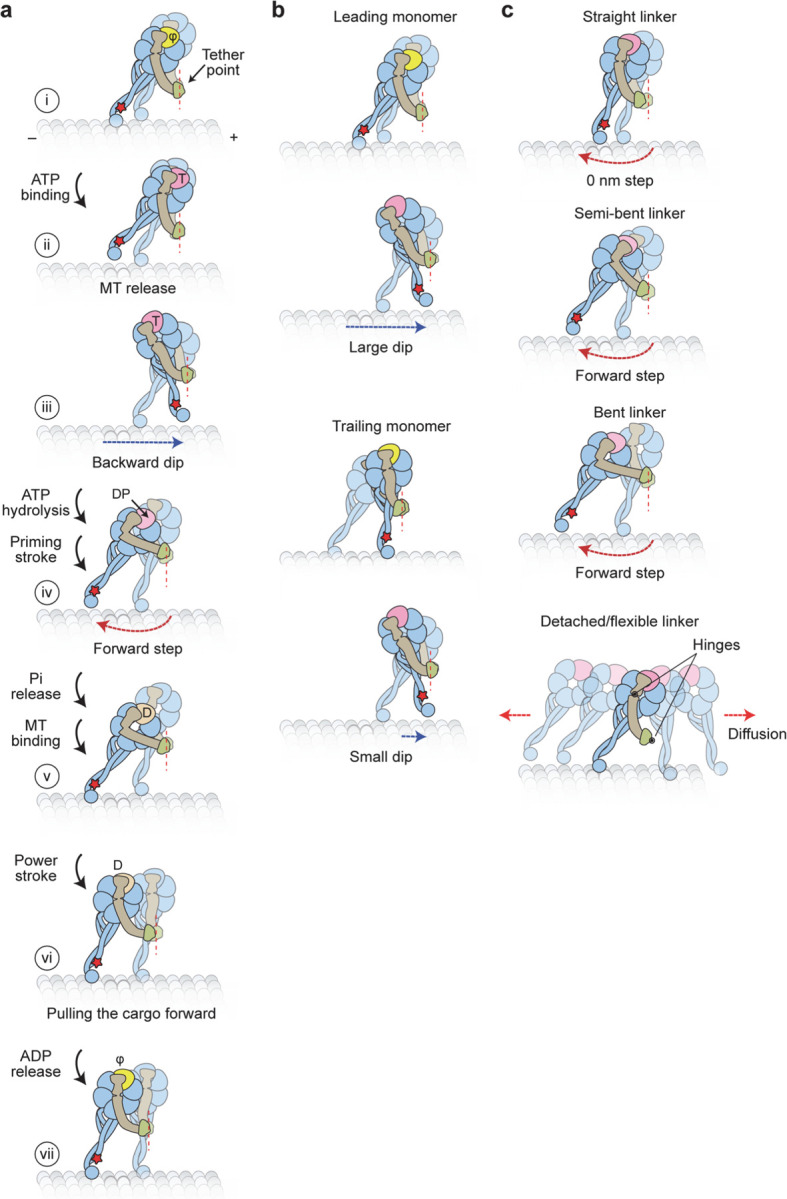
a, Updated model for the mechanochemical cycle of dynein. (i) When AAA1 is in the nucleotide-free state, a dynein monomer is bound to the MT and the linker is in a straight conformation. (ii) ATP binding to AAA1 triggers the release of the motor from the MT. (iii) After MT release, the dynein monomer moves toward the tethering point near the dimerization domain, resulting in dips observed in dynein trajectories. (iv) After ATP hydrolysis, the priming stroke of the linker pushes the MTBD towards the MT minus end. (v) Dynein rebinds the MT and releases inorganic phosphate (Pi). (vi) The Pi release stabilizes the straight conformation of the linker (power stroke) and dynein pulls its cargo in the forward direction. (vii) Upon ADP release, the linker further straightens towards AAA5, and the motor resets for the next ATPase cycle. b, Dip size depends on the relative positions of the stepping monomer and the MT-bound monomer. Dips are oriented backward due to the plus-end-directed tilting of the stalk relative to the MT. If a step is initiated from a monomer in the lead, the stepping monomer takes a large dip in the backward direction to move towards the tether point in the dimer. However, if the trailing monomer steps, the relaxation towards the tether point may be small or directed towards the minus end, resulting in a shorter dip in the backward direction. c, Different conformations of the linker in the ADP.Pi state may contribute to the high variability of the dynein step size. The linker can orient in straight, semi-bent, and bent conformations or detach from the AAA+ ring. Straightening of the linker before MT rebinding may result in no net displacement and account for 0 nm steps. Semi-bent and bent conformations push the MTBD towards the minus-end, which may be responsible for the small minus-end-directed bias in the dynein step size. Detachment of the linker from the AAA+ ring increases the degrees of freedom and results in a larger diffusional search, which may account for the high variability of the dynein step size distribution.
Extended Data Table 1 |. Characterization of cysteines in truncated dynein.
Cysteines of the dynein motor domain were categorized into three groups based on available cryo-EM structures of yeast dynein (PDB ID: 4AKG)23. The surface-exposed cysteines were accessible to surrounding water over 5 Å, partially exposed cysteines were solvent accessible to over 2.5 Å, and buried cysteines to less than 2.5 Å. Surface-exposed cysteines of the GST-dimerization were also removed. SpyCatcher and SpyTag do not contain any cysteines and are therefore omitted from this table.
| Protein | Surface exposed | Partially exposed | Buried |
|---|---|---|---|
| GFP | C48 | C70 | |
| GST | C84 C137 C177 |
C168 | |
| Dyn314kDa | C2037 C2268 C2372 C3499 C4016 |
C1405 C1663 C1718 C2210 C2220 C2724 C2892 C3141 C3206 C3324 C3684 C3704 C3959 |
C1428 C1626 C1793 C1822 C1846 C1960 C1980 C2057 C2078 C2417 C2486 C2535 C2603 C2806 C2814 C2912 C3382 C3652 C3877 C3928 C4011 |
| Total | 8 | 15 | 22 |
Extended Data Table 2 |. Testing of unique cysteine for labeling DynCLM at its MTBD.
Protein expression and motility properties of DynCLM constructs mutated with a single surface-exposed cysteine at the MTBD, compared to WT Dyn314kDa. Assays were performed in 1 mM ATP. The most processive and well-expressing motor (Q3231C) was used for the tracking assays. The denaturing gel used for quantifying protein expression levels is shown in Extended Data Fig. 1.
| Construct Number | Construct Name | Expression Level | Average velocity (nm/s) | % processive motors landed on MTs |
|---|---|---|---|---|
| - | Wildtype | High | 80 | - |
| 1 | Q3147C | Low | 10 | >90 |
| 2 | H3169C | High | 80 | 50 |
| 3 | Q3179C | High | 70 | 50 |
| 4 | K3234C | - | - | - |
| 5 | K3242C | Medium | 50 | 50 |
| 6 | S3097C | Low | 100 | 25 |
| 7 | S3190C | Medium | 78.6 | 25 |
| 8 | K3182C | Medium | 73.3 | 25 |
| 9 | Q3211C | Medium | 76.6 | 25 |
| 10 | Q3231C | Medium | 80 | >90 |
| 11 | H3175C | Low | 83.3 | 50 |
| 12 | E3107C | Low | 58 | 25 |
Extended Data Table 3 |. Yeast strains used in this study.
UPF1 was knocked out of the CY1 genome (MATa; his3–11,15; ura3–1; leu2–3,112; ade2–1; trp1–1) using the KanMX6 marker. The KanMX6 was amplified from pFA6a and inserted into the CY1 genome via homologous recombination. SpyCatcher and SpyTag facilitate heterodimerization of the tail-truncated dynein constructs. HALO (DHA) is a genetic tag used for labeling the protein with a fluorescent dye.
| Name | Description | Genotype | Database | Reference |
|---|---|---|---|---|
| WT Dynein | GFP-GST-Dyn1314kDa | MATa; his3–11,15; ura3–1; leu2–3,112; ade2-1; trp1–1; PEP4::HIS5; PRB1D pDyn-pGAL-ZZ-TEV-GFP-3XHA-GST-DYN1314kDa-gsDHA:Kan | CY31 | (Reck-Peterson et al., 2006)25 |
| Tail-labeled Dynein | DHA-GST-Dyn1314kDa | MATa; his3–11,15; ura3–1; leu2–3,112; ade2-1; trp1–1; PEP4::HIS5; PRB1D pDyn-pGAL-ZZ-TEV-gsDHA-GST- DYN1314kDa | VY268 | (Reck- Peterson et al., 2006)25 |
| GSTCLM- Dyn | CY1 + P354 | MATa; his3–11,15; leu2–3,112; ade2–1; trp1–1; PEP4::HIS5; PRB1D pDyn-pGAL-ZZ-TEV-GFP-3XHA-GSTC84S,C137S,C177S -D6-Dyn1314kDa-gsDHA:Kan | CY392 | This study |
| DynCLM | CY1 + P372 | MATa; his3–11,15; leu2–3,112; ade2–1; trp1–1, PEP4::HIS5; PRB1D pDyn-pGAL-ZZ-TEV-GFP-3XHA-GSTC84S,C137S,C177S-Dyn1314kDa, C2037S, C2268S, C2372S, C3499S, C4016S-gsDHA:Kan | CY393 | This study |
| SpyCatcher- DynCLM | CY1 + P372-SpyCatcher | MATa; his3–11,15; leu2–3,112; ade2–1; trp1–1, PEP4::HIS5; PRB1D pDyn-pGAL-ZZ-TEV-GFP-3XHA-ΔUPF1::KanMX6, SpyCatcher-Dyn1314kDa, C2037S, C2268S, C2372S, C3499S, C4016S | CY483 | This study |
| SpyTag- DynCLM | CY1 + P372-SpyTag | MATa; his3–11,15; leu2–3,112; ade2–1; trp1–1, PEP4::HIS5; PRB1D pDyn-pGAL-ZZ-TEV-GFP-3XHA-ΔUPF1::KanMX6, SpyTag-Dyn1314kDa, C2037S, C2268S, C2372S, C3499S, C4016S | CY487 | This study |
| GST-DynCLM Q3231C | GFP-GST- DynCLM + Q3231C | MATa; his3–11,15; leu2–3,112; ade2–1; trp1–1, PEP4::HIS5; PRB1D pDyn-pGAL-ZZ-TEV-GFP-3XHA-ΔUPF1::KanMX6,GST-C84S,C137S,C177S -Dyn1314kDa, C2037S, C2268S, C2372S, C3499S, C4016S, Q3231C | CY486 | This study |
| SpyTag- DynCLM Q3231C | CY1 + P372-SpyTag- DynCLM + Q3231C | MATa; his3–11,15; leu2–3,112; ade2–1; trp1–1, PEP4::HIS5; PRB1D pDyn-pGAL-ZZ-TEV-GFP-3XHA-ΔUPF1::KanMX6,GSTC84S,C137S,C177S - Dyn1314kDa, C2037S, C2268S, C2372S, C3499S, C4016S, Q3231C | CY488 | This study |
| SpyCatcher- DynCLM Q3231C | CY1 + P372-SpyCatcher- DynCLM,Q3231C | MATa; his3–11,15; leu2–3,112; ade2–1; trp1–1, PEP4::HIS5; PRB1D pDyn-pGAL-ZZ-TEV-GFP-3XHA-ΔUPF1::KanMX6, SpyCatcher-Dyn1314kDa, C2037S, C2268S, C2372S, C3499S, C4016S, Q3231C | CY489 | This study |
Extended Data Table 4 |. List of MINFLUX parameters used to track dynein at different temporal resolution.
10* and 100** represent the minimum photon counts needed for the next iteration. Laser power was optimized to obtain an average number of 30* and 140** photons, respectively. The 0.28 ms temporal resolution algorithm also required increasing the pinhole to 0.97 Airy units.
| 0.28 ms temporal resolution MINFLUX tracking algorithm | |||||
|---|---|---|---|---|---|
| L size (nm) | 284 | 302 | 151 | 75 | 30 |
| Pattern | Hexagon | Hexagon | Hexagon | Hexagon | Hexagon |
| Minimum photons | 40 | 20 | 10 | 10 | 10* |
| Laser power (µW) | 152 | 152 | 304 | 380 | 912 |
| Minimum dwell time (µs) | 400 | 400 | 400 | 400 | 150 |
| Pattern repeat | 1 | 1 | 1 | 1 | 1 |
| Background threshold (kHz) | 70 | 70 | 40 | 40 | 135 |
| 2.5 ms temporal resolution MINFLUX tracking algorithm | |||||
| L size (nm) | 284 | 302 | 151 | 75 | |
| Pattern | Hexagon | Hexagon | Hexagon | Hexagon | |
| Minimum photons | 40 | 20 | 10 | 100** | |
| Laser power (µW) | 47 | 47 | 94 | 118 | |
| Minimum dwell time (µs) | 400 | 400 | 400 | 300 | |
| Pattern repeat | 1 | 1 | 1 | 3 | |
| Background threshold (kHz) | 70 | 70 | 40 | 40 | |
| 8.3 ms temporal resolution MINFLUX tracking algorithm | |||||
| L size (nm) | 284 | 302 | 151 | 75 | |
| Pattern | Hexagon | Hexagon | Hexagon | Hexagon | |
| Minimum photons | 40 | 20 | 10 | 100** | |
| Laser power (µW) | 47 | 47 | 94 | 118 | |
| Minimum dwell time (µs) | 400 | 400 | 400 | 100 | |
| Pattern repeat | 1 | 1 | 1 | 3 | |
| Background threshold (kHz) | 70 | 70 | 40 | 40 | |
| Dead time (ms) | - | - | - | 5 | |
Acknowledgements
We thank M. DeWitt and A. Yonar for preliminary work on FIONA, J. Matthias, K. Boateng, and G. Fried for technical assistance with the MINFLUX microscope in IGB at Univ. of Illinois. This work was funded by grants from the NSF Science and Technology Center for Quantitative Cell Biology (2243257 to P.R.S.), NSF (MCB-1055017 and MCB-1617028 to A.Y., and DGE 2146752 to J.S.), NIH (GM136414 to A.Y.; GM132392 to P.R.S.), and the Medical Research Council (MC_UP_A025_1011 to A.P.C.).
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Reck-Peterson S.L., Redwine W.B., Vale R.D. & Carter A.P. The cytoplasmic dynein transport machinery and its many cargoes. Nature reviews. Molecular cell biology 19, 382–398 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canty J.T., Tan R., Kusakci E., Fernandes J. & Yildiz A. Structure and Mechanics of Dynein Motors. Annual review of biophysics 50, 549–574 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgess S.A., Walker M.L., Sakakibara H., Knight P.J. & Oiwa K. Dynein structure and power stroke. Nature 421, 715–718 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Kon T., Nishiura M., Ohkura R., Toyoshima Y.Y. & Sutoh K. Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein. Biochemistry 43, 11266–11274 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Gibbons I.R. et al. The affinity of the dynein microtubule-binding domain is modulated by the conformation of its coiled-coil stalk. The Journal of biological chemistry 280, 23960–23965 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kon T. et al. Helix sliding in the stalk coiled coil of dynein couples ATPase and microtubule binding. Nature structural & molecular biology 16, 325–333 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt H., Zalyte R., Urnavicius L. & Carter A.P. Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature 518, 435–438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts A.J. et al. AAA+ Ring and linker swing mechanism in the dynein motor. Cell 136, 485–495 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhabha G. et al. Allosteric communication in the dynein motor domain. Cell 159, 857–868 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chai P. et al. Cryo-EM Reveals the Mechanochemical Cycle of Reactive Full-length Human Dynein-1. bioRxiv, 2024.2005.2001.592044 (2024). [Google Scholar]
- 11.DeWitt M.A., Chang A.Y., Combs P.A. & Yildiz A. Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science 335, 221–225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu W. et al. Dynein achieves processive motion using both stochastic and coordinated stepping. Nature structural & molecular biology 19, 193–200 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imai H. et al. Direct observation shows superposition and large scale flexibility within cytoplasmic dynein motors moving along microtubules. Nature communications 6, 8179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lippert L.G. et al. Angular measurements of the dynein ring reveal a stepping mechanism dependent on a flexible stalk. Proceedings of the National Academy of Sciences of the United States of America 114, E4564–E4573 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Can S., Lacey S., Gur M., Carter A.P. & Yildiz A. Directionality of dynein is controlled by the angle and length of its stalk. Nature 566, 407–410 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niekamp S., Stuurman N., Zhang N. & Vale R.D. Three-color single-molecule imaging reveals conformational dynamics of dynein undergoing motility. Proceedings of the National Academy of Sciences of the United States of America 118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yildiz A. et al. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science 300, 2061–2065 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Balzarotti F. et al. Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science 355, 606–612 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Wolff J.O. et al. MINFLUX dissects the unimpeded walking of kinesin-1. Science 379, 1004–1010 (2023). [DOI] [PubMed] [Google Scholar]
- 20.Deguchi T. et al. Direct observation of motor protein stepping in living cells using MINFLUX. Science 379, 1010–1015 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elshenawy M.M. et al. Cargo adaptors regulate stepping and force generation of mammalian dynein-dynactin. Nat Chem Biol 15, 1093–1101 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schleske J.M. et al. MINFLUX Reveals Dynein Stepping in Live Neurons. bioRxiv, 2024.2005.2022.595351 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt H., Gleave E.S. & Carter A.P. Insights into dynein motor domain function from a 3.3-A crystal structure. Nature structural & molecular biology 19, 492–497, S491 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carter A.P. et al. Structure and functional role of dynein’s microtubule-binding domain. Science 322, 1691–1695 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reck-Peterson S.L. et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell 126, 335–348 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altman R.B. et al. Enhanced photostability of cyanine fluorophores across the visible spectrum. Nature methods 9, 428–429 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gennerich A., Carter A.P., Reck-Peterson S.L. & Vale R.D. Force-induced bidirectional stepping of cytoplasmic dynein. Cell 131, 952–965 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Can S., Dewitt M.A. & Yildiz A. Bidirectional helical motility of cytoplasmic dynein around microtubules. eLife 3, e03205 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferro L.S., Can S., Turner M.A., ElShenawy M.M. & Yildiz A. Kinesin and dynein use distinct mechanisms to bypass obstacles. eLife 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho C., Reck-Peterson S.L. & Vale R.D. Regulatory ATPase sites of cytoplasmic dynein affect processivity and force generation. The Journal of biological chemistry 283, 25839–25845 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeWitt M.A., Cypranowska C.A., Cleary F.B., Belyy V. & Yildiz A. The AAA3 domain of cytoplasmic dynein acts as a switch to facilitate microtubule release. Nature structural & molecular biology 22, 73–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng C.S. et al. Nanometer-Resolution Long-term Tracking of Single Cargos Reveals Dynein Motor Mechanisms. bioRxiv, 2022.2001.2005.475120 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Holzbaur E.L. & Johnson K.A. ADP release is rate limiting in steady-state turnover by the dynein adenosinetriphosphatase. Biochemistry 28, 5577–5585 (1989). [DOI] [PubMed] [Google Scholar]
- 34.Belyy V., Hendel N.L., Chien A. & Yildiz A. Cytoplasmic dynein transports cargos via load-sharing between the heads. Nature communications 5, 5544 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts A.J. et al. ATP-driven remodeling of the linker domain in the dynein motor. Structure 20, 1670–1680 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheiderer L., Wirth J.O., Tarnawski M. & Hell S.W. Dual-color MINFLUX: Kinesin-1 takes Chassé-Inchworm steps. bioRxiv, 2024.2003.2005.583551 (2024). [Google Scholar]
- 37.Zhao Y., Oten S. & Yildiz A. Nde1 promotes Lis1-mediated activation of dynein. Nature communications 14, 7221 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gwosch K.C. et al. MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells. Nature methods 17, 217–224 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Ezber Y., Belyy V., Can S. & Yildiz A. Dynein harnesses active fluctuations of microtubules for faster movement. Nature Physics 16, 312–316 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]