Abstract
Proteins which bind intrinsically disordered proteins (IDPs) and intrinsically disordered regions (IDRs) with high affinity and specificity could have considerable utility for therapeutic and diagnostic applications. However, a general methodology for targeting IDPs/IDRs has yet to be developed. Here, we show that starting only from the target sequence of the input, and freely sampling both target and binding protein conformation, RFdiffusion can generate binders to IDPs and IDRs in a wide range of conformations. We use this approach to generate binders to the IDPs Amylin, C-peptide and VP48 in a range of conformations with Kds in the 3 −100nM range. The Amylin binder inhibits amyloid fibril formation and dissociates existing fibers, and enables enrichment of amylin for mass spectrometry-based detection. For the IDRs G3bp1, common gamma chain (IL2RG) and prion, we diffused binders to beta strand conformations of the targets, obtaining 10 to 100 nM affinity. The IL2RG binder colocalizes with the receptor in cells, enabling new approaches to modulating IL2 signaling. Our approach should be widely useful for creating binders to flexible IDPs/IDRs spanning a wide range of intrinsic conformational preferences.
Keywords: Intrinsically disordered protein, intrinsically disordered region, Amyloid fibril dissociation, diagnostics, protein design, RFdiffusion, Rosetta, deep learning
IDPs and IDPRs (structured proteins with intrinsically disordered regions) are abundant in nature and carry out important biological functions without adopting a single well-defined structure, and hence are well established biomarkers in clinical care and biomedical research (Fig. 1a). Designing binders specific for disordered regions could be valuable for clinical diagnosis, therapeutic development, and scientific research1–4. Current methods largely rely on antibodies, which have limitations such as high production costs, reproducibility, and complex engineering requirements5,6; the dynamic nature of disordered proteins can also complicate the elicitation of antibodies7,8. Computational protein design has created binders of peptides in extended beta strand9,10, helical11, and polyproline II conformations12. While powerful, these methods require prespecification of the target peptide geometry, which can be limiting because the optimal conformation given both the intrinsic sequence biases of the peptide, and the opportunities for making high affinity interactions, may be quite irregular.
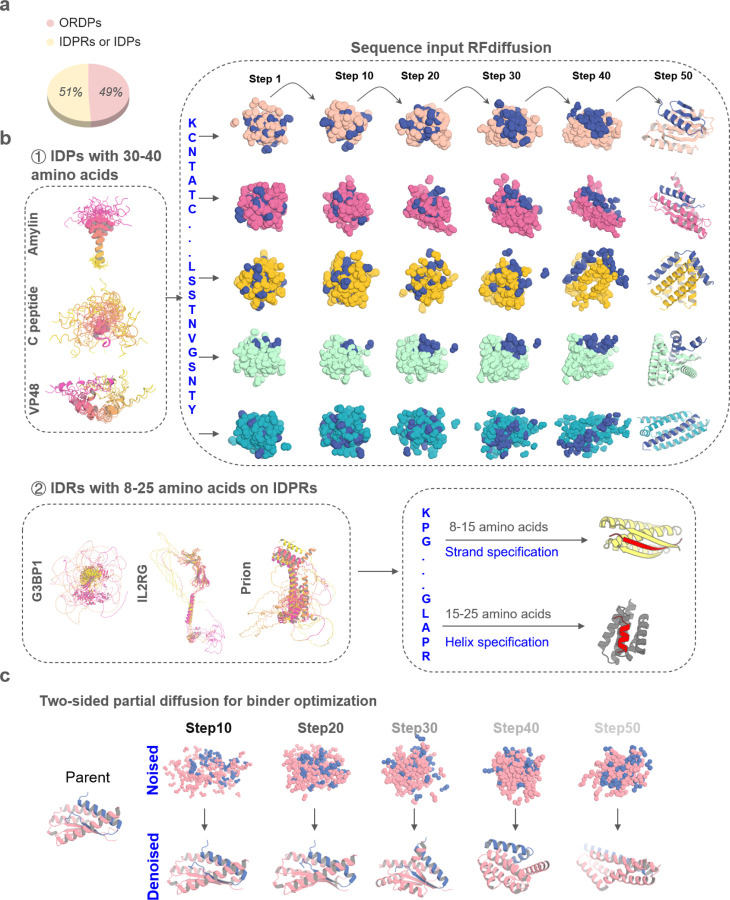
Figure. 1. Design strategies for binding conformational flexible peptides.
a, Frequency of ORDPs (ordered proteins), IDPRs /IDPs (intrinsically disordered proteins) in the human proteome41. b, ① Left, the NMR structure of Amylin (PDBID: 2KB8), C peptide (PDBID: 1T0C), the predicted structures of VP48 by five AlphaFold models22. The 5 predicted structures of VP48 are aligned together, revealing the flexibility of the intrinsically disordered protein. Right, Diffusion models for proteins are trained to recover noised protein structures and to generate new structures by reversing the corruption process through iterative denoising of initially random noise into a realistic structure. Here, A modified version of RFdiffusion was trained on two chain systems from the PDB to permit the design of protein binders to targets, for which only the sequence of the target was specified. The fine-tuned was found to generate binders to peptides in finely varying helix conformationsWith solely sequence input. ② Left, the predicted structures of G3BP1, IL2RG and prion by five AlphaFold models22. Right, A modified version of RFdiffusion was trained, allowing for specification of the secondary structure of a region, along with its sequence (See Method). When provided with the same target sequence input but different secondary structure specifications (helix or strand), the resulting conformations of the target could vary. c, Top: two sided partial diffusion. RFdiffusion is used to denoise a randomly noised starting parent design for both target and binder ; varying the extent by different noised step of initial noising (top row) enables control over the extent of introduced structural variation (bottom row; colours, new designs; grey, parent design).
We sought to develop a general approach to design high-affinity binders for intrinsically disordered proteins that starts from the target sequence alone and does not require prespecification of the target geometry (Fig. 1b ①). We reasoned that a version of RFdiffusion trained on two chain systems from the PDB, noising the structure on one and providing only the sequence on the second, could have such capability. This was used previously to generate binders to bioactive peptide hormones restricted to helical conformations11; here we begin by investigating the application of the approach to IDPs in a much broader range of conformations (the sequences of many targets are not compatible with uninterrupted helical conformations). To target shorter IDRs, we reasoned that strand pairing, as employed by Sahtoe et al using Rosetta13, coupled with RFdiffusion14 to sample the many different possible variations of strand conformation, could provide a general approach to maximizing interactions over a short region since backbone-backbone hydrogen bonds contribute to binding energy in addition to sidechain-sidechain interactions (Fig. 1b ②).
We first experimented with designing binders to the human islet amyloid polypeptide (hIAPP), also known as amylin, a 37-residue hormone co-secreted with insulin by pancreatic islet β-cells to modulate glucose levels15,16. Cysteine residues 2 and 7 form disulfide bridge which is critical for the full biological activity of amylin15. NMR studies conducted in lipid environments or under SDS micelle binding conditions have indicated helical propensity in Amylin fragments17,18; the overall structure appears to be intrinsically disordered19,20.
We employed the flexible target fine-tuned RFdiffusion to design binders against Amylin using only the Amylin sequence as input – the structure of the binding protein, the Amylin conformation, and the binding mode are entirely unspecified. Starting from the amino acid sequence of Amylin, RFdiffusion generated complexes encompassing a variety of conformations for both peptides and binders. Representative design trajectories are shown in Supplementary Video 1; starting from a random distribution of residues of both Amylin and binder; in sequential denoising steps, the Amylin adopts different conformations while the binder residue distribution shifts to surround Amylin and progressively organizes into a folded structure which cradles nearly the entire surface of the peptide (Fig. 1b①). The resulting library of backbones were sequence designed using ProteinMPNN21, and filtered using AlphaFold2 (AF2)22 for the monomer conformation and AF2 initial guess for the complex23.
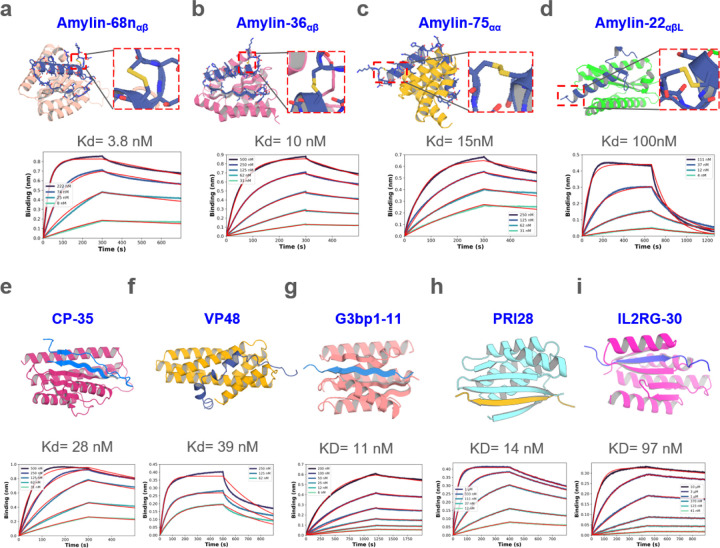
We obtained synthetic genes encoding 96 designs binding amylin in a variety of conformations, expressed the proteins in E.Coli, and purified them using immobilized metal ion affinity chromatography (IMAC). Amylin binding affinities determined using bio-layer interferometry (BLI) ranged from 100 nM to 454 nM (Supplementary Fig. 1a). Since binders to peptides in entirely helical conformations have been studied11, here we focused on other geometries. To optimize the binding affinity of initial hits to αβ, αβL, and αα conformations, we implemented a two sided partial diffusion approach (see Methods; in contrast to one sided partial diffusion which only diversifies the binder conformation and keeps the target fixed, two sided partial diffusion allows simultaneous conformation changes of both target and binder which leads to broader sampling (Fig. 1c, Supplementary Fig. 2a)). We carried out 5,000 two sided diffusion trajectories from initial designs noised over 5 to 20 steps (complete randomization corresponds to 50 steps), and found that this yielded designs with generally better metrics than one sided diffusion likely because the peptide conformation can adapt to that of binder resulting in greater shape complementarity and more extensive interactions (Supplementary Fig. 2). We obtained synthetic genes encoding the 174 resulting designs with the best metrics that span amylin conformations in the αβ, αβL, and αα conformations. 107 out of 174 refined designs bound Amylin; the highest affinity binders (Amylin-68nαβ, Amylin-36αβ, Amylin-75αα and Amylin-22αβL) which bind Amylin in different conformations, have affinities of 3.8 nM, 10 nM, 15 nM and 100 nM, respectively (Fig. 2a-d). While the Amylin adopts very different conformations in different designs, the diffusion process was able to maintain the disulfide bond, key to amylin function, in all designs15 (Fig. 2a-d). Circular dichroism studies showed that all four binders were largely helical as designed and thermostable up to 95 °C (Supplementary Fig. 1b)
Figure. 2. Design of disordered region binder.
a-d, Binder design of Amylin using sequence input diffusion. Top, from left to right, design model of Amylin and its binder Amylin-68nαβ, Amylin-36αβ, Amylin-75αα and Amylin-22αβL, respectively. The secondary structure of Amylin is indicated in the subscript of the binder’s name. For each of the designs, the Amylin disulfide bonds between 2nd Cysteine and 7th Cysteine were retained well. Bottom, from left to right, the BLI measurement indicated that the binding affinity between Amylin-68nαβ, Amylin-36αβ, Amylin-75αα, Amylin-22αβL and Amylin are 3.8, 10, 15, 100 nM respectively. e-f, Binder design of CP and VP48 using sequence input diffusion, the binder affinity of CP and VP48 are 28 and 39 nM, respectively. g-i, Binder design using strand specification. Top, from left to right, design model of G3BP1RBD, prion and IL2RG and their binders G3bp1–11, PRI28 and IL2RG-30. Bottom, the BLI measurement indicated that the binding affinity of G3bp1–11, PRI28 and IL2RG-30 binders are 11, 14 and 97 nM, respectively.
C-peptide is a 31 residue peptide secreted by islet β cells that is made from the same precursor – proinsulin – as insulin24. Measurement of plasma C-peptide levels is important for accurate classification and diagnosis of type I and type II diabetes25. We carried out sequence-input diffusion with C-peptide allowed to sample diverse conformations (Supplementary Fig. 3a). Of 96 designs tested, one in which the C-peptide forms a long strand, followed by a long dynamic loop and a small strand paired with the long strand had weak binding affinity (Supplementary Fig. 3b-c). This design had more hydrogen bonds between target and binder (13) than all but 5 of the 96 designs (Supplementary Fig. 3d), and we hypothesized that this was important for binding. To optimize the initial hit to improve binding affinity, we again used two sided partial diffusion and included the number of hydrogen bonds in filtering. Screening with BLI revealed a much higher success rate, with six designs binding C peptide with better than 100nM binding affinity; the highest affinity binder (CP-35) had a Kd of 28nM (Fig. 2e). Circular dichroism studies showed that CP-35 was largely helical, consistent with the design model, and thermostable up to 95 °C (Supplementary Fig.3e).
We next chose to target VP48 (39 amino acid), a potent activator of transcription26. In a first round of 30,000 unconstrained RFdiffusion trajectories, the most enriched conformations after filtering contained substantial secondary structure as in the above cases. To explore binding to more loop-containing conformations, we filtered these designs based on target backbone conformation and a relatively loose PAE cutoff (PAE <16); within this pool, 20 designs were manually selected and further optimized by iterative partial diffusion and backbone extension (see Methods). Of 95 designs tested, 2 showed binding at 2 μM by BLI with the highest affinity 750nM for a design with the VP48 in a conformation with three short helical fragments connected with relatively long loops. Further partial diffusion optimization yielded a design with a Kd of 39nM (Fig. 2f), that again was thermostable up to 95C (Supplementary Fig. 3f).
Targeting shorter IDRs using beta strand interactions
Consistent with the observations of Sahtoe et al using the non-deep learning Rosetta method13, we found that for targeting shorter segments, the RFdiffusion generated designs with the best metrics often made extensive beta strand interactions to targets adopting beta strand conformations. To increase the efficiency of generating such designs, we incorporated into the RFdiffusion sequence input approach the ability to define the secondary structure of the target (See Methods), to enable the specification of either the entire or a portion of the target sequence in helical, strand, or loop conformation. This is particularly important for strand conformations which can vary considerably in actual 3D coordinates; the coordinate specifying approach used by Vasquez et al11 for helical peptides would be less efficient for targeting strands as many trajectories would have to be carried out for beta strand conformations with different twists, etc. To explore the power of this approach, we used it to design binders to three IDR containing targets.
G3BP1 is a central node within the core stress granule (SG) network27 and plays a crucial role in RNA metabolism and stress response, with a disordered RNA-binding domain (abbreviated as RBD; KPGFGVGRGLAPR, 13 amino acid) mediating interactions with RNA molecules, regulating RNA metabolism, and contributing to the assembly and disassembly of stress granules. A first round of 10,000 RFdiffusion trajectories with sequence only specification of the RBD domain of G3BP1, abbreviated as G3bp1RBD yielded designs with the peptide adopting a roughly 5.7 :3.8 :0.5 ratio for helix:strand:loop, respectively (Supplementary Fig. 4a), but only the 23 strand containing designs had AF2 pae_interaction < 10 and plddt_binder > 90 (Supplementary Fig. 4a-b). Based on these observations, we specified the secondary structure as a strand and conducted 10,000 trajectories. The resulting ratio of G3bp1RBD conformation in the complex was 0.54:8.9:0.6 for helix:strand:loop, respectively, with 1,192 designs meeting the same filtering criteria, a ~51 fold improvement; in all passing designs the target had a strand conformation. We narrowed these down to 78 designs by filtering on structure prediction and Rosetta interaction metrics (monomer plddt, hbonds_count, monomer RMSD, sap_score, ddg, and contact_molecular_surface). The 78 designs were subsequently expressed in E. coli and subjected to initial screening using BLI. 5 out of 78 designs were found to bind to G3bp1RBD, with the tightest exhibiting a binding affinity at 18 nM. Through two-side partial diffusion, we further optimized 4 of the binders (G3bp1–4, G3bp1–45, G3bp1–53, and G3bp1–77; Supplementary Fig. 4c); 40 of the 95 refined designs bound G3bp1RBD, with the tightest G3bp1–11 having an affinity of 11nM.
We next sought to make binders of the prion protein which is primarily found in neuronal cells in mammals. Aggregated forms of this protein are linked to prion diseases, a group of transmissible neurodegenerative disorders28,29. The pathological hallmark of prion diseases is the conformational conversion of the native, monomeric cellular prion protein (PrPC) into a misfolded and aggregated form (PrPSc) characterized by a cross-β structure30–33. To target the amyloid core region of the prion protein, we targeted the amino acid sequence VNITIKQH (positions 180–187), specifying its secondary structure as a β-strand and conducted 20,000 trajectories. Using in silico filtering strategies similar to those employed for G3bp1RBD, we selected 48 designs for further validation via BLI. Among these, the tightest binder, PRI28, had a binding affinity of 14 nM (Fig. 2h) with high stability up to 95 °C (Supplementary Fig. 5a), higher affinity and specificity than generally achieved with our earlier Rosetta based β-strand targeting method13 (Supplementary Fig. 5b). Moreover, we found that specifying the secondary structure of the target region as a β-strand resulted in binders with higher affinity than using the target sequence information alone (14 nM from secondary structure specification (PRI28) vs 1.88 μM sequence input (PRI22), Fig. 2h and Supplementary Fig. 5c-d). after refinement through two-sided partial diffusion, the affinity of PRI22 improved to 80 nM, still weaker than PRI28 (Supplementary Fig. 5c-d).
Signal transduction via cell surface receptors is mediated by their intracellular domains, which contain long disordered regions34,35. Developing binders targeted at these domains would be broadly useful for co-localization imaging applications and for the modulation of receptor activation. The common cytokine receptor γ chain (common gamma chain, IL2RG) is a receptor subunit shared among the interleukin (IL) receptors for IL-2, IL-4, IL-7, IL-9, IL-15 and IL-21. Each receptor within the γc family uniquely contributes to the adaptive immune system, influencing the development of T, B, natural killer, and innate lymphoid cells36. To target the intracellular domain of IL2RG, we selected the amino acid sequence ERLCLVSEIP (positions 327–336) as the target region, specifying its secondary structure as a strand and conducted 40,000 trajectories. Employing in silico filtering strategies similar to those used for G3bp1, we selected 94 designs for further validation via BLI. Among these, one design had a binding affinity of 493 nM. Through two sided partial diffusion, we increased the binding affinity to 97 nM, and we named it IL2RG-30 (Fig. 2i); this optimized design again had high thermal stability (Supplementary Fig. 4e).
Structure analysis of designed complexes
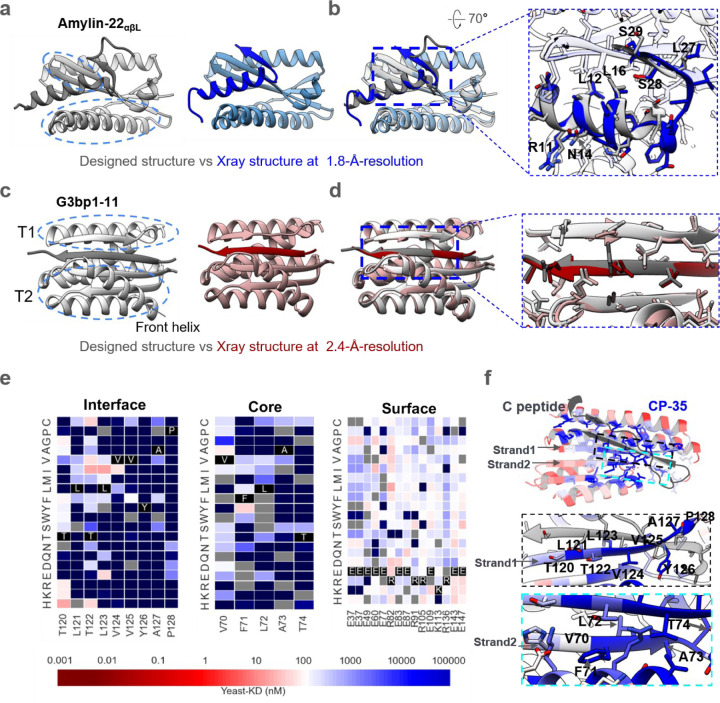
We obtained crystal structures of Amylin-22αβL and G3bp1–11 in complexes with their target at 1.8-Å-resolution and 2.4-Å-resolution, respectively. For Amylin-22αβL, the designed conformation comprises a helix, a strand, and an unstructured loop (Fig. 3a, left). The Amylin helix is embedded within a groove formed by the helix and strand segments of the binder. Adjacent to this, the Amylin strand pairs with a corresponding strand of the binder. The Amylin loop is predicted to be disordered based on the low per-residue AF2 pLDDT (predicted Local Distance Difference Test) (Fig. 3a, left, Supplementary Fig. 1c)22,37. In the crystal structure, the main helix and strand are well resolved, and closely match the computational model; the disordered loop is as anticipated not resolved (Fig. 3a-b). The Ca RMSD between the design model and the crystal structure over the backbone of the binder alone, and over the backbone of the full complex excluding the missing loop of Amylin, are 0.96 and 2.04, respectively. The backbone and sidechains at the designed binder-target interface are also in close agreement between crystal structure and design model (Fig. 3b, interface Ca and sidechain RMSD are 1.33 and 1.87, respectively).
Figure. 3. Structural characterizations.
a, Left, the designed model of Amylin-22αβL, with target and binder proteins rendered in dim gray and gray, respectively. The helical and strand segments that create the groove in the binder, docking the helical segment of Amylin, are highlighted with blue dashed ellipsoid. Right, the crystal structure of Amylin-22αβL at 1.8 Å-resolution, with target and binder proteins rendered in blue and cornflower blue, respectively. b, Left, the overlay of the design model and the crystal structure of Amylin-22αβL. Right, magnified views of the regions indicated with black dotted frames in the left panel are provided to illustrate the detailed interface view of the design and crystal structure. The binder proteins are rendered with 90% transparency to enhance the visibility of the peptide target. The key residues on the Amylin are labeled to illustrate the good alignment of the key residues between designed protein and crystal structure. c, Left, the designed model of G3bp1–11, with target and binder proteins rendered in dim gray and gray, respectively. The two α/β topologies (T1 and T2) of the binders, forming the cleft where the target strand is positioned, are highlighted with blue dashed ellipses. The front helix of T2 is denoted by a black arrow. Right, the crystal structure of G3bp1–11 at 2.4 Å-resolution, with target and binder proteins rendered in dark red and rosy brown, respectively. d, Left, the overlay of the design model and the crystal structure of G3bp1–11. Right, magnified views of the regions indicated with black dotted frames in the left panel. The front helix of T2 has been surface capped to reveal the strand pairing interface. e, Heat maps representing C peptide-binding Kd (nM) values for single mutations in the designed interface (left), core (middle) and the surface (right). Substitutions that are heavily depleted are shown in blue, and beneficial mutations are shown in red, gray color indicates the lost yeast strains. For the interface region, we highlighted and showcased strand 1 (indicated by the arrow), which serves as the primary interaction secondary structure with the C peptide. For the core region, we showcased the right segment of strand 2 (indicated by the arrow), representing a main core region that does not form interactions with the C peptide. For the surface region, we selected the most exposed surface residues that don’t form any connections with other residues (Supplementary Fig. 6c). Full SSM map over all positions for CP35 is provided in Supplementary Fig. 6b. f, Top, designed binding proteins are colored by positional Shannon entropy from site saturation mutagenesis, with blue indicating positions of low entropy (conserved) and red those of high entropy (not conserved). Bottom, zoomed-in views of central regions of the design interface and core with the C peptide.
In the G3bp1–11 design model, the peptide is in a β-strand conformation and lies within a cleft formed by two α/β structures, T1 and T2, in the designed binder, pairing with two adjacent strands (Fig. 3c). An additional helix in T2 also interacts with the target, potentially enhancing binding affinity and specificity (Fig. 3c, Supplementary Fig. 6a). The crystal structure of G3bp1–11 closely recapitulates the design model, with the peptide clamped in a β-strand conformation (Fig. 3c-d, Ca RMSD 0.8 Å for entire complex between design and crystal structure) with the interface residues nearly perfectly aligned with the design model structure (Fig. 3c-d, interface Ca and sidechain RMSD are 0.86 and 2.29, respectively).
We were unable to solve crystal structures of the CP binders, so we instead obtained a lower resolution structural footprint of the binding site by generating a site saturation mutagenesis library (SSMs) for CP-35 in which every residue was substituted with each of the 20 amino acids one at a time. Next generation sequencing before and after FACS sorting for CP binding revealed that residues at the binding interface and protein core were largely conserved (Fig. 3e-f and Supplementary Fig. 6b-c), supporting the design model.
Specificity of designed binders
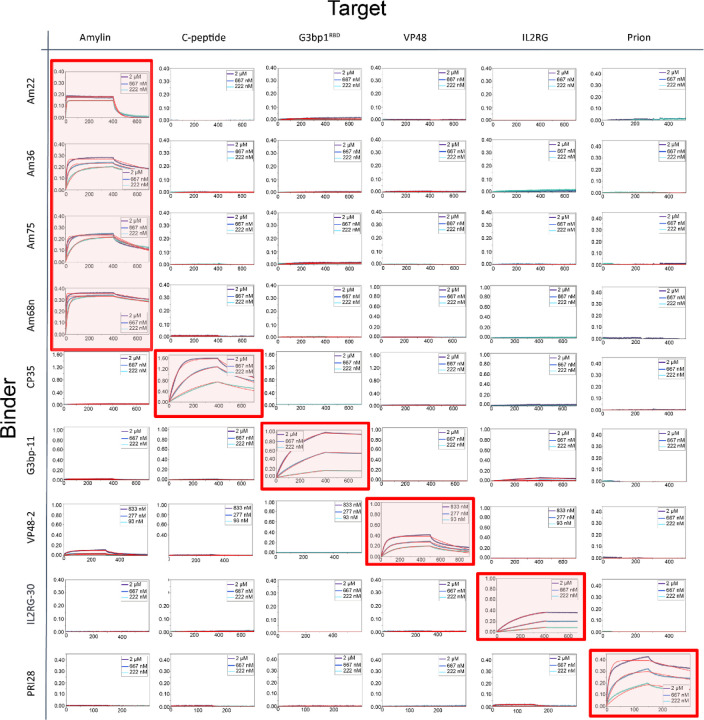
We investigated the specificity of the binders by carrying out all by all binding experiments (Fig. 4). BLI binding characterization of 9 binders against 6 targets showed that the designs had high specificity for their intended peptide targets. Very weak off target binding was observed at high concentrations in two cases: VP48 weakly bound Amylin above 800 nM, perhaps reflecting the ~50% helical content of both peptides (specificity could potentially be further improved through another round of partial diffusion, or decreasing the helical percentage through secondary structure specification) and G3BP1–11 weakly bound IL2RG at 2 uM. Overall, the much higher on-target than off-target binding suggests the binders should be broadly usable as affinity reagents.
Figure. 4. Specificity profile of designed binders in BLI.
Biotinylated peptides were immobilized onto octet streptavidin biosensors at equal densities and incubated with all binders in separate experiments at three concentrations (2, 0.667 and 0.222 μM except VP48 binder at 0.833, 0.277 and 0.093 μM). Amylin-68nαβ, Amylin-36αβ, Amylin-75αα, Amylin-22αβL are abbreviated as Am68n, Am36, Am75 and Am22, respectively. The designed on-target interactions are indicated with a light red background.
Designed binders colocalize with their targets in mammalian cells
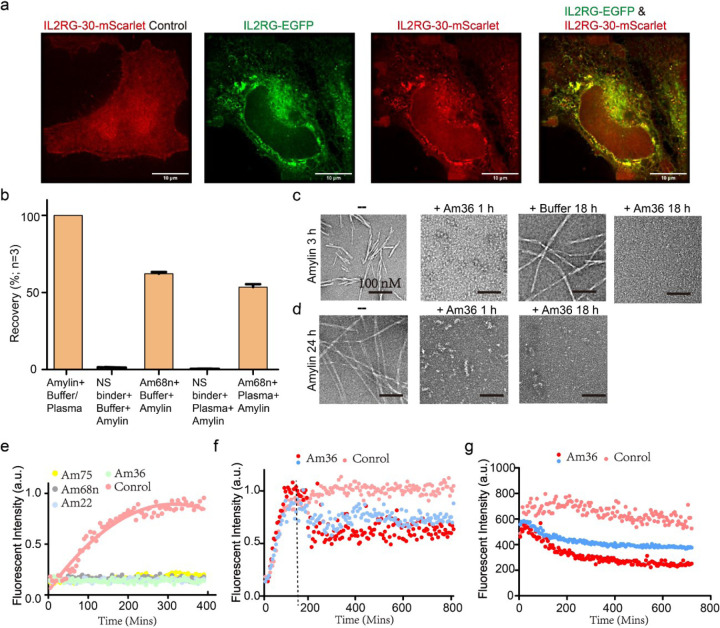
To examine whether the designs could fold properly and bind to the target proteins in mammalian cells, we knocked out the endogenous IL2RG in HeLa cells using CRISPR-Cas9, and then transfected the cells with a construct encoding IL2RG fused to EGFP. When cells were additionally transfected with mScarlet-labeled IL2RG binder IL2RG-30, colocalization of GFP and mScarlet was observed, indicating binding (Fig. 5a). In IL2RG knockout cells transfected only with IL2RG-30-mScarlet, no colocalization was observed (Fig. 5a, left), confirming that the interaction occurs through the designed interface.
Figure. 5. Applications of designed binders.
a, Colocalization of binder IL2RG-30 and target membrane receptor IL2RG in HeLa Cells. Cells with endogenous IL2RG knocked out express only the red fluorescent mScarlet-tagged binder IL2RG-30, which is uniformly distributed throughout the cell (left). In contrast, cells co-expressing green EGFP-tagged IL2RG and red mScarlet-tagged IL2RG-30 show specific colocalization of both proteins. b, The LC–MS/MS recovery percent of Amylin from PBS-0.1% CHAPS buffer and EDTA-anticoagulated plasma was compared between BSA-blocked tosyl-activated bead, an off-target binder, and amylin-targeted binders (Am68n). Percent recovery was calculated using the peak area of a sample of pure amylin peptide in elution solvent as the denominator (i.e., 100% recovery of the peptide). Error bars represent SD (n=3). c-d, Visualization of fibril dissociation by Amylin-36αβ binder using negative staining electron microscopy. panels (c) and (d) demonstrate the dissociation of existing fibrils at elongation phase (c) and mature phase (d) following the addition of Amylin-36αβ. Scale bars, 100 nM. e, Thioflavin T (ThT) assay revealed that all 4 binders could strongly inhibit fibril formation at molar ratio of binder to Amylin 1:4. f, Amylin-36αβ could dissociate fibrils at elongation phase in concentration-dependent manner. The Tht assay was performed since the Amylin monomer, Amylin-36αβ was added at 3h when Amylin fibrils were at elongation phase, marked with a dotted line. Red dot and blue dot indicate that Amylin-36αβ to Amylin is 1:4 and 1:40, respectively. g, Tht assay was performed after the mature Amylin fibrils were formed for 24 h, at the same time, Amylin-36αβ was added, the data revealed that fibril fluorescence decreased in a concentration-dependent manner. Red dot and blue dot indicate that Amylin-36αβ to Amylin is 1:4 and 1:40, respectively.
Enrichment for LC–MS/MS detection
We explored the use of amylin binder Amylin-68n as a capture agent for immunoaffinity enrichment combined with liquid chromatography–tandem mass spectrometry (LC–MS/MS), a general platform for detecting low-abundance protein biomarkers in human serum38. We prepared Amylin-binder-conjugated beads as described in the Methods. Amylin enrichment was calculated based on detection of intact, alkylated amylin in either human plasma or simplified PBS-CHAPS matrix39 (Methods). We found that the designed binder enabled capture of Amylin from buffer and human plasma supplemented with Amylin (the endogenous levels are too low for reliable detection) with recoveries of 62.2% and 53.5%, respectively (Figure. 5b).
Designs inhibit Amylin fibril formation and dissociate existing fibrils
Amylin fibril formation is implicated in type 2 diabetes, where the aggregation of amylin into insoluble fibrils contributes to islet amyloid deposition and β-cell dysfunction40. We investigated the effect of four binders—Amylin-68nαβ, Amylin-36αβ, Amylin-75αα and Amylin-22αβL—on Amylin fibril formation. At a binder to Amylin molar ratio of 1:4, with concentrations of 40 μM for Amylin and 10 μM for binders, all binders completely inhibited fibril formation (Fig. 5e). Further tests with Amylin-22αβL and Amylin-36αβ at binder to Amylin molar ratios of 1:4, 1:40, and 1:400 revealed a concentration-dependent retardation of fibril formation (Supplementary Fig. 7a). Inhibition of fibril formation was also observed by negative stain electron microscopy (NS-EM), with Amylin-22αβL and Amylin-36αβ at binder to Amylin molar ratios of 1:4. Addition of Amylin-36αβ blocked fiber formation at both 1 h and 18 h, whereas some short fibrils were observed 18 hours post-addition of Amylin-22αβL (Supplementary Fig. 7b-c).
We next investigated whether the amylin binders were able to disaggregate pre-formed amylin fibrils. We generated short Amylin fibrils by incubating the peptide at 40 μM for 3 hours at 37 °C, to reach the elongation phase, and then incubated with 10 μM Amylin-36αβ. NS-EM revealed no fibrillar structures after treatment with Amylin-36αβ at both 1 h and 18 h time points (Fig. 5c). Thioflavin T (ThT) assays with Amylin-36αβ added at the 3-hour Amylin fiber stage also showed fiber disassembly in a design concentration-dependent manner (Fig. 5f).
To test whether Amylin-36αβ could dissociate mature fibrils that had formed over 24 hours at 10 μM, we incubated them with 10 μM of the binder. Small oligomers were still observed at 1 hour, but were completely dissociated by 18 hours (Fig. 5d). Fibril ThT fluorescence again decreased in a designed binder concentration-dependent manner (Fig. 5g).
Discussion
Our results demonstrate the utility of RFdiffusion in designing binders for IDPs ranging from 30–40 amino acids in length in diverse conformations, expanding its applicability beyond helical peptides. The ability to target IDPs without specifying the target structure is important as such proteins have no single defined conformation. During the design process, the target protein samples a wide range of possible conformations as the designed binding protein diffuses around it; the co-folding of design and target effectively enables the selection of conformations particularly suitable for binding. The versatility of our approach is highlighted by the design binders for Amylin in diverse conformations while consistently forming the Amylin peptide disulfide.
For shorter peptides which can adopt beta strand like conformations, we show the introduction of a secondary structure type specification feature within the RFdiffusion model enables targeting of peptides in the beta strand conformation. The generated structures resemble previous strand targeting designs generated using Rosetta, but exhibit higher specificity and binding affinity.
The binders and approaches described here could be broadly useful given the current difficulty in targeting IDPs and IDRs, and the important roles these play in both normal physiology and disease. For example, the Amylin binder both inhibits the formation of Amylin fibers and dissociating pre-existing fibers, which could have therapeutic utility. Additionally, it facilitates the enrichment and detection of Amylin using mass spectrometry. The designed binders bind their targets in cells, as illustrated by the colocalization of PRI28 with the intracellular tail of the IL2 receptor gamma subunit, opening up new ways of modulating cytokine signaling in feedback loops for adoptive cell therapies and other applications.
Supplementary Material
Acknowledgements:
This research used resources (FMX/AMX) of the National Synchrotron Light Source II, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. The Center for BioMolecular Structure (CBMS) is primarily supported by the National Institutes of Health, National Institute of General Medical Sciences (NIGMS) through a Center Core P30 Grant (P30GM133893), and by the DOE Office of Biological and Environmental Research (KP1607011). Amylin fibril inhibition and dissociation work is supported by the Chinese Academy of Sciences (CAS) (XDB37010100), and the basic Research Program Based on Major Scientific Infrastructures, CAS JZHKYPT-2021-05). We appreciate the help provided by David Juergens in the training of the RFdiffusion model with Joseph L. Watson.
Footnotes
Code availability:
Code explanation and examples of binder design using RFdiffusion can be found at <github_link>. Partial-diffusion code explanation and examples can be found at <github_link>.
Data Availability:
Crystal structures of Amylin-22αβL and G3bp1–11 have been deposited in Protein Data Bank, with the accession IDs 9CC5 and 9CC6, respectively.
References
- 1.Zhang G. et al. Islet amyloid polypeptide cross-seeds tau and drives the neurofibrillary pathology in Alzheimer’s disease. (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dyson H. J. & Wright P. E. Intrinsically unstructured proteins and their functions. Nature Reviews Molecular Cell Biology 6, 197–208, doi: 10.1038/nrm1589 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett 579, 3346–3354, doi: 10.1016/j.febslet.2005.03.072 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Uversky V. N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol 44, 18–30, doi: 10.1016/j.sbi.2016.10.015 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Bradbury A. & Plückthun A. Reproducibility: Standardize antibodies used in research. Nature 518, 27–29, doi: 10.1038/518027a (2015). [DOI] [PubMed] [Google Scholar]
- 6.Baker M. Reproducibility crisis: Blame it on the antibodies. Nature 521, 274–276, doi: 10.1038/521274a (2015). [DOI] [PubMed] [Google Scholar]
- 7.Olejniczak E. T. et al. Rapid Determination of Antigenic Epitopes in Human NGAL Using NMR. Biopolymers 93, 657–667, doi: 10.1002/bip.21417 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Fernández-Quintero M. L. et al. Conformational selection of allergen-antibody complexes-surface plasticity of paratopes and epitopes. Protein Eng Des Sel 32, 513–523, doi: 10.1093/protein/gzaa014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahtoe D. D. et al. Transferrin receptor targeting by de novo sheet extension. Proc Natl Acad Sci U S A 118, doi: 10.1073/pnas.2021569118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danny D. S. et al. Design of amyloidogenic peptide traps. bioRxiv, 2023.2001.2013.523785, doi: 10.1101/2023.01.13.523785 (2023). [DOI] [Google Scholar]
- 11.Vázquez Torres S. et al. De novo design of high-affinity binders of bioactive helical peptides. Nature 626, 435–442, doi: 10.1038/s41586-023-06953-1 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu K. et al. De novo design of modular peptide-binding proteins by superhelical matching. Nature 616, 581–589, doi: 10.1038/s41586-023-05909-9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sahtoe D. D. et al. Design of amyloidogenic peptide traps. Nature Chemical Biology, doi: 10.1038/s41589-024-01578-5 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watson J. L. et al. De novo design of protein structure and function with RFdiffusion. Nature 620, 1089–1100, doi: 10.1038/s41586-023-06415-8 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts A. N. et al. Molecular and Functional-Characterization of Amylin, a Peptide Associated with Type-2 Diabetes-Mellitus. P Natl Acad Sci USA 86, 9662–9666, doi:DOI 10.1073/pnas.86.24.9662 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westermark P. Amyloid in the islets of Langerhans: Thoughts and some historical aspects. Upsala J Med Sci 116, 81–89, doi: 10.3109/03009734.2011.573884 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLisle C. F., Malooley A. L., Banerjee I. & Lorieau J. L. Pro-islet amyloid polypeptide in micelles contains a helical prohormone segment. The FEBS Journal 287, 4440–4457, doi: 10.1111/febs.15253 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Patil S. M., Xu S., Sheftic S. R. & Alexandrescu A. T. Dynamic α-Helix Structure of Micelle-bound Human Amylin*. Journal of Biological Chemistry 284, 11982–11991, doi: 10.1074/jbc.M809085200 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunker A. K. et al. Intrinsically disordered protein. Journal of Molecular Graphics and Modelling 19, 26–59, doi: 10.1016/S1093-3263(00)00138-8 (2001). [DOI] [PubMed] [Google Scholar]
- 20.He J., Dai J., Li J., Peng X. & Niemi A. J. Aspects of structural landscape of human islet amyloid polypeptide. The Journal of Chemical Physics 142, 045102, doi: 10.1063/1.4905586 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Dauparas J. et al. Robust deep learning-based protein sequence design using ProteinMPNN. Science 378, 49–55, doi: 10.1126/science.add2187 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jumper J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583-+, doi: 10.1038/s41586-021-03819-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett N. R. et al. Improving de novo protein binder design with deep learning. Nat Commun 14, doi:ARTN 2625 10.1038/s41467-023-38328-5 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei Y., Quan L., Zhou T., Du G. & Jiang S. The relationship between different C-peptide level and insulin dose of insulin pump. Nutrition & Diabetes 11, 7, doi: 10.1038/s41387-020-00148-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iqbal S., Jayyab A. A., Alrashdi A. M. & Reverté-Villarroya S. The Predictive Ability of C-Peptide in Distinguishing Type 1 Diabetes From Type 2 Diabetes: A Systematic Review and Meta-Analysis. (2023). [DOI] [PubMed] [Google Scholar]
- 26.Cheng A. W. et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res 23, 1163–1171, doi: 10.1038/cr.2013.122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang P. et al. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 181, 325–345.e328, doi: 10.1016/j.cell.2020.03.046 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aguzzi A., Sigurdson C. & Heikenwaelder M. Molecular mechanisms of prion pathogenesis. Annu Rev Pathol 3, 11–40, doi: 10.1146/annurev.pathmechdis.3.121806.154326 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Scheckel C. & Aguzzi A. Prions, prionoids and protein misfolding disorders. Nat Rev Genet 19, 405–418, doi: 10.1038/s41576-018-0011-4 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Prusiner S. B. Novel Proteinaceous Infectious Particles Cause Scrapie. Science 216, 136–144, doi:DOI 10.1126/science.6801762 (1982). [DOI] [PubMed] [Google Scholar]
- 31.Prusiner S. B., Groth D. F., Bolton D. C., Kent S. B. & Hood L. E. Purification and Structural Studies of a Major Scrapie Prion Protein. Cell 38, 127–134, doi:Doi 10.1016/0092-8674(84)90533-6 (1984). [DOI] [PubMed] [Google Scholar]
- 32.Bolton D. C., Mckinley M. P. & Prusiner S. B. Identification of a Protein That Purifies with the Scrapie Prion. Science 218, 1309–1311, doi:DOI 10.1126/science.6815801 (1982). [DOI] [PubMed] [Google Scholar]
- 33.Thody S. A., Mathew M. K. & Udgaonkar J. B. Mechanism of aggregation and membrane interactions of mammalian prion protein. Bba-Biomembranes 1860, 1927–1935, doi: 10.1016/j.bbamem.2018.02.031 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Minezaki Y., Homma K. & Nishikawa K. Intrinsically disordered regions of human plasma membrane proteins preferentially occur in the cytoplasmic segment. J Mol Biol 368, 902–913, doi: 10.1016/j.jmb.2007.02.033 (2007). [DOI] [PubMed] [Google Scholar]
- 35.De Biasio A. et al. Prevalence of intrinsic disorder in the intracellular region of human single-pass type I proteins: The case of the Notch ligand Delta-4. J Proteome Res 7, 2496–2506, doi: 10.1021/pr800063u (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leonard W. J., Lin J. X. & O’Shea J. J. The gamma(c) Family of Cytokines: Basic Biology to Therapeutic Ramifications. Immunity 50, 832–850, doi: 10.1016/j.immuni.2019.03.028 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Guo H.-B. et al. AlphaFold2 models indicate that protein sequence determines both structure and dynamics. Scientific Reports 12, 10696, doi: 10.1038/s41598-022-14382-9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi J. et al. A distributable LC-MS/MS method for the measurement of serum thyroglobulin. J Mass Spectrom Adv Clin Lab 26, 28–33, doi: 10.1016/j.jmsacl.2022.09.005 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoofnagle A. N., Becker J. O., Wener M. H. & Heinecke J. W. Quantification of thyroglobulin, a low-abundance serum protein, by immunoaffinity peptide enrichment and tandem mass spectrometry. Clin Chem 54, 1796–1804, doi: 10.1373/clinchem.2008.109652 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hull R. L., Westermark G. T., Westermark P. & Kahn S. E. Islet amyloid: a critical entity in the pathogenesis of type 2 diabetes. J Clin Endocrinol Metab 89, 3629–3643, doi: 10.1210/jc.2004-0405 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Deiana A., Forcelloni S., Porrello A. & Giansanti A. Intrinsically disordered proteins and structured proteins with intrinsically disordered regions have different functional roles in the cell. PLoS One 14, e0217889, doi: 10.1371/journal.pone.0217889 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Senior A. W. et al. Improved protein structure prediction using potentials from deep learning. Nature 577, 706-+, doi: 10.1038/s41586-019-1923-7 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Simons K. T., Kooperberg C., Huang E. & Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol Biol 268, 209–225, doi:DOI 10.1006/jmbi.1997.0959 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Bennett N. R. et al. Improving de novo protein binder design with deep learning. Nat Commun 14, 2625, doi: 10.1038/s41467-023-38328-5 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D 66, 133–144, doi: 10.1107/S0907444909047374 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winn M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67, 235–242, doi: 10.1107/S0907444910045749 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCoy A. J. et al. ıt Phaser crystallographic software. Journal of Applied Crystallography 40, 658–674, doi: 10.1107/S0021889807021206 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221, doi: 10.1107/S0907444909052925 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P. & Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132, doi: 10.1107/S0907444904019158 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Williams C. J. et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci 27, 293–315, doi: 10.1002/pro.3330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystal structures of Amylin-22αβL and G3bp1–11 have been deposited in Protein Data Bank, with the accession IDs 9CC5 and 9CC6, respectively.