Abstract
Influenza A viruses possess two glycoprotein spikes on the virion surface: hemagglutinin (HA), which binds to oligosaccharides containing terminal sialic acid, and neuraminidase (NA), which removes terminal sialic acid from oligosaccharides. Hence, the interplay between these receptor-binding and receptor-destroying functions assumes major importance in viral replication. In contrast to the well-characterized role of HA in host range restriction of influenza viruses, there is only limited information on the role of NA substrate specificity in viral replication among different animal species. We therefore investigated the substrate specificities of NA for linkages between N-acetyl sialic acid and galactose (NeuAcα2-3Gal and NeuAcα2-6Gal) and for different molecular species of sialic acids (N-acetyl and N-glycolyl sialic acids) in influenza A viruses isolated from human, avian, and pig hosts. Substrate specificity assays showed that all viruses had similar specificities for NeuAcα2-3Gal, while the activities for NeuAcα2-6Gal ranged from marginal, as represented by avian and early N2 human viruses, to high (although only one-third the activity for NeuAcα2-3Gal), as represented by swine and more recent N2 human viruses. Using site-specific mutagenesis, we identified in the earliest human virus with a detectable increase in NeuAcα2-6Gal specificity a change at position 275 (from isoleucine to valine) that enhanced the specificity for this substrate. Valine at position 275 was maintained in all later human viruses as well as swine viruses. A similar examination of N-glycolylneuraminic acid (NeuGc) specificity showed that avian viruses and most human viruses had low to moderate activity for this substrate, with the exception of most human viruses isolated between 1967 and 1969, whose NeuGc specificity was as high as that of swine viruses. The amino acid at position 431 was found to determine the level of NeuGc specificity of NA: lysine conferred high NeuGc specificity, while proline, glutamine, and glutamic acid were associated with lower NeuGc specificity. Both residues 275 and 431 lie close to the enzymatic active site but are not directly involved in the reaction mechanism. This finding suggests that the adaptation of NA to different substrates occurs by a mechanism of amino acid substitutions that subtly alter the conformation of NA in and around the active site to facilitate the binding of different species of sialic acid.
Influenza virions contain two glycoproteins on their surface, hemagglutinin (HA) and neuraminidase (NA), which recognize sialic acid on host cell glycoconjugates. HA binds to cell surface sialyloligosaccharides and then mediates the entry of the virus into the cell (23), while NA prevents the aggregation of progeny virions by removing sialic acid on the oligosaccharides of newly synthesized HA and NA polypeptides (14). It also facilitates the elution of progeny virions from infected cells by removing sialic acid from host cell glycoconjugates.
The HA receptor specificity of influenza viruses differs according to the host species of origin (3, 15). That is, HA preferentially recognizes the sialic acid-galactose linkages expressed on cells of the host from which the virus was isolated. For example, duck intestinal epithelial cells express primarily N-acetylneuraminic acid (NeuAc) bound to galactose through an α2,3 linkage (NeuAcα2-3Gal) (7), which is preferentially recognized by duck virus HA (15). Likewise, human virus HA preferentially binds NeuAcα2-6Gal (3, 5, 15, 16), the primary linkage of sialic acid expressed on human tracheal epithelial cells (4). Similarly, the NA specificity for NeuAcα2-3Gal and NeuAcα2-6Gal depends on the viral isolate examined (1, 24). The specificity of human virus N2 NA has changed over the years since its introduction from an avian virus (1). The earliest human N2 viruses, isolated in 1957, showed enzymatic activity against only NeuAcα2-3Gal, while N2 viruses isolated in 1967 and 1968 showed limited activity against NeuAcα2-6Gal and still retained primary activity against NeuAcα2-3Gal. By 1972, human virus N2 NA had developed similar specificities for both NeuAcα2-6Gal and NeuAcα2-3Gal. From this finding, it was concluded that the N2 NA of human viruses had acquired the ability to recognize the same linkage of sialic acid that is preferentially recognized by the HA of the viruses, probably to facilitate the efficient release of progeny virus from cells and to prevent self-aggregation of virions.
Influenza viruses also infect animals (e.g., pigs) that express high levels of N-glycolylneuraminic acid (NeuGc) bound to galactose in cellular glycoconjugates. NeuGc has not been detected on human tracheal epithelial cells but is expressed at high levels on both swine and equine tracheal epithelial cells (19, 19a), suggesting that it may play a role in influenza virus infection of these hosts. This notion is supported by the observation that swine influenza viruses efficiently bind glycoconjugates that contain NeuGc, while human viruses do not (19).
In comparison with the HA receptor specificity, information on the NA substrate specificity of influenza A viruses is limited in terms of correlations between the host animals from which the viruses were isolated and the structural basis for substrate recognition. To address these issues, we first compared the NA specificities for linkages between sialic acid and galactose (NeuAcα2-3Gal versus NeuAcα2-6Gal) and for different molecular species of sialic acids (NeuAc versus NeuGc) among avian, swine, and human viruses. We then determined the amino acid residues contributing to these specificities by using chimeric NAs and site-specific mutagenesis.
MATERIALS AND METHODS
Viruses and cells.
Influenza A viruses used in this study were obtained from the repository at St. Jude Children’s Research Hospital and the Istituto Superiore di Sanita. Abbreviations (used on the figures) and subtypes of the viruses used are enclosed in parentheses: A/duck/Hong Kong/7/75 (dk/HK/7/75) (H3N2), A/mallard/New York/6750/78 (mal/NY/6750/78) (H2N2), A/gull/Delaware/2838/87 (gull/Del/2838/87) (H3N2), A/swine/Italy/1850/77 (sw/It/1850/77) (H3N2), A/swine/Belgium/1/79 (sw/Bel/1/79) (H3N2), A/swine/Finestere/55/80 (sw/Fin/55/80) (H3N2), A/swine/Netherlands/3/80 (sw/Ned/3/80) (H3N2), A/swine/Netherlands/12/85 (sw/Ned/12/85) (H3N2), A/swine/Belgium/1/83 (sw/Bel/1/83) (H3N2), A/swine/Ukkel/1/84 (sw/Uk/1/84) (H3N2), A/swine/France/5027/87 (sw/Fr/5027/87) (H3N2), A/swine/Italy/635/87 (sw/It/635/87) (H3N2), A/Singapore/1/57 (Sing/1/57) (H2N2), A/Ann Arbor/6/60 (AA/6/60) (H2N2), A/England/12/62 (Eng/12/62) (H2N2), A/Tokyo/3/67 (Tokyo/3/67) (H2N2), A/Berkeley/1/68 (Berk/1/68) (H3N2), A/Aichi/2/68 (Aichi/2/68) (H3N2), A/Hong Kong/1/68 (HK/1/68) (H3N2), A/Hong Kong/8/68 (HK/8/68) (H3N2), A/Korea/426/68 (Korea/426/68) (H2N2), A/Memphis/1/68 (Mem/1/68) (H3N2), A/Netherlands/20/69 (Ned/20/69) (H3N2), A/Netherlands/84/68 (Ned/84/68) (H3N2), A/Udorn/307/72 (Udorn/307/72) (H3N2), A/Bangkok/1/79 (Bang/1/79) (H3N2), and A/Los Angeles/2/87 (LA/2/87) (H3N2). 293T cells were cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal calf serum.
NA substrate specificity assays.
For the NA substrate specificity assays, a concentrated stock of each virus was prepared. Each virus was grown in 60 11-day-old embryonated chicken eggs. The allantoic fluid was clarified by centrifugation at 5,000 × g for 15 min at 4°C. Each virus was pelleted by centrifugation at 65,000 × g for 1 h at 4°C and resuspended in 1 ml of phosphate-buffered saline. Portions of the solution were stored as aliquots at −20°C.
For the substrate specificity assays, the NA enzymatic activity of each virus preparation was determined. Dilutions of the concentrated viruses were prepared in calcium-saline (CaS) buffer (6.8 mM CaCl2, 154 mM NaCl, 19.5 mM H3BO3, 0.136 mM Na2B4O7). Five microliters of each dilution was incubated in a 96-well plate with 5 μl of 0.2 mM 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (Sigma) in 0.2 mM potassium phosphate (pH 5.9) for 30 min at 37°C. The reaction was stopped by adding 200 μl of 0.1 M glycine–25% ethanol (pH 10.7) per well. The fluorescence of released 4-methylumbelliferone was determined by use of a fluorometer (Labsystems Fluoroskan II) with excitation at 355 nm and emission at 460 nm. One unit of NA enzymatic activity was defined as 1 μmol of released NeuAc, equivalent to the amount of released 4-methylumbelliferone per min at 37°C.
Two types of substrates were used for the NA substrate specificity assays: (i) sialyllactoses containing NeuAc acid bound to the galactose group of lactose through either an α2-3 (NeuAcα2-3Gal) or an α2-6 (NeuAcα2-6Gal) ketosidic linkage (Sigma) and (ii) GM3 gangliosides with either the NeuAc or the NeuGc species of sialic acid. NeuAc-GM3 was purified from human liver (18), and NeuGc-GM3 was purified from equine erythrocytes (6). Reactions were performed by use of 50 μl of CaS buffer with 0.1 mM substrate and 10 mU of viral NA per ml. The NeuAcα2-6Gal specificities of several human viruses were also determined with 100 mU of NA per ml. Reactions with GM3 gangliosides were performed with 0.1% sodium deoxycholate (SDC), required to solubilize the gangliosides. All reactions were performed in duplicate at 37°C and various time points, typically between 15 and 60 min, and were stopped by heating at 100°C for 15 min. The amount of liberated sialic acid was determined by the periodate-thiobarbituric acid assay (22). The colored chromophore was extracted into 750 μl of n-butanol–5% HCl (vol/vol), and the absorbance was measured at 549 nm. Although these assays and subsequent assays described below were done at multiple times, we typically selected results obtained at either 15 or 30 min, depending on the interval that fell within the linear response range of the sialidase activity-versus-time plot. The data reported are representative of two or three separate assays.
Cloning of NA genes.
Full-length cDNAs of the NA genes from A/Singapore/1/57 (H2N2), A/England/12/62 (H2N2), and A/Tokyo/3/67 (H2N2) were amplified by PCR with oligonucleotides 5′-TCTCTTCGAGCAAAAGCAGGAGTGAAAATG-3′ and 5′-ATTAACCCTCACTAAAAGTAGAAACAAGGAGTTTTTTTC-3′ and Pfu DNA polymerase (Stratagene). The full-length PCR products were cloned into the pCRII vector of the TA cloning kit (Invitrogen) according to the provided instructions. The sequence of each gene was determined with N2-specific primers (sequences available upon request) and an automated sequencer (Applied Biosystems Inc., Foster City, Calif.). The NA genes of A/Singapore/1/57, A/England/12/62, and A/Tokyo/3/67 were then subcloned into the EcoRI restriction enzyme site of the plasmid expression vector pCAGGS/MCS (8, 13) to generate pCAT3DKSING57NASAP, pCAT3DKENG62NASAP, and pCAT3TOKYO67NASAP, respectively. Each gene was similarly subcloned into the EcoRI site of pUC19 to generate pUCT3DKSING57NASAP, pUCT3DKENG62NASAP, and pUCT3TOKYO67NASAP, respectively. The sequence of each cloned NA gene was confirmed to be identical to that of the wild-type gene by sequence analysis and thus did not contain errors introduced by PCR.
Generation of chimeric constructs.
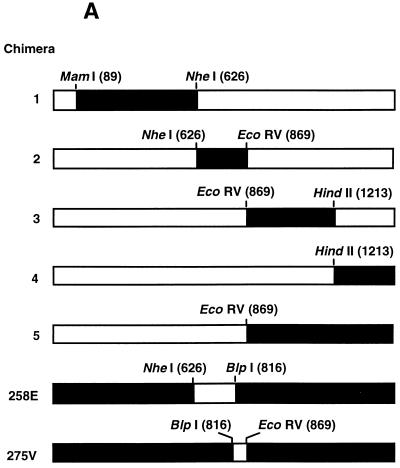
Using shared unique restriction enzyme sites in pUCT3DKSING57NASAP and pUCT3DKENG62NASAP, we generated five chimeric constructs in which portions of the A/England/12/62 NA gene were replaced with the corresponding region of the A/Singapore/1/57 NA gene. Sites for MamI, NheI, EcoRV, and HindII at N2 gene nucleotide positions 98, 626, 869, and 1213, respectively, and Asp718 in the pUC19 multiple cloning site (MCS) were used to generate the chimeras (see Fig. 2A). Two additional chimeras were generated to investigate individual amino acid differences between the NAs of A/Singapore/1/57 and A/England/12/62. To replace Lys258 of A/Singapore/1/57 with Glu258 of A/England/12/62, we substituted the pUTCDKENG62NASAP sequence for the NheI-BlpI fragment of pUCT3DKSING57NASAP, resulting in the pUCT3DKSING57-258E construct. Similarly, Ile275 of A/Singapore/1/57 was replaced with Val275 of A/England/12/62 by replacing the BlpI-EcoRV fragment of pUCT3DKSING57NASAP with pUTCDKENG62NASAP to generate pUCT3DKSING57-275V. Each chimeric NA was then subcloned into the EcoRI site of pCAGGS/MCS to generate pCAT3DKSING57-258E and pCAT3DKSING57-275V, respectively.
FIG. 2.
Determination of amino acid residues involved in NeuAcα2-6Gal specificity by use of A/Singapore/1/57-A/England/12/62 chimeric constructs. (A) In these constructs, the sequences of A/England/12/62 N2 NA (□; pCAT3DKENG62NASAP) were replaced with the sequences of A/Singapore/1/57 N2 NA (■; pCAT3DKSING57NASAP) by use of shared restriction sites to generate chimeras 1 to 5. Amino acid mutations K258→E and I275→V in A/Singapore/1/57 N2 NA were generated in constructs pCAT3DKSING-258E (258E) and pCAT3DKSING-275V (275V) by substitution of the NheI-BlpI and BlpI-EcoRV fragments of pCAT3DKENG62NASAP into the equivalent regions of pCAT3DKSING57NASAP, respectively. (B and C) 293T cells were transfected with 2 μg of each plasmid expression vector containing parental or chimeric NA per well (six-well plate). Cells expressing parental NAs and chimeric constructs 1 to 5 (B) or mutant constructs pCAT3DKSING-258E and pCAT3DKSING-275V (C) were harvested at 40 h posttransfection. Cell-expressed NA (0.5 mU) was assayed for the amount of sialic acid released from 0.1 mM sialyllactose containing the NeuAcα2-6Gal linkage in 30 min at 37°C. Released sialic acid was determined as described in the legend to Fig. 1.
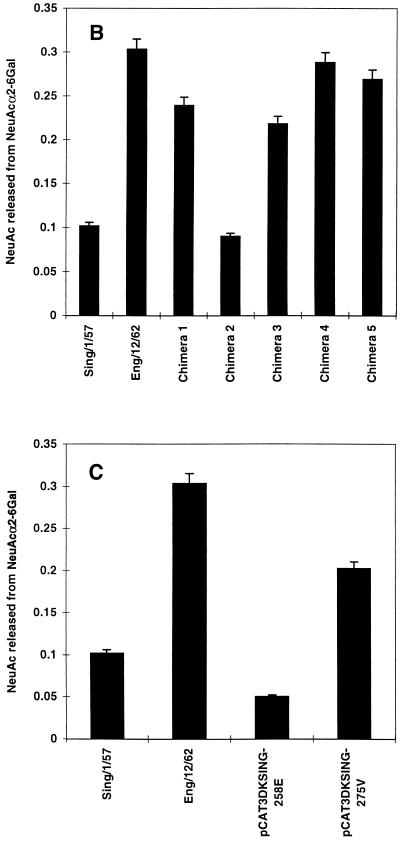
Additional chimeric constructs (designated 6 to 10) in which portions of the A/England/12/62 N2 NA gene were replaced with the corresponding region of the A/Tokyo/3/67 N2 NA gene exactly as described above were generated (see Fig. 4A).
FIG. 4.
Determination of amino acid residues contributing to NeuGc specificity. (A) Chimeras 6 to 10, in which the sequences of A/England/12/62 N2 NA (□; pCAT3DKENG62NASAP) were replaced with the equivalent sequences of A/Tokyo/3/67 N2 NA (■; pCAT3TOKYO67NASAP), were generated as described in the legend to Fig. 2 for chimeras 1 to 5. Parental NAs and chimeras 6 to 10 (B) or A/England/12/62 mutant constructs containing amino acid substitutions N401→D and W403→R (pCAT3DKENG-401D,403R) and Q431→K (pCAT3DKENG-431K) (C) were expressed as described in the legend to Fig. 2. Cell-expressed NA (0.5 mU) was incubated with 0.1 mM GM3 ganglioside substrate containing NeuAc or NeuGc in the presence of 0.1% SDC for 30 min at 37°C. The amount of sialic acid released from NeuGcα2-3Gal was determined as described in the legend to Fig. 1.
Site-directed mutagenesis of specific NA residues.
To introduce three specific point mutations, representing nucleotides in A/Tokyo/3/67, into A/England/12/62, we synthesized two mutagenic primers for PCR amplifications. The first mutagenic oligonucleotide, 5′-GTCATAGTTGACAGCGATAATCGGTCAGG-3′, contains the HindIII site of the NA gene at position 1213. The reverse primer, 5′-ATGACCATGATTACGCCAAGCTTGC-3′, binds nucleotides 445 to 469 of the pUC19 MCS. PCR was done with Pfu DNA polymerase for 30 cycles at an annealing temperature of 60°C and with pUCT3DKENG62NASAP as a template. A PCR fragment containing nucleotide mutations G1221→A and T1227→C was generated, resulting in amino acid mutations Asn401→Asp and Trp403→Arg, respectively. This fragment was subcloned into the HindII site (nucleotide position 1213 of the NA gene) and SalI (MCS) site of pUCT3DKENGNASAP to generate pUCT3DKENG-401D,403R. Using the second mutagenic primer, 5′-GTCATAGTTGACAGTAATAATTGGTCAGG-3′, and pUCT3TOKYO67NASAP as a template, we generated a PCR fragment containing nucleotide mutations G1221→A and C1227→T, resulting in amino acid mutations Asp401→Asn and Arg403→Trp, respectively. This fragment was subcloned into pUCT3DKENG62NASAP and had the net effect of introducing Gln431→Lys into the A/England/12/62 NA protein, generating the mutant NA construct pUCT3DKENG-431K. pUCT3DKENG-401D,403R and pUCT3DKENG-431K were each subcloned into the EcoRI site of pCAGGS/MCS to generate pCAT3DKENG-401D,403R and pCAT3DKENG-431K, respectively. Introduction of only the desired mutation in each final construct was confirmed by sequence analysis of the entire region generated by PCR.
Substrate specificity of cell-expressed NA.
The pCAGGS/MCS expression plasmid construct for each NA gene or chimeric construct was expressed in 293T cells. Cells at 70 to 80% confluency were transfected with 2 μg of each plasmid DNA per well of a six-well tissue culture plate by use of Lipofectamine (Life Technologies Inc.). After incubation for 40 h at 37°C, the cells were washed from the plate, washed once with phosphate-buffered saline, and then resuspended in CaS buffer. Dilutions of the cell suspensions were prepared in CaS buffer and assayed for NA enzymatic activity with the substrate 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid as described above. On the basis of the calculated activity of each expressed NA, 0.5 mU of NA activity per reaction was used to determine the substrate specificity of each expressed NA. The required volume of cell suspension was divided into aliquots, which were placed in 1.5-ml tubes. The cells were then pelleted for 30 s at 16,000 × g. After the supernatant fluids were aspirated, the cells were resuspended at 4°C in 50 μl of CaS buffer containing either 0.1 mM sialyllactose or GM3 ganglioside and 0.1% SDC. The remainder of the assay was performed as described for the concentrated viruses. The reported data represent duplicate assays and duplicate reactions within each assay.
NA amino acid sequence analysis.
The amino acid sequences of avian, swine, and human N2 NAs were compared at positions 275 and 431. For sequences available from GenBank, the accession numbers are reported. Additional human sequences were obtained from a published sequence analysis (11). Sequences not available from GenBank or literature sources were determined by automated sequencing. The identity of amino acid 275, inferred from the nucleotide sequence, was determined with oligonucleotide 5′-GTAATGACTGATGGAAGTGC-3′, which binds nucleotides 737 to 756 (human virus N2 NA sequence numbering). Similarly, the identity of amino acid 431 was determined with oligonucleotide 5′-GTAATGACTGATGGAAGTGC-3′, which binds nucleotides 1148 to 1164.
RESULTS
Specificities of avian, human, and swine virus N2 NAs for NeuAcα2-3Gal and NeuAcα2-6Gal.
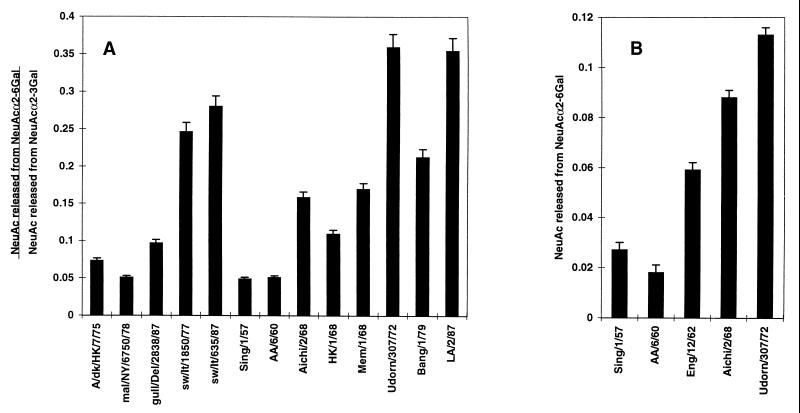
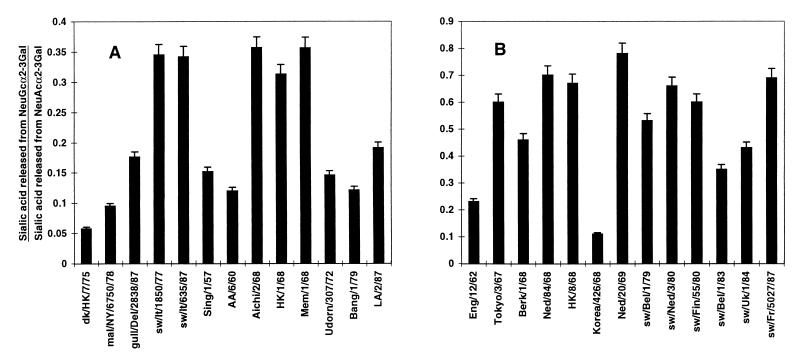
A drift in the substrate specificity of human virus N2 NAs, due to continuous replication of N2 viruses in humans since the introduction of the N2 NA from an avian virus in 1957, has led to a higher specificity for NeuAcα2-6Gal (1). To understand the molecular basis of this change, we examined the substrate specificities of a panel of human N2 viruses isolated between 1957 and 1987 to identify the point at which NA first showed a detectable increase in specificity for NeuAcα2-6Gal. In these studies, we compared avian virus N2 NAs for their ability to recognize the NeuAcα2-3Gal and NeuAcα2-6Gal substrates. Swine virus N2 NAs were also of interest because they were introduced from humans and because pig trachea contains both NeuAcα2-3Gal and NeuAcα2-6Gal (7). Using equal amounts of viral NAs, as determined with the substrate 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid, we found that all of the viral NAs had high activity for NeuAcα2-3Gal. Within the linear response range of the NA activity assay with NeuAcα2-3Gal, the levels of sialic acid released, expressed as the optical density at 549 nm of the sialic acid derivative generated by the Warren assay (22), were in the range of 0.130 to 0.255, with a random distribution of the viruses from all three hosts within this range.
Sialidase activity for the NeuAcα2-6Gal linkage was dependent on the host species and, for the human viruses, on the year of isolation (Fig. 1A). Avian viruses showed very low activity for the NeuAcα2-6Gal linkage, while the two swine viruses showed high activity for that substrate. Human viruses isolated in 1957 and 1960, soon after the first appearance of the N2 NA in human viruses, showed low activity for NeuAcα2-6Gal, similar to that of the avian viruses. To verify and extend these results, we performed additional assays with 100 mU of NA per ml to improve quantitation of the released sialic acid (Fig. 1B) and added A/England/12/62 to the analysis to provide an intermediate isolate. The first detectable increase in specificity for NeuAcα2-6Gal among human virus N2 NAs was observed with A/England/12/62. There was a gradual increase in specificity for NeuAcα2-6Gal that continued until a maximal level of activity was observed by 1972 (Fig. 1). There is some isolate-specific variation in specificity in later isolates, as in the lower specificity of A/Bangkok/1/79 than of A/Udorn/307/72 and A/Los Angeles/2/87, but an overall trend toward higher activity for NeuAcα2-6Gal, compared to that of the 1957 and 1960 isolates, is maintained. These results are similar to those of Baum and Paulson (1), who showed that the human virus N2 NA acquired specificity for NeuAcα2-6Gal, with activities for NeuAcα2-6Gal and NeuAcα2-3Gal becoming approximately equal, by 1972. In contrast to this latter finding, the maximal activity of the N2 NA for NeuAcα2-6Gal in our study was only about 30% that for NeuAcα2-3Gal. In addition, our findings also indicate that NeuAcα2-6Gal specificity was preserved in pigs after the introduction of the N2 NA from humans.
FIG. 1.
NeuAcα2-6Gal substrate specificities of avian, swine, and human virus N2 NAs. Virus strains were compared for their ability to release sialic acid from sialyllactoses containing either the NeuAcα2-3Gal or the NeuAcα2-6Gal linkage. (A) Ratio of sialic acid released from a 0.1 mM concentration (50 μl) of each substrate (NeuAcα2-6Gal and NeuAcα2-3Gal) by 10.0 mU of viral NA per ml. (B) Absolute amount of sialic acid released from a 0.1 mM concentration (50 μl) of NeuAcα2-6Gal-containing sialyllactose by 100.0 mU of NA per ml in 30 min at 37°C. The amount of released sialic acid was determined by the periodate-thiobarbituric acid assay (22) and reported as the absorbance of the colored chromophore derivative at 549 nm.
Region of NA important for NeuAcα2-6Gal substrate specificity.
To identify the region of the human virus N2 NA protein that confers increased specificity for NeuAcα2-6Gal, we studied chimeric constructs representing A/England/12/62, the virus showing the first incremental increase in specificity, and A/Singapore/1/57, one of the first N2 NA-containing human viruses. In the five constructs, sequential portions of A/England/12/62 N2 NA were replaced by equivalent regions from A/Singapore/1/57 N2 NA (Fig. 2A). Each chimera NA and the parental virus NA were individually expressed in 293T cells, and the substrate specificity of the expressed protein was determined. The parental virus and chimeric NAs all exhibited similar specificities for NeuAcα2-3Gal (data not shown). As was observed with the NA from concentrated virus, the N2 NA of A/Singapore/1/57 showed very low specificity for NeuAcα2-6Gal, in contrast to N2 NA of A/England/12/62, whose specificity for this linkage was approximately 2.5- to 3.0-fold higher (Fig. 1B and 3B). Chimeras 1, 3, 4, and 5 had NeuAcα2-6Gal specificities similar to that of parental virus A/England/12/62, although chimera 2 had a lower specificity for NeuAcα2-6Gal, similar to that of parental virus A/Singapore/1/57. This result suggested that the region of A/England/12/62 NA responsible for the higher NeuAcα2-6Gal specificity has been replaced by the corresponding region of A/Singapore/1/57 NA in chimera 2. Conceivably, the lower NeuAcα2-6Gal specificity of chimera 2 could have been caused by a global perturbation in the NA structure due to transfer of an A/Singapore/1/57 NA sequence into A/England/12/62 NA. This possibility is unlikely, as the NA activity for NeuAcα2-3Gal remained similar to that of the parental viruses and the other chimeras.
FIG. 3.
NeuGc substrate specificities of avian, swine, and human virus N2 NAs. The abilities of avian, swine, and human virus N2 NAs (A) and NAs from additional human viruses isolated in 1962 and 1967 to 1969 and swine viruses (B) to release sialic acid from GM3 ganglioside substrates containing NeuAcα2-3Gal or NeuGcα2-3Gal were compared. Viral NA (10 mU/ml) was incubated with 0.1 mM substrate containing 0.1% SDC (50 μl) for 30 min at 37°C, and the amount of liberated sialic acid was determined as described in the legend to Fig. 1. The ratio of sialic acid released from NeuGcα2-3Gal versus that released from NeuAcα2-3Gal is reported.
Amino acid residues affecting NeuAcα2-6Gal specificity.
The region exchanged in chimera 2 (amino acid residues 204 to 283) contained two amino acid differences between the two parental virus NAs (Table 1). In A/England/12/62 NA, a glutamate had replaced lysine at position 258, and a valine had replaced isoleucine at position 275. To determine if one or both of the two residues are responsible for the higher NeuAcα2-6Gal specificity of A/England/12/62 NA, we generated two site-specific A/Singapore/1/57 NA mutants. Residues 258 and 275 of A/Singapore/1/57 NA were separately exchanged with those of A/England/12/62 NA, introducing valine at position 275 and lysine at position 258 to yield constructs pCAT3DKSING57-275V and pCAT3DKSING57-258E (pCAGGS/MCS expression vector constructs), respectively. In 293T cells, the expressed proteins had high levels of sialidase activity for NeuAcα2-3Gal, as did the parental virus NAs (data not shown). Compared with the situation for A/Singapore/1/57 NA, the amino acid substitution at position 258 did not affect NA activity for NeuAcα2-6Gal (Fig. 2C). On the other hand, the substitution of valine for isoleucine at position 275 increased NA activity for NeuAcα2-6Gal to about 65% that of A/England/12/62 NA, indicating that the residue at this position plays a significant role in determining the specificity of the N2 NA for NeuAcα2-6Gal.
TABLE 1.
Comparison of amino acid residues in the NAs of N2 viruses A/England/12/62, A/Singapore/1/57, and A/Tokyo/3/67
| Amino acid position | Amino acida in:
|
||
|---|---|---|---|
| A/Singapore/1/57 | A/England/12/62 | A/Tokyo/3/67 | |
| 30 | Ala | Val | Val |
| 81 | Glu | Glu | Lys |
| 127 | Gly | Gly | Val |
| 147 | Gly | Asp | Asp |
| 149 | Ile | Ile | Val |
| 172 | Lys | Lys | Arg |
| 173 | Glu | Gln | Gln |
| 176 | Val | Val | Ile |
| 194 | Val | Val | Ile |
| 199 | Arg | Lys | Lys |
| 258 | Lys | Glu | Glu |
| 266 | Ile | Ile | Ser |
| 269 | Ser | Ser | Ala |
| 275 | Ile | Val | Val |
| 286 | Asp | Asp | Gly |
| 303 | Ile | Ile | Val |
| 331 | Ser | Arg | Arg |
| 339 | Asp | Asn | Asn |
| 346 | Asn | Asn | Thr |
| 347 | Pro | Pro | Gln |
| 358 | Asp | Asp | Asn |
| 360 | Val | Val | Leu |
| 367 | Asn | Ser | Ser |
| 370 | Ser | Leu | Leu |
| 400 | Asn | Ser | Ser |
| 401 | Asn | Asn | Asp |
| 403 | Trp | Trp | Arg |
| 431 | Pro | Gln | Lys |
Amino acids in A/Singapore/1/57 or A/Tokyo/3/67 that differ from those in A/England/12/62 are shown in bold.
Substrate specificities of avian, swine, and human virus NAs for NeuGc.
Another species of sialic acid that is expressed at high levels on the cells of some host species infected by influenza A viruses is NeuGc attached to galactose by an α2-3 linkage (NeuGcα2-3Gal). This type of sialic acid is found on swine cells but not on human cells at the sites of virus infection (19). To examine the role played by NeuGcα2-3Gal in influenza virus infection, we compared the specificities of avian, swine, and human virus NAs for NeuAc-GM3 and NeuGc-GM3, which contain NeuAcα2-3Gal and NeuGcα2-3Gal, respectively. The N2 NAs of the avian, swine, and human viruses all had similarly high specificities for NeuAc-GM3 (data not shown). This result was expected and mirrors the high specificities shown by the NAs for the NeuAcα2-3Gal-containing sialyllactose substrate (both substrates contain the same species and linkage of sialic acid). In contrast, there were species-dependent differences in the recognition of NeuGc-GM3 by the NAs (Fig. 3A). Avian viruses had low to moderate specificity for NeuGc, whereas swine viruses had high specificity, as previously reported (19). Human virus N2 NAs showed an unexpected trend in specificity. Early human N2 viruses, isolated between 1957 and 1962, exhibited low to moderate specificity for NeuGc, as did avian viruses. However, A/Aichi/2/68, A/Hong Kong/1/68, and A/Memphis/8/68 showed high specificity for NeuGc, much like the results for swine viruses. By 1972, the NeuGc specificity of the human viruses had returned to low to moderate levels like those observed before 1968.
We further investigated the unusual NeuGc specificity of human viruses isolated in 1968 by examining additional H2N2 and H3N2 human viruses isolated between 1967 and 1969. All but one of these human viruses (A/Korea/426/68) showed high specificity for NeuGc (Fig. 3B). Analysis of additional swine viruses supported the earlier finding that such viruses possess high NeuGc specificity (Fig. 3B). A/England/12/62, which had low to moderate NeuGc specificity like those of other early human viruses, was included in the analysis because it represented the nearest available human virus isolate relative to viruses isolated in 1967 and 1968.
Amino acids affecting NeuGc substrate specificity.
A/Tokyo/3/67 had an approximately 2.6-fold higher specificity for NeuGc than did A/England/12/62 (Fig. 3B). This difference was used to identify amino acid residues in NA responsible for the higher NeuGc specificity of A/Tokyo/3/67. Taking advantage of the high homology between the NA genes of A/England/12/62 and A/Tokyo/3/67, we generated five chimeric constructs (chimeras 6 to 10) in which portions of the A/England/12/67 NA gene were replaced with the corresponding regions of the A/Tokyo/3/67 NA gene (Fig. 4A). Each chimeric and parental NA was then expressed in 293T cells, and the substrate specificities of the cell-expressed NAs were determined with NeuAc-GM3 and NeuGc-GM3, using equal amounts of NA activity.
In the cell-expressed NA assays, parental A/Tokyo/3/67 NA consistently showed 1.5 to 2.0 times higher NeuGc specificity than did parental A/England/12/62 NA (Fig. 4B). The NeuGc specificities of chimeras 6 and 7 were about 60% that shown by A/Tokyo/3/67 and were slightly higher than that of parental A/England/12/62. The NeuGc specificity of chimera 8 was similar to that of A/England/12/62. Chimeras 9 and 10 were comparable to A/Tokyo/3/67 in NeuGc specificity, indicating that the determinants of higher NeuGc specificity resided within the region exchanged in these two constructs. Because chimera 10 combines regions from chimeras 8 and 9 and chimera 9 alone showed elevated NeuGc specificity, we reasoned that the determinant of increased specificity lay in the sequences exchanged in chimera 9, which comprise three amino acid differences between the NAs of A/England/12/62 and A/Tokyo/3/67 (Table 1). The changes were asparagine 401 to aspartic acid, tryptophan 403 to arginine, and glutamine 431 to lysine. Since amino acid 431 is located near the edge of the sialic acid-binding pocket of the enzymatic active site (Fig. 5), we generated a mutant of A/England/12/62 NA, pCAT3DKENG-431K, that carried the A/Tokyo/3/67 residue in this position. The roles of the amino acid differences at positions 401 and 403 were investigated by replacing these residues in A/England/12/62 NA with aspartic acid and arginine, respectively, to generate construct pCAT3DKENG-401D,403R. The substrate specificities of these mutants indicated that the primary determinant of the higher NeuGc specificity of A/Tokyo/3/67 was lysine 431. Cell-expressed pCAT3DKENG-431K displayed 94% of the NeuGc specificity of A/Tokyo/3/67, indicating that this single amino acid change was sufficient to confer increased NeuGc specificity to A/England/12/62 (Fig. 4C). The amino acid substitutions Asn401→Asp and Trp403→Arg did not appreciably affect NeuGc specificity.
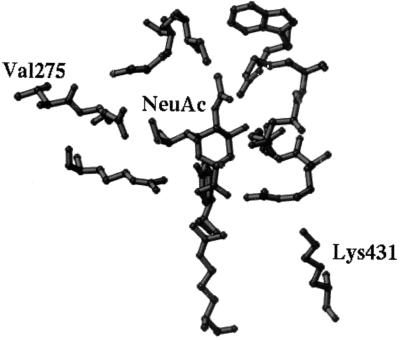
FIG. 5.
Active-site residues of N2 influenza virus NA and bound NeuAc presented as a ball-and-stick model. Valine at residue 275 (Val275) is located below the center of the active site, while lysine at residue 431 (Lys431) lies above the center of the active site (at twice the distance of Val275).
DISCUSSION
We have identified two amino acid residues, at positions 275 and 431, near the enzymatic active center of N2 NA, that are involved in the ability of NA to recognize NeuAcα2-6Gal and NeuGc, respectively. Residue 275 is an isoleucine in avian virus NAs as well as NAs of early human N2 viruses isolated in 1957 (Table 2). Isoleucine at this position is associated with high activity for NeuAcα2-3Gal and very low activity for NeuAcα2-6Gal. A change to valine results in higher NeuAcα2-6Gal activity without affecting NeuAcα2-3Gal activity. In A/England/12/62, valine at position 275 is associated with the first increment in the gradual increase in NeuAcα2-6Gal specificity that characterizes human N2 viruses. This single amino acid change is maintained in the sequences of all human H2N2 and H3N2 viruses isolated after 1962 (Table 2). Hence, the change in the identity of residue 275 may be the first important step in the shift of N2 NA toward increased specificity for NeuAcα2-6Gal. Valine at position 275 is also found in the N2 sequences of swine viruses, which have higher NeuAcα2-6Gal specificity than do avian viruses (Fig. 1A) (24).
TABLE 2.
Comparison of N2 NA amino acid residues at positions 275 and 431 in avian, swine, and human viruses
| Host | Isolate | Subtype | Residue 275 | Residue 431 | Reference or source of sequencea | Relative activity for the following substrateb:
|
||
|---|---|---|---|---|---|---|---|---|
| NeuAcα2-3Gal | NeuAcα2-6Gal | NeuGcα2-3Gal | ||||||
| Avian | A/turkey/Colorado/8/72 | H5N2 | Ile | Pro | This study | |||
| A/duck/Hong Kong/7/75 | H3N2 | Ile | Pro | This study | ||||
| A/duck/Hong Kong/24/76 | H3N2 | Ile | Pro | D21184 | ++++ | + | + | |
| A/goose/Hong Kong/10/76 | H3N2 | Ile | Pro | This study | ||||
| A/duck/Hong Kong/245/77 | H3N2 | Ile | Pro | D21185 | ||||
| A/mallard/New York/6750/78 | H2N2 | Ile | Pro | This study | ++++ | + | + | |
| A/duck/Hong Kong/342/78 | H5N2 | Ile | Pro | This study | ||||
| A/turkey/Minnesota/833/80 | H4N2 | Ile | Pro | This study | ||||
| A/chicken/Pennsylvania/1370/83 | H5N2 | Ile | Pro | M12051 | ||||
| A/gull/Delaware/475/86 | H2N2 | Ile | Pro | This study | ||||
| A/gull/Delaware/2838/87 | H7N2 | Ile | Pro | This study | ++++ | + | ++ | |
| Swine | A/swine/Italy/1850/77 | H3N2 | Val | Lys | This study | ++++ | ||
| A/swine/Italy/635/87 | H3N2 | Val | Gly | This study | ++++ | |||
| Human | A/Singapore/1/57 | H2N2 | Ile | Pro | This study | ++++ | + | ++ |
| A/Rhode Island/5−/57 | H2N2 | Ile | Pro | J02156 | ||||
| A/Leningrad/134/57 | H2N2 | Ile | Pro | L37329 | ||||
| A/Ann Arbor/6/60 | H2N2 | Val | Pro | This study | ++++ | + | + | |
| A/England/12/62 | H2N2 | Val | Gln | This study | ++++ | ++ | ||
| A/Berlin/3/64 | H2N2 | Val | Gln | This study | ||||
| A/Tokyo/3/67 | H2N2 | Val | Lys | This study | ++++ | ++ | +++ | |
| A/Cordoba/522/67 | H2N2 | Val | Lys | 11 | ||||
| A/England/10/67 | H2N2 | Val | Lys | 11 | ||||
| A/Georgia/1/67 | H2N2 | Val | Lys | 11 | ||||
| A/Poland/6/67 | H2N2 | Val | Lys | 11 | ||||
| A/Korea/426/68 | H2N2 | Val | Gln | This study | ++++ | + | ||
| A/Aichi/2/68 | H3N2 | Val | Lys | M55059 | ++++ | ++ | +++ | |
| A/NT/60/68 | H3N2 | Val | Lys | J02136 | ||||
| A/Hong Kong/8/68 | H3N2 | Val | Lys | U42630 | ++++ | +++ | ||
| A/Perg/1/68 | H2N2 | Val | Lys | 11 | ||||
| A/Udorn/307/72 | H3N2 | Val | Glu | J02168 | ++++ | +++ | ++ | |
| A/Victoria/3/75 | H3N2 | Val | Glu | J02173 | ||||
| A/Bangkok/1/79 | H3N2 | Val | Glu | K01150 | ++++ | ++ | ++ | |
| A/Los Angeles/2/87 | H3N2 | Val | Glu | This study | ++++ | +++ | ++ | |
Letter-number designations are GenBank accession numbers.
Relative levels of activity for each substrate from lowest (+) to highest (++++) are indicated for the viruses that were tested and for which sequence data are available.
The increase in NeuAcα2-6Gal specificity in human virus N2 NAs, due to the change at residue 275, was followed by further increases in specificity for that substrate (Fig. 1) (1). The lack of additional mutations at this position suggests that the gradual shift to higher NeuAcα2-6Gal specificity seen in human virus N2 NAs resulted from successive amino acid changes in other key residues that each subtly altered the conformation of the active site to better accommodate this linkage of sialic acid. However, there appears to be a limit to this process of adaptation, as maximal specificity for NeuAcα2-6Gal was apparently attained by 1972 (Fig. 1A) (1). Whether further adaptation is not possible because of an adverse effect on enzymatic activity or whether there is simply insufficient selective pressure from the environment to select variants with still higher levels of NeuAcα2-6Gal specificity is uncertain. Couceiro et al. (4) have suggested that NeuAcα2-6Gal on human epithelial tracheal cells, together with NeuAcα2-3Gal in bronchial mucus secretions, may exert selective pressure to determine human virus HA specificity (4). Similarly, we propose that in addition to acquiring NeuAcα2-6Gal specificity, it would be advantageous for human virus NA to maintain appreciable NeuAcα2-3Gal specificity to support movement of the virus in bronchial mucus.
The mutation at residue 275 may not be the only one required to promote an increase in NeuAcα2-6Gal specificity in early human N2 viruses. The mutation of isoleucine at residue 275 in A/Singapore/1/57 NA to valine resulted in about 65% of the NeuAcα2-6Gal specificity shown by A/England/12/62 NA. This result suggests that an additional mutation(s) may be necessary to maximize binding to the NeuAcα2-6Gal substrate or to compensate for conformational changes induced in the rest of the enzyme as a consequence of the change at residue 275. Analysis of the N2 NA sequences of other viruses showed that A/Ann Arbor/6/60 NA already had valine at position 275, although it showed the same low NeuAcα2-6Gal specificity as A/Singapore/1/57 NA (Fig. 1B and Table 2). Since A/Ann Arbor/6/60 NA has several amino acid differences that are not shared by either A/Singapore/1/57 NA or A/England/12/62 NA, one or more of these differences may counter the effect of valine at position 275 on the activity of the enzyme for NeuAcα2-6Gal.
Site-specific mutagenesis demonstrated that a lysine at position 431 determines the highest NeuGc specificity, although A/swine/Italy/635/87 NA, which contains a glycine at this position, also has high NeuGc specificity (Table 2). That swine viruses have high NeuGc specificity is not unexpected, because swine tracheal epithelial cells express high levels of NeuGc (19). Examination of other swine N2 virus sequences indicated that lysine and glycine are not the only residues that can occupy position 431. Arginine is present at this position in A/swine/Kanagawa/2/78 (12) but represents only a conservative change from lysine, and the substrate specificity of the isolate was not examined. Proline was observed at position 431 in avian and human viruses; it was found in all avian N2 virus sequences examined as well as those of human N2 viruses isolated from 1957 to 1960. Other human viruses contain glutamine or glutamate which, like proline, is associated with low to moderate NeuGc specificity. A close relationship between lysine at position 431 and high NeuGc specificity is further supported by the fact that A/Korea/426/68 has glutamine at this position and is the only human virus isolated from 1967 to 1969 with low activity for this sialic acid.
The mechanism by which amino acid changes affect NA substrate specificity is expected to involve very small shifts in NA structure that subtly affect the enzymatic active site. The high degree of conservation among active-site residues (2, 20, 21) suggests that amino acid substitutions within the active site which could permit the binding of alternative linkages or species of sialic acid are not likely to occur, a prediction substantiated by site-specific mutagenesis (9). Indeed, both of the amino acid substitutions found to affect the specificity of NA for NeuAcα2-6Gal and NeuGc occur at positions close to the enzymatic active site and not at those believed to be involved in the reaction mechanism. Residue 275 is directly adjacent to glutamate 276 and glutamate 277, both of which are highly conserved and directly involved in the binding of sialic acid to the enzymatic active site (21).
The mechanism of NA specificity for the α2-3 or α2-6 glycosidic linkage appears to entail restrictions on accessibility to the active site. In the crystal structure of an intramolecular trans-sialidase which has strict specificity for NeuAcα2-3Gal (10), the side chain of a tryptophan lies above the C-2 position, excluding binding to a substrate with a NeuAcα2-6Gal linkage. Hence, it is important to consider possible interactions with the substrate of side chains in the upper portion of the active site. When there is a valine at position 275 below the active site, as in the structure shown in Fig. 5, the side chain of glutamate 276 in the next position becomes involved in good contacts with the glycerol group in NeuAc. The pyranose ring of NeuAc is horizontal in the active site, allowing the remaining moiety of the α2-3Gal or α2-6Gal oligosaccharide to extend freely to the open region of that site. If valine 275 is replaced with a bulkier residue, such as isoleucine, glutamate 276 will have to be lifted a little to accommodate the substitution. Such a conformational change may cause the pyranose ring of NeuAc to tilt toward the right side of the active site. The side chain of aspartic acid 151 is near the glycosidic oxygen, right above the pyranose ring of NeuAc. Such geometry may allow free access to a substrate with a NeuAcα2-3Gal linkage but may impede access to a substrate with a NeuAcα2-6Gal linkage owing to steric hindrance of the remaining moiety of the α2-6Gal oligosaccharide by the side chain of aspartic acid 151.
The side chain of lysine 431, which confers NeuGc specificity in N2 NA, is far above the active site (Fig. 5). There do not seem to be any direct contacts of this residue with the substrate sialic acid. The only possible interactions would be with the second or third sugar moiety of NeuAc in an oligosaccharide substrate; such interactions would be unlikely to impose any restrictions on NeuGc substrates. A substitution at this position with glycine or perhaps arginine does not seem to change enzyme specificity for NeuGc, compared to that of NAs with lysine at position 431. Glutamate or glutamine can decrease NeuGc specificity, an effect that may result from long-range charge-charge interactions between the negatively charged glutamate (or the partially negatively charged glutamine) and the positively charged arginines in the active site. Substitutions with glutamate or glutamine could also alter the active-site conformation through long-range effects. Proline 431, as found in avian and early (1957 to 1960) human viruses, could restrict the conformational flexibility required for NeuGc binding.
Human tracheal epithelial cells do not possess detectable levels of NeuGc (19). Hence, one would not anticipate any pressure during viral replication in humans for the selection of N2 NA variants with higher NeuGc specificity. An unexpected observation was that most of the H2N2 and H3N2 human viruses isolated between 1967 and 1969 had high NeuGc specificity and possessed lysine at position 431, like swine viruses. The only exception was A/Korea/426/68 (Fig. 3A). One explanation for this finding is that some event occurring between 1967 and 1969 favored the emergence of human viruses which had high NeuGc specificity and which rapidly replaced viruses with glutamine at position 431. A possible scenario (although highly speculative) to explain these observations is the transmission of H2N2 viruses from humans to pigs or some other animal species with high NeuGc content on its cells prior to 1967 such that NA acquired increased specificity for NeuGc in the new host. These viruses were then transmitted back to humans. During this period, the same N2 NA virus was reassorted with an avian H3 virus to generate the lineage of human H3N2 viruses (17) with high NeuGc specificity. Although this scenario would explain our observations, evidence to confirm this scenario (i.e., the presence of N2 viruses in pigs prior to 1967) does not exist due to a lack of surveillance of influenza virus infections in swine at that time. After 1969, high NeuGc specificity was not maintained in human viruses, probably due to a lack of NeuGc on the human cells that are infected by influenza viruses. Alternatively, the appearance of viruses with high NeuGc specificity in humans in 1967 to 1969 might not have had any biologic significance and might have been the result of point mutations selected by immunologic pressure.
Cross-species transfer of influenza viruses will continue to be a source of potentially serious disease in animals, including humans. The active site of the NA molecule is highly conserved among the nine NA subtypes; however, during the 20th century, only two subtypes, N1 and N2, have been identified among human viruses, suggesting that specific requirements must be met before a new subtype can emerge and support influenza virus growth in humans. Understanding the role that NA plays in cross-species transfer of influenza viruses and the adaptive mechanism that it uses to efficiently support virus growth in new hosts will greatly aid in understanding how subtypes of viruses become established and are maintained. Such knowledge will also enhance surveillance efforts by helping to identify avian virus subtypes that potentially pose the most serious threat to humans and domestic animals, such as pigs.
ACKNOWLEDGMENTS
We thank Krisna Wells and Scott Krauss for excellent technical assistance, Clayton Naeve and the St. Jude Children’s Research Hospital for preparing oligonucleotides and for computer support, and John Gilbert for editing the manuscript.
Support for this work was provided by National Institute of Allergy and Infectious Diseases Public Health Service Research grants, a Cancer Center Support (CORE) grant from the National Cancer Institute, and the American Lebanese Syrian Associated Charities (ALSAC).
REFERENCES
- 1.Baum L G, Paulson J C. The N2 neuraminidase of human influenza virus has acquired a substrate specificity complementary to the hemagglutinin receptor specificity. Virology. 1991;180:10–15. doi: 10.1016/0042-6822(91)90003-t. [DOI] [PubMed] [Google Scholar]
- 2.Burmeister W P, Ruigrok R W H, Cusack S. The 2.2Å resolution crystal structure of influenza B neuraminidase and its complex with sialic acid. EMBO J. 1992;11:49–56. doi: 10.1002/j.1460-2075.1992.tb05026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Connor R J, Kawaoka Y, Webster R G, Paulson J C. Receptor specificity in human, avian and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 4.Couceiro J N, Paulson J C, Baum L G. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 1993;29:155–165. doi: 10.1016/0168-1702(93)90056-s. [DOI] [PubMed] [Google Scholar]
- 5.Couceiro J N, Baum L G. Characterization of the hemagglutinin receptor specificity and neuraminidase substrate specificity of clinical isolates of human influenza A viruses. Mem Inst Oswaldo Cruz. 1994;89:587–591. doi: 10.1590/s0074-02761994000400015. [DOI] [PubMed] [Google Scholar]
- 6.Gasa S, Makita A, Kinoshita Y. Further study of the chemical structure of the equine erythrocyte hematoside containing O-acetyl ester. J Biol Chem. 1983;258:876–881. [PubMed] [Google Scholar]
- 7.Ito T, Couceiro J N, Kelm S, Baum L G, Krauss S, Castrucci M R, Donatelli I, Kida H, Paulson J C, Webster R G, Kawaoka Y. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobasa D, Rodgers M E, Wells K, Kawaoka Y. Neuraminidase hemadsorption activity, conserved in avian influenza A viruses, does not influence viral replication in ducks. J Virol. 1997;71:6706–6713. doi: 10.1128/jvi.71.9.6706-6713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lentz M R, Webster R G, Air G M. Site-directed mutation of the active site of influenza neuraminidase and implications for the catalytic mechanism. Biochemistry. 1987;26:5351–5358. doi: 10.1021/bi00391a020. [DOI] [PubMed] [Google Scholar]
- 10.Luo Y, Li S-C, Chou M-Y, Li Y-T, Luo M. The crystal structure of an intramolecular trans-sialidase with a NeuAcα2-3Gal specificity. Structure. 1998;6:521–530. doi: 10.1016/s0969-2126(98)00053-7. [DOI] [PubMed] [Google Scholar]
- 11.Nakao H, Nakajima K, Nakajima S. Location on the evolutionary trees of the nonstructural protein (NS) and neuraminidase (NA) genes of late human influenza A (H2N2) viruses: parental viruses of the NS and NA genes of Hong Kong influenza A (H3N2) viruses. J Gen Virol. 1993;74:1667–1672. doi: 10.1099/0022-1317-74-8-1667. [DOI] [PubMed] [Google Scholar]
- 12.Nerome K, Kanegae Y, Yoshioka Y, Itamura S, Ishida M, Gojobori T, Oya A. Evolutionary pathways of N2 neuraminidases of swine and human influenza A viruses: origin of the neuraminidase genes of two reassortants (H1N2) isolated from pigs. J Gen Virol. 1991;72:693–698. doi: 10.1099/0022-1317-72-3-693. [DOI] [PubMed] [Google Scholar]
- 13.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 14.Palese P, Tobita K, Ueda M, Compans R W. Characterization of temperature-sensitive influenza virus mutants defective in neuraminidase. Virology. 1974;61:397–410. doi: 10.1016/0042-6822(74)90276-1. [DOI] [PubMed] [Google Scholar]
- 15.Rogers G N, Paulson J C. Receptor determinants of human influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983;127:361–373. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- 16.Rogers G N, D’Souza B L. Receptor binding properties of human and animal H1 influenza virus isolates. Virology. 1989;173:317–322. doi: 10.1016/0042-6822(89)90249-3. [DOI] [PubMed] [Google Scholar]
- 17.Scholtissek C, Rohde W, Von Hoyningen V, Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 18.Seyfried T N, Ando S, Yu R K. Isolation and characterization of human liver hematoside. J Lipid Res. 1978;19:538–543. [PubMed] [Google Scholar]
- 19.Suzuki T, Horiike G, Yamazaki Y, Kawabe K, Masuda H, Miyamoto D, Matsuda M, Nishimura S-I, Yamagata T, Ito T, Kida H, Kawaoka Y, Suzuki Y. Swine influenza virus strains recognize sialylsugar chains containing the molecular species of sialic acid predominantly present in the swine tracheal epithelium. FEBS Lett. 1997;404:192–196. doi: 10.1016/s0014-5793(97)00127-0. [DOI] [PubMed] [Google Scholar]
- 19a.Suzuki, Y. Unpublished data.
- 20.Varghese J N, Colman P M. Three-dimensional structure of the neuraminidase of influenza virus A/Tokyo/3/67 at 2.2Å resolution. J Mol Biol. 1991;221:473–486. doi: 10.1016/0022-2836(91)80068-6. [DOI] [PubMed] [Google Scholar]
- 21.Varghese J N, McKimm-Breschkin J L, Caldwell J B, Kortt A A, Colman P M. The structure of the complex between influenza virus neuraminidase and sialic acid, the viral receptor. Proteins. 1992;14:327–332. doi: 10.1002/prot.340140302. [DOI] [PubMed] [Google Scholar]
- 22.Warren L. The thiobarbituric acid assay of sialic acids. J Biol Chem. 1959;234:1971–1975. [PubMed] [Google Scholar]
- 23.Wiley D C, Skehel J J. The structure and function of the HA membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 24.Xu G, Suzuki T, Maejima Y, Mizoguchi T, Tsuchiya M, Kiso M, Hasegawa A, Suzuki Y. Sialidase of swine influenza A viruses: variation of the recognition specificities for sialyl linkages and for the molecular species of sialic acid with the year of isolation. Glycocong J. 1995;12:156–161. doi: 10.1007/BF00731360. [DOI] [PubMed] [Google Scholar]