Abstract
Disulfide bonds provide a convenient method for chemoselective alteration of peptide and protein structure and function. We previously reported that mild oxidation of a p53-derived bisthiol peptide (CTFANLWRLLAQNC) under dilute non-denaturing conditions led to unexpected disulfide-linked dimers as the exclusive product. The dimers were antiparallel, significantly α-helical, resistant to protease degradation, and easily reduced back to the original bisthiol peptide. Here we examine the intrinsic factors influencing peptide dimerization using a combination of amino acid substitution, circular dichroism (CD) spectroscopy, and X-ray crystallography. CD analysis of peptide variants suggests critical roles for Leu6 and Leu10 in the formation of stable disulfide-linked dimers. The 1.0 Å resolution crystal structure of the peptide dimer supports these data, revealing a leucine-rich LxxLL dimer interface with canonical knobs-into-holes packing. Two levels of higher-order oligomerization are also observed in the crystal: an antiparallel “dimer of dimers” mediated by Phe3 and Trp7 residues in the asymmetric unit and a tetramer of dimers mediated by Trp7 and Leu10. In CD spectra of Trp-containing peptide variants, minima at 227 nm provide evidence for the dimer of dimers in dilute aqueous solution. Importantly, and in contrast to the original dimer model, the canonical leucine-rich core and robust dimerization of most peptide variants suggests a tunable molecular architecture to target various proteins and evaluate how folding and oligomerization impact various properties, such as cell permeability.
Keywords: Dimerization, Disulfide, Structure, Folding
Introduction
Disulfide bonds play a critical role in the structures and functions of both natural and designed proteins and peptides. Disulfide bonds covalently link distal regions within a wide range of peptides, including oxytocin (Gimpl & Fahrenholz, 2001; Hammer et al., 2023; Muttenthaler et al., 2010), cyclotides (de Veer et al., 2019; Northfield et al., 2014), disulfide-rich peptides (Singh et al., 2010), insulin (Karas et al., 2021), and proteins (Dombkowski et al., 2014; Mitchinson & Wells, 1989; Pace et al., 1988; Zavodszky et al., 2001), including antibodies (Goto & Hamaguchi, 1979; McAuley et al., 2008; Thies et al., 2002). Three-dimensional folding imposed by disulfide bonds is critical for the structure and function of these molecules, many of which have significant clinical value.
Thus, synthetic strategies for regioselective disulfide bond formation have been the subject of intensive study, especially in peptides and proteins containing multiple cysteine (Cys) residues (Albericio et al., 1991; Andreu et al., 1994; Postma & Albericio, 2014). Oxidative folding conditions can be sufficient in some cases but tend to require long reaction times to proceed to thermodynamic equilibrium and/or avoid kinetic traps (Shi et al., 2022; Welker et al., 2001). Orthogonal protecting groups are frequently used to allow formation of one disulfide bond at a time, including “one-pot” strategies (Laps et al., 2021; Postma et al., 2012; Spears et al., 2021). Alternatively, substitution of cysteine with analogs like penicillamine (Pen) favors selective Cys-Pen disulfide bond formation and can allow regioselective disulfide bond formation (Yao et al., 2022; Zheng et al., 2015).
Recently, Victorio and Sawyer reported the surprising, spontaneous formation of disulfide-linked dimers from a p53-derived bisthiol peptide called CV1 (CTFANLWRLLAQNC) using a mild aqueous buffer and dimethyl sulfoxide (DMSO) as oxidant (Victorio & Sawyer, 2023). This peptide is derived from SAH-p53–8, a stapled variant of the N-terminal segment of the p53 protein that interacts with the proteins Mdm2 and MdmX (Baek et al., 2012; Bernal et al., 2010). The interactions between p53 and Mdm2 and/or MdmX are major targets for drug discovery, especially in cancers with Mdm2 and/or MdmX upregulation (Chandramohan et al., 2024; Chang et al., 2013; Momand et al., 1998; Silvestri et al., 2023; Vassilev et al., 2004; Zhu et al., 2022). Beyond the challenge of creating stable peptides that bind tightly and specifically to Mdm2 and/or MdmX, the delivery of p53-derived peptides into target cells remains a significant challenge. Intracellular delivery is impacted by many factors, including amino acid composition, three-dimensional structure, and oligomerization state (Baek et al., 2012; Bernal et al., 2007; Chang et al., 2013; Dantas de Araujo et al., 2018; Henchey et al., 2010; Yoo et al., 2020).
In initial studies, dimers of the CV1 peptide were determined to be exclusively antiparallel with no detectable formation of competing parallel dimers or intramolecular disulfide species. Dimerization was dependent on α-helical folding of CV1, yielding highly α-helical dimers. To account for an unusual minimum observed at 227 nm by circular dichroism (CD) spectroscopy, a model of the dimer was proposed in which tryptophan residues interact at the dimer interface, which is uncommon but not unprecedented in coiled-coil dimers (Rhys et al., 2021). This model accounted for the fact that interactions between aromatic residues frequently give rise to CD maxima/minima in the 225–230 nm wavelength range (Andersen et al., 2006; Cochran et al., 2001; Nazzaro et al., 2023; Sawyer & Arora, 2018).
Here we describe structural characterization of CV1 dimers using a combination of alanine substitution, CD spectroscopy, and X-ray crystallography. Antiparallel dimers were readily produced from all CV1 variants except the L6A variant, which lacked α-helical character as a bisthiol monomer. Dimers displayed a range of α-helical character, and most dimers retained their maximal α-helical structure up to 70°C. The 1.0 Å crystal structure of the CV1 dimer revealed a knobs-into-holes packing interface between leucine residues at the dimer interface. The formation of a “dimer of dimers” arrangement in the asymmetric unit and a higher order “tetramer of dimers” is facilitated by Phe and Trp residues. These data revise the previous model of CV1 oligomerization and suggest a designable molecular architecture for potential targeting of a wide variety of proteins and studying how peptide folding and oligomerization influence interactions with proteins, nucleic acids, and membranes.
Materials and Methods
General Information.
Commercially purchased solvents and reagents were used without further purification. Nα-Fmoc-protected amino acids and peptide synthesis reagents were purchased from Advanced ChemTech, ChemImpex International, Oakwood Chemical, and Gyros Protein Technologies. Peptides were synthesized manually or using a Gyros Protein Technologies PurePep™ Chorus synthesizer. Peptides were purified on preparative C18 columns using reverse-phase high-performance liquid chromatography (RP-HPLC) on a Shimadzu Nexera HPLC system using gradients of water and acetonitrile (ACN) containing 0.1% trifluoroacetic acid (TFA). Peptide purity was evaluated by analytical HPLC using a Luna 5 μm C18(2) column (150 × 4.6 mm) on an Agilent 1100 HPLC system with a flow rate of 0.3 mL/min (gradient:5–95% solvent B over 30 minutes, solvent A = 0.1% TFA, B = 95% ACN, 5% water, 0.1% TFA, see Figure S1, Table S1). High-resolution mass spectrometry data was collected on a Bruker autoflex maX MALDI-TOF/TOF mass spectrometer using α-cyano-4-hydroxycinnamic acid as the matrix. Proteinase K (P8107S) was purchased from New England Biolabs. Mass spectrometry grade trypsin was purchased from Fisher (PI90057).
Peptide Synthesis.
Peptides were synthesized following standard Fmoc solid-phase approaches on high-loading Rink MBHA resin (0.62 mmol/g resin). Peptides were globally deprotected and cleaved from resin by incubation with Reagent R (90% TFA, 5% thioanisole, 3% 1,2-ethanedithiol, and 2% anisole) for 2 h. After filtration, rotary evaporation of TFA and precipitation with ice-cold diethyl ether yielded crude peptides. A list of bisthiol peptides used for these studies is provided in Table 1.
Table 1.
Peptide Naming and Sequences.
| Peptide | Sequence |
|---|---|
| CV1 | Ac-CTFANLWRLLAQNC-NH2 |
| T2A | Ac-CAFANLWRLLAQNC-NH2 |
| F3A | Ac-CTAANLWRLLAQNC-NH2 |
| N5A | Ac-CTFAALWRLLAQNC-NH2 |
| L6A | Ac-CTFANAWRLLAQNC-NH2 |
| W7Y | Ac-CTFANLYRLLAQNC-NH2 |
| R8A | Ac-CTFANLWALLAQNC-NH2 |
| L9A | Ac-CTFANLWRALAQNC-NH2 |
| L10A | Ac-CTFANLWRLAAQNC-NH2 |
| Q12A | Ac-CTFANLWRLLAANC-NH2 |
| N13A | Ac-CTFANLWRLLAQAC-NH2 |
Thiol Oxidation Reactions.
Thiol oxidation reactions were performed by sequentially adding aqueous NH4HCO3 (pH 6.0), DMSO, and peptide to produce a solution with final concentrations of 0.88 mM peptide, 20% DMSO, 5.5 mM NH4HCO3, pH 6.0 (Tam et al., 1991). Reactions were performed at room temperature for 24 h. After each reaction, dimers were purified by RP-HPLC using a gradient of 25–70% ACN over 30 min.
Circular Dichroism.
Circular dichroism spectra were acquired using a Jasco J-1500 CD spectrometer at a concentration of 30 μM in 10 mM sodium phosphate, pH 7.5 in a 0.1 cm pathlength cell. Unless specified, all spectra were collected at 25°C. For temperature-dependent circular dichroism experiments, CD spectra were acquired at 5°C intervals starting at a minimum temperature of 5°C and ending at a maximum temperature of 95°C. After acquisition of each spectrum, the sample temperature was increased at a rate of 1°C/min and held at the new target temperature for a minimum of 60 s before acquisition of the next spectrum.
Trypsin Digestion.
Trypsin digestion samples were prepared by combining 10 μL of 300 μM peptide in water, 40 μL of 125 μM phosphate buffer (pH 7.2), and 10 μL of 1.2 μM trypsin in phosphate buffer at 37°C. Mass spectrometry data were collected after 15 h.
X-ray Crystallography.
A crystallization stock solution was prepared containing the dimeric CV1 peptide in water at a concentration of 36 mg/mL. Crystals were grown by hanging drop vapor diffusion using a crystallization condition optimized from Molecular Dimensions Morpheus crystallization screen (30 mM sodium nitrate, 30 mM dibasic sodium phosphate, 30 mM ammonium sulfate, 20% (v/v) glycerol, 10% (w/v) PEG4000, 100 mM imidazole/MES monohydrate buffer, pH 6.5. A 2 μL drop comprising a 1:1 mixture of crystallization stock solution and the crystallization condition was placed on a glass cover slide and inverted to seal a well containing 150 μL of the crystallization condition. Crystal morphology is shown in Figure S2. Crystals were looped and vitrified in liquid nitrogen. Diffraction data were collected at the AMX beamline (17-ID-1) at the National Synchrotron Light Source II (NSLS-II) at Brookhaven National Laboratory (Upton, NY). Data were indexed, integrated, and scaled using the program XDS (Kabsch, 2010) and merged using the program Aimless (Evans & Murshudov, 2013) as implemented in the autoPROC processing package (Vonrhein et al., 2011). To determine an initial set of phases, a composite reflection file was generated by merging datasets derived from multiple crystals with KAMO (Yamashita et al., 2018). These multi-dataset reflections were then subjected to molecular replacement with the program Fragon (Jenkins, 2018) using three fragment search models comprised of ten-residue ideal α-helices. Following structure solution, ADP and coordinate model refinements were performed against a single crystal dataset using the program phenix.refine (Adams et al., 2010) in combination with manual real-space model building and refinement in the program Coot (Emsley & Cowtan, 2004). Collection and refinement statistics are available in Table S2. The structure has been deposited as PDB code 9C5S.
Results and Discussion
To understand CV1 dimerization, we first sought to define the contribution of individual amino acid residues to dimerization. To do so, we prepared variants of CV1 in which each non-alanine, non-cysteine residue was individually substituted with alanine. In total, ten variants were prepared: T2A, F3A, N5A, L6A, W7Y, R8A, L9A, L10A, Q12A, and N13A. For Trp7, we opted to substitute with tyrosine instead of alanine to allow determination of all peptide concentrations by UV-Vis spectrophotometry. The R8A variant displayed limited aqueous solubility that prevented further study.
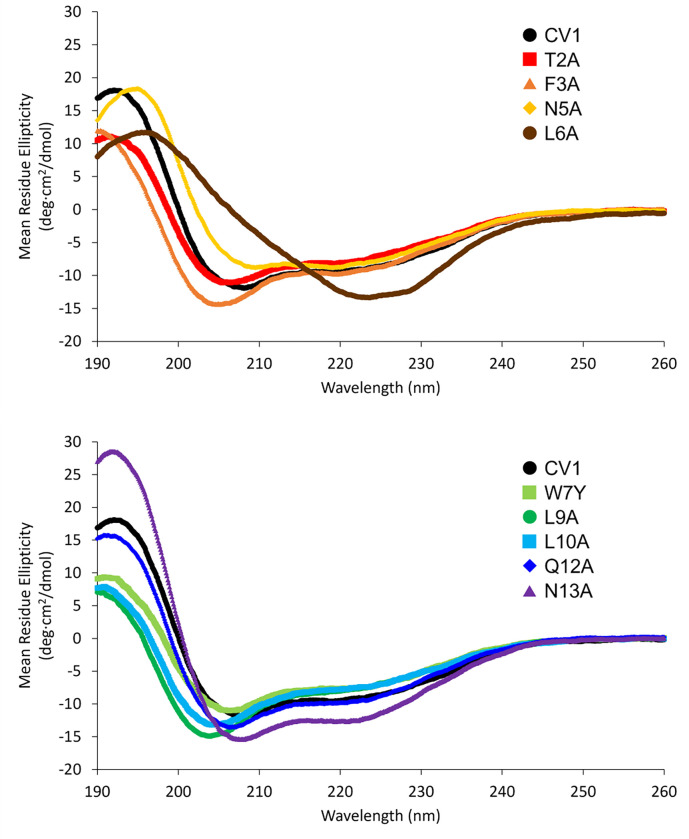
Analysis of bisthiol variants by CD spectroscopy revealed that all variants except L6A exhibited partial α-helical folding in sodium phosphate buffer. Aside from the L6A variant, all CD spectra were consistent with α-helical folding, displaying dual minima at approximately 208 and 222 nm (Fig. 1). Based on a calculated minimum mean residue ellipticity (MRE) of approximately − 27000 deg·cm2/dmol at 222 nm for an ideal 14-residue α-helix (Shepherd et al., 2005; Wang et al., 2006), the original CV1 bisthiol peptide is estimated to be approximately 30% α-helical in 10 mM sodium phosphate buffer at 25 °C, which is consistent with previous results (Victorio & Sawyer, 2023). Estimated α-helix content ranged from 25% for the L10A variant to approximately 45% for the N13A variant at 25°C. In contrast, the L6A variant has a maximum around 196–197 nm and a broad minimum between 220 and 230 nm, indicating a non-α-helical structure that is distinct from a random coil (Bishop et al., 2001).
Figure 1.
Circular dichroism spectra for the CV1 bisthiol peptide and variants. Spectra were acquired using a Jasco J-1500 CD spectrometer at a concentration of 30 μM in 10 mM sodium phosphate, pH 7.5 in a 0.1 cm pathlength cell at 25 °C
Oxidation of bisthiol monomers under mild conditions produced dimers as the major product for many, but not all, variants. Dimerization was performed as previously described using a peptide concentration of 0.88 mM in 5 mM aqueous NH4HCO3 buffer containing 20% (v/v) DMSO as oxidant. Based on HPLC quantification, reactions with CV1 and three variants (N5A, Q12A, N13A) produced the dimer as the exclusive product. The W7Y variant also displayed significant preference for the dimer product (> 90%) over the competing intramolecular disulfide species. For four remaining variants – T2A, F3A, L9A, and L10A – a mixture of dimer and intramolecular disulfide products was observed. For the L6A variant, only the intramolecular disulfide product was observed upon oxidation, which is consistent with previous results suggesting the necessity of α-helical folding of the bisthiol monomer to yield dimers. When corrected for the estimated extinction coefficients of 11,000 M− 1·cm− 1 for the dimer and 5500 M− 1·cm− 1 for the intramolecular disulfide species, the ratios of dimer-to-intramolecular disulfide were 1:1 for the T2A variant, 2:1 for the F3A variant, 3:1 for the L9A variant, and 1:2 for the L10A variant. All dimers were determined to be antiparallel based on trypsin digestion (Figure S3). Based on these data, Leu6 and Leu10 appear to be major contributors to dimerization with additional contributions from Thr2, Phe3, and Leu9. Importantly, the minimal contribution of Trp7 contrasts sharply with the previously proposed model in which tryptophan residues interact at the dimer interface (Victorio & Sawyer, 2023).
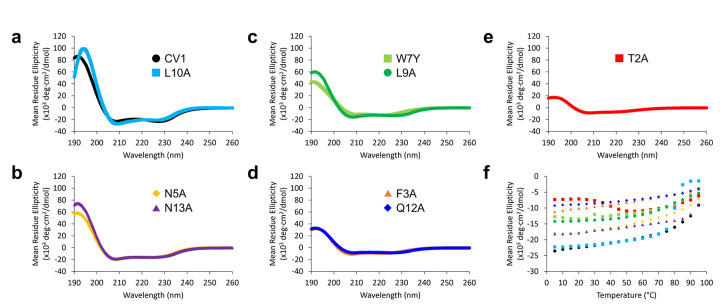
Evaluation of dimers by CD spectroscopy indicated that dimers formed from each variant exhibited different degrees of α-helical folding. At 25°C, the MRE at 222 nm ranged from approximately − 22,000 deg·cm2/dmol (70% α-helical) for CV1 to approximately − 7300 deg·cm2/dmol for the T2A variant (20% α-helical). The nine dimers (CV1 and eight variants) can be grouped into four pairs with one outlier (Fig. 2):
Figure 2.
Circular dichroism spectra for the disulfide-linked CV1 peptide dimer and disulfide-linked dimers of CV1 alanine variants. Spectra were acquired using a Jasco J-1500 CD spectrometer at a concentration of 30 μM in 10 mM sodium phosphate, pH 7.5 in a 0.1 cm pathlength cell. a-e) Spectra at 25 °C. f) Mean residue ellipticity at 222 nm as a function of temperature (5 °C to 95 °C)
The first pair is CV1 and L10A, which were almost 70% α-helical at 25°C (Fig. 2A, black and light blue; Figure S4). Nonetheless, folding stability was starkly different for these peptides. The CV1 dimer shows gradual unfolding with an MRE of −9000 deg·cm2/dmol at 95°C, corresponding to approximately 50% α-helicity at this temperature. In contrast, the dimer of the L10A variant displayed a cooperative loss of α-helical folding between 75°C and 85°C (Fig. 2F, light blue). Thus, consistent with HPLC analysis, Leu10 appears to play a significant role in dimer stability.
Six of the remaining variants can be paired based on their α-helical folding as dimers at 25°C (Fig. 2B–D and Figure S4). At 25 °C, variants N5A and N13A were approximately 50% folded (−17,500 deg·cm2/dmol), variants W7Y and L9A variants were approximately 40% folded (−13,500 deg·cm2/dmol), and variants F3A and Q12A were approximately 30% folded (−9400 deg·cm2/dmol). The relative stability of these variants was maintained across the full temperature range up to 95°C, with retention of greater than 90% of their respective maximum folding up to 70°C.
Dimers of the final T2A variant displayed a highly unusual CD profile as a function of temperature (Fig. 2E–F, red; Figure S4). At 5–25°C, the MRE at 222 nm was relatively constant at approximately 20% α-helical (−7200 deg·cm2/dmol). As the temperature increases from 25°C to 55°C, the MRE at 222 nm decreased to approximately −11,000 deg·cm2/dmol, indicating an unusual increase in α-helical folding to approximately 40% in this temperature range. Beyond 55°C, the MRE for the T2A variant increased to −6200 deg·cm2/dmol at 95°C, approximately 40% of the maximum α-helical folding for this temperature range.
One important caveat to this analysis of relative α-helical folding of dimers is that all but three peptides (T2A, F3A, and W7Y) showed an apparent “shift” in the characteristic 222 nm CD minimum for α-helices to approximately 227 nm (Fig. 2). This shift was not observed for any of the bisthiol peptides, suggesting that the shift is related to dimerization. Aromatic-aromatic interactions, especially those involving tryptophan, frequently give rise to CD minima/maxima within this wavelength range (Andersen et al., 2006; Cochran et al., 2001; Nazzaro et al., 2023; Sawyer & Arora, 2018). This fact contributed to the previously proposed model involving tryptophan interactions at the dimerization interface (Victorio & Sawyer, 2023). The fact that the W7Y variant does not display this CD shift supports the hypothesis that the CD shift arises from interaction between tryptophan residues. The absence of this shift in the T2A and F3A variants was not initially clear from CD analyses but was subsequently rationalized based on the three-dimensional structure of the CV1 dimer (vide infra).
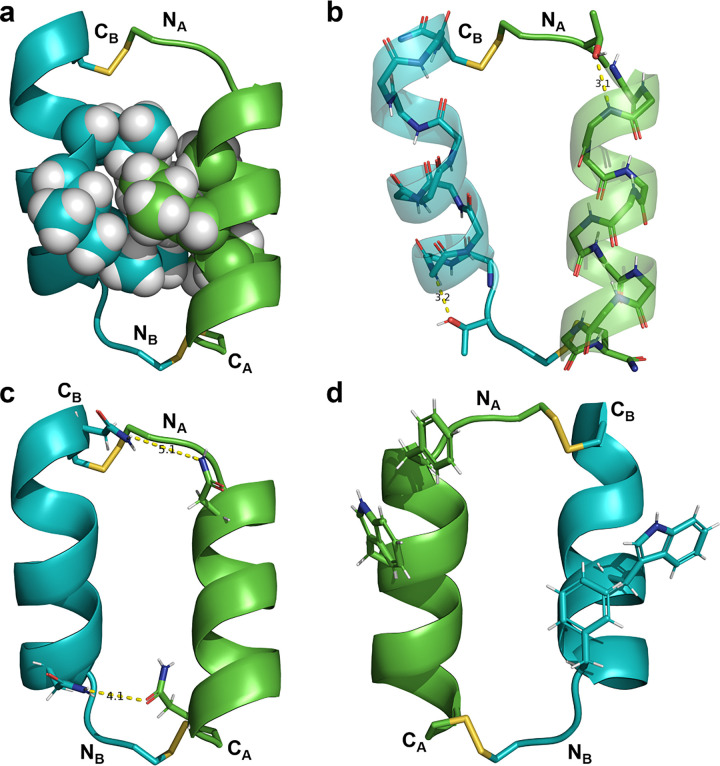
Concurrent with alanine scanning and CD analyses, we also determined the three-dimensional structure of the CV1 dimer at 1.0 Å resolution. The asymmetric unit is composed of two interacting dimers (Fig. 3A). In contrast to the previously proposed dimer interface involving tryptophan residues (Victorio & Sawyer, 2023), the structure of each dimer in the crystal structure reveals a canonical knobs-into-holes packing between Leu6, Leu9, and Leu10 as expected for a dimer of α-helices (Hadley et al., 2008; Keating et al., 2001; Regan & DeGrado, 1988; Tripet et al., 2000). The helical portion of each peptide is capped at the N-terminus by Thr2 (Fig. 3B; (Aurora & Rose, 1998)). Cysteine residues extend beyond the helical segment of each peptide to form bridging disulfide bonds. Asn5 and Asn13 form long-range hydrogen bonding interactions within each dimer (Fig. 3C), which is consistent with the slightly decreased stability of the N5A and N13A dimers relative to CV1 in CD experiments. For each dimer, one of the Phe3 residues and both Trp7 residues are oriented away from the dimer interface, suggesting a limited role for these residues in dimerization/disulfide bond formation (Fig. 3D).
Figure 3.
Crystal structure of disulfide-linked CV1 peptide dimer (PDB 9C5S). In all panels, each helix is represented with the same color (chain A = green, chain B = blue,) and disulfide bonds are represented as yellow sticks. N- and C-termini are labeled as N and C, respectively, with subscripts to indicate the corresponding chain. a) The dimer interface involves interdigitation of the side-chains of Leu6, Leu9, and Leu10 (spheres). b) The oxygen atom of each Thr2 side-chain serves as a hydrogen bond acceptor for Asn5 NH to cap the helix at the N-terminus. c) Asn5 and Asn13 side-chains form long range side-chain-to-side-chain hydrogen bonds between helices. d) Phe3 and Trp7 side-chains (sticks) are peripheral to the helical interface
While not contributing to individual dimers, the Phe3 and Trp7 residues appear to stabilize a dimer of dimers in the asymmetric unit and help to rationalize several observations from previous and current CD data (Victorio & Sawyer, 2023). The individual dimer copies within the dimer of dimers overlay well (Fig. 4A, RMSD = 0.28 Å for all backbone atoms). The arrangement of dimers is antiparallel and highly asymmetrical (Fig. 4B). The interface between dimers resembles a hydrophobic protein core and is composed of four copies each of Phe3, Leu6, Trp7, and Leu10. The Phe3 residues form interacting pairs on either end of the hydrophobic core with a specific packing arrangement in which Hβ atoms from Phe3 of chains B and C form apparent CH-π interactions (Nishio et al., 2014; Zondlo, 2013) with the ring faces of Phe3 from chains D and A, respectively (Fig. 4C). The Trp7 residues form two types of interactions: the Trp7 residues from chains B and C form a parallel displaced stacking arrangement at a distance of approximately 3.2 Å (Figure 4D, left) while the Trp7 residues from chains A and D are parallel to each other but interacting with other hydrophobic core residues instead of each other (Fig. 4D, right).
Figure 4.
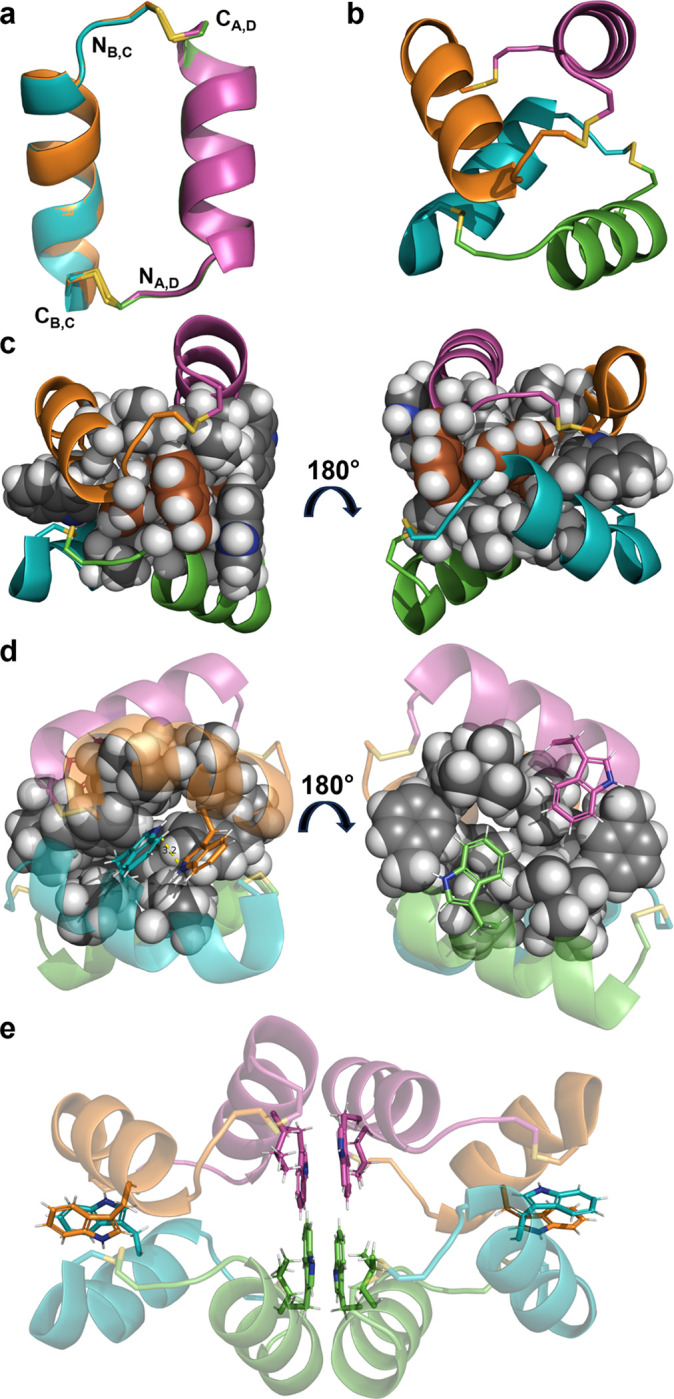
Analysis of crystallographic symmetry. In all panels, each helix is represented with the same color (chain A = green, chain B = blue, chain C = orange, chain D = pink) and disulfide bonds are represented as yellow sticks. N- and C-termini are labeled as N and C, respectively, with subscripts to indicate the corresponding chain. a) An overlay of the two copies of the dimer observed in the asymmetric unit shows high similarity (RMSD = 0.28 Å for all backbone atoms). b) The dimer of dimers is antiparallel with close contact between chains B (blue) and C (orange) and a wider interface between chains A (green) and D (pink). c) Phe3 residues (brown) cap an extended hydrophobic core formed primarily by Phe3, Leu6, Trp7, and Leu10 (spheres). d) Trp7 residues form two different arrangements. Between chains B (blue) and C (orange), Trp7 residues are separated by approximately 3.2 Å in a parallel displaced stacking arrangement. Between chains A (green) and D (pink), Trp7 residues are not interacting but pack in a parallel fashion against other hydrophobic core residues. e) The crystallographic octamer (dimer of dimer of dimers) is formed by extensive hydrophobic and π-π interactions between Trp7 and Leu10 from chains A (green) and D (pink) in adjacent dimer of dimers
In particular, the parallel displaced stacking of Trp residues at the interface between dimers provides a rationale for the apparent shift of the 222 nm CD minimum to 227 nm, which is observed in the spectra of CV1 and many other variants (Grishina & Woody, 1994). As mentioned previously, aromatic-aromatic interactions, especially those involving tryptophan, frequently give rise to CD minima/maxima within the 225–235 nm wavelength range (Andersen et al., 2006; Cochran et al., 2001; Nazzaro et al., 2023; Sawyer & Arora, 2018). The absence of the CD shift for the W7Y dimer supports the idea that the CD shift is related to Trp-Trp interaction. The absence of the 227 nm CD shift for the F3A dimer can be rationalized by the apparent role of the Phe3 residues in stabilizing the dimer of dimers that results in Trp-Trp stacking. The reason for the absence of a CD shift for the T2A dimer is less clear but may be related to the capping function of Thr2, which presumably stabilizes individual helices to promote the formation of the dimer of dimers.
In addition to revealing the interactions driving dimerization of CV1 and its dimer of dimers assembly, the X-ray structure also revealed interactions that drive crystal packing. Residues Trp7 and Leu10 from chains A and D form a relatively flat face that forms hydrophobic and π-π interactions with the same residues from an adjacent dimer of dimers to form an octameric (or dimer of dimer of dimers) structure (Fig. 4E). This hierarchical assembly leads to a porous crystal structure with polar residues (Asn5, Arg8, Gln12, and Asn13) facing the pores, reminiscent of functionalized peptide-derived crystalline frameworks based on π-stacking strategies (Heinz-Kunert et al., 2022; Vijayakanth et al., 2024).
Overall, we report biophysical and crystallographic characterization of spontaneous peptide dimers based on the N-terminus of the p53 protein. CD spectral and structural data highlight major roles for the Leu6 and Leu10 residues in mediating interaction between peptide chains within each dimer. Phe3 and Trp7 residues promote higher-order assemblies of four and eight peptide chains, though such assemblies are not required for efficient dimerization. The four-chain dimer of dimers possesses a protein-like hydrophobic core, and its existence in dilute aqueous solution is supported by the 227 nm minimum observed for almost all Trp-containing variants.
Moreover, structural data suggests a designable molecular architecture beyond the p53 sequence. The all-leucine core at the dimer interface forms a LxxLL pattern, which is a consensus motif through which transcriptional coactivators bind to nuclear receptors in an α-helical conformation (Galande et al., 2005; Heery et al., 1997; Plevin et al., 2005). Beyond its role in transcription, the LxxLL motif is also found in the ubiquitin ligase UBE3A, also known as E6AP, which interacts with the E6 protein of human papillomavirus (HPV) to mediate p53 degradation as a hallmark of the HPV-related carcinogenicity (Ye et al., 2023; Zanier et al., 2013). Thus, it seems highly likely that other LxxLL motif-containing peptides could be dimerized in the same way as the CV1 peptide for potential protein targeting and/or intracellular delivery.
ACKNOWLEDGMENTS
C.G.V. thanks the Henry Luce Foundation for funding support through a Clare Boothe Luce Undergraduate Summer Research Scholarship and the Fordham College Dean’s Office for funding support through a Len Blavatnik STEM Research Fellowship and FCRH Undergraduate Research grants. V.K.O. thanks the University of Missouri for start-up funding. N.S. thanks Fordham University for start-up funding and faculty research grant support. X-ray crystallography data reported here used the AMX beamline of the National Synchrotron Light Source II (NSLS-II), a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. The Center for BioMolecular Structure (CBMS) is primarily supported by the National Institutes of Health, National Institute of General Medical Sciences (NIGMS) through a Center Core P30 Grant (P30GM133893), and by the DOE Office of Biological and Environmental Research (KP1607011).
Footnotes
The authors declare no competing financial interest.
Contributor Information
Magdalena C. DiGiorno, Fordham University
Nisansala Vithanage, University of Missouri.
Clara G. Victorio, Fordham University
Dale F. Kreitler, National Synchrotron Light Source II
Victor K. Outlaw, University of Missouri
Nicholas Sawyer, Fordham University.
References
- 1.Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., & Zwart P. H. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D, 66(2), 213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albericio F., Hammer R. P., GarcÍA-EcheverrÍA C., Molins M. A., Chang J. L., Munson M. C., Pons M., Giralt E., & Barany G. (1991). Cyclization of disulfide-containing peptides in solid-phase synthesis†. International Journal of Peptide and Protein Research, 37(5), 402–413. 10.1111/j.1399-3011.1991.tb00755.x [DOI] [PubMed] [Google Scholar]
- 3.Andersen N. H., Olsen K. A., Fesinmeyer R. M., Tan X., Hudson F. M., Eidenschink L. A., & Farazi S. R. (2006). Minimization and Optimization of Designed β-Hairpin Folds. Journal of the American Chemical Society, 128(18), 6101–6110. 10.1021/ja054971w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreu D., F. A., N.A.. S., M.C. M. M. F., & Barany G. (1994). Formation of disulfide bonds in synthetic peptides and proteins. Methods in Molecular Biology, 35, 91–169. [DOI] [PubMed] [Google Scholar]
- 5.Aurora R., & Rose G. D. (1998). Helix capping. Protein Science, 7(1), 21–38. 10.1002/pro.5560070103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baek S., Kutchukian P. S., Verdine G. L., Huber R., Holak T. A., Lee K. W., & Popowicz G. M. (2012). Structure of the Stapled p53 Peptide Bound to Mdm2. Journal of the American Chemical Society, 134(1), 103–106. 10.1021/ja2090367 [DOI] [PubMed] [Google Scholar]
- 7.Bernal F., Tyler A. F., Korsmeyer S. J., Walensky L. D., & Verdine G. L. (2007). Reactivation of the p53 Tumor Suppressor Pathway by a Stapled p53 Peptide. Journal of the American Chemical Society, 129(9), 2456–2457. 10.1021/ja0693587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernal F., Wade M., Godes M., Davis T. N., Whitehead D. G., Kung A. L., Wahl G. M., & Walensky L. D. (2010). A Stapled p53 Helix Overcomes HDMX-Mediated Suppression of p53. Cancer Cell, 18(5), 411–422. 10.1016/j.ccr.2010.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bishop C. M., Walkenhorst W. F., & Wimley W. C. (2001). Folding of β-sheets in membranes: specificity and promiscuity in peptide model systems11Edited by G. von Heijne. Journal of Molecular Biology, 309(4), 975–988. 10.1006/jmbi.2001.4715 [DOI] [PubMed] [Google Scholar]
- 10.Chandramohan A., Josien H., Yuen T. Y., Duggal R., Spiegelberg D., Yan L., Juang Y.-C. A., Ge L., Aronica P. G., Kaan H. Y. K., Lim Y. H., Peier A., Sherborne B., Hochman J., Lin S., Biswas K., Nestor M., Verma C. S., Lane D. P., Sawyer T. K., Garbaccio R., Henry B., Kannan S., Brown C. J., Johannes C. W., & Partridge A. W. (2024). Design-rules for stapled peptides with in vivo activity and their application to Mdm2/X antagonists. Nature Communications, 15(1), 489. 10.1038/s41467-023-43346-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang Y. S., Graves B., Guerlavais V., Tovar C., Packman K., To K.-H., Olson K. A., Kesavan K., Gangurde P., Mukherjee A., Baker T., Darlak K., Elkin C., Filipovic Z., Qureshi F. Z., Cai H., Berry P., Feyfant E., Shi X. E., Horstick J., Annis D. A., Manning A. M., Fotouhi N., Nash H., Vassilev L. T., & Sawyer T. K. (2013). Stapled α−helical peptide drug development: A potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proceedings of the National Academy of Sciences, 110(36), E3445. 10.1073/pnas.1303002110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cochran A. G., Skelton N. J., & Starovasnik M. A. (2001). Tryptophan zippers: Stable, monomeric β-hairpins. Proceedings of the National Academy of Sciences, 98(10), 5578–5583. 10.1073/pnas.091100898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dantas de Araujo A., Perry S. R., & Fairlie D. P. (2018). Chemically Diverse Helix-Constrained Peptides Using Selenocysteine Crosslinking. Organic Letters, 20(5), 1453–1456. 10.1021/acs.orglett.8b00233 [DOI] [PubMed] [Google Scholar]
- 14.de Veer S. J., Kan M.-W., & Craik D. J. (2019). Cyclotides: From Structure to Function. Chemical Reviews, 119(24), 12375–12421. 10.1021/acs.chemrev.9b00402 [DOI] [PubMed] [Google Scholar]
- 15.Dombkowski A. A., Sultana K. Z., & Craig D. B. (2014). Protein disulfide engineering. FEBS Letters, 588(2), 206–212. 10.1016/j.febslet.2013.11.024 [DOI] [PubMed] [Google Scholar]
- 16.Emsley P., & Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallographica Section D, 60(12 Part 1), 2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 17.Evans P. R., & Murshudov G. N. (2013). How good are my data and what is the resolution? Acta Crystallographica Section D, 69(7), 1204–1214. 10.1107/S0907444913000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galande A. K., Bramlett K. S., Trent J. O., Burris T. P., Wittliff J. L., & Spatola A. F. (2005). Potent Inhibitors of LXXLL-Based Protein–Protein Interactions. ChemBioChem, 6(11), 1991–1998. 10.1002/cbic.200500083 [DOI] [PubMed] [Google Scholar]
- 19.Gimpl G., & Fahrenholz F. (2001). The Oxytocin Receptor System: Structure, Function, and Regulation. Physiological Reviews, 81(2), 629–683. 10.1152/physrev.2001.81.2.629 [DOI] [PubMed] [Google Scholar]
- 20.Goto Y., & Hamaguchi K. (1979). The Role of the Intrachain Disulfide Bond in the Conformation and Stability of the Constant Fragment of the Immunoglobulin Light Chain. The Journal of Biochemistry, 86(5), 1433–1441. [DOI] [PubMed] [Google Scholar]
- 21.Grishina I. B., & Woody R. W. (1994). Contributions of tryptophan side chains to the circular dichroism of globular proteins: exciton couplets and coupled oscillators [ 10.1039/FD9949900245]. Faraday Discussions, 99(0), 245–262. 10.1039/FD9949900245 [DOI] [PubMed] [Google Scholar]
- 22.Hadley E. B., Testa O. D., Woolfson D. N., & Gellman S. H. (2008). Preferred side-chain constellations at antiparallel coiled-coil interfaces. Proceedings of the National Academy of Sciences, 105(2), 530–535. 10.1073/pnas.0709068105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammer R. P., Butrie M. A., Davidson K., Goldblatt P. T., Schrader A. M., Dalluge J. J., Becker A., & Barany G. (2023). Scaled-up Synthesis and Characterization of Oxytocin Trisulfide. International Journal of Peptide Research and Therapeutics, 30(1), 5. 10.1007/s10989-023-10580-9 [DOI] [Google Scholar]
- 24.Heery D. M., Kalkhoven E., Hoare S., & Parker M. G. (1997). A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature, 387(6634), 733–736. 10.1038/42750 [DOI] [PubMed] [Google Scholar]
- 25.Heinz-Kunert S. L., Pandya A., Dang V. T., Tran P. N., Ghosh S., McElheny D., Santarsiero B. D., Ren Z., & Nguyen A. I. (2022). Assembly of π-Stacking Helical Peptides into a Porous and Multivariable Proteomimetic Framework. Journal of the American Chemical Society, 144(15), 7001–7009. 10.1021/jacs.2c02146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henchey L. K., Porter J. R., Ghosh I., & Arora P. S. (2010). High Specificity in Protein Recognition by Hydrogen-Bond-Surrogate α-Helices: Selective Inhibition of the p53/MDM2 Complex. ChemBioChem, 11(15), 2104–2107. 10.1002/cbic.201000378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jenkins H. (2018). Fragon: rapid high-resolution structure determination from ideal protein fragments. Acta Crystallographica Section D, 74(3), 205–214. 10.1107/S2059798318002292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabsch W. (2010). Integration, scaling, space-group assignment and post-refinement. Acta Crystallographica Section D, 66(2), 133–144. 10.1107/S0907444909047374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karas J. A., Wade J. D., & Hossain M. A. (2021). The Chemical Synthesis of Insulin: An Enduring Challenge. Chemical Reviews, 121(8), 4531–4560. 10.1021/acs.chemrev.0c01251 [DOI] [PubMed] [Google Scholar]
- 30.Keating A. E., Malashkevich V. N., Tidor B., & Kim P. S. (2001). Side-chain repacking calculations for predicting structures and stabilities of heterodimeric coiled coils. Proceedings of the National Academy of Sciences, 98(26), 14825–14830. 10.1073/pnas.261563398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laps S., Atamleh F., Kamnesky G., Sun H., & Brik A. (2021). General synthetic strategy for regioselective ultrafast formation of disulfide bonds in peptides and proteins. Nature Communications, 12(1), 870. 10.1038/s41467-021-21209-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McAuley A., Jacob J., Kolvenbach C. G., Westland K., Lee H. J., Brych S. R., Rehder D., Kleemann G. R., Brems D. N., & Matsumura M. (2008). Contributions of a disulfide bond to the structure, stability, and dimerization of human IgG1 antibody CH3 domain. Protein Science, 17(1), 95–106. 10.1110/ps.073134408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchinson C., & Wells J. A. (1989). Protein engineering of disulfide bonds in subtilisin BPN’. Biochemistry, 28(11), 4807–4815. 10.1021/bi00437a043 [DOI] [PubMed] [Google Scholar]
- 34.Momand J., Jung D., Wilczynski S., & Niland J. (1998). The MDM2 gene amplification database. Nucleic Acids Research, 26(15), 3453–3459. 10.1093/nar/26.15.3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muttenthaler M., Andersson A., de Araujo A. D., Dekan Z., Lewis R. J., & Alewood P. F. (2010). Modulating Oxytocin Activity and Plasma Stability by Disulfide Bond Engineering. Journal of Medicinal Chemistry, 53(24), 8585–8596. 10.1021/jm100989w [DOI] [PubMed] [Google Scholar]
- 36.Nazzaro A., Lu B., Sawyer N., Watkins A. M., & Arora P. S. (2023). Macrocyclic β-Sheets Stabilized by Hydrogen Bond Surrogates. Angewandte Chemie International Edition, 62(41), e202303943. 10.1002/anie.202303943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishio M., Umezawa Y., Fantini J., Weiss M. S., & Chakrabarti P. (2014). CH–π hydrogen bonds in biological macromolecules [ 10.1039/C4CP00099D]. Physical Chemistry Chemical Physics, 16(25), 12648–12683. 10.1039/C4CP00099D [DOI] [PubMed] [Google Scholar]
- 38.Northfield S. E., Wang C. K., Schroeder C. I., Durek T., Kan M.-W., Swedberg J. E., & Craik D. J. (2014). Disulfide-rich macrocyclic peptides as templates in drug design. European Journal of Medicinal Chemistry, 77, 248–257. 10.1016/j.ejmech.2014.03.011 [DOI] [PubMed] [Google Scholar]
- 39.Pace C. N., Grimsley G. R., Thomson J. A., & Barnett B. J. (1988). Conformational stability and activity of ribonuclease T1 with zero, one, and two intact disulfide bonds. Journal of Biological Chemistry, 263(24), 11820–11825. 10.1016/S0021-9258(18)37859-1 [DOI] [PubMed] [Google Scholar]
- 40.Plevin M. J., Mills M. M., & Ikura M. (2005). The LxxLL motif: a multifunctional binding sequence in transcriptional regulation. Trends in Biochemical Sciences, 30(2), 66–69. 10.1016/j.tibs.2004.12.001 [DOI] [PubMed] [Google Scholar]
- 41.Postma T. M., & Albericio F. (2014). Disulfide Formation Strategies in Peptide Synthesis [ 10.1002/ejoc.201402149]. European Journal of Organic Chemistry, 2014(17), 3519–3530. 10.1002/ejoc.201402149 [DOI] [Google Scholar]
- 42.Postma T. M., Giraud M., & Albericio F. (2012). Trimethoxyphenylthio as a Highly Labile Replacement for tert-Butylthio Cysteine Protection in Fmoc Solid Phase Synthesis. Organic Letters, 14(21), 5468–5471. 10.1021/ol3025499 [DOI] [PubMed] [Google Scholar]
- 43.Regan L., & DeGrado W. F. (1988). Characterization of a Helical Protein Designed from First Principles. Science, 241(4868), 976–978. 10.1126/science.3043666 [DOI] [PubMed] [Google Scholar]
- 44.Rhys G. G., Dawson W. M., Beesley J. L., Martin F. J. O., Brady R. L., Thomson A. R., & Woolfson D. N. (2021). How Coiled-Coil Assemblies Accommodate Multiple Aromatic Residues. Biomacromolecules, 22(5), 2010–2019. 10.1021/acs.biomac.1c00131 [DOI] [PubMed] [Google Scholar]
- 45.Sawyer N., & Arora P. S. (2018). Hydrogen Bond Surrogate Stabilization of β-Hairpins. ACS Chemical Biology, 13(8), 2027–2032. 10.1021/acschembio.8b00641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shepherd N. E., Hoang H. N., Abbenante G., & Fairlie D. P. (2005). Single Turn Peptide Alpha Helices with Exceptional Stability in Water. Journal of the American Chemical Society, 127(9), 2974–2983. 10.1021/ja0456003 [DOI] [PubMed] [Google Scholar]
- 47.Shi W.-W., Shi C., Wang T.-Y., Li Y.-L., Zhou Y.-K., Zhang X.-H., Bierer D., Zheng J.-S., & Liu L. (2022). Total Chemical Synthesis of Correctly Folded Disulfide-Rich Proteins Using a Removable O-Linked β-N-Acetylglucosamine Strategy. Journal of the American Chemical Society, 144(1), 349–357. 10.1021/jacs.1c10091 [DOI] [PubMed] [Google Scholar]
- 48.Silvestri A. P., Zhang Q., Ping Y., Muir E. W., Zhao J., Chakka S. K., Wang G., Bray W. M., Chen W., Fribourgh J. L., Tripathi S., He Y., Rubin S. M., Satz A. L., Pye C. R., Kuai L., Su W., & Schwochert J. A. (2023). DNA-Encoded Macrocyclic Peptide Libraries Enable the Discovery of a Neutral MDM2–p53 Inhibitor. ACS Medicinal Chemistry Letters, 14(6), 820–826. 10.1021/acsmedchemlett.3c00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh A., Haslach E. M., & Haskell-Luevano C. (2010). Structure-Activity Relationships (SAR) of Melanocortin and Agouti-Related (AGRP) Peptides. In Catania A. (Ed.), Melanocortins: Multiple Actions and Therapeutic Potential (pp. 1–18). Springer New York. 10.1007/978-1-4419-6354-3_1 [DOI] [PubMed] [Google Scholar]
- 50.Spears R. J., McMahon C., & Chudasama V. (2021). Cysteine protecting groups: applications in peptide and protein science [ 10.1039/D1CS00271F]. Chemical Society Reviews, 50(19), 11098–11155. 10.1039/D1CS00271F [DOI] [PubMed] [Google Scholar]
- 51.Tam J. P., Wu C. R., Liu W., & Zhang J. W. (1991). Disulfide bond formation in peptides by dimethyl sulfoxide. Scope and applications. Journal of the American Chemical Society, 113(17), 6657–6662. 10.1021/ja00017a044 [DOI] [Google Scholar]
- 52.Thies M. J. W., Talamo F., Mayer M., Bell S., Ruoppolo M., Marino G., & Buchner J. (2002). Folding and Oxidation of the Antibody Domain CH3. Journal of Molecular Biology, 319(5), 1267–1277. 10.1016/S0022-2836(02)00375-3 [DOI] [PubMed] [Google Scholar]
- 53.Tripet B., Wagschal K., Lavigne P., Mant C. T., & Hodges R. S. (2000). Effects of side-chain characteristics on stability and oligomerization state of a de Novo-designed model coiled-coil: 20 amino acid substitutions in position “d”11Edited by P. E. Wright. Journal of Molecular Biology, 300(2), 377–402. 10.1006/jmbi.2000.3866 [DOI] [PubMed] [Google Scholar]
- 54.Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., & Liu E. A. (2004). In Vivo Activation of the p53 Pathway by Small-Molecule Antagonists of MDM2. Science, 303(5659), 844–848. 10.1126/science.1092472 [DOI] [PubMed] [Google Scholar]
- 55.Victorio C. G., & Sawyer N. (2023). Folding-Assisted Peptide Disulfide Formation and Dimerization. ACS Chemical Biology, 18(7), 1480–1486. 10.1021/acschembio.3c00268 [DOI] [PubMed] [Google Scholar]
- 56.Vijayakanth T., Dasgupta S., Ganatra P., Rencus-Lazar S., Desai A. V., Nandi S., Jain R., Bera S., Nguyen A. I., Gazit E., & Misra R. (2024). Peptide hydrogen-bonded organic frameworks [ 10.1039/D3CS00648D]. Chemical Society Reviews, 53(8), 3640–3655. 10.1039/D3CS00648D [DOI] [PubMed] [Google Scholar]
- 57.Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., Womack T., & Bricogne G. (2011). Data processing and analysis with the autoPROC toolbox. Acta Crystallographica Section D, 67(4), 293–302. 10.1107/S0907444911007773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang D., Chen K., Kulp J. L., & Arora P. S. (2006). Evaluation of Biologically Relevant Short α-Helices Stabilized by a Main-Chain Hydrogen-Bond Surrogate. Journal of the American Chemical Society, 128(28), 9248–9256. 10.1021/ja062710w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Welker E., Wedemeyer W. J., Narayan M., & Scheraga H. A. (2001). Coupling of Conformational Folding and Disulfide-Bond Reactions in Oxidative Folding of Proteins. Biochemistry, 40(31), 9059–9064. 10.1021/bi010409g [DOI] [PubMed] [Google Scholar]
- 60.Yamashita K., Hirata K., & Yamamoto M. (2018). KAMO: towards automated data processing for microcrystals. Acta Crystallographica Section D, 74(5), 441–449. 10.1107/S2059798318004576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yao S., Moyer A., Zheng Y., Shen Y., Meng X., Yuan C., Zhao Y., Yao H., Baker D., & Wu C. (2022). De novo design and directed folding of disulfide-bridged peptide heterodimers. Nature Communications, 13(1), 1539. 10.1038/s41467-022-29210-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ye X., Zhang P., Tao J., Wang J. C. K., Mafi A., Grob N. M., Quartararo A. J., Baddock H. T., Chan L. J. G., McAllister F. E., Foe I., Loas A., Eaton D. L., Hao Q., Nile A. H., & Pentelute B. L. (2023). Discovery of reactive peptide inhibitors of human papillomavirus oncoprotein E6 [ 10.1039/D3SC02782A]. Chemical Science, 14(44), 12484–12497. 10.1039/D3SC02782A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoo D. Y., Barros S. A., Brown G. C., Rabot C., Bar-Sagi D., & Arora P. S. (2020). Macropinocytosis as a Key Determinant of Peptidomimetic Uptake in Cancer Cells. Journal of the American Chemical Society, 142(34), 14461–14471. 10.1021/jacs.0c02109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zanier K., Charbonnier S., Sidi A. O. M. h. O., McEwen A. G., Ferrario M. G., Poussin-Courmontagne P., Cura V., Brimer N., Babah K. O., Ansari T., Muller I., Stote R. H., Cavarelli J., Vande Pol S., & Travé G. (2013). Structural Basis for Hijacking of Cellular LxxLL Motifs by Papillomavirus E6 Oncoproteins. Science, 339(6120), 694–698. 10.1126/science.1229934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zavodszky M., Chen C.-W., Huang J.-K., Zolkiewski M., Wen L., & Krishnamoorthi R. (2001). Disulfide bond effects on protein stability: Designed variants of Cucurbita maxima trypsin inhibitor-V. Protein Science, 10(1), 149–160. 10.1110/ps.26801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng Y., Zhai L., Zhao Y., & Wu C. (2015). Orthogonal Cysteine–Penicillamine Disulfide Pairing for Directing the Oxidative Folding of Peptides. Journal of the American Chemical Society, 137(48), 15094–15097. 10.1021/jacs.5b10779 [DOI] [PubMed] [Google Scholar]
- 67.Zhu H., Gao H., Ji Y., Zhou Q., Du Z., Tian L., Jiang Y., Yao K., & Zhou Z. (2022). Targeting p53–MDM2 interaction by small-molecule inhibitors: learning from MDM2 inhibitors in clinical trials. Journal of Hematology & Oncology, 15(1), 91. 10.1186/s13045-022-01314-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zondlo N. J. (2013). Aromatic–Proline Interactions: Electronically Tunable CH/π Interactions. Accounts of Chemical Research, 46(4), 1039–1049. 10.1021/ar300087y [DOI] [PMC free article] [PubMed] [Google Scholar]