Abstract
Intracellular aggregation of repeat expanded RNA has been implicated in many neurological disorders. Here, we study the role of biomolecular condensates on irreversible RNA clustering. We find that physiologically relevant and disease-associated repeat RNAs spontaneously undergo an age-dependent percolation transition inside multi-component protein-nucleic acid condensates to form nanoscale clusters. Homotypic RNA clusters drive the emergence of multiphasic condensate structures with an RNA-rich solid core surrounded by an RNA-depleted fluid shell. The timescale of the RNA clustering, which drives a liquid-to-solid transition of biomolecular condensates, is determined by the sequence features, stability of RNA secondary structure, and repeat length. Importantly, G3BP1, the core scaffold of stress granules, introduces heterotypic buffering to homotypic RNA-RNA interactions and impedes intra-condensate RNA clustering in an ATP-independent manner. Our work suggests that biomolecular condensates can act as sites for RNA aggregation. It also highlights the functional role of RNA-binding proteins in suppressing aberrant RNA phase transitions.
Introduction
Macromolecular phase separation is a segregative mechanism utilized by living cells to form mesoscale compartments, henceforth referred to as biomolecular condensates1, 2, 3, 4, 5, 6. A common feature of many ribonucleoprotein (RNP) condensates, such as the cytoplasmic stress granules, is their dynamic fluid-like material properties, which enable their facile on-demand formation, dissolution, and rapid macromolecular transport7. Experimental and computational approaches probing intra-condensate rheology have revealed that biomolecular condensates are viscoelastic fluids8, 9, 10, 11 with material and structural properties that can change over time, termed physical aging 12. Age-dependent loss in fluidity of condensates can lead to a liquid-to-solid phase transition12, 13, 14, 15, 16. Both protein and RNA components can contribute to the physical aging of condensates. In the protein-centric model of condensate aging17, 18, 19, 20, phase separation of RNA-binding proteins (RBPs) such as hnRNPA1, FUS, and TDP43 is considered metastable9, 12, 14, 21, 22, 23, 24. Hence, condensates formed by these RBPs are susceptible to transition into stable solids, which can either be a glassy material9, viscoelastic Kelvin-Voigt solid12, or amyloid fiber23, 25. Clinically relevant mutations in these RBPs linked to numerous neurodegenerative disorders including amyotrophic lateral sclerosis (ALS)13, 15, 22, 24 can accelerate the physical aging of condensates.
Multiple lines of recent evidence also suggest that RNA-driven condensation plays a central role in the formation and regulation of RNP condensates including stress granules26, 27, 28 and paraspeckles29, 30. Furthermore, aberrant intracellular aggregation of GC-rich RNAs with expanded repeats is a hallmark of numerous neurological disorders including Huntington’s disease and ALS31, 32, 33, 34, 35, 36, 37. These RNAs form pathological intracellular condensates, which are sensitively dependent on the repeat length and likely involve sequence and structure-specific homotypic RNA-RNA interactions38, 39, 40. Homotypic RNA clustering has also been reported to drive mRNA self-assembly in germ granules41, 42 and in vitro for RNA homopolymers26, indicating condensation of RNA molecules may be ubiquitous in cells. A recent study showed that RNAs have an intrinsic propensity to undergo protein-free phase separation in vitro upon heating with lower critical solution temperatures (LCSTs), driven by desolvation entropy and Mg2+-dependent physical cross-linking of RNA molecules43. The thermo-responsive phase separation of RNAs can be coupled to an intra-condensate networking transition within the dense phase, referred to as percolation44, 45, 46, which engenders long-lived physical cross-linking of RNA chains through nucleobase-specific interactions. Importantly, phase separation and percolation of RNA chains are two distinct transitions, the former being an entropy-driven density transition leading to the formation of phase-separated RNA condensates and the latter being an associative transition mediated by multivalent RNA-RNA interactions. Percolation coupled to phase separation can result in the dynamical arrest of RNAs in the dense phase rendering RNA condensation irreversible. RNA percolation could potentially be exploited in disease conditions to perturb RNP granule dynamics via aberrant RNA clustering, and hence, provides a conceptual framework to model the aggregation landscape of repeat expanded RNAs. These outstanding insights lead to two important questions: (a) do multicomponent RNP condensate microenvironments enhance or suppress RNA percolation, and (b) does intra-condensate RNA percolation contribute to the age-dependent condensate transition from a predominantly liquid to a solid state?
To address these questions, here we employ a designed multi-component condensate system and quantitative microscopy with nanorheology, focusing our attempt to capture key elements of intra-condensate RNA percolation. Our experiments reveal that percolation transitions of physiologically relevant and disease-associated repeat RNAs engender the formation of viscoelastic RNA-rich sub-phases embedded within a fluid-like condensate matrix in an age-dependent manner. The timescale of RNA percolation is tuned by the RNA sequence, secondary structure, and repeat length. Importantly, multivalent RBPs such as G3BP1 can buffer intra-condensate RNA-RNA homotypic interactions, thereby enhancing condensate metastability and delaying the onset of RNA clustering. Overall, our findings suggest that biomolecular condensates can act as sites for RNA clustering and highlight the chaperone-like function of RBPs to buffer the intrinsic capacity of some RNAs to undergo irreversible percolation.
Results
RNA aggregation is enhanced in multi-component biomolecular condensates
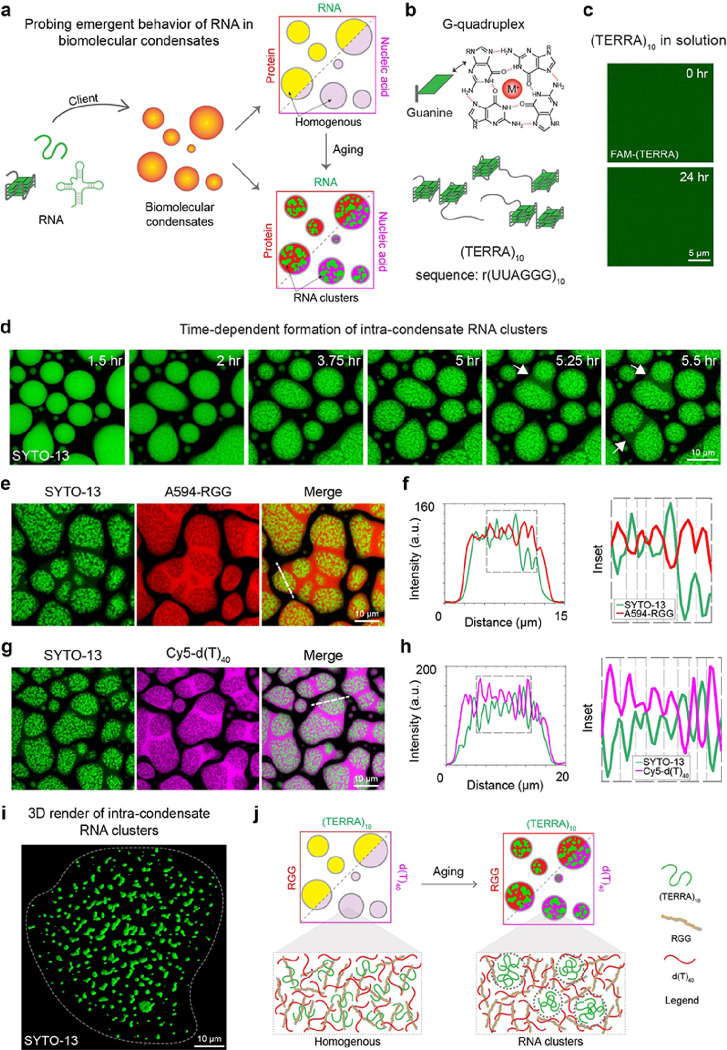
To study the emergent role of homotypic RNA-RNA interactions within heterotypic protein-nucleic acid condensates, we utilized a model condensate system amenable to quantitative experimental interrogation. This model system consists of an RNA binding motif-inspired multivalent disordered polypeptide [RGG; sequence: (RGRGG)5] and a single-stranded nucleic acid [poly-thymine DNA, d(T)40]. Phase separation in RGG-d(T)40 mixtures is driven by obligate cation-π and electrostatic protein-nucleic acid interactions8. Furthermore, the material and physical properties of RGG-nucleic acid condensates are fully tunable via peptide and nucleic acid sequence and length8, 47, providing a robust means to tune the condensate microenvironment. Introducing additional components, such as client RNAs featuring specific primary sequence and secondary structures, to this model condensate allows us to systematically probe the effects of compositional complexity and RNA-driven changes on condensate physical properties (Fig. 1a). We hypothesize that the introduction of RNAs capable of forming homotypic intra- and inter-molecular contacts within RGG-d(T)40 condensates would result in one of two outcomes: the formation of a homogenous ternary condensate where the RNA fully mixes with the two primary condensate-forming components, RGG and d(T)40, or a multiphasic condensate with RNA preferentially partitioning into one of the two co-existing phases. A possible third outcome would be a homogenous ternary condensate undergoing age-dependent transformation into a multiphasic condensate due to RNA demixing (Fig. 1a). In our first set of experiments, we put this idea to the test by utilizing the naturally occurring telomeric repeat-containing RNA48, 49 [TERRA, sequence: (UUAGGG)n] (Fig. 1b), which is known to form G-quadruplex (GQ) structure50, 51, 52.
Figure 1. RNA aggregation is enhanced within multi-component biomolecular condensates.
(a) Depiction of the experimental approach to probing the emergent properties of a client RNA in a heterotypic biomolecular condensate system comprised of RGG and d(T)40. (a) A top-view schematic of Hoogsteen base-pairing in a G-quartet that forms the structural foundation of (TERRA)10. (c) In dilute solution, (TERRA)10 at 10 mg/ml remains soluble and does not show age-dependent aggregation, as probed by FAM-labeled (TERRA)4, over a period of 24 hours at room temperature. (d) In multicomponent condensates containing 1.0 mg/ml (TERRA)10, 5.0 mg/ml RGG, 1.5 mg/ml d(T)40 [buffer = 25 mM Tris-HCl (pH 7.5), 25 mM NaCl, 20 mM DTT], (TERRA)10 undergoes an age-dependent demixing transition into fractal-like clusters, visualized with SYTO-13 (green). RNA demixing results in a core-shell architecture of the condensates. Fusion of the shell phase of neighboring condensates is indicated by white arrows (see SupplementaryVideo 1). Localization of (TERRA)10 in 18 hours aged condensates [same composition as (d)] is negatively correlated with the localizations of RGG (e, f) and d(T)40 (g, h) as visualized by Alexa594-RGG and Cy5-d(T)40. (i) Three-dimensional rendering of super-resolution Z-stack images of (TERRA)10 clusters from an 18-hour-aged sample [same composition as (d)]. (j)Schematic of age-dependent intra-condensate RNA clustering. In experiments utilizing fluorescently labeled components, the concentration is 250 nM to 500 nM. Each experiment was independently repeated at least three times.
Without multivalent cofactors such as Mg2+ ions, (TERRA)10 alone remains fully soluble over a period of 24 hours at room temperature (Fig. 1c). When introduced as a client to RGG-d(T)40 condensates, (TERRA)10 preferentially partitions in the dense phase and remains homogeneously distributed at initial time points (Supplementary Fig. 1). Using a nucleic acid-binding dye (SYTO-13), we next tracked whether intra-condensate spatial distribution of TERRA changes with condensate age. Remarkably, we find that TERRA undergoes age-dependent demixing inside RGG-d(T)40 condensates leading to the formation of RNA clusters (Fig. 1d). As a result, a distinct multiphasic architecture emerges that is defined by an RNA-rich inner core surrounded by an RNA-deplete shell. Utilizing fluorescently labeled anti-sense TERRA, we confirmed that the inner SYTO-13-positive sub-phases in aged condensates are indeed formed by TERRA clusters (Supplementary Fig. 1). The outer phase of aged condensates displays liquid-like dynamics such as fusion with the outer phase of nearby condensates, which is not observed for the RNA-rich core (Fig. 1d; Supplementary Video 1). To elucidate the molecular compositions of these distinct phases, we employ a pairwise imaging approach and identify the localization of each component. The TERRA-enriched clusters within the condensate are significantly depleted of RGG (probed by Alexa594-labeled RGG) and d(T)40 [probed by Cy5-labeled d(T)40] (Fig. 1e–h). Line profiles illustrate the anti-correlation between TERRA and either of these two components (Fig. 1f, h). Moreover, the RNA clusters are shown to be quite irregular in morphology (Fig. 1i). Replacing d(T)40 with r(U)40 as a condensate forming component in the ternary system did not alter the age-dependent clustering of TERRA (Supplementary Fig. 2). However, we do not observe this time-dependent appearance of RNA clusters in the dense phase of binary condensates composed of TERRA and RGG (Supplementary Fig. 3). These observations suggest that age-dependent RNA demixing is an emergent property of the ternary condensates. Overall, these results demonstrate that RNA partitioning into multi-component biomolecular condensates can drive age-dependent homotypic RNA clustering (Fig. 1j). Next, we attempt to dissect which features of the RNA molecules enable this emergent behavior.
Intra-condensate RNA aggregation is driven by RNA percolation transition
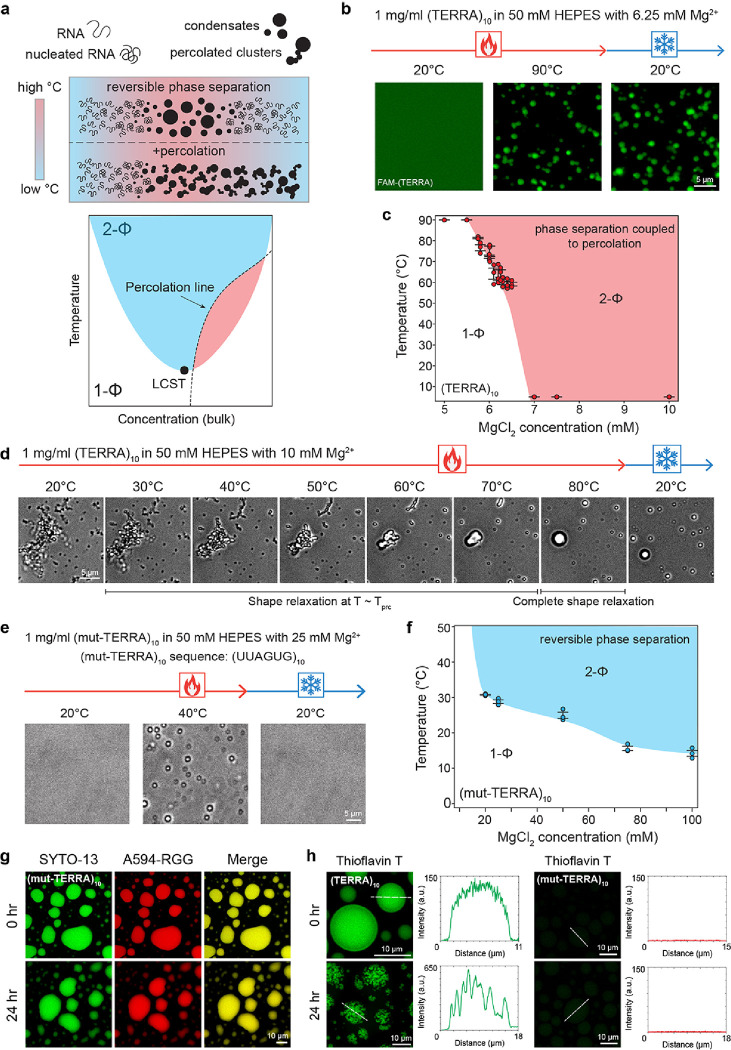
In the absence of proteins, RNA can undergo reversible temperature-dependent phase separation with a secondary percolation transition43 (Fig. 2a). The percolation transition, which is manifested by homotypic RNA-RNA interactions and sensitively depends on the RNA sequence and secondary structure43, dynamically arrests RNA in the dense phase and makes RNA phase separation irreversible. Therefore, RNAs with a strong percolation propensity tend to form irreversible condensates in contrast to RNAs with a weak or no percolation propensity (Fig. 2a). We hypothesized that the demixing of (TERRA)10 in the dense phase of the multicomponent protein-nucleic acid condensates (Fig. 1; Supplementary Fig. 2) stems from a strong percolation propensity of the RNA. To test this idea, we performed temperature-controlled microscopy of (TERRA)10. We observed that (TERRA)10 [1.0 mg/ml RNA in 50 mM HEPES (pH 7.5), 6.25 mM Mg+ 2] remains homogenous at 20°C but undergoes an irreversible phase transition with a lower cloud point temperature (LCPT) of T = 60.2 ± 1.3°C upon heating. The irreversibility of the (TERRA)10 condensates upon cooling to 20°C suggests the formation of a strongly percolated RNA network in the dense phase (Fig. 2b, c; Supplementary Video 2). Using a series of Mg2+ titrations, we mapped the state diagram of (TERRA)10 and observed that the phase separation coupled to percolation behavior of (TERRA)10 is extremely sensitive to Mg2+ concentration with a very narrow range of Mg2+ concentrations (5.75 to 6.5 mM) where thermo-responsive phase separation is experimentally observable (Fig. 2c). At Mg2+concentrations higher than 6.5 mM, we found that (TERRA)10 always exists as percolated clusters even at the lowest temperature tested, 2.0°C (Fig. 2c). At temperatures above percolation temperature (Tprc), (TERRA)10 clusters underwent shape relaxation into energetically favorable spherical condensates that persist after cooling, thereby irreversibly trapping the RNA in the condensed state (Fig. 2d; Supplementary Video 3). These results therefore reveal a strong percolation propensity of (TERRA)10, which is likely to drive its age-dependent aggregation within RGG-d(T)40 condensates.
Figure 2. TERRA undergoes phase separation coupled to percolation that can be perturbed by mutations.
(a) (top) A schematic representing reversible phase separation to form RNA condensates as well as the phase separation coupled to percolation behavior to form irreversible condensates, in response to heating/cooling ramps. (bottom) Depiction of an RNA state diagram, highlighting regions of reversible LCST-type phase separation (blue) and irreversible percolation (red) demarcated by the percolation line (black dashed line). (b) (TERRA)10 with 6.25 mM Mg2+ remains homogenous in solution at 20°C, as visualized using FAM-(TERRA)4. At an elevated temperature, (TERRA)10 phase separates into condensates and does not revert to solution upon cooling, indicative of percolated network formation (Supplementary Video 2) (c) State diagram of (TERRA)10 for a set of experiments similar to (b) with titrations of Mg2+ concentration. (d)Temperature-controlled microscopy shows that (TERRA)10 with 10 mM Mg2+ forms percolated clusters at 20°C, which upon heating above the percolation temperature (Tprc), undergo shape relaxation into spherical condensates that persist when cooled to 20°C (Supplementary Video 3). (e) Temperature-controlled microscopy of 1 mg/ml (mut-TERRA)10 in 50 mM HEPES with 25 mM Mg2+ shows reversible RNA phase separation (Supplementary Video 4).(f) State diagram of (mut-TERRA)10 for a set of experiments similar to (e) with titrations of Mg2+ concentration. (mut-TERRA)10 in RGG-d(T)40 condensates do not show intra-condensate RNA percolation. (h) Thioflavin T (ThT) staining of (TERRA)10 containing RGG-d(T)40 condensates shows homogenous ThT fluorescence at 0 hours and ThT fluorescence within intra-condensate RNA clusters at 24 hours after sample preparation. ThT staining of (mut-TERRA)10 containing RGG-d(T)40 condensates shows the absence of ThT fluorescence at 0 hours and 24 hours after sample preparation. The concentration of ThT used is 50 μM. The concentration of RNA used for temperature-controlled microscopy measurements is 1 mg/ml in 50 mM HEPES (pH 7.5) with the specified Mg2+ concentrations. The composition of the ternary condensate system is 1 mg/ml RNA, 5 mg/ml RGG, and 1.5 mg/ml d(T)40 in a buffer containing 25 mM Tris-HCl (pH 7.5), 25 mM NaCl, and 20 mM DTT. In experiments utilizing fluorescently labeled components, the concentration range is 250 nM to 500 nM. Each experiment was independently repeated at least three times.
An important molecular feature of RNA percolation is the contribution of purine-mediated base-pairing and stacking interactions43. It is also known that G-rich sequences of RNA can form aggregates in solution26. Many of these sequences are also known to form GQ structures with high stability in the presence of monovalent ions53. We reasoned that the propensity of TERRA to form GQ structures50, 52 may contribute to homotypic RNA clustering via percolation. If true, this can be modulated by TERRA sequence perturbations. To test this idea, we employed a mutant (TERRA)10 sequence [(mut-TERRA)10] with a G-to-U substitution e.g., (UUAGUG)10, which is expected to disrupt the stability of the GQ state54,55. We performed temperature-controlled microscopy experiments with (mut-TERRA)10 samples and observed that the LCST transition of (mut-TERRA)10 requires a substantially higher concentration of Mg2+ ions (Fig. 2b, c vs. 2e, f). At 25 mM Mg2+, (mut-TERRA)10 undergoes reversible phase separation upon heating with an LCPT of 28.8 ± 0.42°C (Fig. 2e; Supplementary Video 4). We note that under the same condition, wild-type (WT) (TERRA)10 would exist as percolated clusters at all experimentally accessible temperatures. Percolation of (mut-TERRA)10 was not observed at any conditions tested (Fig. 2f) demonstrating that the G-to-U substitution attenuates the intermolecular RNA-RNA interactions between TERRA chains. Consistent with the absence of RNA percolation in RNA-only condensates, we find that (mut-TERRA)10 does not form age-dependent RNA aggregates in RGG-d(T)40 condensates (Fig. 2g; Supplementary Fig. 4, 5). This data suggests that intra-condensate RNA percolation is RNA sequence- and structure-specific.
Does the intra-condensate aggregation of TERRA, as reported in Fig. 1d–i, proceed via a transition from intra-molecular to inter-molecular GQ structures? We tested this idea by utilizing a GQ-selective fluorescent probe Thioflavin T (ThT), which has been previously shown to display fluorescence activation upon binding to GQ structures with high specificity56, 57, 58. At an early time-point after the preparation of RGG-d(T)40 condensates containing (TERRA)10, ThT fluorescence was observed to be uniformly distributed throughout the condensate suggesting that TERRA is likely forming intramolecular GQs within the homogenous condensate (Fig. 2h). Importantly, RGG-d(T)40 condensates with (TERRA)10 clusters, formed upon aging, displayed ThT positivity, with fluorescence intensities of the clusters being ~ 4-fold higher than nascent homogeneous condensates. This observation indicates that the TERRA in these aggregates is also likely to form GQs (Fig. 2h). In the case of the (mut-TERRA)10, there was no detectable ThT signal under identical imaging conditions at all time points (Fig. 2h). This is consistent with the previous reports that this particular G-to-U substitution disrupts the GQ structure of TERRA54, 55. Negative control of RGG-d(T)40 and RGG-d(T)40 condensates containing a non-percolating RNA43, poly-uridylic acid (polyU), did not show any measurable ThT signal under identical experimental conditions (Supplementary Fig. 6). Finally, intra-condensate (TERRA)10 clusters were observed ubiquitously in the presence of a wide variety of monovalent salts (Supplementary Fig. 7) that differentially impact the stability of RNA GQ structures, suggesting observed RNA percolation may directly stem from the clustering propensity of purine-rich RNAs43.
Timescale of RNA clustering depends on the TERRA repeat number
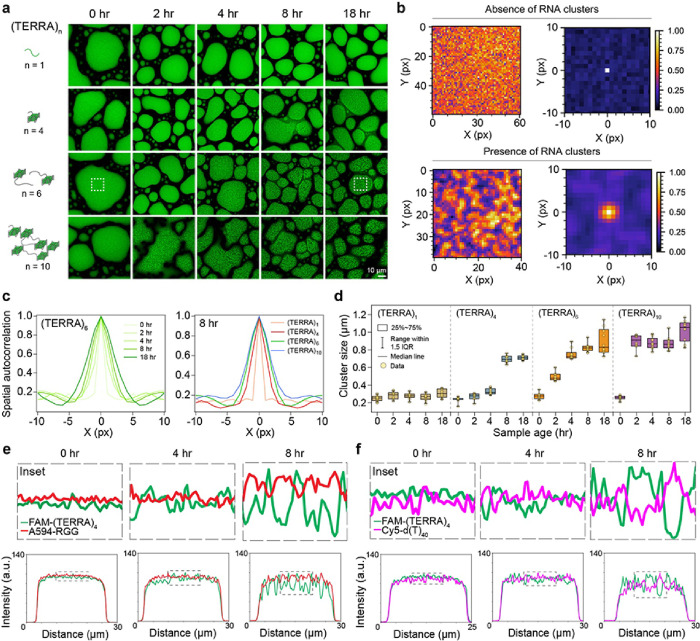
The number of guanine tracts (G-tracts) plays important roles in monomeric RNA GQ and multimeric RNA structure formation59, 60, 61. We reasoned that time-dependent TERRA aggregation in RGG-d(T)40 condensates (Fig. 1d) can be tuned by the ability of TERRA to form multi-molecular GQ structures, which is expected to depend on the number of repeat units of TERRA59, 62. To test this idea, we titrated the number of TERRA repeat units, [UUAGGG]n, where n = 1, 4, 6, and 10; far below the number of repeat units transcribed in cells, 100 to 9000 nucleotides49, 63. We observed that all RNAs that could form intramolecular GQs, e.g., n = 4, 6, and 10, showed age-dependent intra-condensate cluster formation with a corresponding timescale correlated inversely with the number of repeats (Fig. 3a). (TERRA)10 forms microscale aggregates the fastest with a timescale of ~ 2 hours, whereas (TERRA)4 form clusters at substantially slower rate (~ 8 hours) and the clusters are less distinct (Fig. 3a). To quantify the relative intra-condensate cluster sizes as a function of time, we employed spatial autocorrelation analysis (SAC) of the confocal fluorescence images of condensates (Fig. 3a insets, white-dotted boxes; Supplementary Fig. 8). In the absence of clusters larger than the detection limit of the microscope, which is the case for (TERRA)6 at 15 minutes after sample preparation, SAC returns the size of spatial fluctuations, 0.28 ± 0.01 μm, which closely corresponds to the image resolution (1 pixel = 0.2196 μm). However, at 18 hours time point when the RNA clusters in the condensate are detectable, SAC reveals the characteristic size of the RNA clusters being 0.89 ± 0.04 μm (Fig. 3b). Employing SAC, we find that the size of RNA clusters increases, and the timescale of RNA cluster formation decreases with increasing number of repeat units of TERRA (Fig. 3c, d). Simultaneously, we see increased negative correlation between the intensities of TERRA and the primary components, RGG and d(T)40, as a function of condensate age (Fig. 3e, f; Supplementary Fig. 9, 10). As clusters become more pronounced with the aging of condensates, the anti-correlation of intensities becomes more apparent (Fig. 3e, f; Supplementary Fig. 9, 10). These data suggest that percolated TERRA clusters demix from the RGG and d(T)40 in a time-dependent manner. Together with the data presented in the previous section on (mut-TERRA)10 failing to form homotypic intra-condensate clusters, these data suggest that repeat number and RNA sequence variations are molecular regulators of age-dependent RNA percolation within biomolecular condensates.
Figure 3. TERRA repeat numbers dictate the timescale of RNA clustering.
(a) The effect of (TERRA)n repeat number (n) on the timescale of RNA clustering in RGG-d(T)40 condensates. TERRA clusters are visualized with SYTO-13 (green). The white box corresponds to analyses done in (b). (b) Color map images of (TERRA)6 containing RGG-d(T)40 condensates as shown in (a) at 15 minutes (above; absence of RNA clustering) and 18 hours (below; presence of RNA clustering) after sample preparation along with corresponding x-y spatial autocorrelation functions from SAC. (c) SAC line profiles of (TERRA)6 at various time points as shown in (b) (left) and SAC line profiles of (TERRA)n (n= 1, 4, 6, 10) at a sample age of 8 hours (right). (d) Cluster sizes derived from SAC analysis of (TERRA)n at increasing sample age. Pairwise line profile analyses of (TERRA)4 images with respect to RGG (e) and d(T)40 (f) as a function of time (for corresponding images, see Supplementary Fig. 9). Each line profile shown here is normalized with respect to the maximum intensity value, wherein all values were first offset by the minimum intensity value. The composition of the ternary RGG-d(T)40 condensate system is 1 mg/ml RNA, 5 mg/ml RGG, and 1.5 mg/ml d(T)40 in a buffer containing 25 mM Tris-HCl (pH 7.5), 25 mM NaCl, and 20 mM DTT. In experiments utilizing fluorescently labeled components, the concentration range is 250 nM to 500 nM. Each experiment was independently repeated at least three times.
Repeat expanded RNAs form intra-condensate clusters in a length-dependent manner
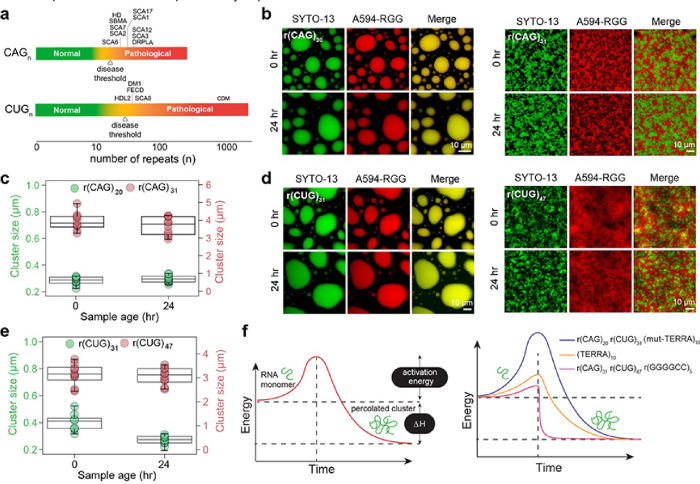
Intracellular RNA aggregation has been widely implicated in several diseases primarily in the category of trinucleotide repeat expansion disorders such as Huntington’s disease (CAG repeat), Fragile-X syndrome (CGG repeat), and myotonic dystrophy (CUG repeat)37, 64, 65, 66, 67, 68, 69. One characteristic of these disorders is that pathology occurs at a number of repeats beyond what is found in healthy individuals. Further, the threshold number of repeats associated with pathological outcomes varies with the repeat RNA sequence. For example, the number of r(CAG) repeats required for disease onset is ~ 15 to 20 repeats less than that required for r(CUG) (Fig. 4a). This implies that the cellular machinery can better tolerate r(CUG) repeats than it can with r(CAG) repeats64, 65, 66, 67, 68.
Figure 4. Repeat expanded RNAs form intra-condensate clusters in a length-dependent manner.
(a) Schematic of disease-associated triplet RNA repeat expansions highlighting the threshold number of RNA repeats (n) that correspond to healthy states versus pathological states. Diseases are listed in an order according to the minimum RNA repeat length required for disease onset. The disease threshold for each GC-rich repeat RNA is marked according to the lowest repeat length linked to disease onset64, 65, 66. Disease abbreviations are as follows. SCA6: Spinocerebellar Ataxia (SCA) Type 6, HD: Huntington’s disease, SBMA: Spinal and Bulbar Muscular Atrophy, SCA7: SCA Type 7, SCA2: SCA Type 2, SCA17: SCA Type 17, SCA1: SCA Type 1, SCA12: SCA Type 12, SCA3: SCA Type 3, DRPLA: Dentatorubral-Pallidoluysian Atrophy, HDL2: Huntington’s Disease-Like 2, DM1: Myotonic Dystrophy Type 1, FECD: Fuchs’ Endothelial Corneal Dystrophy, SCA8: SCA Type 8, CDM: Congenital Myotonic Dystrophy. (b)r(CAG)20 containing RGG-d(T)40 condensates remain homogenous and do not form clusters with time (left) whereas r(CAG)31 containing condensates form microscale RNA clusters. The 0-hour image was acquired after 15 minutes of sample preparation (right). (c) Cluster sizes derived from SAC analysis corresponding to (b). (d) r(CUG)31 containing RGG-d(T)40 condensates remain homogenous for 24 hours whereas r(CUG)47 containing condensates form microscale RNA clusters. The 0-hour image was acquired after 15 minutes of sample preparation. (e) Cluster sizes derived from SAC analysis corresponding to (d). (f)A schematic showing the hierarchy of activation energy barrier for an RNA monomer to form percolated RNA clusters. The composition of the ternary RGG-d(T)40 condensate system is 1 mg/ml RNA [0.45 mg/ml in the case of r(CUG)47], 5 mg/ml RGG, and 1.5 mg/ml d(T)40 in a buffer containing 25 mM Tris-HCl (pH 7.5), 25 mM NaCl, and 20 mM DTT. In experiments utilizing fluorescently labeled components, the concentration range is 250 nM to 500 nM. Each experiment was independently repeated at least three times.
Motivated by our results of enhanced TERRA clustering as a function of increasing repeat number, we interrogated the clustering propensity of two repeat RNAs, r(CAG)n and r(CUG)n, with repeat lengths higher and lower than the pathological limit using our model condensate system. r(CAG) repeat lengths lower than 21 are found in healthy individuals, however, repeat lengths exceeding this number are considered to be at the intermediate-high risk scale of being most likely to attain diseases such as spinocerebellar ataxia and Huntington’s disease (Fig. 4a). We observed that r(CAG)20 stays predominantly homogeneous in the dense phase of RGG-d(T)40 condensates at all ages till 24 hours, the end time of our experiments. The mean RNA cluster size is estimated to be 0.28 ± 0.01 μm, which closely corresponds to the image resolution (1 pixel = 0.2196 μm) (Fig. 4b, c; Supplementary Fig. 11). Strikingly, r(CAG)31, which corresponds to a pathological number of repeats, spontaneously demixes into fractal-like clusters prior to sample imaging (15 minutes post-preparation) (Fig. 4b, c; Supplementary Fig. 11). In these samples, r(CAG)31 clusters are extensively percolated with cluster sizes that are nearly 4-fold larger (3.94 ± 0.14 μm) than those formed by TERRA at the same concentration (Fig. 4c). Reduced r(CAG)31 concentration reduces the intra-condensate cluster size, but the RNA aggregation timescale appears to be independent of r(CAG)31 concentration (Supplementary Fig. 12).
We next examined intra-condensate percolation of r(CUG)n for n = 31, which represents a healthy number of repeats. Similar to r(CAG)20, we did not observe intra-condensate r(CUG)31 clustering. The estimated cluster size is 0.40 ± 0.02 μm, which closely corresponds to the image resolution (1 pixel = 0.2196 μm) (Fig. 4d, e; Supplementary Fig. 11). In stark contrast, r(CUG)47 forms microscale clusters within 15 minutes after sample preparation (Fig. 4d, e; Supplementary Fig. 11). We also tested intra-condensate clustering behavior of a hexanucleotide repeat expanded RNA, r(GGGGCC)n, that is associated with ALS and frontotemporal dementia (FTD)59, 70, 71. We observed spontaneous r(GGGGCC)5 clustering in RGG-d(T)40 within 15 minutes after sample preparation (Supplementary Fig. 13). Importantly, r(GGGGCC)5 clusters showed ThT positivity but not r(CAG)20 or r(CAG)31 clusters (Supplementary Fig. 14). This is consistent with the ability of r(GGGGCC)5 to form GQ structures59. Overall, the distinct differences in the intra-condensate percolation behavior of each of these RNAs and TERRA of various lengths suggest a heuristic framework where a sequence and length-specific activation energy barrier dictates the timescale of intra-condensate RNA clustering (Fig. 4f, left). In this picture, r(CAG)20, r(CUG)31, and (mut-TERRA)10 molecules have a substantially higher energy barrier to undergo intra-condensate percolation as compared to (CAG)31, r(CUG)47, and r(GGGGCC)5, whereas (TERRA)10 features an intermediate activation energy barrier (Fig. 4f, right).
Intra-condensate RNA clustering drives a liquid-to-solid phase transition
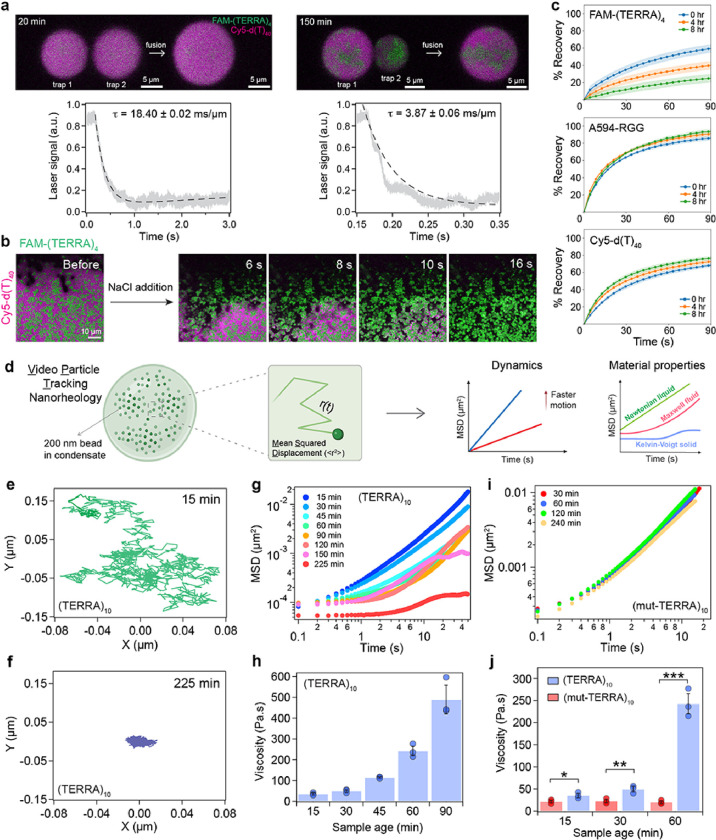
What are the material properties of condensates containing percolated RNA clusters? Time-lapse microscopy suggests that the shell phase of the aged condensates containing TERRA clusters can undergo fusion, signifying liquid-like properties (Fig. 1d, Supplementary Video 1). However, these condensates are in contact with the glass surface, which can influence their fusion kinetics substantially72. To examine the dynamical behavior of (TERRA)10 containing RGG-d(T)40 condensates quantitatively as a function of age, we employed controlled condensate fusion using optical tweezers73, 74. We trapped two condensates using a dual-trap optical tweezer and initiated condensate fusion while recording the fluorescence of FAM-(TERRA)4 and Cy5-d(T)40 simultaneously. At 20 minutes after the condensate preparation, when both TERRA and d(T)40 are uniformly distributed throughout the droplets, they undergo fusion with a fusion relaxation time (τ) of 18.40 ± 0.02 ms/μm. The components were homogeneously mixed after the condensate coalescence was completed (Fig. 5a; Supplementary Video 5). However, for condensates aged for 150 minutes, distinct RNA clusters were visible within condensates (Fig. 5a; Supplementary Video 6). These aged condensates containing RNA clusters were still able to fuse indicating that the condensate shell is dynamic and behaves as a terminally viscous liquid. Force relaxation analysis showed that fusion of the outer shell of aged condensates occurs more rapidly with a fusion relaxation time of 3.87 ± 0.06 ms/μm (Fig. 5a). However, the irregular RNA clusters within the condensate remained demixed at all conditions, indicating that they are in a different material state than RGG-d(T)40 rich phase (Fig. 5a). We tested this directly by designing a condensate dissolution assay, which would preferentially dissolve the non-percolated components (Supplementary Fig. 15a). We observed that addition of 0.5 μl of 5 M NaCl is sufficient to rapidly dissolve condensates formed by RGG-d(T)40 (Supplementary Fig. 15b). However, NaCl addition to aged RGG-d(T)40 condensates containing TERRA clusters showed only partial dissolution where the RGG-d(T)40 rich shell phase dissolved immediately but the RNA clusters persisted (Fig. 5b; Supplementary Videos 7, 8). Notably, freshly prepared (TERRA)10 containing RGG-d(T)40 condensates lacking RNA clusters (15 minutes post preparation) were observed to dissolve completely by NaCl treatment (Supplementary Fig. 15c; Supplementary Video 9). We also probed the translational mobility of each component in the condensate as a function of age by fluorescence recovery after photobleaching (FRAP; Fig. 5c). We chose (TERRA)4 as a representative RNA for these measurements which forms intra-condensate clusters by 8 hours (Fig. 3a, c). FRAP analysis reveals that at early time points before the onset of visible RNA clusters (< 4 hours), all three components are relatively dynamic (Fig. 5c; Supplementary Fig. 16; Supplementary Videos 10, 11, 12). However, FRAP traces of TERRA, but not RGG and d(T)40, show a progressive drop in recovery with condensate age (Fig. 5c; Supplementary Fig. 16; Supplementary Videos 13, 14, 15).
Figure 5. Intra-condensate RNA clustering drives a liquid-to-solid transition.
(a) (top) Optical tweezer-mediated fusion of (TERRA)10 containing RGG-d(T)40 condensates at two different time points, as indicated, after sample preparation (see Supplementary Videos 5, 6). (bottom) Corresponding force relaxation profiles and the estimated fusion relaxation times (error represents ±1 standard deviation). (b) The addition of 0.5 μl of 5 M NaCl to (TERRA)10 containing RGG-d(T)40 condensates at 6 hours after sample preparation shows the dissolution of Cy5-d(T)40-rich shell phase (magenta) but not the (TERRA)10 clusters (green) (see Supplementary Fig. 15; Supplementary Video 7). (c) FRAP recovery profiles of FAM-(TERRA)4, Alexa 594-RGG, and Cy5-d(T)40 in (TERRA)4 containing RGG-d(T)40 condensate system with age (Supplementary Fig. 16; Supplementary Videos 10–15). Shaded regions in each plot signify the standard error. (d) Schematic of video particle tracking (VPT) nanorheology using beads passively embedded inside condensates, which is used to estimate their mean squared displacements (MSD) to ascertain condensate material properties. Created with BioRender.com. Individual bead trajectories at 15 minutes (e) and 225 minutes (f) inside (TERRA)10 containing RGG-d(T)40 condensates. (g) MSDs of beads inside (TERRA)10 containing RGG-d(T)40 condensates as a function of condensate age. (h) The corresponding terminal viscosities are reported. (i) MSDs of beads inside (mut-TERRA)10 containing RGG-d(T)40 condensates as a function of condensate age. (j) The corresponding terminal viscosities are reported in comparison to that of (TERRA)10 containing RGG-d(T)40 condensates as a function of condensate age. Statistical significance was determined by performing a two-sided Student’s t-test (* means p<0.05, ** means p<0.01, *** means p<0.001) between viscosities of (TERRA)10 and (mut-TERRA)10 condensates; the p-values determined at sample age of 15 minutes, 30 minutes, and 60 minutes are 0.0423, 0.0100, and 0.0002, respectively. The composition of the ternary condensate system is 1 mg/ml RNA, 5 mg/ml RGG, and 1.5 mg/ml d(T)40 for the optical tweezer and FRAP experiments; 0.5 mg/ml RNA, 2.5 mg/ml RGG, and 0.75 mg/ml d(T)40 for the condensate dissolution experiments; and 2 mg/ml RNA, 10 mg/ml RGG, and 3.0 mg/ml d(T)40 for the VPT measurements. Buffer composition for all experiments is 25 mM Tris-HCl (pH 7.5), 25 mM NaCl, and 20 mM DTT. In experiments utilizing fluorescently labeled components, the concentration range is 250 nM. The left panel in (a) is representative of three samples, and the right panel in (a) is representative of a single sample. All other measurements were independently repeated at least three times.
The reduced translational mobility of TERRA could result from the formation of dynamically arrested intra-condensate RNA networks, driving a liquid-to-solid phase transition. To directly probe the material properties of condensates, we employed video particle tracking (VPT) nanorheology using 200 nm fluorescently labeled beads8 (Fig. 5d). The mean squared displacement (MSD) profiles of the probe particles have distinct characteristics for Newtonian liquids, viscoelastic fluids with terminal viscous behavior, and Kelvin-Voigt solids with terminal elastic behavior12, 75 (Fig. 5d, right). VPT measurements reveal that freshly prepared RGG-d(T)40 condensates containing (TERRA)10 display material properties similar to a Maxwell fluid9 with a terminal viscosity of 35.2 ± 4.85 Pa.s (Fig. 5e–h; Supplementary Video 16). Upon physical aging, the probe particles within condensates are caged as evidenced by a narrower spread of the particle trajectory (Fig. 5f). Correspondingly, the ensemble-averaged MSD profiles showed dramatic differences at times beyond the emergence of RNA clusters, e.g., t > 60 minutes (Fig. 5g). We find that this is an emergent property of the (TERRA)10 containing RGG-d(T)40 condensate system since the two-component condensate system composed of RGG and d(T)40 does not show time-dependent changes in MSD profiles (Supplementary Fig. 17). Further measurements of MSD profiles in RGG-d(T)40 condensates containing (TERRA)10 revealed a dramatic arrest of the probe particles at time points, t = 30 and 45 minutes, at which the microscale RNA clusters were not clearly visible (Fig. 5g; Fig. 18; Supplementary Video 17). The dynamical slowdown of probe particles prior to the emergence of microscale RNA aggregates may indicate the formation of nanoscale pre-percolation clusters of TERRA76. Upon further aging, the ensemble-averaged MSDs show a plateauing behavior at longer lag times indicating a terminally elastic response reminiscent of Kelvin-Voigt solids12 (Fig. 5g; observation time ≥ 150 minutes). This may stem from the caging of beads due to the onset of percolated RNA clusters (Fig. 5f). The estimated terminal viscosity of the condensates at 90 minutes is 489.3 ± 69.3 Pa.s (Fig. 5h), which is an order of magnitude higher than condensates at 15 minutes. At 150 minutes after sample preparation, the bead motions were completely arrested, which is a characteristic property of terminally solid material12 (Supplementary Video 18).
We further performed nano-rheology measurements with (mut-TERRA)10 (Fig. 2e, f), which lacks the ability to undergo intra-condensate percolation (Fig. 2g; Supplementary Fig. 4, 5). We observed that (observation time ~ 30 minutes) RGG-d(T)40 condensates containing (mut-TERRA)10 behave as a Maxwell fluid with viscosity 21.5 ± 3.1 Pa.s, which is slightly lower than WT (TERRA)10 (Fig. 5i, j; Supplementary Fig. 19). Importantly, no discernable changes in viscoelastic properties was observed for these condensates over the same period of time (4 hours; Fig. 5i, j; Supplementary Fig. 19) where the WT TERRA containing condensates undergo complete dynamical arrest. These results show that selectively inhibiting the percolation ability of the RNA through sequence perturbations abrogates RNA percolation-driven condensate dynamical arrest.
RNA-binding protein G3BP1 inhibits intra-condensate RNA clustering
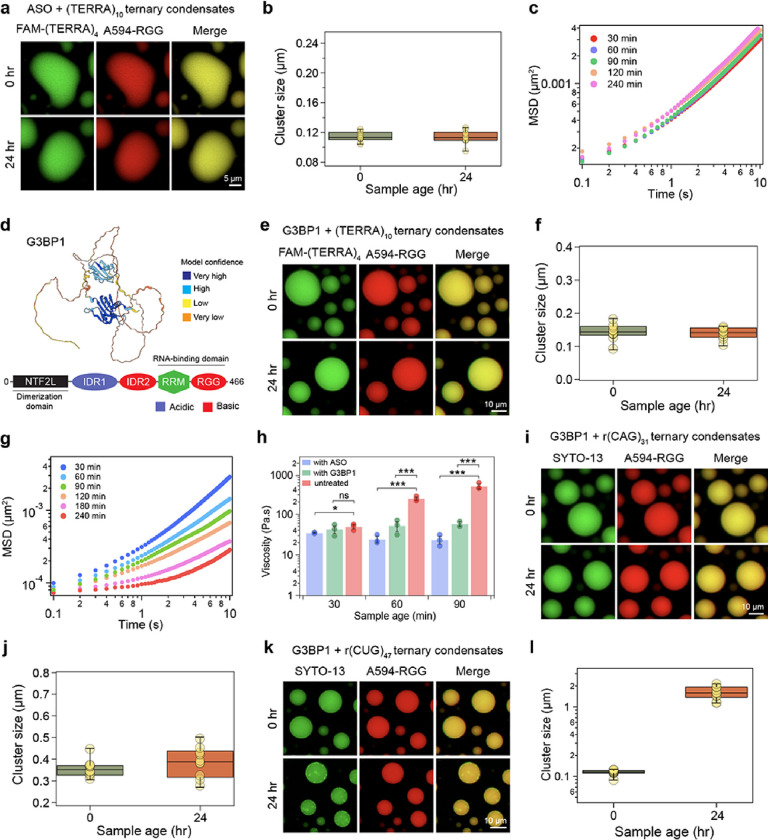
In cells, RNP granules are comprised of diverse RNA and protein species with an assortment of sequence-specific and non-specific interactions77, 78, 79, 80. Based on our results on age-dependent intra-condensate RNA clustering, we reasoned that introducing additional components in these condensates that can compete with homotypic RNA-RNA interactions may inhibit RNA cluster formation. We tested this idea first by employing an anti-sense oligonucleotide [ASO; sequence: r(CCCUAA)] targeting (TERRA)10. ASO-treated condensates did not show any signs of RNA clusters even after 24 hours in our fluorescence microscopy experiments (Fig. 6a, b; Supplementary Fig. 20). VPT-based nanorheology confirmed the absence of nanoscale clusters (Fig. 6c; Supplementary Fig. 21) as no substantial change in condensate viscosity was observed during 24 hours of aging (Fig. 5f). These results suggest that selectively targeting homotypic RNA-RNA interactions can buffer against RNA clustering-mediated liquid- to-solid phase transition. Could such buffering effects be also imparted by multivalent RBPs with broad specificity for a diverse repertoire of RNA? We tested this idea utilizing G3BP1, which is a core scaffolding RBP of cytoplasmic stress granules81, 82, 83. G3BP1 binds to RNAs through a folded RNA recognition motif and disordered Arg/Gly-rich domain (Fig. 6d). In the presence of 10 μM G3BP1, we observed that (TERRA)10 containing RGG-d(T)40 condensates do not show the emergence of RNA clusters after aging (within 24 hours). All components remain homogenous and the cluster sizes derived from SAC analysis show no substantial change in their spatial distribution (0.14 ± 0.01 μm at 15 minutes and 0.14 ± 0.01 μm at 24 hours; 1 pixel = 0.09765 μm) (Fig. 6e, f; Supplementary Fig. 23).
Figure 6. Heterotypic buffering by ASO and G3BP1 can prevent homotypic RNA clustering in biomolecular condensates.
(a) (TERRA)10 containing RGG-d(T)40 condensates treated with TERRA antisense oligonucleotide [ASO; sequence: r(CCCUAA)] does not form age-dependent RNA clusters. (b) The corresponding cluster sizes derived from SAC analysis are reported. (c) Age-dependent MSDs of 200 nm beads inside (TERRA)10 containing RGG-d(T)40 condensates treated with ASO (also see Supplementary Fig. 21). (d) Alphafold285 predicted structure of G3BP1 (identifier: AF-Q13283-F1) with color coding corresponding to model confidence. Domain architecture of G3BP1 (NTF2L, nuclear transport factor 2-like domain; IDR, intrinsically disordered region; RRM, RNA recognition motif; RGG, Arg/Gly-rich domain). (e) (TERRA)10 containing RGG-d(T)40 condensates with G3BP1 do not show age-dependent morphological changes. (f).The corresponding cluster sizes derived from SAC analysis indicate a lack of microscale RNA clustering (g) Age-dependent MSDs of 200 nm beads inside (TERRA)10 containing RGG-d(T)40 condensates with G3BP1 (also see Supplementary Fig. 24). (h) Comparison of terminal viscosities of (TERRA)10 containing RGG-d(T)40 condensates with or without the ASO or G3BP1 [“untreated” data taken from Fig. 5h]. Statistical significance was determined by performing a two-sided Student’s t-test (* means p<0.05, ** means p<0.01, *** means p<0.001, ns means ‘not significant’) between viscosities of untreated condensates versus condensates containing either the ASO or G3BP1. The p-values, between ‘untreated’ and ‘with ASO’ condition at 30 minutes, 60 minutes, and 90 minutes sample age, are 0.041, 0.0003, and 0.0008, respectively. The p-values between ‘untreated’ and ‘with G3BP1’ conditions at 30 minutes, 60 minutes, and 90 minutes sample age are 0.4766, 0.0009, and 0.0004, respectively. (i) r(CAG)31 containing RGG-d(T)40 condensates with G3BP1 do not show age-dependent morphological (j) changes. The corresponding cluster size analysis indicates an absence of RNA clustering. (k) r(CUG)47 containing RGG-d(T)40 condensates with G3BP1 do not show visible RNA clusters at 15 minutes after sample preparation but show some RNA clusters at an age of 24 hours. (l) The corresponding cluster size analysis is reported. The concentrations of the ASO and G3BP1 are 1 mg/ml and 10 μM, respectively. The composition of the condensate system used for imaging is 1 mg/ml RNA [0.45 mg/ml in the case of r(CUG)47], 5 mg/ml RGG, and 1.5 mg/ml d(T)40. For the nanorheology measurements, the relative proportion of the condensate components was kept the same, but the overall concentration of each component was doubled to achieve a higher volume fraction of the dense phase. Buffer composition for all experiments is 25 mM Tris-HCl (pH 7.5), 25 mM NaCl, and 20 mM DTT. In experiments utilizing fluorescently labeled components, the concentration range is 250 nM to 500 nM. Each experiment was independently repeated at least three times, except for the sample with G3BP1 in (h) that aged for 90 minutes, which was independently repeated two times.
Earlier, our nano-rheology experiments on (TERRA)10 containing RGG-d(T)40 condensates (Fig. 5g, h) revealed signs of dynamical arrest and a substantial increase in viscosity even before microscale RNA clusters were visible. To quantify the effects of G3BP1 on the rheology of these condensates, we probed the time-dependent changes in condensate material properties using VPT. MSD profiles and viscosity measurements reveal that despite the absence of microscale RNA clusters, G3BP1 containing condensates undergo a progressive dynamical slowdown (Fig. 6g; Supplementary Fig. 24). There is a concomitant increase in viscosity from 42.3 ± 9.0 Pa.s for condensates at 30 minutes to 154 ± 22.3 Pa.s within 3 hours of condensate preparation (Supplementary Fig. 24). However, condensate aging is substantially slower in the presence of G3BP1 (Fig. 6h). Therefore, although microscale RNA clusters are disfavored in the presence of G3BP1, our rheology measurements suggest that there may be nanoscale RNA clustering that contributes to increased viscoelasticity of aged condensates. Nonetheless, G3BP1 can increase the activation energy barrier of intra-condensate (TERRA)10 clustering.
We further tested the generalizability of the observed buffering effect of G3BP1 with GC-rich repeat RNAs, r(CAG)31 and r(CUG)47, that spontaneously form microscale clusters in RGG-d(T)40 condensates (Fig. 4). In the presence of G3BP1, both of these RNAs formed homogeneous condensates (Fig. 6i–l; Supplementary Fig. 23). Furthermore, in the case of (CAG)31, no microscale clusters were observed even at 24 hours age where the cluster size = 0.39 ± 0.02 μm, close to the detection limit (1 pixel = 0.2196 μm) (Fig. 6i, j; Supplementary Fig. 23). In the case of r(CUG)47 we observed the formation of a few RNA foci at 24 hours (Fig. 6k, l; Supplementary Fig. 23). However, the total fraction of RNA, determined by intensity-based analysis, in these clusters were only ~ 6.65 ± 2.31% (Supplementary Fig. 25), whereas the same RNA was present almost exclusively (~ 100%) in the intra-condensate clusters in the absence of G3BP1 (Fig. 4d). Together, these results signify that G3BP1 can frustrate84 RNA-RNA homotypic interactions in these condensates thereby disfavoring RNA clustering and preserving intra-condensate solubility of repeats RNAs.
Conclusions
Recently, it has been shown that biomolecular condensates can act as sites for pathological protein aggregation21, 22, 23, 24, 25, 86. We now demonstrate that irreversible clustering of repeat expanded RNA molecules, a process widely implicated in many neurological disorders37, 64, 65, 66, 67, 68, 69, can be nucleated in multi-component protein-nucleic acid condensates. The underlying mechanism is sequence-specific percolation transitions of RNA chains driven by homotypic RNA-RNA interactions. RNA percolation engenders an age-dependent liquid-to-solid phase transition of condensates. Multivalent co-factors, such as ASO and RBPs, that compete with homotypic RNA-RNA interactions can increase the activation energy barrier of RNA clustering, thereby acting as inhibitors of this process.
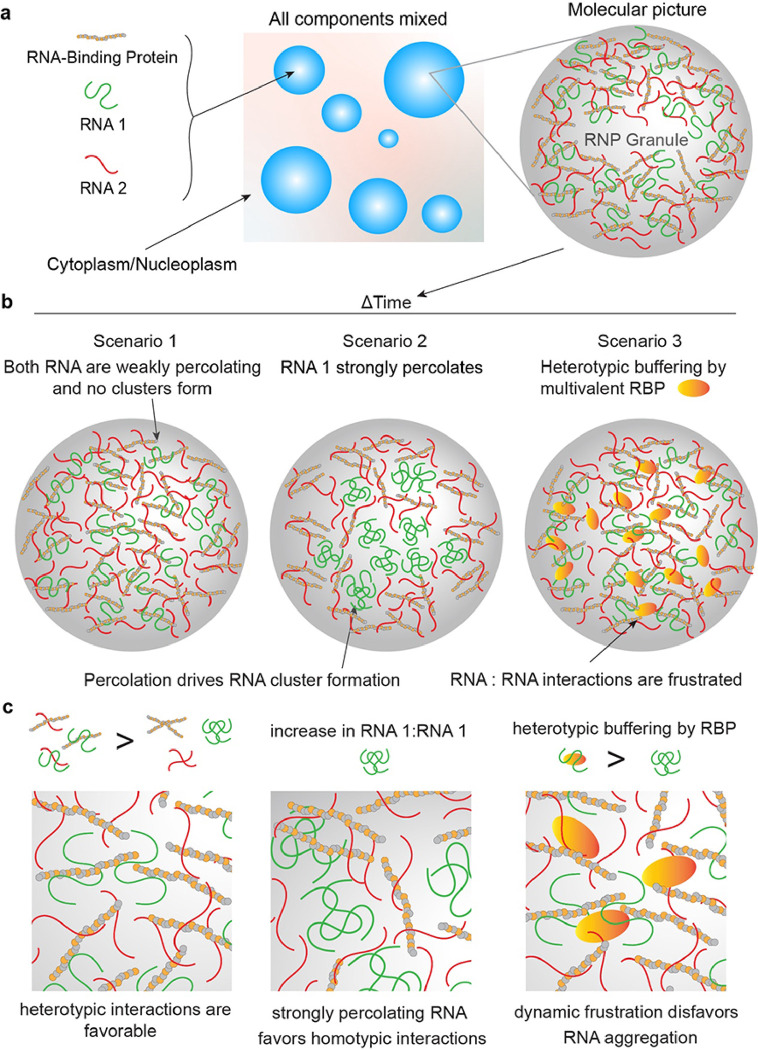
Two important implications, as outlined in Fig. 7, stem from our experiments reported in this study. Firstly, dynamic frustration of homotypic interactions between RNA chains can increase the range of metastability of biomolecular condensates that are poised to undergo RNA percolation-driven physical aging. In this model, a multi-component biomolecular system is frustrated if the probability of minimizing its global free energy through coordinated optimization of all possible interaction modalities of constituent macromolecules is kinetically sluggish due to overlapping inter-molecular interactions84, 87, 88, 89, 90. For RNA containing RGG-d(T)40 condensates, the RNA [e.g., (TERRA)10 or r(CAG)31 or r(CUG)47] is initially kept frustrated through heterotypic interactions with the primary condensate components, despite having a strong propensity to self-associate as evidenced by its hysteretic phase behavior and irreversible clustering43 (Fig. 2). However, with time, RNA-RNA homotypic interactions dominate, leading to age-dependent RNA clusters that demix from the fluid phase formed by RGG and d(T)40. The onset of RNA clustering is determined by the driving force for RNA percolation, which is modulated by repeat RNA length and/or sequence perturbations that weaken base pairing and stacking interactions. Reducing the homotypic RNA-RNA interactions, and hence, the percolation propensity shifts the balance towards heterotypic interactions between RNA and condensate components over RNA self-assembly. Consequently, the cross-play of interactions between all components as opposed to the dominance of a singular intermolecular, homotypic interaction node may be essential for enhancing the metastability of multi-component biomolecular condensates.
Figure 7. schematic showing the proposed model of intra-condensate RNA percolation and heterotypic buffering.
(a) A model of multi-component condensates formed by two RNAs with strong (as shown in green) and weak (as shown in red) percolation propensity, respectively, and an RBP. (b) Three possible scenarios of RNA percolation-driven condensate aging or a lack thereof in the presence of a multivalent RBP. (c) Zoomed-in views of the panels shown above.
Second, in the cases of strongly percolating RNA molecules [e.g., (TERRA)10 or r(CAG)31 or r(CUG)47], G3BP1 reduces the propensity of RNA clustering suggesting that the solubility of RNAs in condensates is enhanced. Based on these observations, we propose that RNA binding proteins in complex biomolecular condensates may employ heterotypic RNA-protein interactions91 as a regulatory mechanism to prevent RNAs from aberrant homotypic self-assembly. This phenomenon, termed heterotypic buffering91, was previously proposed as a mechanism to enhance the solubility of aggregation-prone proteins, which possess a strong preference for homotypic interactions and amyloid fiber formation linked to diseases. Our study extends this thermodynamic framework to rationalize the biological roles of RBPs in regulating intra-condensate RNA aggregation. During cellular stress, polysomes are disassembled and polyA-tailed mRNAs are sequestered in stress granules, thereby globally inhibiting translation92, 93. Intra-stress granule RNA percolation can compromise the disassembly of stress granules upon removal of stress, leading to the accumulation of irreversible granules that are cytotoxic22, 94. Our results indicate that multivalent RNA binding proteins can effectively provide the first line of defense against irreversible RNA clustering before ATP-dependent RNA helicases can actively engage in remodeling these assemblies78.
In summary, we report that percolation-driven irreversible RNA clustering can be enhanced in biomolecular condensates, leading to their liquid-to-solid phase transitions. This can be buffered by multivalent RBPs supporting liquid-phase condensate homeostasis. The insights gained from our study provide a complementary perspective on the role of RBPs in regulating aberrant RNA self-assembly in living cells.
Acknowledgments
This work was supported by the US National Institutes of Health through grants R35 GM138186 (P.R.B) and the St. Jude Children’s Research Collaborative on the Biology and Biophysics of RNP Granules (P.R.B.). We gratefully acknowledge Dr. Paul Taylor’s lab at St. Jude Children’s Research Hospital for providing purified G3BP1 protein. We also gratefully acknowledge Dr. Ibraheem Alshareedah, currently at Boston Children’s Hospital, Harvard Medical School for providing the initial SAC analysis codes. The authors deeply appreciate critical feedback from Drs. Rohit Pappu and Tanja Mittag and the group members of Banerjee laboratory.
Footnotes
Competing interests
P.R.B. is a member of the Biophysics Reviews (AIP Publishing) editorial board. This affiliation did not influence the work reported here. All other authors have no conflicts to report.
Additional Declarations: There is NO Competing Interest.
Contributor Information
Priya Banerjee, University at Buffalo, SUNY.
Tharun Selvam Mahendran, University at Buffalo, SUNY.
Gable Wadsworth, University at Buffalo, SUNY.
Anurag Singh, University at Buffalo.
Data and materials availability
All data are available in the manuscript or the supplementary materials. Codes for nanorheology, FRAP, SAC analysis, and RNA state diagram data analysis are available on GitHub (see https://github.com/BanerjeeLab-repertoire/Biomolecular-Condensates-Can-Enhance-Pathological-RNA-Clustering).
References
- 1.Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science 2017, 357(6357): eaaf4382. [DOI] [PubMed] [Google Scholar]
- 2.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry - Nature Reviews Molecular Cell Biology. Nat Rev Mol Cell Biol 2017, 18(5): 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, et al. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol 2018, 28(6): 420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, et al. Germline P Granules Are Liquid Droplets That Localize by Controlled Dissolution/Condensation. Science 2009, 324(5935): 1729–1732. [DOI] [PubMed] [Google Scholar]
- 5.Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012, 483(7389): 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pappu RV, Cohen SR, Dar F, Farag M, Kar M. Phase Transitions of Associative Biomacromolecules. Chemical Reviews 2023, 123(14): 8945–8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyon AS, Peeples WB, Rosen MK. A framework for understanding the functions of biomolecular condensates across scales. Nature Reviews Molecular Cell Biology 2021, 22(3): 215–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alshareedah I, Moosa MM, Pham M, Potoyan DA, Banerjee PR. Programmable viscoelasticity in protein-RNA condensates with disordered sticker-spacer polypeptides. Nat Commun 2021, 12(6620): 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jawerth L, Fischer-Friedrich E, Saha S, Wang J, Franzmann T, Zhang X, et al. Protein condensates as aging Maxwell fluids. Science 2020, 370(6522): 1317–1323. [DOI] [PubMed] [Google Scholar]
- 10.Riback JA, Eeftens JM, Lee DSW, Quinodoz SA, Donlic A, Orlovsky N, et al. Viscoelasticity and advective flow of RNA underlies nucleolar form and function. Mol Cell 2023, 83(17): 3095–3107.e3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh A, Kota D, Zhou H-X. Shear relaxation governs fusion dynamics of biomolecular condensates. Nature Communications 2021, 12(1): 5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alshareedah I, Borcherds WM, Cohen SR, Singh A, Posey AE, Farag M, et al. Sequence-specific interactions determine viscoelasticity and aging dynamics of protein condensates. bioRxiv 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 2015, 162(5): 1066–1077. [DOI] [PubMed] [Google Scholar]
- 14.St George-Hyslop P, Lin JQ, Miyashita A, Phillips EC, Qamar S, Randle SJ, et al. The physiological and pathological biophysics of phase separation and gelation of RNA binding proteins in amyotrophic lateral sclerosis and fronto-temporal lobar degeneration. Brain Res 2018, 1693(Pt A): 11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X, Sumrow L, Tashiro K, Sutherland L, Liu D, Qin T, et al. Mutations linked to neurological disease enhance self-association of low-complexity protein sequences. Science 2022, 377(6601): eabn5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zbinden A, Pérez-Berlanga M, De Rossi P, Polymenidou M. Phase Separation and Neurodegenerative Diseases: A Disturbance in the Force. Dev Cell 2020, 55(1): 45–68. [DOI] [PubMed] [Google Scholar]
- 17.Alberti S, Carra S. Quality Control of Membraneless Organelles. Journal of Molecular Biology 2018, 430(23): 4711–4729. [DOI] [PubMed] [Google Scholar]
- 18.Alberti S, Gladfelter A, Mittag T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176(3): 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alberti S, Hyman AA. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Biol 2021, 22(3): 196–213. [DOI] [PubMed] [Google Scholar]
- 20.Babinchak WM, Surewicz WK. Liquid–Liquid Phase Separation and Its Mechanistic Role in Pathological Protein Aggregation. J Mol Biol 2020, 432(7): 1910–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin Y, Protter DS, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell 2015, 60(2): 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, et al. Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell 2015, 163(1): 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012, 149(4): 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murakami T, Qamar S, Lin JQ, Schierle GS, Rees E, Miyashita A, et al. ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron 2015, 88(4): 678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linsenmeier M, Faltova L, Morelli C, Capasso Palmiero U, Seiffert C, Küffner AM, et al. The interface of condensates of the hnRNPA1 low-complexity domain promotes formation of amyloid fibrils. Nat Chem 2023, 15(10): 1340–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, Parker R. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proceedings of the National Academy of Sciences 2018, 115(11): 2734–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Protter DSW, Parker R. Principles and Properties of Stress Granules. Trends Cell Biol 2016, 26(9): 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tauber D, Tauber G, Khong A, Van Treeck B, Pelletier J, Parker R. Modulation of RNA Condensation by the DEAD-Box Protein eIF4A. Cell 2020, 180(3): 411–426.e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y, Schmidt BF, Bruchez MP, McManus CJ. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res 2018, 46(7): 3742–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.West JA, Mito M, Kurosaka S, Takumi T, Tanegashima C, Chujo T, et al. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J Cell Biol 2016, 214(7): 817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nature Reviews Genetics 2010, 11(4): 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malik I, Kelley CP, Wang ET, Todd PK. Molecular mechanisms underlying nucleotide repeat expansion disorders. Nature Reviews Molecular Cell Biology 2021, 22(9): 589–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gatchel JR, Zoghbi HY. Diseases of Unstable Repeat Expansion: Mechanisms and Common Principles. Nature Reviews Genetics 2005, 6(10): 743–755. [DOI] [PubMed] [Google Scholar]
- 34.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011, 72(2): 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011, 72(2): 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science 2000, 289(5485): 1769–1773. [DOI] [PubMed] [Google Scholar]
- 37.CAG Repeat Not Polyglutamine Length Determines Timing of Huntington’s Disease Onset. Cell 2019, 178(4): 887–900.e814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jain A, Vale RD. RNA phase transitions in repeat expansion disorders. Nature 2017, 546(7657): 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep 2013, 5(5): 1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimchi O, King EM, Brenner MP. Uncovering the mechanism for aggregation in repeat expanded RNA reveals a reentrant transition. Nature Communications 2023, 14(1): 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trcek T, Douglas TE, Grosch M, Yin Y, Eagle WVI, Gavis ER, et al. Sequence-Independent Self-Assembly of Germ Granule mRNAs into Homotypic Clusters. Mol Cell 2020, 78(5): 941–950 e912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trcek T, Grosch M, York A, Shroff H, Lionnet T, Lehmann R. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat Commun 2015, 6: 7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wadsworth GM, Zahurancik WJ, Zeng X, Pullara P, Lai LB, Sidharthan V, et al. RNAs undergo phase transitions with lower critical solution temperatures. Nat Chem 2023, 15(12): 1693–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mittag T, Pappu RV. A conceptual framework for understanding phase separation and addressing open questions and challenges. Molecular Cell 2022, 82(12): 2201–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harmon TS, Holehouse AS, Rosen MK, Pappu RV. Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. eLife 2017, 6: e30294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dar F, Cohen SR, Mitrea DM, Phillips AH, Nagy G, Leite WC, et al. Biomolecular condensates form spatially inhomogeneous network fluids. Nature Communications 2024, 15(1): 3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alshareedah I, Singh A, Yang S, Ramachandran V, Quinn A, Potoyan DA, et al. Determinants of viscoelasticity and flow activation energy in biomolecular condensates. Science Advances 2024, 10(7): eadi6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cusanelli E, Chartrand P. Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity. Front Genet 2015, 6: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007, 318(5851): 798–801. [DOI] [PubMed] [Google Scholar]
- 50.Xu Y, Suzuki Y, Ito K, Komiyama M. Telomeric repeat-containing RNA structure in living cells. Proceedings of the National Academy of Sciences 2010, 107(33): 14579–14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collie GW, Haider SM, Neidle S, Parkinson GN. A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex. Nucleic Acids Res 2010, 38(16): 5569–5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martadinata H, Phan AT. Structure of Propeller-Type Parallel-Stranded RNA G-Quadruplexes, Formed by Human Telomeric RNA Sequences in K + Solution. Journal of the American Chemical Society 2009, 131(7): 2570–2578. [DOI] [PubMed] [Google Scholar]
- 53.Bhattacharyya D, Mirihana Arachchilage G, Basu S. Metal Cations in G-Quadruplex Folding and Stability. Front Chem 2016, 4: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahama K, Kino K, Arai S, Kurokawa R, Oyoshi T. Identification of Ewing’s sarcoma protein as a G-quadruplex DNA- and RNA-binding protein. The FEBS Journal 2011, 278(6): 988–998. [DOI] [PubMed] [Google Scholar]
- 55.Takahama K, Oyoshi T. Specific Binding of Modified RGG Domain in TLS/FUS to G-Quadruplex RNA: Tyrosines in RGG Domain Recognize 2′-OH of the Riboses of Loops in G-Quadruplex. Journal of the American Chemical Society 2013, 135(48): 18016–18019. [DOI] [PubMed] [Google Scholar]
- 56.Renaud de la Faverie A, Guédin A, Bedrat A, Yatsunyk LA, Mergny JL. Thioflavin T as a fluorescence light-up probe for G4 formation. Nucleic Acids Res 2014, 42(8): e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu S, Li Q, Xiang J, Yang Q, Sun H, Guan A, et al. Thioflavin T as an efficient fluorescence sensor for selective recognition of RNA G-quadruplexes. Sci Rep 2016, 6(1): 24793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yeasmin Khusbu F, Zhou X, Chen H, Ma C, Wang K. Thioflavin T as a fluorescence probe for biosensing applications. TrAC Trends in Analytical Chemistry 2018, 109: 1–18. [Google Scholar]
- 59.Reddy K, Zamiri B, Stanley SYR, Macgregor RB Jr., Pearson CE. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem 2013, 288(14): 9860–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol 2009, 19(8): 414–422. [DOI] [PubMed] [Google Scholar]
- 61.Randall A, Griffith JD. Structure of long telomeric RNA transcripts: the G-rich RNA forms a compact repeating structure containing G-quartets. J Biol Chem 2009, 284(21): 13980–13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martadinata H, Heddi B, Lim KW, Phan AT. Structure of long human telomeric RNA (TERRA): G-quadruplexes formed by four and eight UUAGGG repeats are stable building blocks. Biochemistry 2011, 50(29): 6455–6461. [DOI] [PubMed] [Google Scholar]
- 63.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 2008, 10(2): 228–236. [DOI] [PubMed] [Google Scholar]
- 64.Usdin K. The biological effects of simple tandem repeats: lessons from the repeat expansion diseases. Genome Res 2008, 18(7): 1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fiszer A, Krzyzosiak WJ. RNA toxicity in polyglutamine disorders: concepts, models, and progress of research. J Mol Med (Berl) 2013, 91(6): 683–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sznajder Ł J, Swanson MS. Short Tandem Repeat Expansions and RNA-Mediated Pathogenesis in Myotonic Dystrophy. Int J Mol Sci 2019, 20(13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Orr HT, Chung MY, Banfi S, Kwiatkowski TJ Jr., Servadio A, Beaudet AL, et al. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet 1993, 4(3): 221–226. [DOI] [PubMed] [Google Scholar]
- 68.Davis BM, McCurrach ME, Taneja KL, Singer RH, Housman DE. Expansion of a CUG trinucleotide repeat in the 3’ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc Natl Acad Sci U S A 1997, 94(14): 7388–7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jin P, Zarnescu DC, Zhang F, Pearson CE, Lucchesi JC, Moses K, et al. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron 2003, 39(5): 739–747. [DOI] [PubMed] [Google Scholar]
- 70.Gendron TF, Petrucelli L. Disease Mechanisms of C9ORF72 Repeat Expansions. Cold Spring Harb Perspect Med 2018, 8(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumar V, Hasan GM, Hassan MI. Unraveling the Role of RNA Mediated Toxicity of C9orf72 Repeats in C9-FTD/ALS. Front Neurosci 2017, 11: 711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alshareedah I, Thurston GM, Banerjee PR. Quantifying viscosity and surface tension of multicomponent protein-nucleic acid condensates. Biophys J 2021, 120(7): 1161–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alshareedah I, Kaur T, Banerjee PR. Chapter Six - Methods for characterizing the material properties of biomolecular condensates. In: Keating CD (ed). Methods in Enzymology, vol. 646. Academic Press, 2021, pp 143–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ghosh A, Kota D, Zhou HX. Determining Thermodynamic and Material Properties of Biomolecular Condensates by Confocal Microscopy and Optical Tweezers. Methods Mol Biol 2023, 2563: 237–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Azevedo TN, Rizzi LG. Microrheology of filament networks from Brownian dynamics simulations. Journal of Physics: Conference Series 2020, 1483(1): 012001. [Google Scholar]
- 76.Kar M, Dar F, Welsh TJ, Vogel LT, Kühnemuth R, Majumdar A, et al. Phase-separating RNA-binding proteins form heterogeneous distributions of clusters in subsaturated solutions. Proceedings of the National Academy of Sciences 2022, 119(28): e2202222119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mittag T, Parker R. Multiple Modes of Protein-Protein Interactions Promote RNP Granule Assembly. J Mol Biol 2018, 430(23): 4636–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tauber D, Tauber G, Parker R. Mechanisms and Regulation of RNA Condensation in RNP Granule Formation. Trends Biochem Sci 2020, 45(9): 764–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Protter DSW, Rao BS, Van Treeck B, Lin Y, Mizoue L, Rosen MK, et al. Intrinsically Disordered Regions Can Contribute Promiscuous Interactions to RNP Granule Assembly. Cell Rep 2018, 22(6): 1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Youn JY, Dyakov BJA, Zhang J, Knight JDR, Vernon RM, Forman-Kay JD, et al. Properties of Stress Granule and P-Body Proteomes. Mol Cell 2019, 76(2): 286–294. [DOI] [PubMed] [Google Scholar]
- 81.Yang P, Mathieu C, Kolaitis RM, Zhang P, Messing J, Yurtsever U, et al. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 2020, 181(2): 325–345.e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, et al. Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell 2020, 181(2): 306–324.e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guillén-Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlüßler R, et al. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 2020, 181(2): 346–361.e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ferreiro DU, Komives EA, Wolynes PG. Frustration, function and folding. Current Opinion in Structural Biology 2018, 48: 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596(7873): 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shen Y, Chen A, Wang W, Shen Y, Ruggeri FS, Aime S, et al. The liquid-to-solid transition of FUS is promoted by the condensate surface. Proc Natl Acad Sci U S A 2023, 120(33): e2301366120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ferreiro DU, Komives EA, Wolynes PG. Frustration in biomolecules. Q Rev Biophys 2014, 47(4): 285–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gianni S, Camilloni C, Giri R, Toto A, Bonetti D, Morrone A, et al. Understanding the frustration arising from the competition between function, misfolding, and aggregation in a globular protein. Proceedings of the National Academy of Sciences 2014, 111(39): 14141–14146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jemth P, Karlsson E, Vögeli B, Guzovsky B, Andersson E, Hultqvist G, et al. Structure and dynamics conspire in the evolution of affinity between intrinsically disordered proteins. Science Advances 2018, 4(10): eaau4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vannimenus J, Toulouse G. Theory of the frustration effect. II. Ising spins on a square lattice. Journal of Physics C: Solid State Physics 1977, 10(18): L537. [Google Scholar]
- 91.Mathieu C, Pappu RV, Taylor JP. Beyond aggregation: Pathological phase transitions in neurodegenerative disease. Science 2020, 370(6512): 56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hofmann S, Kedersha N, Anderson P, Ivanov P. Molecular mechanisms of stress granule assembly and disassembly. Biochim Biophys Acta Mol Cell Res 2021, 1868(1): 118876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R. Distinct stages in stress granule assembly and disassembly. eLife 2016, 5: e18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang P, Fan B, Yang P, Temirov J, Messing J, Kim HJ, et al. Chronic optogenetic induction of stress granules is cytotoxic and reveals the evolution of ALS-FTD pathology. eLife 2019, 8: e39578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are available in the manuscript or the supplementary materials. Codes for nanorheology, FRAP, SAC analysis, and RNA state diagram data analysis are available on GitHub (see https://github.com/BanerjeeLab-repertoire/Biomolecular-Condensates-Can-Enhance-Pathological-RNA-Clustering).