Abstract
Background
Lower respiratory tract infection (LRTI) is a leading cause of infant morbidity and mortality globally. LRTI may be caused by viral or bacterial infections, individually or in combination. We investigated associations between LRTI and infant nasopharyngeal (NP) viruses and bacteria in a South African birth cohort.
Methods
In a case-control study of infants enrolled in the Drakenstein Child Health Study (DCHS), LRTI cases were identified prospectively and age-matched with controls from the cohort. NP swabs were tested using quantitative real-time polymerase chain reaction (qPCR) and 16S rRNA gene amplicon sequencing. We calculated adjusted Conditional Odds Ratios (aORs) for qPCR targets and used mixed effects models to identify differentially abundant taxa between LRTI cases and controls and explore viral-bacterial interactions.
Results
Respiratory Syncytial Virus (RSV) [aOR: 5.69, 95% CI: 3.03–10.69], human rhinovirus (HRV) [1.47, 1.03–2.09], parainfluenza virus [3.46, 1.64–7.26], adenovirus [1.99, 1.08–3.68], enterovirus [2.32, 1.20–4.46], Haemophilus influenzae [1.72, 1.25–2.37], Klebsiella pneumoniae [2.66, 1.59–4.46], or high-density (> 6.9 log10 copies/mL) Streptococcus pneumoniae [1.53, 1.01–2.32] were associated with LRTI. Using 16S sequencing, LRTI was associated with increased relative abundance of Haemophilus (q = 0.0003) and decreased relative abundance of Dolosigranulum (q = 0.001), Corynebacterium (q = 0.091) and Neisseria (q = 0.004). In samples positive for RSV, Staphylococcus and Alloprevotella were present at lower relative abundance in cases than controls. In samples positive for parainfluenza virus or HRV, Haemophilus was present at higher relative abundance in cases.
Conclusions
The associations between bacterial taxa and LRTI are strikingly similar to those identified in high-income countries, suggesting a conserved phenotype. RSV was the major virus associated with LRTI. H. influenzae appears to be the major bacterial driver of LRTI, acting synergistically with viruses. The Gram-positive bacteria Dolosigranulum and Corynebacteria may protect against LRTI, while Staphylococcus was associated with reduced risk of RSV-related LRTI.
Funding
National Institutes of Health of the USA, Bill and Melinda Gates Foundation, National Research Foundation South Africa, South African Medical Research Council, L’Oréal-UNESCO For Women in Science South Africa, Australian National Health and Medical Research Council.
Keywords: 16S rRNA gene, bacterial, infant, low- and middle-income country, lower respiratory tract infection, microbiome, nasopharyngeal, qPCR, viral
BACKGROUND
Lower respiratory tract infection (LRTI) remains the leading cause of child death outside of the neonatal period worldwide [1]. The burden of LRTI disproportionately affects low- and middle-income countries (LMICs), with more than 200 deaths per 100,000 children under the age of five years in 2019 in several countries in Sub-Saharan Africa [2]. Viruses, particularly Respiratory Syncytial Virus (RSV), and bacteria not included in vaccines, particularly non-type b Haemophilus influenzae, are considered to play an increasingly important role [1].
Infection of the upper respiratory tract with pathogenic viruses and bacteria frequently precedes LRTI; with local replication and subsequent translocation to the lower respiratory tract [3]. Dysbiotic (imbalanced) NP bacterial community profiles may have reduced capability to resist pathogen overgrowth and invasion. For example, LRTI caused by RSV has been associated with increased abundance of H. influenzae and Streptococcus species and decreased abundance of Staphylococcus aureus [4]. H. influenzae and Streptococcus-dominated profiles have also been associated with an exaggerated inflammatory response and more severe RSV disease [4].
Risk factors for LRTI, such as malnutrition, maternal HIV-infection, lack of exclusive breastfeeding and indoor air pollution, are more prevalent in LMICs compared to high-income countries, which may influence aetiology and pathogen interactions. However, studies investigating the association between infant NP bacterial community profiles, viral infection and LRTI have primarily been performed in high-income settings. We therefore performed a case-control study, nested within a South African birth cohort, to comprehensively investigate associations between NP bacterial community profiles, viral infection and LRTI within the first year of life.
METHODS
Study design, participants, and specimen matching:
We conducted a case-control study of infants enrolled in the Drakenstein Child Health Study (DCHS), a birth cohort study in a peri-urban area in South Africa [5]. Ethical approval was received from the Human Research Ethics Committee (HREC) of the University of Cape Town, South Africa (401/2009 and 585/2015). Mothers were enrolled in their 2nd trimester of pregnancy; all births and hospital care occurred at a central public hospital [5]. Mother-infant pairs were followed from birth, with study visits at 6, 10 and 14 weeks, and 6, 9 and 12 months. Participants who gave additional consent participated in intensive fortnightly NP sampling, across the first year of life. Demographic and clinical data were recorded antenatally at birth, and postnatally during scheduled study visits (Appendix p. 2).
LRTI episodes during the first year of life were identified by active surveillance at local clinics and hospital using World Health Organisation (WHO) criteria [6]. Mothers were counselled about key respiratory symptoms and advised to contact study staff whenever their infant developed cough or difficulty breathing [6]. Infants were followed up through hospitalization or ambulatory illness. LRTI cases and non-LRTI controls were matched 1:1 by birth date and study site.
NP flocked swabs (Copan Diagnostics, CA, USA) were collected fortnightly from infants across the first year of life (0–365 + 14 days) (Figure S1). A window of +/−14 days was allowed between LRTI episode date and specimen collection date for inclusion as a LRTI case specimen. Additional information regarding the categorization of case-control specimens is provided in Appendix (pp. 3–4; Figure S1).
Laboratory procedures
NP specimens were immediately suspended in PrimeStore® Molecular Transport medium (Longhorn Vaccines & Diagnostics, MD, USA), transported on ice and subsequently stored at −80°C. Nucleic acid extracts (Appendix p. 4) were screened for viral and bacterial species using a quantitative real-time polymerase chain reaction (qPCR) [Fast-Track Diagnostics Respiratory Pathogens 33 assay, Luxembourg] [7]. We applied a predefined threshold (> 6.9 log10 copies/mL) for defining high density Streptococcus pneumoniae that best differentiates case-control status, as previously described [8].
We performed short read 16S rRNA gene amplicon sequencing on NP specimens from a subset of case-control specimens from whom sufficient sample was available. Each sequencing run included a comprehensive set of sequencing controls alongside NP specimens (Appendix pp. 4–5; Figure S2) [9]. We measured total 16S rRNA gene copy numbers (16S rRNA gene copies/μl) from nucleic acid extracts via qPCR [9]. A two-step PCR protocol targeting the V4 hypervariable region of the 16S rRNA gene was used to generate 16S rRNA gene amplicons. Pooled libraries were sequenced on the Illumina® MiSeq™ platform using V3 chemistry with 2 × 251 cycles [MiSeq Reagent Kit v3 (600-cycle) Reagent Cartridge (Illumina, CA, USA)] (Appendix pp. 5–7).
We used the DADA2 pipeline (wrapped in the Nextflow algorithm) to filter and trim reads and infer amplicon sequence variants (ASVs) (Appendix p. 7). Taxonomy was assigned using the RDP classifier implementation for DADA2 and SILVA v138. We used R software v4.1.2 [10] and RStudio v2021.09.2 to remove ASVs classified as Eukaryota and ASVs unclassified at Kingdom-level. We applied a step-wise in-silico quality control approach to remove low quality amplicon data [9] and potential contaminant ASVs (Appendix p. 8).
Outcomes
The primary outcome was the association between viral or bacterial species (qPCR), or bacterial taxa (16S rRNA gene amplicon sequencing), in cases with LRTI compared to age-matched controls.
Statistical analysis
We calculated Conditional Odds Ratios (ORs) and Population Attributable Fractions (PAFs) using Miettinen’s formula for each viral and bacterial organism included in the qPCR panel. We further stratified results within three age categories: 0–3 months, > 3–6 months, and > 6–12 months. Findings were adjusted for age and antibiotic use.
Associations between LRTI and NP bacterial load or alpha diversity (Shannon and Chao1) were assessed using linear regression. PERMANOVA (Bray-Curtis dissimilarity assessed using adonis2 function from the vegan package) was used to determine if overall microbial composition differed between cases and controls [11]. Regression and PERMANOVA variables included case status, age and antibiotic exposure.
K means clustering was used to identify specimen clusters, based on the profiles of the 25 most abundant ASVs. We used logistic regression to explore associations between cluster membership and case status. The model included cluster membership, absolute abundances of RSV (A/B), parainfluenza virus, enterovirus, adenovirus, HRV, cytomegalovirus (CMV), age and bacterial load.
We used the Microbiome Multivariable Associations with Linear Model (MaAsLin2) v1.8.0 [12] to identify differentially abundant ASVs between cases and controls. Mixed effects models, with participant identifier as a random effect and case-control group, age, and antibiotic exposure as fixed effects were used. Total sum scaled (TSS) normalization and log transformation were applied. The results were visualized using Seaborn v0.12 [13] in Python. Subsequently, using identical settings, we chose four subsets comprising individuals infected with RSV, HRV, parainfluenza virus or CMV, and applied the MaAsLin2 model to identify bacterial taxa associated with LRTI.
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
RESULTS
Demographics and clinical characteristics
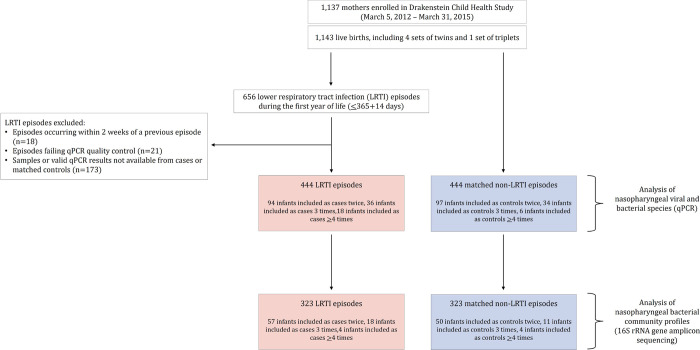
From 29 May 2012 to 3 September 2015, 1,137 mothers were enrolled with 1,143 live births. Cohort retention was high [88.8% (1,015/1,143) infants at 1 year] [14]. A total of 656 LRTI episodes were identified during the first year of life (Fig. 1). Results from qPCR were available for 444 case specimens and 444 matched control specimens (Fig. 1). A subset of these specimens [323 LRTI specimens and 323 control specimens] had high-quality 16S rRNA gene amplicon sequence data available (Appendix pp. 13–17).
Figure 1.
Study flow diagram
Maternal smoking was identified in 27% (151/544) participants. Poor socio-economic status was reflected by low maternal educational attainment (67% of mothers had primary level education only), any parent employment rate (49.6%), and monthly income [< 5,000 ZAR per month for 88% of households]. The median household size was 4 people [interquartile range (IQR): 3–6]. Most infants were delivered by vaginal delivery (80%, 442/542) with 17% (93/544) born prematurely (< 37 weeks’ gestation); most [58/93 (62%)] late preterm. Twenty seven percent (148/544) of infants were HIV-exposed but none were HIV-infected, and duration of exclusive breastfeeding was short [1.4 months (IQR: 0.5–3.0)]. Immunization coverage for all expanded programme on immunization vaccines was high (> 98%) including for H. influenzae type B conjugate vaccine and 13-valent pneumococcal conjugate vaccine.
The median age of NP specimen collection from age-matched cases and controls at the time of LRTI was 139 days (IQR: 81–220 days) (Table 1). Nineteen percent (85/444) of case specimens were collected from cases who received antibiotics prior to specimen collection; none of the controls received antibiotics (p < 0.001).
Table 1.
Characteristics of DCHS study population included in the LRTI case control analysis.
| qPCR data | 16S rRNA gene amplicon dataϕ | |
|---|---|---|
| N = 544 | N = 479 | |
| n (%) | n (%) | |
| Maternal smoking (self-report): | ||
| Yes | 151 (27.3) | 127 (26.5) |
| Maternal education: | ||
| Primary level | 52 (9.4) | 43 (9.0) |
| Started secondary level | 319 (57.6) | 277 (57.8) |
| Completed secondary level | 151 (27.3) | 132 (27.6) |
| Any tertiary level | 32 (5.8) | 27 (5.6) |
| Parent employed: | ||
| Yes | 275 (49.6) | 237 (49.5) |
| Household income (per month): | ||
| < 1,000 ZARi | 187 (33.8) | 160 (33.4) |
| 1,000–5,000 ZAR | 302 (54.5) | 264 (55.1) |
| > 5,000 ZAR | 65 (11.7) | 55 (11.5) |
| Household density, median (IQRii) | 4 (3–6) | 4 (3–6) |
| Mode of delivery: | ||
| Vaginal delivery | 442 (80.1)2 | 382 (79.9)1 |
| Season of birth: | ||
| Summer | 152 (27.4) | 129 (26.9) |
| Autumn | 152 (27.4) | 136 (28.4) |
| Winter | 134 (24.2) | 114 (23.8) |
| Spring | 116 (20.9) | 100 (20.9) |
| WAZiii at birth, median (IQR) | −0.3 (−0.0–0.4) | −0.2 (−0.9–0.4) |
| Gestational age: | ||
| Premature (< 37 weeks’ gestation) | 93 (16.8) | 72 (15.0) |
| Sex: | ||
| Male | 294 (53.1) | 252 (52.6) |
| HIViv-exposure: | ||
| HIV-exposed, uninfected | 148 (26.7) | 127 (26.5) |
| Exclusive breastfeeding (months), median (IQR) | 1.4 (0.5–3.0)10 | 1.1 (0.5–2.8) |
| Vaccine coverage | ||
| Birth (BCG, OPV) | 545 (99.1)4 | 472 (99.2)3 |
| 6 weeks (OPV, RV, DTaP-IPV-Hib-HepB, PCV13) | 548 (99.6)4 | 474 (99.6)3 |
| 10 weeks (DTaP-IPV-Hib-HepB) | 548 (99.6)4 | 474 (99.6)3 |
| 14 weeks (RV, DTaP-IPV-Hib-HepB, PCV13) | 545 (99.5)6 | 471 (99.4)5 |
| 9 months (DTaP-IPV-Hib-HepB, PCV13) | 523 (97.8)19 | 452 (97.8)17 |
| Specimen-level data | ||
| qPCR data | 16S rRNA gene amplicon dataϕ | |
| N = 888 | N = 646 | |
| n (%) | n (%) | |
| Age at specimen collection (days), median (IQR) | 139 (81–220) | 128 (75–211) |
| Antibiotic use at specimen collection | ||
| No | 799 (90.0) | 581 (89.9) |
| Antibiotic commenced < 24 hours prior to specimen collection | 55 (6.2) | 41 (6.4) |
| Antibiotic commenced > 24 hours and < 7 days prior to specimen collection | 30 (3.4) | 24 (3.7) |
| Unknown | 4 (0.4) | 0 (0.0) |
Superscript values represent missing values (n)
Subset of qPCR data
ZAR: South African Rand
IQR: interquartile range
WAZ: Weight-for-age z-score
HIV: Human immunodeficiency virus
BCG: Bacillus Calmette-Guerin
OPV: Oral polio vaccine
RV: Rotavirus vaccine
DTaP-IPV-Hib-HepB: Diphtheria-tetanus-acellular pertussis-injectable polio-Haemophilus influenzae b-Hepatitis B vaccine
PCV13: 13-valent Pneumococcal conjugate vaccine
qPCR of nasopharyngeal pathogens associated with LRTI
qPCR (n = 888 specimens) identified bacteria and viruses which were associated with LRTI (Fig. 2). Bacterial species associated with LRTI included H. influenzae [adjusted odds ratio (aOR): 1.72, 95% confidence interval (95% CI): 1.25–2.37], Klebsiella pneumoniae [aOR: 2.66, 95% CI: 1.59–4.46], and high-density Streptococcus pneumoniae [aOR: 1.53, 95% CI: 1.01–2.32] (Fig. 2).
Figure 2.
Associations between bacteria and viruses detected from nasopharyngeal (NP) specimens and lower respiratory tract infection (LRTI) during the first year of life Adjusted odds ratios (aORs) and population attributable fractions (PAFs) calculated for each of the NP pathogens screened using the Fast-Track Diagnostics Respiratory Pathogens 33 (FTDResp33) assay for 544 participants (444 LRTI and 444 non-LRTI specimens). Significant associations are denoted by an asterisk.
Adjusted odds ratios (aORs) and population attributable fractions (PAFs) calculated for each of the NP pathogens screened using the Fast-Track Diagnostics Respiratory Pathogens 33 (FTDResp33) assay for 544 participants (444 LRTI and 444 non-LRTI specimens). Significant associations are denoted by an asterisk.
Among infants aged 0–3 months, bacteria significantly associated with LRTI were H. influenzae, K. pneumoniae and S. pneumoniae (Figure S4 A). Among children aged > 3–6 months, H. influenzae and Moraxella catarrhalis were associated with LRTI (Figure S5 A), while among children aged > 6–12 months, K. pneumoniae was associated with LRTI (Figure S6 A).
Viruses associated with LRTI during the first year of life included RSV [aOR: 5.69, 95% CI: 3.03–10.69], HRV [aOR: 1.47, 95% CI: 1.03–2.09], parainfluenza virus [aOR: 3.46, 95% CI: 1.64–7.26], adenovirus [aOR: 1.99, 95% CI: 1.08–3.68], and enterovirus [aOR: 2.32, 95% CI: 1.20–4.46] (Fig. 2). RSV, CMV and parainfluenza virus were significantly associated with LRTI among children aged 0–3 months (Figure S4 A), while HRV and RSV were associated with LRTI among children > 3–6 months (Figure S5 A) and RSV was associated with LRTI among children > 6–12 months (Figure S6 A-B). Results of qPCR in the subset of participants from whom 16S rRNA gene amplicon sequence data were available (n = 646 specimens) corresponded with the full dataset (Figure S3).
The population attributable fraction (PAF) reflects the proportion of LRTI which may be attributed (alone or in combination) to a particular pathogen. For RSV, which was both prevalent and strongly associated with LRTI, the PAF was 17.3%, 95% CI: 14.0–19.0 (Fig. 2). Other organisms which contributed substantially to LRTI aetiology were H. influenzae [PAF 19.2%, 95% CI: 9.1–26.6], K. pneumoniae [PAF 9.7%, 95% CI: 5.8–12.1], and HRV [PAF 9.9%, 95% CI: 1.0–16.2].
Investigation of bacterial community profiles associated with LRTI
Following quality control steps (Fig. 1; Appendix p. 13–17), a total of 323 age- and site-matched case-control sets (n = 646 specimens) were included. The median read count per specimen was 19,283 (IQR: 13,022–25,220). Following the removal of potential contaminant amplicon sequence variants (ASVs) (Table S1), a total of 826 ASVs were included, 98% of which were classifiable at genus-level.
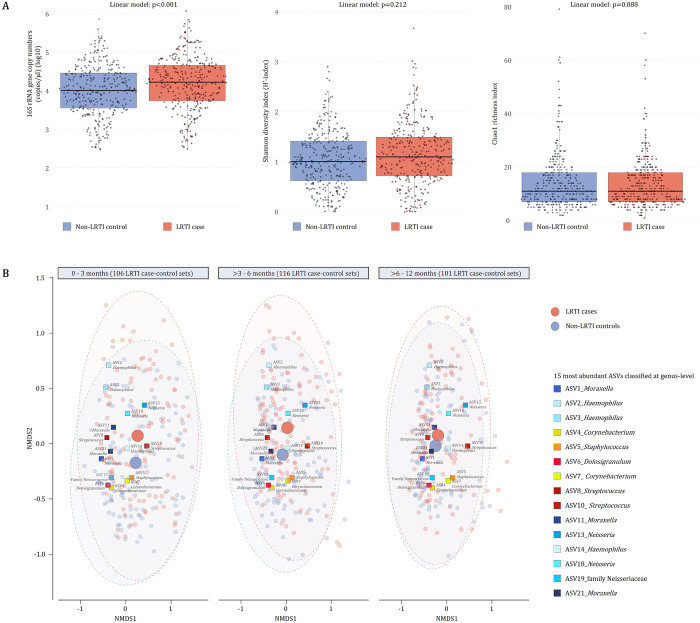
NP bacterial load (16S rRNA gene copies/μl) was higher in specimens from cases compared with controls (p < 0.0001) (Fig. 3A). Lower NP bacterial load was observed in specimens from infants who had antibiotic therapy prior to specimen collection, compared with infants with no antibiotic therapy (≤ 24 hours: p = 0.004, > 24 hours ≤ 7 days: p < 0.001). NP microbiome alpha diversity (Shannon: p = 0.213, Chao1: p = 0.088) was not significantly different when comparing cases with controls (Fig. 3A). Higher alpha diversity was observed in specimens from infants who had received antibiotic therapy prior to specimen collection compared with those who had not (Shannon: p = 0.036 and Chao1: p = 0.011 for antibiotic therapy > 24 hours ≤ 7 days prior to specimen collection vs no antibiotics; Chao1: p = 0.026 for antibiotic therapy < 24 hours of collection vs no antibiotics). There were significant differences in overall NP microbiome composition between cases and controls (PERMANOVA: p < 0.001, PERMDISP: p = < 0.001) (Fig. 3B).
Figure 3.
Bacterial density and diversity measured from NP specimens collected from LRTI cases and controls
A) Bacterial density (16S rRNA gene copies/μl) (left panel) and within-specimen bacterial diversity [Shannon diversity (middle panel) and Chao1 richness (right panel)] compared between nasopharyngeal (NP) specimens collected from lower respiratory tract infection (LRTI) cases and controls. P-values are derived from linear models adjusted for specimen collection age and antibiotic therapy prior to specimen collection. Median values are presented by horizontal lines within each of the boxplots while upper and lower ranges of the boxplots represent the 75% and 25% quartiles, respectively. Maximum and minimum values, excluding outliers, are shown by whiskers. B) Non-metric Multi-Dimensional Scaling (NMDS) plots of Bray-Curtis dissimilarity showing loadings for the 15 most abundant amplicon sequence variants (ASVs) present in the dataset. ASVs are denoted using multicoloured squares. Shades of multicoloured squares are used to present phylum-level classification of each of the ASVs [shades of blue: Proteobacteria, shades of yellow: Actinobacteria, shades of red: Firmicutes]. The three NMDS plots (left, middle and right) show NP specimens collected at 0–3 months, >3–6 months, and >6–12 months, respectively. Small red and blue circles represent NP specimens collected from LRTI cases and controls, respectively. Large red and blue circles represent centroids of LRTI cases and controls, respectively. The number of LRTI case-control sets included in each age interval are shown at the top of each NMDS plot. Alpha bags enclosing 90% of samples are shown for LRTI cases and controls.
Composition of the nasopharyngeal microbiome varies with age, antibiotic therapy and case-status
Compositional mean relative abundances of the top 15 ASVs in each age category are summarised in Fig. 4 and Table S2. Differential abundance testing [12] showed that participant age (Table S3), was strongly associated with relative abundance of several ASVs (including negative association between age and Staphylococcus, Corynebacterium, Streptococcus-ASV10 and positive association between age and Moraxella, Haemophilus, Dolosigranulum and Streptococcus-ASV8). Antibiotic therapy prior to specimen collection was associated with reduced relative abundance of Dolosigranulum and Moraxella and increased relative abundance of anaerobes, including Alloprevotella, Porphyromonas, and Gemella, as well as Streptococcus ASV10 and Family Neisseriaceae ASV19 (Table S4).
Figure 4.
Barplots of compositional mean relative abundances of the top 15 amplicon sequence variants (ASVs) detected from nasopharyngeal (NP) specimens collected at 0–3 months (n=212), >3–6 months (n=232) and >6–12 months (n=202)
The 15 most abundant ASVs from each age group are shown using colours representing phylum-level classification (Shades of blue: Proteobacteria; shades of yellow: Actinobacteria; shades of red: Firmicutes, shades of green: Bacteroidetes).
When adjusting for age and antibiotic therapy, specimens from cases had significantly higher relative abundances of Haemophilus (ASV2) when compared to controls (Fig. 5A). NP specimens with both high bacterial load (> 50,000 16S rRNA gene copies/μl) and high relative abundances (> 40%) of Haemophilus (ASV2) were primarily collected from LRTI cases (77%, 27/35) (Fig. 5A) (McNemar test: p < 0.0001). In contrast, Corynebacterium (ASV4), Dolosigranulum (ASV6), and an unclassified genus of the Family Neisseriaceae (ASV19) were detected at significantly higher relative abundances in specimens from controls compared to cases (Fig. 5B–D). NP specimens with both low bacterial load (< 3,000 16S rRNA gene copies/μl) and high relative abundances (> 20%) of Corynebacterium (ASV4) or Dolosigranulum (ASV6) were primarily collected from controls (81%, 13/16, McNemar test: p < 0.0001 or 81%, 9/11, McNemar test: p < 0.0001 respectively).
Figure 5.
Relative abundances of amplicon sequence variants (ASVs) associated with LRTI case or control status
Relative abundances of A) Haemophilus (ASV2), B) Corynebacterium(ASV4), C) Dolosigranulum (ASV6), and D) an unclassified ASV from the family Neisseriaceae (ASV19) detected from nasopharyngeal (NP) specimens collected from lower respiratory tract infection (LRTI) cases and controls. Violin plots on the left of each plot show the distribution of relative abundances for each of the ASVs detected from NP specimens collected from LRTI cases and controls. Differential abundance testing results (q values and coefficients) using Microbiome Multivariable Associations with Linear Models (MaAsLin2) are shown for LRTI case-control status. ASVs with p values <0.05 and q values <0.25 were considered significantly differentially abundant. MaAsLin2 linear models were adjusted for age at specimen collection and antibiotic therapy prior to specimen collection. Scatter plots of relative abundances of each ASV plotted against bacterial load (16S rRNA gene copies/μl) are shown on the right of each plot, with specimens collected from LRTI cases shown in red and controls in blue. Trendlines represent the trends estimated by LOESS (locally estimated scatterplot smoothing). Shaded areas represent 95% confidence intervals.
Relative abundances of A) Haemophilus (ASV2), B) Corynebacterium (ASV4), C) Dolosigranulum (ASV6), and D) an unclassified ASV from the family Neisseriaceae (ASV19) detected from nasopharyngeal (NP) specimens collected from lower respiratory tract infection (LRTI) cases and controls. Violin plots on the left of each plot show the distribution of relative abundances for each of the ASVs detected from NP specimens collected from LRTI cases and controls. Differential abundance testing results (q values and coefficients) using Microbiome Multivariable Associations with Linear Models (MaAsLin2) are shown for LRTI case-control status. ASVs with p values < 0.05 and q values < 0.25 were considered significantly differentially abundant. MaAsLin2 linear models were adjusted for age at specimen collection and antibiotic therapy prior to specimen collection. Scatter plots of relative abundances of each ASV plotted against bacterial load (16S rRNA gene copies/μl) are shown on the right of each plot, with specimens collected from LRTI cases shown in red and controls in blue. Trendlines represent the trends estimated by LOESS (locally estimated scatterplot smoothing). Shaded areas represent 95% confidence intervals.
Analysis of possible viral-bacterial interactions using qPCR and short read 16S rRNA gene amplicon sequencing
K-means clustering was used to group NP specimens (n = 646) into five clusters (Fig. 6). “MOR” cluster (n = 278) included NP specimens dominated by ASV1 (Moraxella). “STA_COR” cluster (n = 50) included NP specimens dominated by ASV5 (Staphylococcus) or ASV7 (Corynebacterium). “HAE_II” cluster (n = 94) included NP specimens dominated by ASV2 (Haemophilus), with variable contribution from ASV1 or ASV3 (Haemophilus). “HAE_III” cluster (n = 56) included specimens dominated by ASV3 with variable contribution from ASV1. “MIX” cluster (n = 168) was dominated by a range of other ASVs.
Figure 6.
K-means clustering of relative abundances of the top 25 amplicon sequence variants (ASVs) detected from LRTI case-control nasopharyngeal (NP) specimens (n=646)
A) Dendogram representing unsupervised hierarchical clustering distances of nasopharyngeal (NP) specimens based on relative abundances of the top 25 amplicon sequence variants (ASVs) in the dataset (n=646). NP specimens were grouped into five clusters: “MOR”, “STA_COR”, “HAE_II”, “HAE_III” and “MIX”. The most abundant ASVs from each specimen are shown using colours representing phylum-level classification (Shades of yellow: Actinobacteria; shades of green: Bacteroidetes; shades of red: Firmicutes, shades of blue: Proteobacteria). B) NP specimen collection age (in days). C) 16S rRNA gene amplicon copy numbers (copies/μl) measured from each NP specimen. D) Heatmap of relative abundances of the top 25 ASVs present in the dataset. E) Participant status (hospitalized LRTI case, ambulatory LRTI case or control). F) Presence/absence data of viruses screened using multiplex qPCR.
The distribution of age, case status and viral detection by cluster membership are shown in Table S6. We modelled the association between cluster membership and case status, controlling for age, bacterial load, and viral detection (Table S7). Compared with membership of cluster HAE_II, membership of cluster STA_COR was negatively associated with case status (estimate − 0.72, p = 0.001).
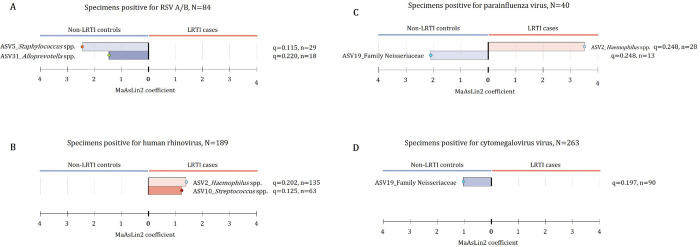
To explore whether the microbiome is associated with development of LRTI in infants infected with a respiratory virus, we investigated which bacterial taxa were associated with case status among infants infected with a particular virus. Among the subset of infants in whom RSV was detected (n = 84, 68 cases and 16 controls), we observed significantly lower relative abundances of Staphylococcus spp. (ASV5) and Alloprevotella spp. (ASV31) in cases when compared to controls (Fig. 7A). Among infants in whom HRV was detected (n = 189, 103 cases and 86 controls), we observed lower abundances of Moraxella spp. (ASV1) in cases than controls, and higher abundances of Haemophilus spp. (ASV_2) and Streptococcus spp. (ASV10) in cases (Fig. 7B). Similarly, in infants with parainfluenza virus (n = 40, 30 cases and 10 controls), Haemophilus spp. (ASV_2) relative abundance was higher among cases compared to controls.
Figure 7.
Differential abundance testing for viral and bacterial pathogens detected using multiplex qPCR
Bacterial taxa which were associated with case-status (LRTI case vs. control, using linear regression models adjusted for age and antibiotic therapy prior to specimen collection) are shown for subsets of NP specimens positive on multiplex qPCR for A) respiratory syncytial virus (RSV), B) human rhinovirus (HRV), C) parainfluenza, and D) cytomegalovirus (CMV).
Bacterial taxa which were associated with case-status (LRTI case vs. control, using linear regression models adjusted for age and antibiotic therapy prior to specimen collection) are shown for subsets of NP specimens positive on multiplex qPCR for A) respiratory syncytial virus (RSV), B) human rhinovirus (HRV), C) parainfluenza, and D) cytomegalovirus (CMV).
DISCUSSION
We investigated associations between NP bacterial community profiles, viral infection and LRTI in South African infants enrolled in a birth cohort through the first year of life. Using qPCR, we found that viruses RSV, HRV, parainfluenza virus, adenovirus, and enterovirus, and the bacteria Haemophilus influenzae, K. pneumoniae and S. pneumoniae (high density colonization) were associated with LRTI. Using 16S rRNA gene amplicon quantification and sequencing, LRTI was associated with increased bacterial load, higher relative abundances of Haemophilus, and lower relative abundances of the commensal taxa Corynebacterium, Dolosigranulum and an unclassified genus within the family Neisseriaceae. Interactions between viruses and bacteria positively associated with LRTI included HRV with Haemophilus or Streptococcus and parainfluenza virus with Haemophilus, while the interaction between RSV and Staphylococcus or Alloprevotella was negatively associated with LRTI.
Our findings of a positive association of LRTI with Haemophilus and a negative association with Corynebacterium and Dolosigranulum via 16S rRNA gene amplicon sequencing are strikingly similar to those observed in children with upper or lower respiratory infection in the United States [15], Australia [16], Europe [17] and Botswana [18]. This finding therefore represents a conserved phenotype associated with respiratory infection across both high and middle-income settings and identifies key bacterial targets for prophylactic or therapeutic intervention. We did not find evidence that NP bacterial diversity was associated with LRTI. This is in contrast to previous findings that acute upper respiratory infections such as acute otitis media [19] and mucosal inflammation in chronic rhinosinusitis [20] are associated with a decrease in local bacterial diversity.
We have previously reported on the association between the detection of microbes, using qPCR, in the NP of children in this cohort and LRTI [21, 22]. Here, we report on a larger, partially overlapping cohort, which enabled age-stratification of findings. In keeping with our previous findings,[6, 7] we detected significant associations between LRTI and detection of viruses (RSV, HRV, parainfluenza virus and adenovirus) and bacteria (H. influenzae, S. pneumoniae and K. pneumoniae). Our findings are also similar to those previously reported in the multicenter PERCH study, but 16S microbiome analysis was not done [23]. A new finding from this study is that CMV was strongly associated with LRTI in the first three months of life, while M. catarrhalis was associated with LRTI between three and six months.
RSV contributed to a substantial proportion of LRTI cases (PAF = 17%) and was strongly enriched among cases. In contrast, the contribution of the highly prevalent H. influenzae, M. catarrhalis and HRV to LRTI is less straightforward to interpret. These three organisms were only weakly (although significantly) associated with LRTI, and were commonly detected in healthy children, suggesting that interactions between these organisms and other pathogens or host factors may be required for progression to LRTI. We therefore explored possible interactions between viruses and bacteria.
Among children in whom RSV was detected, the relative abundance of Staphylococcus or Alloprevotella was lower in children with LRTI compared with controls. Several reports have identified lower prevalence or abundance of Staphylococcus in children with RSV infection, compared with controls [4, 24]. Neutrophilic inflammation in the respiratory tract has been shown to predict symptomatic RSV disease [25], and S. aureus nasal colonization impairs neutrophil recruitment [26], suggesting that S. aureus colonization may reduce the risk of symptomatic RSV infection.
Among those infants infected with HRV or parainfluenza virus, Haemophilus relative abundance was higher among children with LRTI compared with controls. This is in keeping with the hypothesis that for viruses with relatively low pathogenicity, co-infection with bacteria, particularly H. influenzae, is important in driving progression to LRTI. Previous reports have identified that H. influenzae or S. pneumoniae infection may drive more severe illness in children with RSV LRTI, [4, 27], however our cohort included primarily children with mild LRTI, and we did not identify associations between the major Haemophilus ASVs (ASV2 or ASV3) or Streptococcus and RSV disease.
The role of Moraxella species remains unclear. Moraxella-ASV1 was the most abundant ASV detected, in keeping with our previous finding that Moraxella dominates the NP microbiota of infants [28]. qPCR targeting the species M. catarrhalis showed a positive association with LRTI, however, the genus Moraxella was not associated with LRTI overall, and, in children with HRV infection, was more abundant in controls than in cases. ASV1 also includes the understudied species M. nonliquefasciens, and it is possible that the balance between these two species of Moraxella in the NP influences associations with LRTI.
Although we excluded congenital pneumonia cases from our analysis, we identified an association between nasopharyngeal detection of CMV and LRTI in the first three months of life. CMV viraemia has been described in infants with hypoxic pneumonia [29], although whether CMV contributes to respiratory illness or whether illness triggers CMV viraemia is unclear. Autopsy studies have also identified CMV as an important contributory cause to death in children under the age of five years [30].
The combination of high bacterial load and high relative abundance of Haemophilus was strongly predictive of LRTI, while low bacterial load and high relative abundance of Corynebacterium or Dolosigranulum was associated with health. This supports the hypothesis that translocation of large numbers of opportunistic pathogens from the nasopharynx to the lower respiratory tract may be key to pathogenesis of LRTI, and that a low density microbiome dominated by these Gram-positive species may be protective.
Our study has several key strengths, including a robust birth cohort study design which reduces the risk of bias in identifying cases and controls, high cohort retention, strong surveillance for LRTI, large sample size and detection of viruses and bacteria using highly multiplexed qPCR as well as 16S rRNA gene amplicon sequencing. There are some limitations. First, although this is among the largest studies of its kind, our ability to study virus-bacteria interactions was limited for less prevalent viruses. Second, due to the cross-sectional study design, causal inferences are not possible. Third, while the NP niche is a reasonable proxy for the lower respiratory tract [31], we were unable to directly sample the lung due to the need for invasive sampling methods. A limitation of short-read 16S rRNA gene amplicon sequencing is taxonomic resolution. Finally, host responses including immune and metabolic profiles are required to understand host-microbe interactions in LRTI.
CONCLUSIONS
The associations between bacterial taxa and LRTI are strikingly similar to those identified in high-income countries, suggesting a conserved phenotype. RSV was the major virus associated with LRTI. H. influenzae appears to be the major bacterial driver of LRTI, acting synergistically with viruses. The Gram-positive bacteria Dolosigranulum and Corynebacteria may protect against LRTI, while Staphylococcus was associated with reduced risk of RSV-related LRTI. Since patterns of microbial detection associated with LRTI are conserved across different continents and across varying income levels, interventions to reduce the global burden of LRTI should address these conserved patterns. With the implementation of new passive immunisation prevention against RSV in the first six months of life, active immunisation strategies and new interventions targeting non-typeable H. influenzae should be prioritized to reduce the burden of LRTI. Treatments, such as probiotics, should be explored to enhance natural protection associated with Gram-positive commensals. Mechanisms through which Gram-positive bacterial colonization of the nasopharynx may reduce the risk of viral LRTI require further study.
Research in context.
Evidence before this study
We searched PubMed on 8 February 2024, using the search terms “child” AND (“pneumonia” OR “lower respiratory tract infection”) and “microbiome” and “virus”. We identified case-control or cohort studies from high-income countries and a study from Botswana studying the bacterial component of the microbiome with or without associated viral detection. Most studies identified that the bacterial genera Haemophilus and Streptococcus were associated with lower respiratory tract infection (LRTI), while Corynebacterium and Dolosigranulum were associated with health. Viruses, including RSV, influenza, parainfluenza, HRV and human metapneumovirus were frequently associated with infection.
Added value of this study
To our knowledge, this is the largest study incorporating both multiplex viral detection and 16S rRNA gene sequencing in infants with LRTI and age-matched controls, and only the second in a low-or middle-income country (LMIC). We used a cohort design with nested case-control analysis to minimize bias. Our data confirm that the pattern of bacterial taxa associated with LRTI (high Haemophilus, low Corynebacterium, Dolosigranulum) is similar to that in high-income countries (HICs) and that the viral pathogens associated with LRTI are similarly conserved. Our analysis of bacterial-viral associations, enabled by the large sample size, suggests that specific interactions are associated with increased risk of LRTI (Haemophilus with parainfluenza virus or RSV) or decreased risk of LRTI (Staphylococcus with RSV).
Implications of all the available evidence
Patterns of microbial detection associated with LRTI are similar in a LMIC to that in high-income countries, indicating that these microbes should be targeted to reduce the global burden of LRTI. Preventive interventions should aim to reduce bacterial infection with Haemophilusinfluenzae and enhance the growth of the beneficial genera Corynebacterium and Dolosigranulum. Mechanisms through which Gram-positive bacterial colonization of the nasopharynx may reduce the risk of viral LRTI require further study.
Acknowledgements
We thank DCHS participants and their families. We thank the DCHS clinical, laboratory, and data team staff. We thank the staff of the Western Cape Government Health Department at Paarl Hospital and at the clinics for support of the study. We acknowledge facilities provided by the University of Cape Town’s ICTS High Performance Computing team (http://hpc.uct.ac.za). We thank Prof. Debby Bogaert’s team at the Centre for Inflammation Research, University of Edinburgh, for guidance related to their qPCR protocol.
Funding
This work was supported by an H3Africa U01 award from the National Institutes of Health of the USA (1U01AI110466–01A1), the Bill and Melinda Gates Foundation (OPP1017641; OPP1017579), the National Research Foundation South Africa, and the South African Medical Research Council. SC was supported by the Drakenstein Child Health Study, funded by Bill and Melinda Gates Foundation (OPP1017641), the National Research Foundation South Africa, and L’Oréal-UNESCO For Women in Science (South African Young Talents Award). MPN was supported by an Australian National Health and Medical Research Council Investigator Grant (APP1174455). HJZ was supported by the South African Medical Research Council. Funding bodies had no role in the design of the study and collection, analysis, and interpretation of the data and in writing the manuscript and the decision to publish this report.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Ethical approval was received from the Human Research Ethics Committee (HREC) of the University of Cape Town, South Africa (401/2009 and 585/2015).
Contributor Information
Shantelle Claassen-Weitz, University of Cape Town.
Yao Xia, University of Western Australia.
Lesley Workman, Red Cross War Memorial Children’s Hospital.
Luke Hannan, University of Cape Town.
Sugnet Gardner-Lubbe, Stellenbosch University.
Kilaza S Mwaikono, Dar es Salaam Institute of Technology.
Stephanie Harris Mounaud, J. Craig Venter Institute.
William C. Nierman, J. Craig Venter Institute
Samantha Africa, University of Cape Town.
Fadheela Patel, University of Cape Town.
Felix Sizwe Dube, University of Cape Town.
Veronica Allen, University of Cape Town.
Lemese Ah Tow Edries, University of Cape Town.
Heather J. Zar, University of Cape Town
Mark Patrick Nicol, University of Western Australia.
Availability of data and material
qPCR data is available at https://github.com/xy-repo/Drakenstein-Nested-Case-Control. The 16S rRNA gene sequencing data supporting the conclusions of this article will be made available in the National Centre for Biotechnology Information (NCBI) Sequence Read Archive (SRA). Metadata included in this study is available at https://github.com/xy-repo/Drakenstein-Nested-Case-Control/raw_data/metadata_323sets_final.csv. This paper reports original code for MaAsLin2 analyses, which is available in https://github.com/xy-repo/Drakenstein-Nested-Case-Control.
References
- 1.Troeger CE, Khalil IA, Blacker BF, Biehl MH, Albertson SB, Zimsen SRM, et al. Quantifying risks and interventions that have affected the burden of lower respiratory infections among children younger than 5 years: an analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis. 2020;20:60–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Globally F, Bill F. Age-sex differences in the global burden of lower respiratory infections and risk factors, 1990–2019: results from the Global Burden of Disease Study 2019. Lancet Infect Dis. 2022;22:1626–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel SJ, Weiser JN. Mechanisms of Bacterial Colonization of the Respiratory Tract. Annu Rev Microbiol. 2015;69:425–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Steenhuijsen Piters WAA, Heinonen S, Hasrat R, Bunsow E, Smith B, Suarez-Arrabal MC, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med. 2016;194:1104–15. doi: 10.1164/rccm.201602-0220OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zar HJ, Barnett W, Myer L, Stein DJ, Nicol MP. Investigating the early-life determinants of illness in Africa: the Drakenstein Child Health Study. Thorax. 2014;0:1–3. doi: 10.1136/thoraxjnl-2014-206242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zar HJ, Barnett W, Stadler A, Gardner-Lubbe S, Myer L, Nicol MP. Aetiology of childhood pneumonia in a well vaccinated South African birth cohort: A nested case-control study of the Drakenstein Child Health Study. Lancet Respir Med. 2016;4:463–72. doi: 10.1016/S2213-2600(16)00096-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zar HJ, MacGinty R, Workman L, Burd T, Smith G, Myer L, et al. Klebsiella pneumoniae Lower Respiratory Tract Infection in a South African Birth Cohort: a Longitudinal Study. Int J Infect Dis. 2022;121:31–8. doi: 10.1016/j.ijid.2022.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baggett HC, Watson NL, Knoll MD, Brooks WA, Feikin DR, Hammitt LL, et al. Density of upper respiratory colonization with Streptococcus pneumoniae and its role in the diagnosis of pneumococcal pneumonia among children aged < 5 years in the PERCH study. Clin Infect Dis. 2017;64 Suppl 3:S317–27. doi: 10.1093/cid/cix100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claassen-Weitz S, Gardner-Lubbe S, Mwaikono KS, du Toit E, Zar HJ, Nicol MP. Optimizing 16S rRNA gene profile analysis from low biomass nasopharyngeal and induced sputum specimens. BMC Microbiol. 2020;20:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.R Core Team. R Foundation for Statistical Computing. R: A language and environment for statistical computing. 2018. https://www.r-project.org/. [Google Scholar]
- 11.Oksanen J, Blanchet F., Kindt R, Legendre P, Minchin P., O’Hara R., et al. vegan: Community Ecology Package. 2019. https://cran.r-project.org/package=vegan. [Google Scholar]
- 12.Mallick H, Rahnavard A, McIver LJ, Ma S, Zhang Y, Nguyen LH, et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput Biol. 2021;17:1–27. doi: 10.1371/journal.pcbi.1009442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waskom M, Botvinnik O, O’Kane D, Hobson P, Lukauskas S, Gemperline D, et al. Seaborn. 2017. 10.5281/zenodo.883859. [DOI] [Google Scholar]
- 14.McCready C, Haider S, Little F, Nicol MP, Workman L, Gray DM, et al. Early childhood wheezing phenotypes and determinants in a South African birth cohort: longitudinal analysis of the Drakenstein Child Health Study. Lancet Child Adolesc Heal. 2023;7:127–35. doi: 10.1016/S2352-4642(22)00304-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laufer AS, Metlay JP, Gent JF, Fennie KP, Kong Y, Pettigrew MM. Microbial Communities of the Upper Respiratory Tract and Otitis Media in Children. MBio. 2011;2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The Infant Nasopharyngeal Microbiome Impacts Severity of Lower Respiratory Infection and Risk of Asthma Development. Cell Host Microbe. 2015;17:1–12. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Steenhuijsen Piters W, Watson R, de Koff E, Hasrat R, Ar K, Chu M, et al. Early-life viral infections are associated with disadvantageous immune and microbiota profiles and recurrent respiratory infections. Nat Microbiol. 2022;7:224–37. [DOI] [PubMed] [Google Scholar]
- 18.Kelly MS, Surette MG, Smieja M, Pernica JM, Rossi L, Luinstra K, et al. The Nasopharyngeal Microbiota of Children with Respiratory Infections in Botswana. Pediatr Infect Dis J. 2017;36:e211–8. doi: 10.1097/INF.0000000000001607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chonmaitree T, Jennings K, Golovko G, Khanipov K, Pimenova M, Patel JA, et al. Nasopharyngeal microbiota in infants and changes during viral upper respiratory tract infection and acute otitis media. PLoS One. 2017;12:e0180630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abreu N, Nagalingam N, Song J, Roediger F, Pletcher SD Goldberg A, Lynch S. Sinus Microbiome Diversity Depletion and Corynebacterium tuberculostearicum Enrichment Mediates Rhinosinusitis. Sci Transl Med. 2012;4:51ra124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zar HJ, MacGinty R, Workman L, Burd T, Smith G, Myer L, et al. Klebsiella pneumoniae Lower Respiratory Tract Infection in a South African Birth Cohort: a Longitudinal Study. Int J Infect Dis. 2022;121:31–8. doi: 10.1016/j.ijid.2022.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zar HJ, Nduru P, Stadler JAM, Gray D, Barnett W, Lesosky M, et al. Early-life respiratory syncytial virus lower respiratory tract infection in a South African birth cohort: epidemiology and effect on lung health. Lancet Glob Heal. 2020;8:e1316–25. doi: 10.1016/S2214-109X(20)30251-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The Pneumonia Etiology Research for Child Health (PERCH) Study Group. Causes of severe pneumonia requiring hospital admission in children without HIV infection from Africa and Asia: the PERCH multi-country case-control study. Lancet. 2019;394:757–79. doi: 10.1016/S0140-6736(19)30721-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McClintock J, Odom-Mabey A, Kebere N, Ismail A, Mwananyanda L, Gill C, et al. Postmortem Nasopharyngeal Microbiome Analysis of Zambian Infants With and Without Respiratory Syncytial Virus Disease. Pediatr Infect Dis J. 2023;42:637–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Habibi MS, Thwaites RS, Chang M, Jozwik A, Paras A, Kirsebom F, et al. Neutrophilic inflammation in the respiratory mucosa predisposes to RSV infection. Science (80-). 2020;370:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly AM, Leech JM, Doyle SL, McLoughlin RM. Staphylococcus aureus-induced immunosuppression mediated by IL-10 and IL-27 facilitates nasal colonisation. PLoS Pathog. 2022;18:1–23. doi: 10.1371/journal.ppat.1010647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diaz-Diaz A, Bunsow E, Garcia-Maurino C, Moore-Clingenpeel M, Naples J, Juergensen A, et al. Nasopharyngeal Codetection of Haemophilus influenzae and Streptococcus pneumoniae Shapes Respiratory Syncytial Virus Disease Outcomes in Children. J Infect Dis. 2022;225:912–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Claassen-Weitz S, Gardner-Lubbe S, Xia Y, Mwaikono KS, Mounaud SH, Nierman WC, et al. Succession and determinants of the early life nasopharyngeal microbiota in a South African birth cohort. Microbiome. 2023;11:1–21. doi: 10.1186/s40168-023-01563-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lakhan A, Gie A, Rhode D, Mfingwana L, Parker N, Goussard P. Cytomegalovirus viral load as predictor of the clinical course of hypoxic pneumonia in children. Int J Tuberc Lung Dis. 2023;27:49–54. [DOI] [PubMed] [Google Scholar]
- 30.Taylor AW, Blau DM, Bassat Q, Onyango D, Kotloff KL, Arifeen S El, et al. Initial findings from a novel population-based child mortality surveillance approach: a descriptive study. Lancet Glob Heal. 2020;8:e909–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zar HJ, Workman L, Isaacs W, Munro J, Black F, Eley B, et al. Rapid Molecular Diagnosis of Pulmonary Tuberculosis in Children Using Nasopharyngeal Specimens. Clin Infect Dis. 2012;55:1088–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
qPCR data is available at https://github.com/xy-repo/Drakenstein-Nested-Case-Control. The 16S rRNA gene sequencing data supporting the conclusions of this article will be made available in the National Centre for Biotechnology Information (NCBI) Sequence Read Archive (SRA). Metadata included in this study is available at https://github.com/xy-repo/Drakenstein-Nested-Case-Control/raw_data/metadata_323sets_final.csv. This paper reports original code for MaAsLin2 analyses, which is available in https://github.com/xy-repo/Drakenstein-Nested-Case-Control.