Abstract
The three-dimensional structure of soluble recombinant integrin αvβ5 bound to human adenovirus types 2 and 12 (Ad2 and -12) has been determined at ∼21-Å resolution by cryoelectron microscopy (cryo-EM). The αvβ5 integrin is known to promote Ad cell entry. Cryo-EM has shown that the integrin-binding RGD (Arg-Gly-Asp) protrusion of the Ad2 penton base protein is highly mobile (P. L. Stewart, C. Y. Chiu, S. Huang, T. Muir, Y. Zhao, B. Chait, P. Mathias, and G. R. Nemerow, EMBO J. 16:1189–1198, 1997). Sequence analysis indicated that the Ad12 RGD surface loop is shorter than that of Ad2 and probably less flexible, hence more suitable for structural characterization of the Ad-integrin complex. The cryo-EM structures of the two virus-receptor complexes revealed a ring of integrin density above the penton base of each virus serotype. As expected, the integrin density in the Ad2 complex was diffuse while that in the Ad12 complex was better defined. The integrin consists of two discrete subdomains, a globular domain with an RGD-binding cleft ∼20 Å in diameter and a distal domain with extended, flexible tails. Kinetic analysis of Ad2 interactions with αvβ5 indicated ∼4.2 integrin molecules bound per penton base at close to saturation. These results suggest that the precise spatial arrangement of five RGD protrusions on the penton base promotes integrin clustering and the signaling events required for virus internalization.
The integrins comprise a large family of heterodimeric cell surface receptors that mediate biological processes as diverse as cell-cell communication, cell movement and adhesion, leukocyte trafficking, and even short-term memory in Drosophila (20, 25). Because of their prominent role in cellular adhesive and promigratory interactions, integrins are also involved in a number of disease states, including tumor growth and metastasis, osteoporosis, and the development of atherosclerotic lesions (21). There are 16 known α and 8 known β subunits that combine to form at least 22 distinct α-β heterodimers (24). Integrins are able to recognize both cell surface and extracellular matrix ligands, with ligand-binding specificity conferred by the pairing of different subunits.
The regulation of integrin-mediated signal transduction is complex and is governed by a number of factors, including ligand occupancy, receptor conformation and/or aggregation, and interaction with divalent metal cations (22, 23, 33). Cell signaling by integrins results in the colocalization of cytoskeleton-associated proteins, such as talin and α-actinin, as well as tyrosine phosphorylation and activation of downstream effector kinases (27). Efficient signal transduction requires both receptor occupancy and clustering (36), which can be induced by multivalent ligands, such as many extracellular matrix proteins or the penton base protein of adenovirus type 2 (Ad2) (31, 32).
The two subunits of the integrin heterodimer are known to noncovalently interact with each other at their N termini to form a large extracellular head. This globular domain is connected to two separate tails that cross the plasma membrane and possess short cytoplasmic signal transduction sequences at their C termini. The large size of the integrin heterodimer (∼250 kDa), coupled with the presence of multiple domains and numerous glycosylation sites, has made high-resolution structural studies difficult. To date, the only crystal structures of integrin solved are those of the α-chain “inserted” subdomains (i.e., the I domain or A domain) of αLβ2, αMβ2, and α2β1, which reveal a novel metal cation binding region and a potential ligand-receptor coordination site (18, 30, 39). Crystallographic characterization of an RGD ligand-integrin complex may be hindered by the observed flexibility in integrin-binding RGD loops (4, 17, 43). Overall dimensions of the entire integrin heterodimer have been estimated from rotary-shadow, negative-stain, and freeze-fracture electron microscopy (EM) (12, 37, 41, 48). The reported dimensions of the globular head are in the range of 80 by 80 to 80 by 120 Å.
In addition to their normal role in mediating cellular adhesion, integrins have been usurped by a number of viral and bacterial pathogens in order to gain entrance into host cells. For example, the vitronectin binding αvβ3 and αvβ5 integrins are internalization receptors for human Ad (8, 49). Following initial Ad attachment via the fiber receptor (9), there is rapid internalization of the virus into clathrin-coated vesicles mediated by penton base association with integrin αvβ3 or αvβ5. Ad endocytosis also requires cell-signaling events mediated by the activation of PI3-kinase (32) and the Rho family of small GTPases (31). These signaling molecules stimulate polymerization of cortical actin filaments needed for virus uptake into cells.
The Ad penton base protein consists of five polypeptide subunits, each containing an integrin-binding RGD (Arg-Gly-Asp) peptide sequence (49). The RGD motif is highly conserved in multiple Ad serotypes and is found in a number of cell matrix and adhesion proteins, such as fibronectin and vitronectin, that also bind integrins. The presence of a conserved RGD motif in different Ad serotypes, as well as competition studies with function-blocking monoclonal antibodies (MAbs) to αv integrins, indicate that different Ad serotypes use a common pathway for entry into host cells (35). Sequence alignment of four different Ad penton base proteins reveals that the RGD residues are positioned in the middle of a highly variable stretch, with a length ranging from ∼20 residues for Ad12 to more than 80 residues for Ad2. A cryo-EM reconstruction of human Ad2 complexed with a neutralizing Fab fragment from an RGD-specific MAb (named DAV-1) localizes the RGD sequence to a highly mobile surface protrusion on the penton base protein (43). The mobility of RGD loops has been postulated to be a structural feature that facilitates integrin binding.
The ectodomain, or extracellular portion, of αvβ5 integrin has been expressed as a soluble recombinant molecule (34), enabling structural studies of Ad-integrin interactions by cryo-EM. Here we present the structures of the Ad2-integrin and Ad12-integrin complexes at ∼21-Å resolution. We hypothesized that the smaller and less flexible RGD protrusion of Ad12 would be advantageous for structural characterization of the complex. Our results reveal a well-defined ring of integrin bound to the Ad12 penton base and shed further light on the mechanisms of integrin interactions.
MATERIALS AND METHODS
Production and kinetic analyses.
Recombinant baculovirus vectors encoding soluble αvFos and β5Jun integrin subunits were constructed as previously described (34). The secreted integrin heterodimer was isolated by immunoaffinity chromatography with the P1F6 anti-αvβ5 MAb. Eluted fractions containing the ∼155- to 100-kDa integrin heterodimer were pooled and concentrated (C-100 microconcentrator; Amicon) to 2 mg/ml (Bio-Rad protein assay with bovine serum albumin as a standard) and stored at −80°C. Real-time measurements of integrin association with intact Ad2 particles were carried out with an automated biosensor (BIAcore 2000; Pharmacia). A similar approach was used to measure the precise kinetics of soluble intercellular adhesion molecule-1 interaction with human rhinovirus (13). The A18 MAb, which recognizes the distal-domain portion of the Ad2 fiber protein, was used to “capture” intact virus particles onto biosensor chips. Carboxymethyl-dextran coated biosensor chips (CM5 research grade; Pharmacia) were activated with N-hydroxysuccimide and N-ethyl-N′-(dimethylaminopropyl)carbodiimide for 7 min, and a 135-μg/ml solution of A18 MAb in 10 mM sodium acetate buffer, pH 4.4, was passed over the surface at a flow rate of 5 μl/min for 4 min. Nonspecific sites were quenched by addition of 1 M ethanolamine-HCL, pH 8.5, for 7 min. Following covalent coupling of the A18 MAb, 10 μl of 140 μg of purified Ad2/ml in 10 mM Tris-HCl-buffered saline, pH 7.4, was passed over the biosensor chip at a flow rate of 1 μl/min and allowed to equilibrate. Various concentrations of the DAV-1 anti-penton base MAb (6 to 500 nM) or the purified αvβ5 integrin (0.1 to 8.0 μM) in the presence or absence of divalent metal cations were passed at 40 μl/min over a blank biosensor chip or a chip containing the immobilized Ad2 particles. The surface was regenerated after each binding interval by using 15 μl of 50 mM diethylamine, pH 10.0, at a flow rate of 100 μl/min. Kinetic binding data, including association (ka) and dissociation (kd) rate constants, were determined with BIAevaluation software version 3.0. Stoichiometric data were obtained by observing changes in surface plasmon resonance at saturation. Molecular masses of 250 kDa for integrins, 150 kDa for DAV-1, and 1.5 × 105 kDa for Ad2 particles were used in calculations for stoichiometry.
Cryo-EM and negative-stain EM.
For negative-stain EM, the soluble recombinant αvβ5 integrin was concentrated to ∼275 μg/ml, negatively stained with 2% uranyl acetate, and examined with a Philips CM120 transmission EM. For cryo-EM, the volumes of the purified Ad2 and Ad12 virus preparations were adjusted to achieve a concentration of ∼200 μg/ml, which we have found to be optimal for well-spaced particles in the cryogrid. To generate the Ad-integrin complexes, recombinant αvβ5 and the Ad particles were incubated at a molar ratio ranging from 5:1 to 7:1 integrins per viral binding site in Tris (pH 8.1)–1 mM Ca2+–1 mM Mg2+ buffer for 12 h on ice (0°C). Cryoplunging of Ad2-integrin complexes, Ad12-integrin complexes, and the corresponding unbound particles was performed according to well-established procedures (3). Holey carbon grids were prepared by layering Triafoil plastic film with 0.1- to 1-μm-sized holes on 400-mesh copper grids. A 3-μl droplet of concentrated sample was placed on a glow-discharged holey carbon grid, blotted for 10 s, and plunged immediately into ethane slush chilled by liquid nitrogen. The frozen grid was then transferred under liquid nitrogen to a prechilled Gatan 626 cryotransfer holder and inserted into a CM120 transmission EM equipped with cryoaccessories. Digital micrographs of the virus particles were collected with a Gatan slow-scan charge-coupled device camera (1,024 by 1,024 pixels) under low-dose conditions (<20 electrons/Å2) at three discrete levels of defocus (−0.5, −1.0, and −1.5 μm). The variation in defocus within a set of micrographs collected with the same nominal value is approximately ±5%, as estimated from the reproducibility of setting the focus visually. Experimentally, we have found that 17-Å-resolution reconstructions can be achieved even with slight variations in defocus value within a set of images taken with the same nominal defocus value (42). The Ad images were collected with a nominal magnification of ×45,000, corresponding to an effective magnification of ×59,000 at the level of the charge-coupled device camera. The image pixel size, considering the effective magnification, was 4.1 Å and was confirmed by calibration with a catalase crystal.
Image processing.
The QVIEW software package was used to extract individual particle images as 400- by 400-pixel fields (40). Preliminary image processing with QVIEW involved exclusion of density from nearby particles and the carbon edge, planar background subtraction, and application of a circular mask. All subsequent processing steps were performed in the context of the IMAGIC image-processing system (47). Four sets of 396-, 436-, 243-, and 285-total-particle images of Ad2, Ad12, Ad2-integrin, and Ad12-integrin, respectively, were collected. The number of Ad-integrin particle images was limited by the amount of soluble recombinant αvβ5 available. The sets were partitioned according to defocus value, and each subset was then normalized as well as translationally aligned to a calculated reference (the rotationally averaged sum of the input images). Next, the initial Euler orientational angles for the particle images were determined via the technique of angular reconstitution, assuming icosahedral symmetry.
Correction for the microscope contrast transfer function (CTF) was carried out for each subset with a deconvolution program, 2D-DECON, written in FORTRAN (15, 42). The CTF was modeled in Fourier space as a sinusoidal function multiplied by an exponentially decaying “envelope” (44). The parameters used for the modeled CTF equation (spherical abberation constant [Cs], 2 mm; fraction of amplitude contrast, 0.1; kV, 120; decay constant, 20 nm2; Fermi filter resolution cutoff, 8.2 Å; filter width, 3 Å; defocus, −0.5 to −1.5 μm) were selected to minimize CTF ringing effects as observed in the particle images. These parameters also best removed the defocus ringing effects as observed in the central slice of an Ad reconstruction calculated from images of a single defocus value. Deconvolution was performed by dividing the Fourier amplitudes of the particle images by the CTF, assuming a threshold of 0.05 to prevent division by zero at the CTF nodes. The deconvolved particle images were then recombined, and preliminary three-dimensional reconstructions were calculated with IMAGIC by exact filtered back projection (Hamming filter factor, 0.75).
Anchor sets of projections from initial particle reconstructions were used to progressively refine the Euler angles of the input images. In anchor set refinement, the sinograms of projections spanning the asymmetric unit are compared with the sinogram of the current projection and the sinogram correlation function is calculated to find a better particle orientation (38, 47). We have modified this procedure slightly by considering only nearby orientations within the asymmetric unit (nearest-neighbor anchor set refinement) when calculating the sinogram correlation function. This conserves processing time and allows a finer-step search for particles whose orientations are nearly correct. Further translational refinement was also carried out by translating the input images to maximize the cross-correlation coefficients with their corresponding projections. The cycles of orientational and translational refinement were continued until there was no longer any improvement in the resolution of the final structure as assessed by Fourier shell correlation (FSC). Typically, convergence occurred after two to three iterations.
Particle images showing the largest percent error were removed from the Ad2 and Ad12 sets to match the number in the corresponding Ad2-integrin and Ad12-integrin complex sets. Thus, the final reconstructions were calculated from 243 particle images for Ad2 and Ad2-integrin and 285 particle images for Ad12 and Ad12-integrin with a 4.1-Å voxel. The resolution of the final reconstructions was evaluated by first applying a “soft” mask with a Gaussian fall off at the edges to select just the ordered viral capsid (42). The resolution of the reconstructions was assessed by FSC (10, 16) and the Fourier shell phase residual (FSPR) (38, 46). The resolution indicated by the 0.5 FSC criterion and the 45° FSPR threshold was 20 to 22 Å for all four reconstructions. For purposes of difference imaging, all reconstructions were first filtered to 22-Å resolution and then normalized based on the strong capsid density. Difference imaging of the filtered and scaled maps isolated the density corresponding to αvβ5 integrin bound to Ad2 and Ad12. Specifically, the uncomplexed Ad reconstruction was masked at the isosurface threshold chosen for the viral capsid and subtracted from the corresponding Ad-integrin complex reconstruction.
Due to computational limitations, the maximum size of the reconstructed density map was limited to (256 pixels)3. This corresponded to an outer diameter of (1,050 Å)3 with a pixel size of 4.1 Å and truncated a portion of the integrin density. Additional (200-pixel)3 maps were thus calculated, with an interpolated pixel size of 8.2 Å and a maximum diameter of (1,640 Å)3 to encompass all of the integrin. The 2003 interpolated maps were found by FSC to have resolutions of ∼24 Å.
Structural analysis.
Isosurface representations of the scaled density maps were displayed with the AVS visualization software package (Advanced Visualization Systems, Inc.). The isosurface value for the viral capsid was selected to yield a continuous, nonholey capsid surface. The weak density in the Ad2 and Ad12 penton base protrusions was contoured at a level just above noise. For the αvβ5 density in the Ad12 complex reconstruction which included both integrin domains, the isosurface value was chosen to correspond to the molecular mass of five bound integrin heterodimers (250 kDa × 5 = 1,250 kDa). For the αvβ5 density in the Ad12 complex reconstruction with just the integrin proximal domain, the integrin isosurface was chosen to match the appearance of the proximal domain in the full integrin. For the αvβ5 density in the Ad2 complex reconstruction, two isosurface values were selected: one chosen to match the level of the better-defined Ad12 integrin density and the other set just above the noise level.
An occupancy estimate for the integrin in the Ad12 complex was determined from the average of three measurements, assuming 100% occupancy for the penton base protein. The first method involved summing up all of the density within the chosen contour for the integrin, dividing by the molecular mass of five bound soluble integrin molecules, and comparing this ratio with that found for the penton base. Since the penton base is difficult to isolate from the viral capsid, we estimated the sum of the density by multiplying the mean density within the approximately cropped penton base by the expected volume. The second approach involved the ratio of the average density for these two protein components, and the third was based on the ratio of the maximum density. Structural flexibility in the Ad2 RGD loop resulted in αvβ5 density that was diffuse and spread out over a larger volume. Thus, only the first measurement was used to estimate the integrin occupancy in the Ad2 complex. The length of the variable region flanking the RGD motif in the Ad2 and Ad12 penton base sequences was determined by a BLAST search multiple-alignment procedure (5, 11). The alignment was carried out on the penton base sequences of five different Ad serotypes obtained from the SWISS-PROT and TREMBL databases.
RESULTS
Kinetic analysis of integrin and antibody associations with Ad particles.
Structural analysis of integrins has been limited by an inability to purify sufficient amounts of the native receptor from mammalian cells. In addition, integrins isolated from normal human tissues frequently aggregate via their hydrophobic transmembrane domains, thus hindering precise measurement of integrin-binding properties. To circumvent these problems, we expressed the entire ectodomain of integrin αvβ5 as a soluble protein in insect cells. The intact integrin heterodimer retains the ability to bind to its natural ligand, vitronectin, as well as to the penton base protein of several different Ad serotypes (34). Negative-stain EM of the soluble recombinant αvβ5 integrin revealed >99% of the heterodimers to be in a monodispersed form in the absence of ligand.
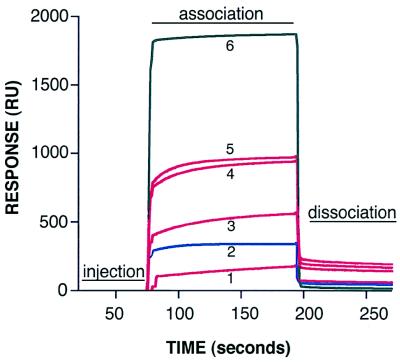
In order to determine the stoichiometry and kinetics of soluble integrin association with intact virus particles, we performed binding studies with an automated biosensor (BIAcore 2000; Pharmacia). Ad2 particles were indirectly linked to a biosensor chip via a MAb (A18) specific for the distal portion (knob) of the Ad2 fiber protein. Various amounts of soluble αvβ5 integrin were passed over immobilized virus particles (Fig. 1). The association (ka) and dissociation (kd) rate constants and stoichiometry were determined by monitoring the change in surface plasmon resonance over time (Table 1). Analysis of the binding site at close to saturating conditions indicated ∼50 integrin molecules bound to each adenovirus particle, or ∼4.2 integrin molecules bound per penton base protein. Binding of the integrin also required the presence of divalent metal cations, as its association with virus particles was completely abolished in the presence of 20 mM EDTA.
FIG. 1.
BIAcore sensorgrams of soluble αvβ5 integrin or DAV-1 MAb binding to Ad2 particles. Various amounts of soluble αvβ5 integrin or antibody (1, 500 nM integrin; 2, 50 nM DAV-1 MAb; 3, 2 μM integrin; 4, 4 μM integrin; 5, 8 μM integrin, 6, 500 nM integrin plus 20 mM EDTA) were passed over a biosensor chip containing immobilized Ad2 particles, and the association and dissociation rates were measured in real time with a BIAcore 2000 biosensor as described in Materials and Methods. For clarity, only the sensorgram of DAV-1 MAb binding (blue line) at saturating conditions is shown. The relatively large bulk-flow response seen in sensorgram 6 is due to the presence of 20 mM EDTA. This response does not represent specific integrin binding, as indicated by the absence of resonance units (RUs) following dissociation.
TABLE 1.
Association and dissociation rate constants, affinity, and stoichiometry of antibody and αvβ5 integrin interaction with Ad2 particlesa
| Ligand | ka (M−1 S−1) | kd (103 S−1) | KD (nM) | Stoichiometry |
|---|---|---|---|---|
| DAV-1 MAb | 745,000 | 1.09 | 1.45 | 1.7 |
| αvβ5 | 7,780 | 0.56 | 73.20 | 4.2 |
Binding constants, including the association rate (ka), dissociation rate (kd), and affinity (KD), and stoichiometry were determined by analysis on a BIAcore biosensor as described in Materials and Methods. Stoichiometry for MAb and integrin binding to Ad2 was determined by the change in surface plasmon resonance at or close to saturation, respectively. The data are representative of three separate experiments.
This experiment was repeated with a penton base MAb (DAV-1) directed against the integrin-binding site (IRGDTFATR) (43). Only ∼1.7 molecules of the DAV-1 MAb were calculated to bind to each penton base protein of Ad2. Due to the 50-fold-higher affinity of the DAV-1 MAb (1.4 nM) over the integrin (73 nM), it was much easier to reach saturation with the MAb. It is unknown whether the DAV-1 MAb binds monovalently or bivalently to the penton base; however, it is known that it does not neutralize the virus (43). This is consistent with our current stoichiometry measurement, which shows the MAb does not bind to all five RGD sites on the penton base. In contrast, four to five αvβ5 integrin molecules can clearly associate with a single penton base, as shown by our stoichiometry measurements.
Cryo-EM structures of Ad2 and Ad12.
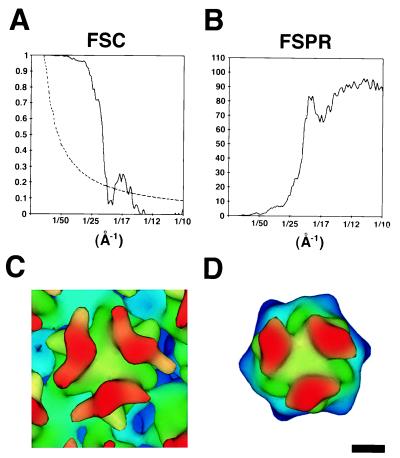
Three-dimensional image reconstructions of Ad2 and Ad12 were calculated to perform a detailed comparison of the RGD-containing penton base protrusions as well as to allow difference imaging with the Ad-integrin complex structures. The resolution of the Ad2 reconstruction was 21 Å as assessed by the commonly used FSC (10, 16) and FSPR methods (38, 46) (Fig. 2A and B). The resolution of the Ad12 reconstruction was similar at 22 Å. An independent resolution estimate was also made for Ad2 by comparison of the cryo-EM density with the filtered crystallographic structure of the Ad2 major capsid protein hexon (6). This comparison confirms that the cryo-EM resolution is at least 21 Å (Fig. 2C and D). The three towers of the cryo-EM hexon appear broader (65 Å) than the filtered crystallographic hexon (30 Å). The broad towers represent a real structural feature that is apparent when the crystallographic hexon is filtered to 15- to 17-Å resolution (42).
FIG. 2.
Resolution assessment of the Ad2 cryo-EM reconstruction. (A) Plot of the FSC (solid line) and the corresponding 23.2ς (3ς, adjusted for icosahedral symmetry) significance threshold curve (dashed line) (38). (B) Plot of the FSPR. Both the FSC and FSPR were calculated from soft masked reconstructions that only included the ordered protein capsid without the disordered fiber and core density. (C) One hexon from the Ad2 cryo-EM reconstruction filtered to 21 Å, the resolution indicated by the 0.5 correlation point of the FSC and the 45° criterion of the FSPR. (D) The crystallographic structure of the Ad2 hexon (6) filtered to 21-Å resolution. The density maps shown in panels C and D are color coded by height. The magnification of the cryo-EM structure was adjusted by ∼5% for the best match with the filtered crystallographic hexon. The scale bar is 25 Å.
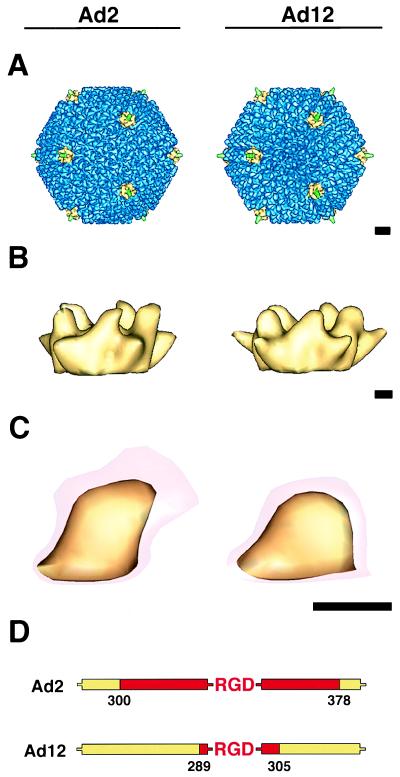
Overall, the reconstructions showed that Ad2 and Ad12 particles were similar, with only minor structural variations observed at the tops of the hexon towers and penton base protrusions (Fig. 3A to C). A BLAST alignment of the Ad2 and Ad12 hexon sequences indicates that there are 45 fewer residues in the tower region of the Ad12 hexon, consistent with the smaller appearance of the Ad12 hexon towers. The fiber is truncated in both reconstructions as only the lower portion of the flexible shaft follows icosahedral symmetry. Close inspection of the penton base protrusions in the two serotypes revealed a noticeable size difference. When the penton base was contoured at a level to show only the strong, well-defined capsid density, the protrusions of the two serotypes appeared similar. However, when contoured at a level just above noise, the Ad2 protrusion showed much more weak density (Fig. 3C). For Ad2, the weak density extended ∼15 Å outwards from the well-defined portion of the protrusion, whereas for Ad12, the weak density extended only ∼6 Å. Moreover, the total volume encompassed by a single Ad2 protrusion, including the weak density, is ∼26,100 Å3, while the corresponding volume for Ad12 is only ∼17,500 Å3. Interestingly, this difference in volume corresponds to the volume of ∼58 amino acid residues. This is in close agreement with the observation that there are 62 additional residues flanking the RGD motif in Ad2 (Fig. 3D). The finding of additional weak density above the Ad2 penton base protrusion implies the presence of a more flexible RGD loop.
FIG. 3.
Cryo-EM reconstructions of Ad2 (left) and Ad12 (right) at ∼21-Å resolution. (A) Ad capsids viewed along an icosahedral threefold axis. The penton base proteins at the icosahedral vertices are shown in yellow, the reconstructed portion of the flexible fibers are in green, and the remaining capsid density is in blue. (B) Side views of the external portion of the penton base contoured at a level corresponding to the strong capsid density. (C) Enlargements of a single penton base protrusion at two isosurface levels, one just above noise (transparent red) and the other showing well-defined density (yellow). (D) The lengths of the variable regions flanking the RGD sequence (red) in Ad2 and Ad12 are obtained from sequence alignment of five different Ad serotypes. The scale bars are 100 (A) and 25 (B and C) Å.
Cryo-EM structures of Ad complexed with soluble αvβ5 integrin.
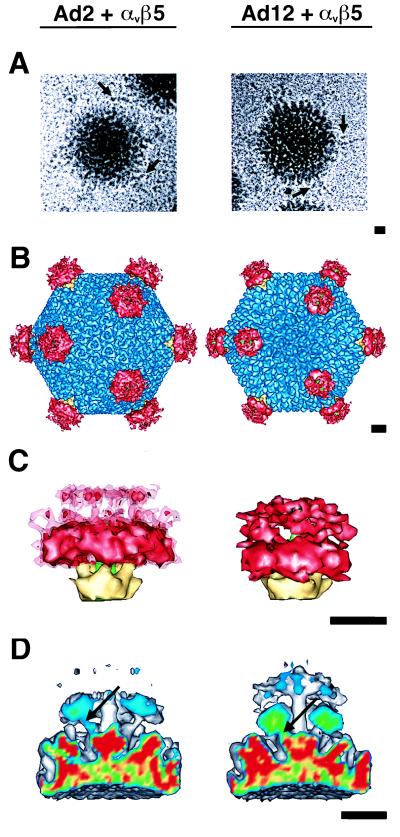
To ensure maximal occupancy of the viral RGD-binding sites by integrin molecules, an ∼6-fold molar excess of αvβ5 integrin per RGD-binding site was mixed with concentrated Ad2 or Ad12 virus preparations. Ca2+ and Mg2+ ions were added to promote and stabilize ligand-receptor complex formation. Occasionally, cryo-EM images of the virus-receptor complexes revealed extra density near the Ad fibers (Fig. 4A). Given the low signal-to-noise ratio in cryo-electron micrographs, it was difficult to discern any differences in integrin binding between Ad2 and Ad12 in the raw particle images. Three-dimensional reconstructions of both Ad2 and Ad12 complexed with integrin were calculated to resolutions of 20 and 21 Å, respectively. Due to computational limitations, it was necessary to interpolate the density maps in order to visualize the full extent of the integrin. These interpolated maps (Fig. 4B to D) spanned the maximum diameter of the Ad-integrin complexes, ∼1,200 Å, and had resolutions of ∼24 Å.
FIG. 4.
Cryo-EM reconstruction of the Ad2-integrin (left) and Ad12-integrin (right) complexes at ∼24-Å resolution. (A) Representative particle images with arrows indicating regions of density attributed to αvβ5 integrin. (B) Ad complexes viewed along an icosahedral threefold axis. The color scheme is the same as in Fig. 3A, with αvβ5 integrin density shown in red. (C) Side views of the penton base, fiber, and integrin. The Ad12-integrin density is contoured at a level corresponding to five bound αvβ5 heterodimers. The Ad2-integrin density is contoured at the same level (solid red) and just above noise (transparent red). (D) A cropped view of the vertex regions of the Ad complexes showing connections between the integrin and the penton base via the RGD protrusions (arrows). The slice plane is color coded on the basis of density, with the strongest density values shown in red and the weakest in blue. The scale bars are 100 Å.
The structure of soluble αvβ5 integrin bound to the Ad penton base.
Difference imaging between the Ad-integrin complexes and the corresponding uncomplexed reconstructions revealed that bound integrin molecules form a ring-like cluster above each penton base protein (Fig. 4B). Enlarged views of the vertex region show that the integrin density in the Ad2 complex extends farther from the surface of the viral capsid and has a larger volume than in the Ad12 complex (Fig. 4C). The αvβ5 integrin in the Ad12 complex was contoured to enclose a volume equivalent to five bound molecules, assuming a molecular mass of 250 kDa for an integrin heterodimer. The integrin density in the Ad12 complex consists of a globular proximal domain and a tail-like distal domain, where proximal and distal are defined with respect to the virus rather than the cell surface. While the Ad12 proximal domain displayed well-defined density, the distal domain showed weaker, more diffuse density suggestive of flexible tails. The proximal and distal integrin domains in the Ad2 complex are disconnected at an equivalent isosurface (Fig. 4C). Connections were only observed when the isosurface level was further lowered to just above noise (Fig. 4C). At this lowered isosurface level, the volume of the integrin in the Ad2 complex is 150% of that expected for five bound integrin heterodimers. The integrin in the Ad2 complex also has weaker density values than that in the Ad12 complex, as shown in crop planes through the vertex regions (Fig. 4D). In both complex structures, the integrin is clearly connected to the penton base via the protrusions and appears to bind at an ∼45° angle with respect to the fivefold fiber axis.
The finding of stronger, more compact integrin density in the Ad12 complex cannot be explained by differences in ligand-receptor affinities. Measurements of αvβ5 interaction with Ad2 and Ad12 in solid-phase binding assays indicated that integrin has a higher binding capacity for Ad2 than for Ad12 (34). Integrated density measurements of the reconstructions showed nearly 100% occupancy of the integrin receptor for Ad2 but only ∼73% occupancy for Ad12. We surmise that the longer and more mobile RGD region in Ad2 allows greater flexibility for the bound integrin than does the shorter RGD region in Ad12, thus explaining the weaker, less compact integrin density in the Ad2 complex despite higher occupancy. The flexibility of the RGD-binding site in Ad2 prevents a detailed structural analysis of the integrin in the Ad2 complex, as the diffuse integrin density is a result of averaging over multiple orientations. In contrast, the well-defined integrin density in the Ad12 complex is consistent with a constrained orientation for at least the globular proximal domains of the bound integrin.
Ring structure formed by integrin-proximal domains.
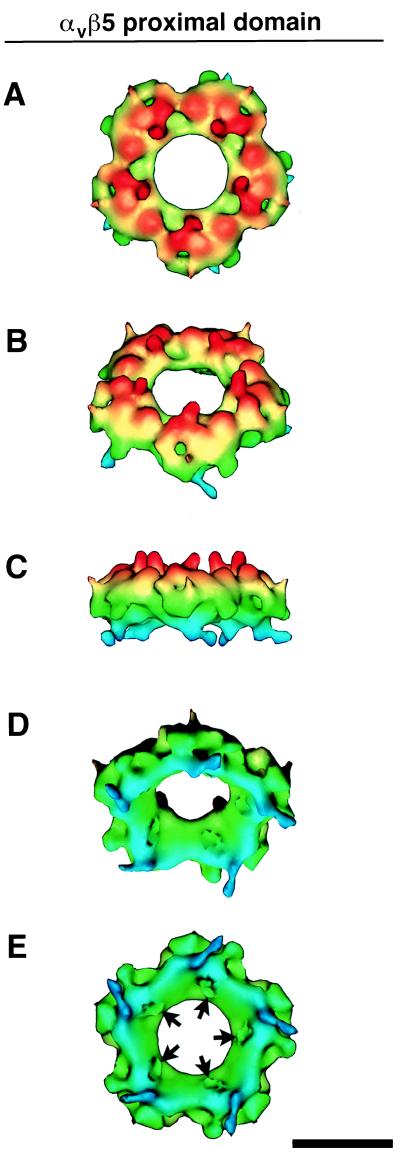
Further structural analysis of the αvβ5 integrin was carried out on the Ad12 complex reconstruction. The integrin proximal domains were observed to form a ring of density, shown color coded by height in Fig. 5. The integrin ring has a height of 80 Å and a maximum outer diameter of 200 Å and appears to be segmented into five regions (Fig. 5A). The side view in Fig. 5C shows five columns of density that connect the proximal domain to the more diffuse density in the distal domain. The tilted view in Fig. 5D reveals that the inner surface of the integrin ring is sloped from an inner diameter ranging from 70 Å at the top to 130 Å at the base. The bottom view in Fig. 5E displays five clefts, ∼20 Å in diameter, where the penton base protrusions bind (see Fig. 7).
FIG. 5.
Integrin ring from the Ad12-αvβ5 complex at ∼21-Å resolution. The ring is formed by associations between the RGD-binding proximal domains. (A to E) Five views of the height-color-coded ring from top to bottom. Note that the top surface has five columns of density (red) that connect the proximal domains to the more flexible distal domains (not shown). The bottom-surface view shows five clefts (arrows), each ∼20 Å in diameter, that bind the RGD-containing protrusions of the penton base protein. The scale bar is 100 Å.
FIG. 7.
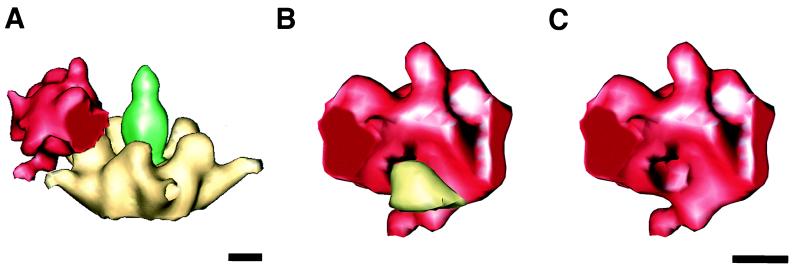
Model for the interaction between the integrin proximal domain and the Ad12 penton base protein. One-fifth of the integrin ring density is shown extracted along estimated boundaries to model the proximal domain of a single αvβ5 heterodimer. (A) The modeled proximal domain shown in association with the penton base protein. (B) The modeled proximal domain is rotated ∼90 degrees with respect to the view in panel A to show the interaction with a single penton base protrusion. (C) The same view as in panel B but with the protrusion removed to reveal the RGD-binding cleft on the inner surface. The scale bars are 25 Å.
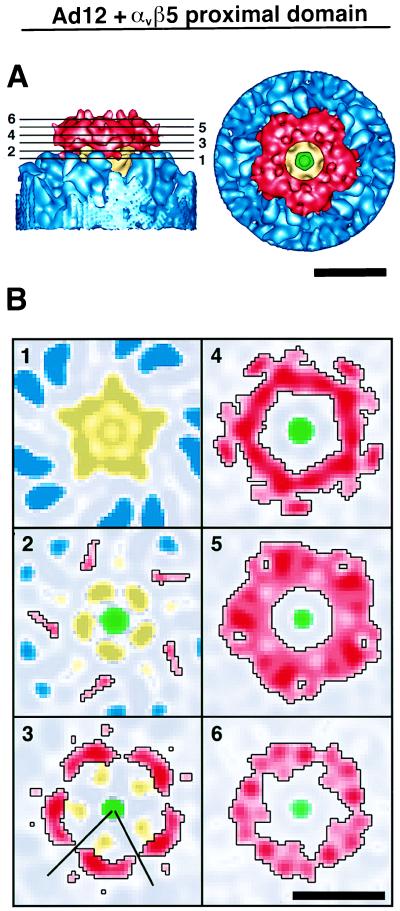
Successive slices through the vertex region of the Ad12-integrin complex show the interaction between the integrin and the penton base (Fig. 6). In slice plane 1, only density corresponding to the penton base and hexon towers is observed. The remaining slice planes show the molecular edge of the integrin outlined in black. In slice plane 3, the five penton base protrusions are observed to contact the sloping inner surface of the integrin ring. Note also that in this slice there is a clear separation of the integrin ring into five regions. However, at planes further from the viral surface, there are large regions of contact between neighboring integrin molecules (planes 4 to 6). Close apposition of adjacent receptor molecules is also noticeable in the top-surface view of the integrin ring (Fig. 6A).
FIG. 6.
Integrin ring in association with the Ad12 capsid. (A) Side- and top-surface views. (B) Slice planes through the integrin density perpendicular to an icosahedral fivefold symmetry axis. The heights of the slice planes are indicated by numbered lines in panel A. The color scheme for the individual proteins is as shown in Fig. 4B. Stronger density values are represented by darker shades, and weaker density values are represented by lighter shades. The black lines in slice 3 designate the boundary for the extracted model of one integrin proximal domain displayed in Fig. 7. The scale bars are 100 Å.
One-fifth of the integrin ring formed by the proximal domains is shown extracted and in context with the Ad12 penton base in Fig. 7A. We present this as a model for the proximal domain of a single integrin heterodimer. The BIAcore measurements indicate that division of the density into five parts is reasonable given that integrin molecules were capable of binding to all five RGD sites on the Ad2 penton base. The extracted integrin density enables visualization of a single penton base protrusion binding within a cleft on the inner sloping surface of the proximal domain (Fig. 7B and C). The protrusion is known to contain the RGD motif from previous cryo-EM studies of Ad2 complexed with an RGD-specific Fab fragment derived from a monoclonal antibody (DAV-1) (43).
DISCUSSION
Comparison of integrin binding to Ad2 and Ad12.
We report the structure of Ad2 and Ad12 complexed with its internalization receptor, αvβ5 integrin. Detailed structural and functional analyses of integrin have been hindered by the inherent difficulties in isolating and purifying αv integrins from human tissue, as well as in generating amounts large enough for structural analysis. Recent expression of the ectodomain of integrin αvβ5 as a soluble recombinant protein (34) has allowed us to characterize the structural interactions of αvβ5 integrin with human Ads by cryo-EM image reconstruction. Previous studies have shown that the Ad2 penton base protrusion, like several native RGD ligands, binds integrin via a highly flexible RGD loop (4, 17, 43). We reasoned that the flexibility of the Ad2 RGD loop would not sufficiently constrain the position of the bound integrin to allow structural characterization. Thus, we also chose to examine another Ad serotype (Ad12) with a much shorter variable RGD flanking sequence, as it has been reported that virus cell entry by both Ad2 and Ad12 is mediated via αv integrins (7). We hypothesized that a more rigid, well-defined integrin structure would be observed for the Ad12 complex.
Consistent with this hypothesis, the structures of Ad12-integrin and Ad2-integrin, solved in parallel and scaled equivalently, reveal the integrin density in the Ad12 complex to be compact and well defined, whereas that in the Ad2 complex is weak and diffuse. Differences in receptor occupancy are unlikely to account for the more diffuse integrin density of the Ad2 complex, as soluble αvβ5 has been shown by solid-phase binding assays to have a higher binding capacity for Ad2 than for Ad12 (34). Moreover, integrated density measurements on the cryo-EM structures indicate that there is actually an ∼30% greater occupancy of the integrin receptor on Ad2 than on Ad12. We also noted that the penton base protrusion in the uncomplexed Ad2 has more weak, diffuse density than the Ad12 protrusion. Taken together, these observations indicate that the αvβ5 integrin in the Ad2 complex is bound via a highly flexible RGD surface loop and hence does not have a well-constrained orientation. This explains the observation of diffuse, disconnected integrin density for the Ad2 complex and the measurement of an apparent integrin volume that is 150% of that expected. In contrast, the αvβ5 integrin is rigidly bound to the smaller, less mobile Ad12 RGD protrusion, making a detailed structural analysis possible.
Although the total height of the soluble integrin was the same for both reconstructions (145 Å), the integrin in the Ad2 complex was located farther from the surface of the viral capsid. This suggests that the Ad2 loop is more extended than the Ad12 loop. A more extended and flexible RGD loop may facilitate αvβ5 integrin binding, accounting for both the higher measured binding capacity of the Ad2 penton base for αvβ5 and the higher integrin occupancy in the Ad2 complex.
Integrin structural features.
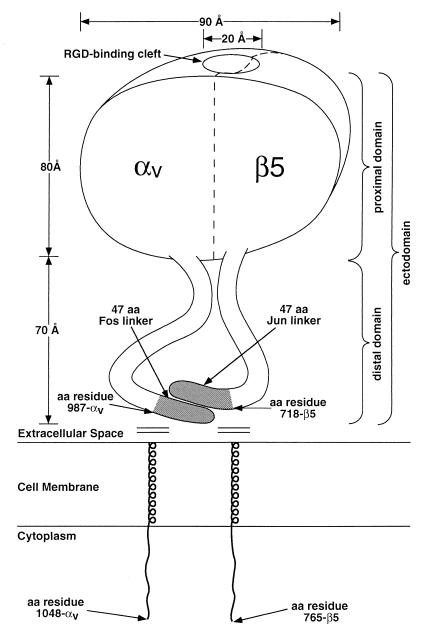
The cryo-EM reconstruction of soluble recombinant αvβ5 integrin bound to Ad12 reveals a ring-like integrin structure above the penton base protein. The integrin is observed to consist of two discrete domains, a globular proximal domain and a flexible, tail-like distal domain (Fig. 8). The visualization of a two-domain structure is in agreement with that seen previously by rotary-shadow, negative-stain, and freeze-fracture EM. The cryo-EM integrin density extracted in Fig. 7C, which is estimated to correspond to a proximal domain from one αvβ5 heterodimer, has dimensions of 65 Å in depth, 80 Å in height, and 90 Å in width (between the estimated boundaries). Previous measurements of 80 by 80 Å (41), 80 by 100 Å (12), and 80 by 120 Å (37, 48) are in rough agreement with the cryo-EM dimensions. The reconstructed integrin density presented here also reveals an ∼20-Å-diameter RGD-binding cleft on the inner surface of the integrin proximal domain, a structural feature not previously observed. Most integrin-binding RGD motifs have been found on extended surface loops (2, 28, 29) and would likely fit within a cleft of this size.
FIG. 8.
Schematic diagram of the soluble αvβ5 integrin heterodimer. Approximate dimensions are derived from the cryo-EM reconstruction of the Ad12-integrin complex. Amino acid residue numbers are from Mathias et al. (34). The proximal and distal domains are defined with respect to the viral surface.
Comparison of integrin and antibody binding to the Ad RGD loop.
Surface plasmon resonance was used to measure antibody and integrin associations with Ad particles in real time. We sought to minimize steric hindrance as well as alteration of the native virus structure during the immobilization step by indirectly linking Ad2 particles to a biosensor chip via a MAb directed against the distal domain of the elongated fiber protein. Under these conditions, ∼4.2 integrins but only ∼1.7 antibody molecules were found to bind on average to each penton base protein in the intact virus particle. The stoichiometry of antibody binding to purified penton base protein was previously reported to be ∼2.8 (43). The lower value obtained for intact virus particles might be due to steric hindrance from the fiber protein. In the present study, we also report that the apparent affinity of integrin was markedly lower than that of the DAV-1 antibody for the penton base. The measured affinity of αvβ5 integrin obtained in the BIAcore studies (KD = 73 nM) is very close to that previously determined in Scatchard analyses of soluble penton base binding to cells (KD = 55 nM) (49). The slower on-rate (ka) for integrin versus antibody molecules may be due to a number of factors, including their larger mass and the possibility that the number of ligand-binding sites becomes limiting as more integrin molecules are bound over time.
Mass considerations alone would suggest that fewer molecules of αvβ5 integrin (250 kDa) would bind to penton base relative to DAV-1 MAb (150 kDa). However, the linear, two-domain shape of the integrin, unlike the Y-shape of the immunoglobulin G antibody, appears to be more conducive for binding to the closely spaced penton base protrusions (∼60 Å apart). In addition, intermolecular associations between bound integrin molecules, but not antibodies, may promote full occupancy on the penton base protein.
Implications for Ad-induced integrin complex formation.
In the Ad12-integrin reconstruction, the bound integrin molecules were observed to form a continuous ring, exhibiting close contact between neighboring receptors. The interactions between adjacent receptors are likely to constrain the position and orientation of the individual integrin heterodimers. In the case of Ad2, the more diffuse integrin density observed in the complex may arise from the integrin ring moving as a single unit of five interlocked heterodimers. As the Ad12 RGD protrusion is less flexible, the bound integrin ring in the Ad12 complex has a more constrained position and hence better-defined density. Since integrin-mediated signaling pathways are activated in response to receptor occupancy and/or clustering, the αvβ5 ring structure visualized by cryo-EM may represent a conformation capable of inducing signals involved in virus endocytosis (31, 32). In support of this idea, the Ad penton base, but not a monomeric RGD peptide derived from the penton base, activates p72 Syk kinase and also promotes adhesion of B lymphoblastoid cells (45). These structural and biochemical findings are consistent with the possibility that integrin clustering in the plane of the cell membrane following interaction with multivalent ligands plays a role in facilitating signaling events.
Receptor redistribution and clustering at adhesion sites play crucial roles in signal transduction events mediated by integrins. In vitro binding experiments have shown that integrin occupancy and integrin aggregation can each mediate separate biological signaling events (36). The synergistic action of both stimuli was required to induce the accumulation of cytoskeletal proteins that mediate cell adhesion. In addition, adhesion-dependent apoptosis and cell spreading by integrins were found to be stimulated by multimeric rather than monomeric ligands (14). These studies also suggest that transmembrane signaling by integrin not only occurs as a result of receptor clustering following ligand binding but is also dependent on the precise spatial arrangement of the bound receptor molecules. Our structural results with the Ad-integrin complex suggest that such integrin clustering may occur via interactions between the globular, RGD-binding domains.
During the course of evolution, various signaling pathways have been exploited by viruses and other pathogens to gain entry into host cells (19). The icosahedral structure of many viruses, with two-, three-, and fivefold axes, readily enables the mimicking of native multimeric ligands. Interestingly, the spacing of the integrin-binding RGD sites on Ad, ∼60 Å, is virtually identical to that on foot-and-mouth disease virus, which is evolutionarily unrelated (1, 26). The fivefold symmetry of the Ad penton base promotes the formation of an integrin ring structure that may play a key role in mediating virus internalization. It is possible that integrin interactions with their natural ligands, extracellular matrix proteins, also involves direct associations between receptor ectodomains, allowing regulation of integrin-mediated cell functions.
ACKNOWLEDGMENTS
We express our gratitude to Mike Galleno (Invitrogen) for helpful suggestions on the expression of soluble αvβ5. We also thank Jack Johnson for reading the manuscript and David Cheresh and members of his laboratory for helpful comments.
This work was supported by NIH grants HL54352 and EY11431 to G.R.N. and AI42929 to P.L.S. C.Y.C. was supported by a fellowship from the Life and Health Insurance Medical Research Fund, a NIH-MSTP training grant (GM08042), and the Aesculapians fund of the UCLA School of Medicine.
REFERENCES
- 1.Acharya R, Fry E, Stuart D, Fox G, Rowlands D, Brown F. The three-dimensional structure of foot-and-mouth disease virus at 2.9 Å resolution. Nature. 1989;337:709–716. doi: 10.1038/337709a0. [DOI] [PubMed] [Google Scholar]
- 2.Adler M, Lazarus R A, Dennis M S, Wagner G. Solution structure of kistrin, a potent platelet aggregation inhibitor and GP IIb-IIIa antagonist. Science. 1991;253:445–448. doi: 10.1126/science.1862345. [DOI] [PubMed] [Google Scholar]
- 3.Adrian M, Dubochet J, Lepault J, McDowall A W. Cryo-electron microscopy of viruses. Nature. 1984;308:32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- 4.Akke M, Liu J, Cavanagh J, Erickson H P, Palmer A G I. Pervasive conformational fluctuations on microsecond time scales in a fibronectin type III domain. Nat Struct Biol. 1998;5:55–59. doi: 10.1038/nsb0198-55. [DOI] [PubMed] [Google Scholar]
- 5.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 6.Athappilly F K, Murali R, Rux J J, Cai Z, Burnett R M. The refined crystal structure of hexon, the major capsid protein of adenovirus type 2, at 2.9 Å resolution. J Mol Biol. 1994;242:430–455. doi: 10.1006/jmbi.1994.1593. [DOI] [PubMed] [Google Scholar]
- 7.Bai M, Campisi L, Freimuth P. Vitronectin receptor antibodies inhibit infection of HeLa and A549 cells by adenovirus type 12 but not by adenovirus type 2. J Virol. 1994;68:5925–5932. doi: 10.1128/jvi.68.9.5925-5932.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai M, Harfe B, Freimuth P. Mutations that alter an Arg-Gly-Asp (RGD) sequence in the adenovirus type 2 penton base protein abolish its cell-rounding activity and delay virus reproduction in flat cells. J Virol. 1993;67:5198–5205. doi: 10.1128/jvi.67.9.5198-5205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 10.Böttcher B, Wynne S A, Crowther R A. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 11.Brown N P, Leroy C, Sander C. MView: a web-compatible database search or multiple alignment viewer. Bioinformatics. 1998;14:380–381. doi: 10.1093/bioinformatics/14.4.380. [DOI] [PubMed] [Google Scholar]
- 12.Carrell N A, Fitzgerald L A, Steiner B, Erickson H P, Phillips D R. Structure of human platelet membrane glycoproteins IIb and IIIa as determined by electron microscopy. J Biol Chem. 1985;260:1743–1749. [PubMed] [Google Scholar]
- 13.Casasnovas J M, Springer T A. Kinetics and thermodynamics of virus binding to receptor. Studies with rhinovirus, intercellular adhesion molecule-1 (ICAM-1), and surface plasmon resonance. J Biol Chem. 1995;270:13216–13224. doi: 10.1074/jbc.270.22.13216. [DOI] [PubMed] [Google Scholar]
- 14.Chen C S, Mrksich M, Huang S, Whitesides G M, Ingber D E. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 15.Chiu C Y, Cary R B, Chen D J, Peterson S R, Stewart P L. Cryo-EM imaging of the catalytic subunit of the DNA-dependent protein kinase. J Mol Biol. 1998;284:1075–1081. doi: 10.1006/jmbi.1998.2212. [DOI] [PubMed] [Google Scholar]
- 16.Conway J F, Cheng N, Zlotnick A, Wingfield P T, Stahl S J, Steven A C. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 17.Copie V, Tomita Y, Akiyama S K, Aota S, Yamada K M, Venable R M, Pastor R W, Krueger S, Torchia D A. Solution structure and dynamics of linked cell attachment modules of mouse fibronectin containing the RGD and synergy regions: comparison with the human fibronectin crystal structure. J Mol Biol. 1998;277:663–682. doi: 10.1006/jmbi.1998.1616. [DOI] [PubMed] [Google Scholar]
- 18.Emsley J, King S L, Bergelson J M, Liddington R C. Crystal structure of the I domain from integrin alpha2beta1. J Biol Chem. 1997;272:28512–28517. doi: 10.1074/jbc.272.45.28512. [DOI] [PubMed] [Google Scholar]
- 19.Finlay B B, Cossart P. Exploitation of mammalian host cell functions by bacterial pathogens. Science. 1997;276:718–725. doi: 10.1126/science.276.5313.718. [DOI] [PubMed] [Google Scholar]
- 20.Grotewiel M S, Beck C D, Wu K H, Zhu X R, Davis R L. Integrin-mediated short-term memory in Drosophila. Nature. 1998;391:455–460. doi: 10.1038/35079. [DOI] [PubMed] [Google Scholar]
- 21.Horwitz A F. Integrins and health. Sci Am. 1997;276:68–75. doi: 10.1038/scientificamerican0597-68. [DOI] [PubMed] [Google Scholar]
- 22.Hughes P E, Pfaff M. Integrin affinity modulation. Trends Cell Biol. 1998;8:359–364. doi: 10.1016/s0962-8924(98)01339-7. [DOI] [PubMed] [Google Scholar]
- 23.Humphries M J. Integrin activation: the link between ligand binding and signal transduction. Curr Opin Cell Biol. 1996;8:632–640. doi: 10.1016/s0955-0674(96)80104-9. [DOI] [PubMed] [Google Scholar]
- 24.Humphries M J, Newham P. The structure of cell-adhesion molecules. Trends Cell Biol. 1998;8:78–83. [PubMed] [Google Scholar]
- 25.Hynes R O. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 26.Jackson T, Sharma A, Ghazaleh R A, Blakemore W E, Ellard F M, Simmons D L, Newman J W, Stuart D I, King A M. Arginine-glycine-aspartic acid-specific binding by foot-and-mouth disease viruses to the purified integrin alpha(v)beta3 in vitro. J Virol. 1997;71:8357–8361. doi: 10.1128/jvi.71.11.8357-8361.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jenkins A L, Nannizzi-Alaimo L, Silver D, Sellers J R, Ginsberg M H, Law D A, Phillips D R. Tyrosine phosphorylation of the beta3 cytoplasmic domain mediates integrin-cytoskeletal interactions. J Biol Chem. 1998;273:13878–13885. doi: 10.1074/jbc.273.22.13878. [DOI] [PubMed] [Google Scholar]
- 28.Leahy D J, Aukhil I, Erickson H P. 2.0 A crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- 29.Leahy D J, Hendrickson W A, Aukhil I, Erickson H P. Structure of a fibronectin type III domain from tenascin phased by MAD analysis of the selenomethionyl protein. Science. 1992;258:987–991. doi: 10.1126/science.1279805. [DOI] [PubMed] [Google Scholar]
- 30.Lee J O, Rieu P, Arnaout M A, Liddington R. Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18) Cell. 1995;80:631–638. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- 31.Li E, Stupack D, Bokoch G M, Nemerow G R. Adenovirus endocytosis requires actin cytoskeleton reorganization mediated by Rho family GTPases. J Virol. 1998;72:8806–8812. doi: 10.1128/jvi.72.11.8806-8812.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li E, Stupack D, Klemke R, Cheresh D A, Nemerow G R. Adenovirus endocytosis via alpha(v) integrins requires phosphoinositide-3-OH kinase. J Virol. 1998;72:2055–2061. doi: 10.1128/jvi.72.3.2055-2061.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Longhurst C M, Jennings L K. Integrin-mediated signal transduction. Cell Mol Life Sci. 1998;54:514–526. doi: 10.1007/s000180050180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathias P, Galleno M, Nemerow G R. Interactions of soluble recombinant integrin αvβ5 with human adenoviruses. J Virol. 1998;72:8669–8675. doi: 10.1128/jvi.72.11.8669-8675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathias P, Wickham T, Moore M, Nemerow G. Multiple adenovirus serotypes use alpha v integrins for infection. J Virol. 1994;68:6811–6814. doi: 10.1128/jvi.68.10.6811-6814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyamoto S, Akiyama S K, Yamada K M. Synergistic roles for receptor occupancy and aggregation in integrin transmembrane function. Science. 1995;267:883–885. doi: 10.1126/science.7846531. [DOI] [PubMed] [Google Scholar]
- 37.Nermut M V, Green N M, Eason P, Yamada S S, Yamada K M. Electron microscopy and structural model of human fibronectin receptor. EMBO J. 1988;7:4093–4099. doi: 10.1002/j.1460-2075.1988.tb03303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orlova E V, Dube P, Harris J R, Beckman E, Zemlin F, Markl J, van Heel M. Structure of keyhole limpet hemocyanin type 1 (KLH1) at 15 Å resolution by electron cryomicroscopy and angular reconstitution. J Mol Biol. 1997;271:417–437. doi: 10.1006/jmbi.1997.1182. [DOI] [PubMed] [Google Scholar]
- 39.Qu A, Leahy D J. Crystal structure of the I-domain from the CD11a/CD18 (LFA-1, alpha L beta 2) integrin. Proc Natl Acad Sci USA. 1995;92:10277–10281. doi: 10.1073/pnas.92.22.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah A K, Stewart P L. QVIEW: software for rapid selection of particles from digital electron micrographs. J Struct Biol. 1998;123:17–21. doi: 10.1006/jsbi.1998.4011. [DOI] [PubMed] [Google Scholar]
- 41.Sharma A, Rao Z, Fry E, Booth T, Jones E Y, Rowlands D J, Simmons D L, Stuart D I. Specific interactions between human integrin alpha v beta 3 and chimeric hepatitis B virus core particles bearing the receptor-binding epitope of foot-and-mouth disease virus. Virology. 1997;239:150–157. doi: 10.1006/viro.1997.8833. [DOI] [PubMed] [Google Scholar]
- 42.Stewart, P. L., R. B. Cary, S. R. Peterson, and C. Y. Chiu. Digitally collected cryo-electron micrographs for single particle reconstruction. Microsc. Res. Tech., in press. [DOI] [PubMed]
- 43.Stewart P L, Chiu C Y, Huang S, Muir T, Zhao Y, Chait B, Mathias P, Nemerow G R. Cryo-EM visualization of an exposed RGD epitope on adenovirus that escapes antibody neutralization. EMBO J. 1997;16:1189–1198. doi: 10.1093/emboj/16.6.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart P L, Fuller S D, Burnett R M. Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 1993;12:2589–2599. doi: 10.1002/j.1460-2075.1993.tb05919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stupack D G, Li E, Silletti S A, Kehler J A, Geahlen R L, Hahn K, Nemerow G R, Cheresh D A. Matrix valency regulates integrin-mediated lymphoid adhesion via Syk kinase. J Cell Biol. 1999;144:777–788. doi: 10.1083/jcb.144.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Heel M. Similarity measures between images. Ultramicroscopy. 1987;21:95–100. [Google Scholar]
- 47.van Heel M, Harauz G, Orlova E V, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 48.Weisel J W, Nagaswami C, Vilaire G, Bennett J S. Examination of the platelet membrane glycoprotein IIb-IIIa complex and its interaction with fibrinogen and other ligands by electron microscopy. J Biol Chem. 1992;267:16637–16643. [PubMed] [Google Scholar]
- 49.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]