Abstract
The UL49 gene product (VP22) of herpes simplex virus types 1 and 2 (HSV-1 and HSV-2) is a virion phosphoprotein which accumulates inside infected cells at late stages of infection. We previously (J. A. Blaho, C. Mitchell, and B. Roizman, J. Biol. Chem. 269:17401–17410, 1994) discovered that the form of VP22 packaged into infectious virions differed from VP22 extracted from infected-cell nuclei in that the virion-associated form had a higher electrophoretic mobility in denaturing gels. Based on these results, we proposed that VP22 in virions was “undermodified” in some way. The goal of this study is to document the biological and biochemical properties of VP22 throughout the entire course of a productive HSV-1 infection. We now report the following. (i) VP22 found in infected cells is distributed in at least three distinct subcellular localizations, which we define as cytoplasmic, diffuse, and nuclear, as measured by indirect immunofluorescence. (ii) Using a synchronized infection system, we determined that VP22 exists predominantly in the cytoplasm early in infection and accumulates in the nucleus late in infection. (iii) While cytoplasmic VP22 colocalizes with the HSV-1 glycoprotein D early in infection, the nuclear form of VP22 is not restricted to replication compartments which accumulate ICP4. (iv) VP22 migrates as at least three unique electrophoretic species in denaturing sodium dodecyl sulfate-DATD-polyacrylamide gels. VP22a, VP22b, and VP22c have high, intermediate, and low mobility, respectively. (v) The relative distribution of the various forms of VP22 derived from infected whole-cell extracts varies during the course of infection such that low-mobility species predominate at early times and high-mobility forms accumulate later. (vi) The highest-mobility forms of VP22 partition with the cytoplasmic fraction of infected cells, while the lowest-mobility forms are associated with the nuclear fraction. (vii) Finally, full-length VP22 which partitions in the nucleus incorporates radiolabel from [32P]orthophosphate whereas cytoplasmic VP22 does not. Based on these results, we conclude that modification of VP22 coincides with its appearance in the nucleus during the course of productive HSV-1 infection.
The synthesis of viral proteins during herpes simplex virus type 1 (HSV-1) infection can be divided into at least three different temporal classes: the immediate-early (IE) phase, when α genes are expressed, the early (E) phase, when β genes are expressed, and the late (L) phase, when γ genes are expressed (23, 24). During the IE phase of infection, several α proteins, including ICP22 and ICP4, localize to the nucleus, where they serve to regulate the expression of later viral genes, whose products are responsible for viral DNA replication and virion assembly (35). Proteins synthesized during the E phase of infection drive the replication of the viral genome (reviewed in references 7 and 35). The localization of these proteins to specific subnuclear compartments within infected cells has been well documented (10, 29, 34, 39, 42). Viral proteins synthesized late in infection are associated with virion particle assembly (32, 38), exit (9), and entry (36) during subsequent cycles of infection. L proteins involved in capsid assembly accumulate in the nuclei of infected cells (8), while components of the virion envelope, such as glycoprotein gD, are found predominantly in cytoplasmic compartments (36, 37). Late in infection, when viral proteins accumulate, these nuclear and cytoplasmic structures differ morphologically from those found in uninfected cells (1, 11, 40, 41).
Changes in cell morphology, generally defined as cytopathic effects, observed late in infection probably represented manifestations of a reorganization of subcellular compartments. Examples of such reorganizations upon infection include the (i) formation of subnuclear replication compartments (10, 13, 25, 41), (ii) fragmentation of the Golgi apparatus in certain cell types and virus strains (40), and (iii) restructuring of the microtubule network (1). While the regulation of viral gene expression is an important factor defining the continuum of the infectious cycle, the subcellular location of HSV-1 proteins in infected cells also plays an equally important role in determining viral function during each phase of replication.
In contrast to the attention focused on the subcellular localizations of IE and E proteins, the localizations of L proteins, particularly those of the tegument, have not been extensively studied. The tegument is defined as the amorphous region, located between the virion capsid and envelope, containing at least nine viral gene products (reviewed in reference 35). While IE and E proteins accumulate predominantly in the nucleus, proteins of the tegument are found distributed in numerous subcellular compartments (30). One component of the tegument, VP22, has been of particular interest to our laboratory (4). VP22 is the protein product of the UL49 gene (19), which is expressed late in infection (21). Although the function of VP22 during viral infection is unclear, several observations have sparked significant interest in this gene product.
Recently, Elliott and O’Hare reported that VP22 is capable of intercellular transport (17), and later they presented data which suggests that this movement of VP22 between cells may involve actin microfilaments (16). They further demonstrated that VP22 colocalizes with microtubules and proposed that one function of VP22 may be the stabilization of microtubule bundles (16). VP22 has also been implicated in the recruitment of other tegument proteins since it directs the relocalization of VP16 when plasmids encoding the two proteins are cotransfected into cells in the absence of other viral proteins (15). Together, these findings suggest that VP22 has the ability to redirect both cellular and viral proteins and may play a role in the modification of microtubule morphologies.
Earlier studies have shown that during productive HSV-1 infection, VP22 exists as a virion phosphoprotein (4). In addition, VP22 is highly posttranslationally modified during infection, and these modifications include nucleotidylation and ADP-ribosylation (4). In vitro, VP22 accepts phosphorylation from at least two cellular kinases, casein kinase II and protein kinase C (18, 31). VP22 in infected cell extracts can be resolved into at least two differently migrating forms in denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The slowest-migrating form is phosphorylated, while the faster-migrating form is found incorporated into purified virions (4, 18). It is noteworthy that while the bovine herpes type 1 virus UL49 homologue is dispensable for viral growth (28), a recombinant HSV-1 with the UL49 gene deleted has not been reported to date (2, 4). Currently, the significance of posttranslational modification of VP22 during productive infection is unknown.
Work in our laboratory has focused on examining the role of VP22 during wild-type HSV-1 infection. Here, we report the relationship between changes in the subcellular localization of VP22 and its modification during synchronized HSV-1 infection of Vero cells. Our research has led to the following observations. First, the localization of VP22 in the infected cell changes over the course of infection from predominantly cytoplasmic early in infection to predominantly nuclear late in infection as observed by indirect immunofluorescence microscopy. During infection, VP22 exhibits three distinct patterns of subcellular localization. We have defined these patterns as cytoplasmic, nuclear, and diffuse. The second observation is that VP22 exists in at least three different forms (VP22a, VP22b, and VP22c), which can be distinguished based on their migrations in denaturing gels. In addition, the distribution of these forms varies during the course of the infection cycle, such that slower-migrating (low-mobility) forms are observed early in infection while the faster-migrating (high-mobility) forms accumulate later. The third observation is derived from fractionation experiments which confirm that slower-migrating forms of VP22 are associated with its presence in the nucleus as well as with its modification by incorporation of radiolabel from [32P]orthophosphate. The modified, slowest-migrating forms of VP22 are not observed in the cytoplasmic fractions. From these results, we conclude that (i) the subcellular location of VP22 is regulated during the infection cycle and (ii) modification of VP22 which results in slower electrophoretic forms is associated with the translocation of VP22 into the nucleus of infected cells.
MATERIALS AND METHODS
Cells and virus.
African green monkey kidney (Vero) cells were obtained from the American Type Culture Collection and passaged in Dulbecco’s modified Eagle’s medium supplemented with 5% fetal bovine serum. The virus strain used in all experiments was the prototype HSV-1(F) (14) provided by Bernard Roizman, University of Chicago. To obtain viral stocks, subconfluent monolayer Vero cultures (approximately 3 × 106 cells) were inoculated with HSV-1(F) at a multiplicity of infection (MOI) of 0.01 for 2 h at 37°C in 199V medium (Life Technologies) supplemented with 2% serum (199V), the inoculum was then removed, fresh medium was added, and the cells were incubated at 37°C in 5% CO2. Viral stocks were prepared once the infection reached a cytopathic effect of 100%, viral titers were determined on Vero cells, and aliquots were stored at −80°C. All MOIs were derived from the number of PFU of virus on Vero cells.
Viral plaque formation.
For experiments involving viral plaques, approximately 106 Vero cells were seeded onto 22-mm2 glass coverslips in six-well dishes 1 day before infection. The following day, cells were infected at an MOI of 0.001 by diluting the virus stock in 199V medium and adsorbing at 37°C for at least 1 h. The viral inoculum was then aspirated, 199V medium containing 10 μg of human immunoglobulin (Ig) (Sigma) per ml was added, and the cells were incubated at 37°C until cytopathic effects indicating plaque formation were observed (about 2 days later). Cells were fixed for immunofluorescence at this point as described below.
Synchronized infections.
Vero cells were seeded the day before infection in either six-well dishes containing 22-mm2 coverslips for indirect immunofluorescence or 25-cm2 flasks for infected cell extracts. For synchronized infections (22), the cells were incubated on ice on an orbital shaker at 4°C for 15 min before the addition of virus. The cells were then inoculated while still on ice with an MOI of 30 and returned to the shaker at 4°C. After the virus had adsorbed for 1 h, the cells were rinsed with 4°C phosphate-buffered saline (PBS). The cells were then removed from the ice, 37°C 199V medium was added immediately, and the cells were returned to a 37°C incubator. The time point at which the 37°C medium was added was defined as T = 1 in our experiments. Synchronous infections were defined as uniform staining in all cells in a microscopic field at a given time postinfection (p.i.), as determined by indirect immunofluorescence with a single antibody specific for a unique HSV-1 polypeptide. In control experiments, it was determined that an HSV-1(F) MOI of 30 was necessary to ensure that all cells were synchronously infected when viewed by immunofluorescence (data not shown). Lower MOIs tested (0.001, 0.1, 1.0, and 10) resulted in nonsynchronized infections as evidenced by indirect immunofluorescent staining. In these control studies, cells were stained with an antibody against the immediate-early protein ICP4 (1101) to determine whether all the cells were infected (data not shown). When we performed a titer determination under the adsorption conditions for synchronized infection (data not shown), we observed an effective viral titer of 7 × 107 PFU/ml compared to 3 × 108 PFU/ml with the standard 37°C-only titer control. This result indicates that the addition of virus at an MOI of 30 under our low-temperature adsorption conditions results in an effective MOI of 7 PFU/cell. In addition, we previously reported (3) that similar infections at an MOI of 50 did not elicit any toxic effects.
Induction of synchronous infection by adsorption of the inoculum at 4°C is routine and has little or no effect on cells in culture. However, it should noted that there was some evidence that the 4°C incubation used to synchronize the infections may have had a transient effect on the monolayer cells. We compared mock- and HSV-1-infected Vero cells which were incubated on ice at 4°C for 1 h with cells maintained at 37°C throughout the infection. Cells fixed immediately after the temperature shift to 37°C (T = 1), were formaldehyde fixed and acetone permeabilized and stained with a monoclonal antibody specific for the cytoskeletal protein α-tubulin. The α-tubulin staining pattern (data not shown) in the cells incubated at 4°C appeared slightly more diffuse than that observed in the cells held at 37°C (33). Consistent with the fact that the dynamics of polymerization-depolymerization of microtubules are energy dependent and would be expected to be reduced at the lower (4°C) temperature, this apparent change in the α-tubulin organization was not observable at 4 h after the temperature shift (33). In fact, by 4 h p.i., the two sets of cells (4 and 37°C) were indistinguishable (data not shown) based on α-tubulin staining (33). Therefore, we believe that the cells had recovered from the 4°C shock by this time.
Immunological reagents.
The generation of the RGST49 rabbit polyclonal antibody against a glutathione S-transferase (GST)-VP22 fusion protein was described previously (4). Affinity-purified RGST49 antibody was generated as follows. Anti-GST antibodies were first removed from RGST49 polyclonal sera by affinity chromatography with purified GST protein cross-linked to agarose by using dimethyl pimelimidate (Pierce) as specified by the manufacturer. Next, the flowthrough from the GST-agarose column was applied to a GST-VP22 affinity column (agarose cross-linked with GST-VP22 as described for GST-agarose) and low-pH elutions were tested for anti-VP22 immunoreactivity by immunoblotting with HSV-1(F)-infected Vero cell extracts. All GST fusion proteins were purified from Escherichia coli cells as described previously (reviewed in reference 5). The G49 antibody is a mouse monoclonal antibody which was raised (Mount Sinai Department of Microbiology Hybridoma Center) against a GST-VP22 fusion protein (4). Growth medium from a resulting hybridoma cell line following 5 to 6 days of incubation at 37°C was used directly for immunoblotting experiments, and this antibody is referred to as G49. RGST22 is a polyclonal antibody against a GST-ICP22 fusion protein (6). Monoclonal antibodies 1101 and 1114 against ICP4 and 1103 against glycoprotein D (gD) were purchased from the Goodwin Institute for Cancer Research, Inc. (Plantation, Fla.) and used at a dilution of 1:1,000 in 1% bovine serum albumin (BSA) for immunofluorescence. Monoclonal antibodies specific for the Golgi 58,000-molecular-weight (58K) protein and α-tubulin were obtained from Sigma. During all indirect immunofluorescence experiments described in this paper and in many additional experiments (data not shown), no significant differences between the staining patterns of monoclonal antibodies 1114 and 1101 were observed. Fluorescein isothiocyanate-conjugated anti-rabbit IgG heavy plus light chains (H+L), tetramethylrhodamine isothiocyanate (Texas Red)-conjugated anti-rabbit IgG (H+L), and Texas Red-conjugated anti-mouse IgG (H+L) were purchased from Vector Laboratories (Santa Cruz, Calif.) and were used at a dilution of 1:100 in 1% BSA as secondary antibodies for indirect immunofluorescence. FITC-conjugated anti-mouse IgG (H+L) was purchased from Boehringer Mannheim (Indianapolis, Ind.) and was used at a dilution of 1:500 in 1% BSA.
Indirect immunofluorescence and microscopy.
Both standard and synchronized infections were terminated, after cells were rinsed twice in PBS, by fixing in 2% methanol-free formaldehyde (Polysciences, Inc.) for 20 min at room temperature. Next, the cells were rinsed twice again with PBS and permeabilized with 100% acetone at −20°C for 3 to 5 min, rinsed twice again in PBS, and then blocked for at least 8.5 h at 4°C in 1% BSA containing 10 μg of pooled human Ig (mainly IgG) (Sigma) per ml. The cells were then rinsed twice in PBS, and each primary antibody was added for 1 h. The primary antibodies used for immunofluorescence studies were diluted in 1% BSA as follows: RGST22, 1:500; RGST49, 1:500; affinity-purified RGST49, 1:10; anti-ICP4, 1101 and 1114, 1:1,000; anti-gD 1103, 1:1,000. After extensive rinsing with PBS, the appropriate secondary antibody was added and incubated for an additional 1 h. Finally, the cells were preserved in a 0.1% solution of Mowiol (Sigma) with 2.5% DABCO (Sigma) used as an antibleaching agent under a fresh coverslip and sealed with nail polish. No staining was observed (data not shown) with any secondary antibody in the absence of primary antibody (33). No cross-reactivity was observed (data not shown) between anti-rabbit secondary antibody and mouse primary antibody and vice versa (33). We cannot exclude the possibility that avidity differences exist between the antibodies used in these studies. Cells were visualized on either a Zeiss Axiophot fluorescence microscope or a Leica (Heidelberg) confocal laser-scanning microscope as indicated in the Results. In the later case, the thinnest possible sections (0.5 μm) were confocally imaged by using a 40× objective with a pinhole size of 40.
Infected whole-cell extracts and isolation of cytoplasmic and nuclear fractions.
Synchronized infections were terminated by scraping cells (∼106) into 140 mM NaCl–3 mM KCl–10 mM Na2HPO4–1.5 mM KH2PO4 (pH 7.5) (PBS) containing protease inhibitors [10 mM l-1-chlor-3-(4-tosylamido)-7-amino-2-heptanon-hydrochloride (TLCK), 10 mM l-1-chlor-3-(4-tosylamido)-4-phenyl-2-butanone (TPCK), 100 mM phenylmethylsulfonyl fluoride (Sigma)]. Whole extracts of the infected cells were prepared after pelleting by low-speed centrifugation and resuspending the pellet in PBS containing 1.0% Triton X-100 plus protease inhibitors. Lysis by sonication was performed with a Branson Sonifier.
For experiments involving the fractionation of cytoplasmic and nuclear compartments (5), infected cells (∼106) were scraped into PBS containing protease inhibitors, pelleted, resuspended in the same buffer containing 0.4% Nonidet P-40 (NP-40), and pelleted again. The supernatant from this centrifugation step was considered the cytoplasmic fraction. The pellet was washed once in PBS containing protease inhibitors plus 0.1% NP-40, pelleted, and resuspended in PBS containing protease inhibitors and 0.4% NP-40. This fraction was sonicated as above and pelleted, and the supernatant was considered the nuclear fraction. The pellet remaining after the nuclear supernatant was collected was resuspended in PBS containing protease inhibitors and 0.4% NP-40, sonicated, and designated the nuclear matrix (12).
Denaturing gel electrophoresis and immunoblotting.
The protein concentrations of all extracts were determined by a modified Bradford assay (Bio-Rad, Richmond, Calif.) as specified by the manufacturer. Equal amounts of infected-cell protein were separated in a sodium dodecyl sulfate (SDS)–15% polyacrylamide gel cross-linked with N,N′-diallyltartdiamide (DATD; Sigma) (5), electrically transferred to nitrocellulose, and probed with anti-VP22 polyclonal antibody RGST49 (1:500 in 1% BSA), anti-ICP22 polyclonal antibody RGST22 (1:500 in 1% BSA), or anti-VP22 monoclonal antibody G49 (hybridoma culture medium) as indicated in the figure legends. Horseradish peroxidase-conjugated anti-rabbit or anti-mouse (Amersham) secondary antibodies were diluted 1:1,000 in PBS and incubated with the blots for 1 h. Specific viral bands were detected following development with chemiluminescence reagents (Amersham) and autoradiography at 25°C with X-OMAT film (Kodak, Rochester, N.Y.). Alkaline phosphatase-conjugated goat anti-rabbit antibody used at 1:500 in PBS was purchased from Southern Biotech (Birmingham, Ala.).
[32P]orthophosphate labeling of cells and immunoprecipitation reactions.
The method for specifically immunoprecipitating [32P]orthophosphate-labeled VP22 was a slight modification of that described previously (4). Approximately 3 × 106 Vero cells were synchronously infected with HSV-1(F) (MOI = 30) as described above, except that 100 mCi of carrier-free 32P-orthophosphate (NEN) was added to the 199V medium at 5 h p.i. The labeled medium was removed from the cells at 7 h p.i. and replaced with fresh 199V medium, and the infections were stopped at the time points indicated in the Results. The whole, cytoplasmic, nuclear, and matrix fractions were isolated as described above.
For the immunoprecipitation reactions, exactly equal amounts (500 μg) of infected-cell protein from each extract or fraction were added to 0.5 ml of 70 mM Tris-HCl (pH 7.5)–150 mM NaCl–5 mM EDTA–1% deoxycholate–1% Triton X-100 (RIPA buffer) plus phenylmethylsulfonyl fluoride, TPCK, and TPLK protease inhibitors. Antibody RGST49, specific for VP22, was added to a final dilution of 1:500, and the mixture was rotated for 18 h at 4°C. By using a snipped micropipet tip, 40 μl of a 50% protein A-Sepharose slurry (Repligen) was added to each reaction mixture and incubated at 4°C for an additional 4 h. By using brief microcentrifugations, the protein A-Sepharose beads were rinsed three times with 1 ml of RIPA buffer plus protease inhibitors and once finally with PBS plus protease inhibitors. Infected-cell protein was eluted from the protein A-Sepharose beads by boiling in protein gel disruption buffer containing 0.1% SDS before being loaded on a 15% DATD-acrylamide denaturing gel. Autoradiography was carried out at 25°C using X-OMAT film.
Computer imaging and Web-posted electronic data.
Immunoblots, autoradiograms, and 35-mm slides were digitized at 600 to 1,200 dots per in. (d.p.i.) resolution by using an AGFA Arcus II scanner linked to a Macintosh G3 PowerPC workstation. Raw digital images, saved as tagged image files (TIF) with Adobe Photoshop version 5.0, were organized into figures by using Adobe Illustrator version 7.1. Grayscale or color prints of figures were obtained by using a Codonics dye sublimation printer. Certain results referred to in the text as “data not shown” may be accessed through our Website (33a).
RESULTS
We previously reported that VP22 is a virion component which is extensively posttranslationally modified and that these modifications include [32P]orthophosphorylation, mono-ADP-ribosylation, and nucleotidylylation (4). In this earlier report, we focused exclusively on VP22 derived from either purified HSV-1 and HSV-2 virions or high-salt nuclear extracts of infected cells. One of the significant findings of that study was that the virion-associated form of VP22 migrates faster in denaturing gels than do the nuclear-derived forms, suggesting that the virion form is “undermodified” (4). The implications of these findings are twofold. First, virion-derived VP22, which could potentially enter target cells following viral envelope and cell membrane fusion, is the “undermodified” form. Second, a novel gatekeeping process probably acts to partition the “undermodified” virion forms from the modified nuclear forms of the protein. The goals of the studies presented in this report are to document the intracellular localization of VP22 during productive infection and to determine whether modifications of the protein occur in the cytoplasm or the nucleus of the infected cell.
Three distinct patterns of VP22 subcellular localization are observed by indirect immunofluorescence of HSV-1(F) plaques.
The first set of experiments used indirect immunofluorescence of viral plaques to detect changes in the subcellular localization of VP22 during different stages of infection. Plaque formation represents a situation in which, at a given point in time, monolayer cells are at different stages of infection and, generally, internal cells of the plaque are at late stages of infection and peripheral cells are at early stages. Plaques were produced by infecting Vero cells at a low multiplicity (MOI = 0.001) and allowing the infection to progress for approximately 2 days. Infection of neighboring cells by virus released into medium was minimized by the addition of neutralizing human Ig. Cells were fixed with formaldehyde, permeabilized with acetone, and stained with either RGST49 or affinity-purified RGST49 antibody specific for VP22, RGST22 antibody specific for ICP22, or 1114 antibody specific for ICP4 as described in Materials and Methods. Two experiments were performed. Initially, plaques were singly stained with antibody specific for VP22 to define the subcellular location of this protein during infection or with antibody specific for ICP22 as a control. Next, double staining with antibodies specific for VP22 or ICP4 was performed to compare the locations of these two proteins within the same cell. Antibodies against the α or IE proteins ICP22 (for single labeling) and ICP4 (for double labeling) were specifically chosen as markers for cells in the earliest phase of infection. The IE proteins, while synthesized in the cytoplasm, localized quickly and almost exclusively within the nuclei of infected cells. The results (Fig. 1) were as follows.
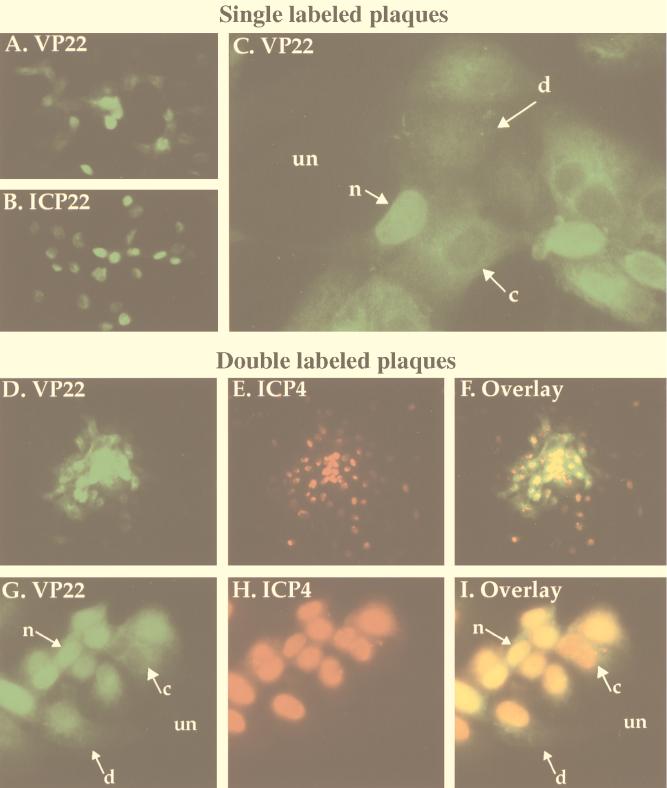
FIG. 1.
Indirect immunofluorescence of HSV-1(F)-infected cell plaques singly labeled with antibodies specific for VP22 (A and C) and ICP22 (B) and doubly labeled with antibodies specific for VP22 and ICP4 (D to I). Vero cells were infected at an MOI of 0.001 with HSV-1(F) for 51 h, fixed with formaldehyde, permeabilized with acetone, and stained with antibodies RGST49 (A and C), RGST22 (B), affinity-purified RGST49 (D, F, G, and I), or 1114 (E, F, H, and I) as described in Materials and Methods. Abbreviations: un, uninfected cells; n, c, and d nuclear, cytoplasmic, and diffuse subcellular localizations of VP22, respectively. Low magnifications were ×10 (D to F) and ×20 (A and B); high magnifications were ×40 (G to I) and ×63 (C).
Single viral plaques were apparent at low magnification following staining with antibodies specific for VP22 or ICP22 (Fig. 1A and B). While ICP22 was uniformly nuclear in all cells in a plaque (Fig. 1B), distinct subcellular distributions of VP22 were observed in different cells. When anti-VP22-labeled plaques were viewed at a higher magnification, at least three unique subcellular distributions of VP22 were observed in different cells (Fig. 1C). VP22 was predominantly nuclear in most infected cells in the plaque. However, in some cells, VP22 appeared to be concentrated in the cytoplasm, and in a few cells, it exhibited a diffuse staining pattern seen throughout the cell.
To directly compare the localization of VP22 with a protein synthesized early in infection, HSV-1 plaques were doubly labeled for indirect immunofluorescence with affinity-purified RGST49 antibody specific for VP22 and a monoclonal antibody specific for ICP4. At low magnification, plaques demonstrated positive staining for both VP22 and ICP4, and a single representative plaque is shown (Fig. 1D to F). While the majority of VP22 staining appeared at the center of the plaque (Fig. 1D and F), ICP4 staining could be observed in the center and at the edges of the plaque (Fig. 1E and F). In another representative plaque viewed at higher magnification, the previously defined (Fig. 1C) three localizations of VP22 were observed (Fig. 1G and I). As expected, ICP4 in the same cells was concentrated in the nucleus (Fig. 1H), although a few cells showed diffuse staining for ICP4. In cases where VP22 is predominantly nuclear, staining with antibodies to both VP22 and ICP4 caused the nuclei of these cells to appear yellow in the overlay (Fig. 1I). In cells where VP22 localization was cytoplasmic, exclusion of VP22 from the nucleus led to a clear visualization of the Texas Red signal on ICP4 antibodies in the nuclei of these cells, while the diffuse VP22 localizations yielded a green cytoplasmic image.
From these results we conclude the following. (i) At least three unique subcellular localizations of VP22 occur during the HSV-1 infection cycle. We refer to these staining patterns as cytoplasmic, nuclear, and diffuse. (ii) Essentially all ICP22 and ICP4 localized to the nucleus. (iii) VP22 was observed only in cells that already expressed ICP4, inasmuch as all cells which stained for VP22 also stained for ICP4. ICP4 was observed in cells at the periphery of plaques in which we did not find detectable levels of VP22, indicating that cells at the edges of plaques which stained only for ICP4 are cells in the earliest stages of viral replication. We conclude from this experiment that VP22 exhibits different subcellular localizations in infected cells during the course of productive infection. Cells that exhibit these forms are observed in viral plaques.
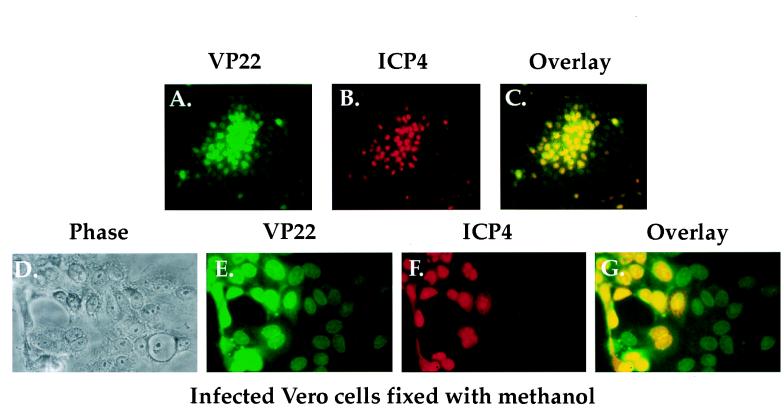
Detection of VP22, but not ICP4, in the nuclei of some cells in HSV-1(F) plaques fixed and permeabilized with methanol.
The results presented in Fig. 1 confirm that viral IE proteins like ICP4 are detected in all infected cells in a plaque while L proteins like VP22 predominate in cells at the center of a plaque. These results are consistent with the temporal regulation of HSV-1 protein synthesis (23). However, based on the report that after infection by a replication defective virus, VP22 is observed in the nuclei of cells that are devoid of a β-galactosidase marker for infection (17), we were intrigued by the fact that we did not observe cells which stained for VP22 alone and not ICP4. To investigate this further, Vero cells were infected at low MOI with HSV-1(F) in parallel with those shown in Fig. 1D to I. Next, the cells were fixed and permeabilized only by the addition of 100% methanol as described previously (17). Plaques were then doubly labeled for immunofluorescence with affinity-purified RGST49 antibody and monoclonal antibody 1114 specific for ICP4 as described in Materials and Methods. As a control, phase contrast images of the cells were also documented.
The results (Fig. 2) demonstrated that at low magnification, many of the cells at the edges of plaques, which exhibited weak staining with the anti-VP22 antibody (Fig. 2A), showed no detectable staining with the antibody specific for ICP4 (compare Fig. 2A with Fig. 2B and C). At a higher magnification, labeling for both VP22 and ICP4 was observed in the nuclei of many cells (Fig. 2G). Some cells also showed a diffuse staining of ICP4 (Fig. 2F). Overlay of the VP22 and ICP4 images (Fig. 2G) demonstrated that many cells had nuclear staining only for VP22 and not ICP4. The intensity of the unique VP22 staining was substantially lower than that observed for VP22 in the cells which also stained for ICP4 (compare Fig. 2E with Fig. 2F and G).
FIG. 2.
Phase-contrast (D) and indirect immunofluorescence (A to C and E to G) of HSV-1(F) plaques doubly labeled with antibodies specific for VP22 and ICP4. Vero cells were infected at an MOI of 0.001 with HSV-1(F) for 51 h in parallel with those in Fig. 1D to I, fixed and permeabilized with 100% methanol (17), and stained with affinity-purified RGST49 (A, C, E, and G) and 1114 (B, C, F, and G) as described in Materials and Methods.
From these results, we conclude the following. (i) Since the experiments in Fig. 1 and 2 were performed in parallel under identical infection conditions, the relative distributions of ICP4 and VP22 were identical in each set of infected cells prior to fixing and permeabilizing, independent of the chemical treatments performed later. (ii) The use of the different fixing and permeabilizing conditions had little or no effect on ICP4 since the staining pattern of ICP4 did not differ between Fig. 1 and 2. (iii) The differences observed in the staining patterns of VP22 are due solely to the use of different conditions for fixing and permeabilizing the cells. In keeping with our above-mentioned understanding of the temporal regulation of protein expression in HSV-1, we conclude that the formaldehyde fixation and acetone permeabilization technique is the method of choice for indirect immunofluorescence experiments presented in this report. Accordingly, this method was used in all subsequent experiments involving indirect immunofluorescence.
The plaque-staining assay proved valuable for observing cells undergoing the full range of the infectious cycle in a single microscopic field. In the next series of experiments, we sought to determine whether the different localizations of VP22 corresponded to different points during the infection cycle by using a coordinated infection system.
Migration of VP22 to the nucleus during HSV-1(F) infection.
The next experiment was designed to determine whether any of the subcellular localizations of VP22 presented in Fig. 1 predominated at a particular time during the course of HSV-1 infection. In an attempt to coordinate the time of infection, Vero cells were infected with HSV-1(F) at an MOI of 0.1 on ice at 4°C for 1 h. At 1 h p.i., fresh 37°C medium was added, and the infections were stopped at regular intervals after the temperature shift. At 7, 9, 13, and 25 h p.i., cells were fixed and permeabilized with formaldehyde and acetone as described in Materials and Methods. The cells were stained with both monoclonal antibody 1101 specific for ICP4 and affinity-purified RGST49 antibody specific for VP22 and visualized by indirect immunofluorescence. This infection strategy was designed to examine the subcellular distribution of VP22 during an entire low-multiplicity replication cycle. The results (Fig. 3) were as follows.
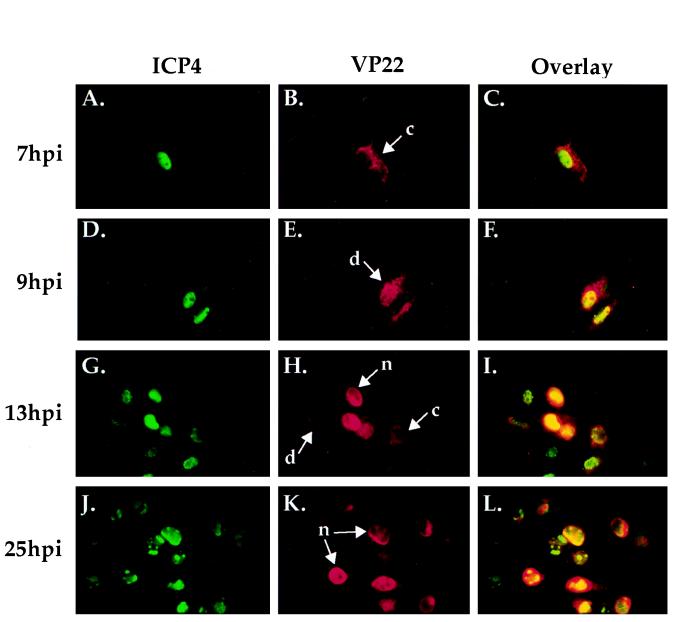
FIG. 3.
Indirect immunofluorescence of HSV-1(F)-infected cells doubly labeled with antibodies specific for VP22 (B, E, H, and K) and ICP4 (A, D, G, and J). Vero cells were infected with 0.1 PFU of HSV-1(F) per ml on ice at 4°C for 1 h. 1 h p.i. refers to the point at which fresh (37°C) medium was added back to the flasks. Infections were terminated at 7, 9, 13, and 25 h p.i. prior to formaldehyde-acetone fixation followed by immunostaining with affinity-purified RGST49 and 1101 as described in Materials and Methods, n, c, and d refer to the nuclear, cytoplasmic, and diffuse subcellular localizations of VP22, respectively (Fig. 1). Overlays are shown in panels C, F, I, and L.
The localization of ICP4 was observed to be nuclear throughout infection (Fig. 3A, D, G, and J). The presence of ICP4 in globular replication compartments (10; reviewed in reference 13) was consistent with earlier reports (25). At 7 h p.i., VP22 exhibited a cytoplasmic distribution in the infected cells (Fig. 3B). Only ICP4 was observed in the nucleus at 7 h p.i. in the overlay of the two images (Fig. 3C), further supporting the idea VP22-specific staining was limited to the cytoplasm. At 9 h p.i. (Fig. 3E), VP22 was detected in both the cytoplasm and nuclei of infected cells, yielding the staining pattern defined as diffuse in the plaque-staining assay. At 13 h p.i., a combination of all three VP22 staining patterns was observed (Fig. 3H), while at 25 h p.i., VP22 localization was almost exclusively nuclear in all infected cells (Fig. 3K). Overlay of VP22 and ICP4 at 25 h p.i. demonstrated that although VP22 accumulated in the nucleus at late times postinfection, its nuclear distribution was not limited to replication compartments as was that of ICP4 (Fig. 3L). Finally, there was a marked increase in the number of infected cells as the time course progressed.
From these results, we conclude that VP22 is distributed in the cytoplasm early in infection while its nuclear localization occurs later in infection. The diffuse pattern appears to represent an intermediate phase of accumulation of VP22 in the nucleus. The results in Fig. 3 are consistent with cell-to-cell asynchronous spread in which additional rounds of replication have occurred. For this reason, high-multiplicity infections were used to obtain synchronous infection.
Differing subcellular localizations of VP22 and gD during a synchronized HSV-1(F) infection.
We have now compared the subcellular localizations of VP22 with two HSV-1 IE proteins, ICP4 and ICP22, which are targeted to the nucleus. During this analysis, we discovered that VP22 possessed a demonstrative cytoplasmic phase. To accurately define the stage of infection at which VP22 accumulates predominantly in the cytoplasm, we directly compared its localization with that of a well-defined viral glycoprotein, gD. Vero cell monolayers were synchronously infected with HSV-1(F) by using low-temperature (4°C) adsorption followed by a temperature shift to 37°C as described in Materials and Methods, and infection was stopped by fixing cells at specific times p.i. The cells were subsequently stained for indirect immunofluorescence with monoclonal antibody 1103, specific for gD, and affinity-purified RGST49 as described in Materials and Methods. The results (Fig. 4) showed the following.
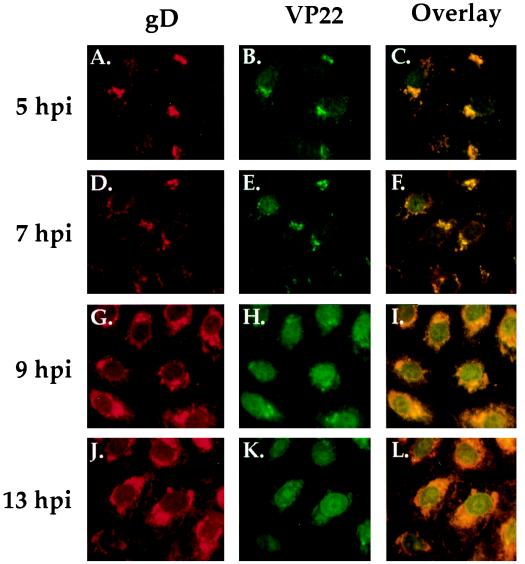
FIG. 4.
Double-label indirect immunofluorescence of synchronously infected cells stained with antibodies specific for VP22 (B, E, H, and K) and gD (A, D, G, and J). Vero cells were synchronously infected with HSV-1(F) (MOI = 30) and at 5, 7, 9, 13 h p.i. were formaldehyde fixed and acetone permeabilized before being immunostained with affinity-purified RGST49 and 1103 as described in Materials and Methods. 1 h p.i. refers to the point at which fresh (37°C) medium was added back to the flasks following a 1-h absorption at 4°C. Overlays are shown in panels C, F, I, and L.
Labeling with either antibody at each time point (gD in Fig. 4A, D, G, and J or VP22 in Fig. 4B, E, H, and K) showed a uniform staining pattern in all cells at each time point, indicating that the infection was synchronized. At 5 and 7 h p.i., VP22 staining was restricted mainly to the cytoplasm adjacent to the cell nucleus (Fig. 4B and E). gD also localized to this structure at 5 and 7 h p.i. (Fig. 4A, C, and F). At 7 h p.i. (Fig. 4E), VP22 was still distributed mainly in the cytoplasm but low levels could be detected in the nucleus. By 9 h p.i., VP22 was observed in the nuclei of all cells (Fig. 4H). At this point in infection, VP22 still exhibited some staining in the cytoplasm around the nucleus (Fig. 4H), and this distribution of VP22 was constant up to 13 h p.i. (Fig. 4K). As expected, gD accumulated in the cytoplasm during the course of infection and was never observed to accumulate to significant levels in the nuclei of infected cells (Fig. 4D, G, and J). VP22 which colocalized with gD in the cytoplasm appeared yellow in the overlay (Fig. 4C, F, I, and L), while VP22 which accumulated in the nucleus at 9 and 13 h p.i. appeared green in the overlay (Fig. 4I and L).
The conclusions from this experiment (Fig. 4) were as follows. (i) Anti-gD labeling, measured by indirect immunofluorescence, was uniform in all cells at each time point. This indicates that the criteria for synchronized infection were met by the technique of shifting the temperature of cells preadsorbed with HSV-1(F) (MOI = 30) from 4 to 37°C. (ii) Double labeling of synchronously infected Vero cells for gD and VP22 enabled us to clearly differentiate between time points when VP22 is predominantly cytoplasmic (<7 h p.i.) and those when it is predominantly nuclear (>7 h p.i.). (iii) Based on our findings, we also conclude that the major influx of VP22 into nucleus initiates between 7 and 9 h p.i.
Uniform accumulation of VP22 in the nuclei of synchronously infected cells at 9 h p.i. detected by confocal immunofluorescence.
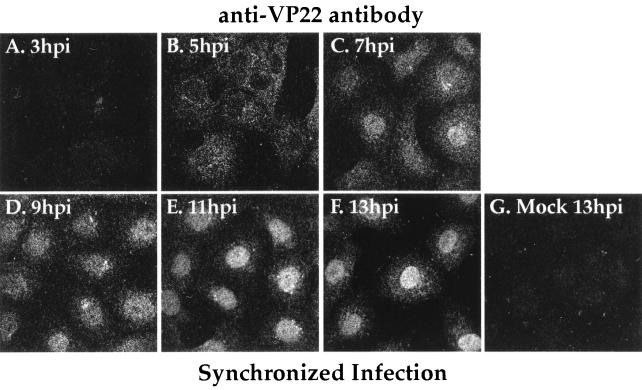
Double labeling (Fig. 4) with antibodies specific for VP22 and gD indicated that VP22 exhibited nuclear localization late in infection. However, since all of the previous data was obtained with a standard fluorescence microscope, we could not eliminate the possibility that VP22 which appeared to be within the nucleus represented VP22 in the cytoplasm above the nucleus. Thus, to precisely determine the subcellular location of VP22 at specific points during infection, Vero cells were synchronously infected with HSV-1(F) and indirect immunofluorescence of VP22 was examined by confocal laser-scanning microscopy as described in Materials and Methods. The pinhole size was adjusted to obtain the thinnest possible sections (0.5 μm) so that we could conclusively determine whether VP22 was nuclear at each time point.
The results (Fig. 5) showed that essentially no VP22 was observed by indirect immunofluorescence at 3 h p.i. (Fig. 5A). At 5 h p.i., VP22 was observed throughout the cytoplasm with areas of stronger staining adjacent to the nucleus (Fig. 4) while little or none was present within the nucleus (Fig. 5B). At 7 h p.i., VP22 was still predominantly distributed throughout the cytoplasm, although a demonstrable amount was also observed in the nuclei of infected cells (Fig. 5C). This combination of cytoplasmic and nuclear staining corresponds to the pattern previously (Fig. 1) referred to as diffuse. At 9 h p.i., VP22 had accumulated within the nuclei of infected cells, with some staining observed in the cytoplasm (Fig. 5D). Areas of decreased immunoreactivity observed in the nuclei of some cells (Fig. 5C and D) indicated that VP22 was either absent from the nucleolus or present in this structure at lower levels than in other areas of the nucleus. At later time points (11 and 13 h p.i.), the vast majority of VP22 was observed throughout the nuclei (Fig. 5E and F). No VP22 signal was observed in mock-infected cells at 13 h p.i. (Fig. 5G). The results presented here are consistent with those presented above (Fig. 1, 3, and 4). In summary, we conclude that during HSV-1(F) infection, VP22 is cytoplasmic prior to 5 h p.i., begins to enter the nucleus between 5 and 7 h p.i., and is located predominantly inside the nucleus after 9 h p.i.
FIG. 5.
Confocal indirect immunofluorescence of synchronously infected cells stained with antibodies specific for VP22. Vero cells were synchronously infected with HSV-1(F) (MOI = 30) or mock infected and at 3, 5, 7, 9, 13 h p.i. were formaldehyde fixed and acetone permeabilized before being immunostained with affinity-purified RGST49 as described in Materials and Methods. Confocal images were obtained with a Leica laser-scanning microscope. The mock control at 13 h p.i. is shown. 1 h p.i. refers to the point at which fresh (37°C) medium was added to the flasks following a 1-h adsorption at 4°C.
Changes in the electrophoretic migrations of VP22 in a denaturing gel during the course of a synchronized HSV-1(F) infection.
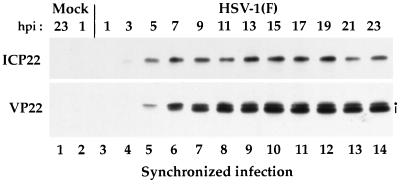
As described in the introduction, VP22 is known to undergo a variety of modifications, both in vitro and in vivo. During our initial analysis, we reported that the form of VP22 which is packaged in the virion migrates faster in a denaturing gel than does its nonpackaged form (4). The next experiment was designed to determine whether the different forms of VP22 observed in denaturing gels accumulated at different rates during the course of a synchronized infection. As described in Materials and Methods, Vero cells were synchronously infected with HSV-1(F) and whole extracts of the infected cells were prepared every 2 h over a 25-h period. Equal amounts of infected-cell protein from each extract were loaded on a 0.1% SDS–15% polyacrylamide gel cross-linked with DATD (5), transferred to nitrocellulose, and probed with antibodies RGST22 and RGST49. ICP22 was specifically chosen as a control marker for the IE (α) phase of infection.
The results (Fig. 6) of this Western blot analysis showed that low levels of ICP22 could be detected at 3 h p.i. (lane 4). The amount of ICP22 remained relatively constant after 5 h p.i. until 19 h p.i. The reduction of ICP22 and VP22 protein observed at 21 and 23 h p.i. may represent the loss of infected-cell protein as a result of cell lysis. VP22 was first observed at 5 h p.i. as a single band (lane 5). An additional, faster-migrating VP22 band was observed in extracts prepared at 7 h p.i. and thereafter (lanes 6 to 14). While both the slow- and fast-migrating forms of VP22 appeared to represent equal amounts of VP22 protein at 7 h p.i., the faster-migrating forms of VP22 predominated from 9 to 23 h p.i. (compare lanes 7 to 14 with lane 6). The distribution pattern in which the faster-migrating form of VP22 predominates appeared to remain constant from 11 to 23 h p.i. As expected, no ICP22 or VP22 was observed in the mock-infected extracts (lanes 1 and 2) or at 1 h p.i. (lane 3). From this experiment, we conclude that (i) VP22 exists in at least two forms in extracts derived from synchronously infected Vero cells and (ii) the distribution of these forms appears to be regulated during the course of infection inasmuch as the low-mobility forms predominate early in infection (<7 h) while the high-mobility forms accumulate later.
FIG. 6.
Immunoreactivities of infected cell proteins extracted during the course of a synchronized HSV-1(F) infection. Vero cells were synchronously infected with HSV-1(F) (MOI = 30) or mock infected. At 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, and 23 h p.i., whole-cell extracts were prepared and polypeptides were separated in a denaturing gel and transferred to nitrocellulose prior to chemiluminescent staining with antibodies RGST22 and RGST49 as described in Materials and Methods. The dot marks the slowest-migrating form of VP22, and the vertical line marks the faster form(s). 1 h p.i. refers to the point at which fresh (37°C) medium was added back to the flasks following a 1-h absorption at 4°C.
Slowest-migrating (low-mobility) forms of VP22 derived from the nuclear fraction of HSV-1(F)-synchronously infected Vero cells.
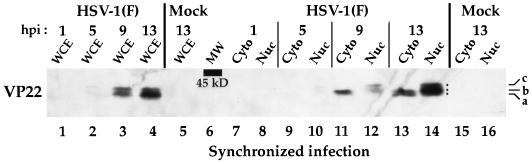
The next set of experiments was designed to determine whether the differently migrating forms of VP22 correlated with changes in subcellular localization of VP22 during a synchronized infection as observed by indirect immunofluorescence (Fig. 5). Nuclear and cytoplasmic fractions were prepared from Vero cells at 1, 5, 9, and 13 h during a synchronized infection with HSV-1 and infected-cell polypeptides were separated in a denaturing gel and transferred to nitrocellulose prior to immunoblotting with G49 antibody specific for VP22 as described in Materials and Methods. As a control, a portion of the infected cells was reserved and whole-cell extracts were prepared from this population. The results of this experiment are shown in Fig. 7.
FIG. 7.
Immunoreactivities of polypeptides derived from either whole-cell extracts (WCE) or nuclear (Nuc) and cytoplasmic (Cyto) fractions during the course of a synchronized HSV-1(F) infection. Vero cells were synchronously infected with HSV-1(F) (MOI = 30) or mock infected. At 1, 5, 9, and 13 h p.i., either whole-cell extracts or nuclear and cytoplasmic fractions were prepared and polypeptides were separated in a denaturing gel and transferred to nitrocellulose before undergoing chemiluminescent staining with antibody G49 as described in Materials and Methods. Dots mark the locations of the three observed electrophoretic forms of VP22 (a, b, and c). 1 h p.i. refers to the point at which fresh (37°C) medium was added back to the flasks following a 1-h adsorption at 4°C. The location of the 45-kDa molecular mass marker is indicated (MW).
The results with control whole-cell extracts confirmed the observations (Fig. 6) that the slowest-migrating form of VP22 was observed at 5 h p.i. (Fig. 7, lane 2), the faster form of VP22 was seen at 9 h p.i. (lane 3), and this faster form predominated at 13 h p.i. (lane 4). This finding indicates that the RGST49 and G49 antibodies yield identical results. No VP22 could be detected at 1 h p.i. in either the whole-cell extracts (lane 1) or the cytoplasmic (lane 7) and nuclear (lane 8) fractions. Comparison of the infected whole-cell extract (lane 2) with the fractions prepared at 5 h p.i. (lane 9 and 10) showed that all of the detectable VP22 was present in the nuclear fraction (lane 10). At 9 h p.i., the majority of VP22 present in the whole-cell extract (lane 3) was represented as a faster-migrating form observed in the cytoplasmic fraction (lane 11), while at least two slower-migrating forms of VP22 were seen in the nuclear fraction (lane 12). By 13 h p.i., the majority of VP22 present in the whole-cell extract (lane 4) was represented as at least three forms observed in the nuclear fraction (lane 14) while two, less abundant, faster-migrating forms of VP22 were seen in the cytoplasmic fraction (lane 13). Comparison of the nuclear and cytoplasmic fractions at 9 and 13 h p.i. indicated that the low-mobility forms of VP22 partitioned exclusively in the nuclear fraction. As expected, no VP22 was present in the mock-infected cells (lane 5, 15, and 16).
Based on the results presented in Fig. 6 and 7, we conclude that (i) during the course of a synchronized infection, at least three different forms of VP22 can be resolved in a denaturing gel. We have designated these forms as VP22a, VP22b, and VP22c (VP22a migrates fastest, and VP22c migrates slowest). (ii) The slowest-migrating form, VP22c, is exclusively nuclear and is detected earliest during infection (5 h p.i.), while forms VP22b and VP22a are detected later in infection. VP22b appears to be the major form of VP22 at 9 h p.i., and this form predominates in the cytoplasm. Although both VP22b and VP22a are present in the cytoplasm at 13 h p.i., VP22 at this time is predominantly nuclear (Fig. 7, compare lanes 13 and 14), consistent with the cell staining presented in Fig. 5. Interestingly, all three forms of VP22 (VP22a to VP22c) are found in the nucleus late in infection. An alternative possibility exists that the fastest-migrating VP22 is a proteolytically processed form, as we have suggested previously (4), and that this processing of VP22 occurs at late times in infection.
[32P]orthophosphate-modified VP22 localizes to the nucleus during a synchronized HSV-1(F) infection.
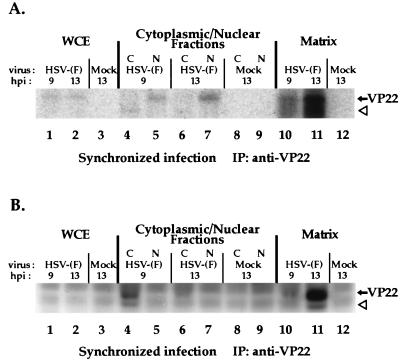
The results above (Fig. 7) demonstrated that VP22c was the slowest-migrating form of VP22 observed in a denaturing gel and that this form was found exclusively in nuclear fractions of infected cells. Since a slower-migrating form of VP22 has been shown to be phosphorylated (4, 18), a likely hypothesis is that VP22c represents one of the posttranslationally modified forms of VP22 described in the introduction. To confirm that the slowest-migrating, nuclear forms of VP22 were, in fact, phosphorylated, Vero cells were synchronously infected with HSV-1(F) or mock infected in the presence of [32P]orthophosphate and total VP22 protein derived from whole-cell extracts or cytoplasmic and nuclear fractions was immunoprecipitated with the RGST49 antibody as described in Materials and Methods. Immunoprecipitations were also performed from the matrix, which remained following the isolation of the nuclear fraction. Immunoprecipitated polypeptides were separated in a denaturing gel, transferred to nitrocellulose, and tested for immunoreactivity with antibody RGST49 prior to autoradiography. The results (Fig. 8) were as follows.
FIG. 8.
Autoradiographic images (A) and immunoreactivities (B) of 32P-orthophosphate-labeled infected-cell polypeptides immunoprecipitated with an anti-VP22 antibody during the course of a synchronized HSV-1(F) infection. Vero cells were synchronously infected with HSV-1(F) (MOI = 30) or mock infected in radiolabel-free medium. At 5 h p.i., medium containing 32P-orthophosphate was added, and after a 2-h incubation, radiolabel was removed. At 9 and 13 h p.i., whole-cell extracts (WCE), nuclear (N) and cytoplasmic (C) fractions, or the residual matrices were precipitated with the RGST49 antibody as described in Materials and Methods. Immunoprecipitated (IP) infected-cell polypeptides were separated in a denaturing 15% DATD-acrylamide gel, transferred to nitrocellulose, and reacted with antibody RGST49. An alkaline phosphatase-conjugated secondary antibody was used for immunostaining prior to autoradiography. The solid arrows mark the location of VP22, while the open arrowheads mark the potential degradation product referred to in the text.
Since the autoradiogram (Fig. 8A) showed labeled bands only in the region where VP22 was detected, only this portion is presented. Weak but detectable levels of radiolabeled protein were precipitated from infected but not mock-infected whole-cell extracts at 9 and 13 h p.i. (lanes 1 and 2). At 9 h p.i., the radiolabeled band precipitated from the nuclear fraction (lane 5) migrated with those observed in the whole-cell extracts (lane 1 and 2). Unexpectedly, a low-molecular-weight radiolabeled band was precipitated from the cytoplasmic fraction (lane 4 and 6). This band probably represents a degradation product of VP22. At 13 h p.i., a radiolabeled band was precipitated from the nuclear fraction (lane 7) and the intensity of the nucleus-derived band increased from 9 to 13 h p.i. When the nuclear matrices were solubilized by sonication and added to immunoprecipitation reaction mixtures, radiolabeled bands (lane 10 and 11) were observed at 9 and 13 h p.i. migrating with those observed in the whole-cell extracts (lane 1 and 2). The radiolabel signal was most intense in the nuclear matrix-derived band present at 13 h p.i. (lane 11).
The portion of the immunoblot (Fig. 8B) containing the whole-cell extracts and the cytoplasmic and nuclear fractions (lanes 1 to 9) showed demonstrable levels of immunostaining of VP22 in the 9 h p.i. cytoplasmic lane. Two immunoreactive species were observed: (i) an abundant form that had an apparent molecular weight similar to that of the radiolabeled bands observed in whole-cell extracts in Fig. 8A, and (ii) the minor, low-molecular-weight protein (lane 4) believed to be a degradation product. Direct alignment of the autoradiogram with the immunoblot at 9 h p.i. showed that the abundant immunoreactive band in the cytoplasmic fraction (Fig. 8B, lane 4) was not radiolabeled (Fig. 8A, lane 4). However, the location of the low-molecular-weight immunoreactive protein (Fig. 8B, lane 4) coincided with the low-molecular-weight radiolabeled protein (Fig. 8A) observed in the cytoplasmic fraction. These results indicate that phosphorylation of full-length VP22 in the cytoplasm is undetectable although a degradation product of VP22 present in the cytoplasm is phosphorylated. Immunoblotting revealed an abundant amount of VP22 present in the nuclear matrix at 13 h p.i. along with a minor amount of the low-molecular-weight species (Fig. 8B, lane 11). VP22 present in the nuclear fraction, which incorporated significantly more radiolabel than did VP22 in the cytoplasm (Fig. 8A, compare lane 4 with lane 5 and compare lane 6 with lane 7), was undetectable by immunoblotting (Fig. 8B, lanes 5 and 7). This indicates that the limited amount of VP22 in the nuclear fraction incorporated a significant amount of radiolabel. However, the majority of VP22 present in the nucleus was associated with the nuclear matrix (Fig. 8B, lanes 10 and 11). VP22 present in whole-cell extracts was not detected by immunoblotting because equal amounts of infected cell protein were used in each precipitation reaction, subsequently diluting the concentration of VP22 in the whole-cell extract relative to the subcellular fractions. Nevertheless, the [32P]orthophosphate label on this small amount of VP22 was detectable in the autoradiogram.
From these results, we conclude that under the conditions of our fractionations, the majority of VP22 at 9 h p.i. is partitioned to the cytoplasm and the majority at 13 h p.i. is located in the nucleus and may be associated with chromatin. In addition, a portion of the VP22 which partitions to the nucleus is modified by the incorporation of phosphate from [32P]orthophosphate since both the nuclear fractions and nuclear matrices accumulated radiolabeled VP22 during the course of infection. At this time, we do not know the significance of the low-molecular-weight form of VP22 observed in the cytoplasmic (at 9 h p.i.) and matrix (at 13 h p.i.) fractions, although we believe that it is a degradation product of VP22. This species has been observed previously under conditions of excess polypeptide loading on denaturing gels (4), and its high electrophoretic mobility indicates that it is not related to the VP22a to VP22c forms defined in Fig. 7. Since the majority of the cytoplasmic form of VP22 was not radiolabeled, we conclude that modification of VP22 correlates with its presence in the nucleus.
DISCUSSION
The goal of this study was to document the biological and biochemical properties of the HSV-1 VP22 protein during the course of productive infection. To accomplish this task, we developed infection protocols which would enable us to dissect the subcellular localization and compartmentalization of the protein. The key findings of our study are summarized as follows.
(i) A low-temperature adsorption step and subsequent shift to 37°C resulted in a synchronized HSV-1 infection. It was our desire to develop an infection system in which all cells in a monolayer were at the same stage of infection at any given time. Using a modification of a previously published report (22), we determined that a MOI of 30 PFU per cell was necessary to achieve synchronized infection. Indirect immunofluorescence of cells fixed at regular intervals p.i. indicated that staining for the HSV-1 protein gD was uniform in all cells at each time point examined. Therefore, this technique presents an infection system in which all cells in a monolayer are at the same stage of infection at any given time and is suitable for studies involving indirect immunofluorescence. Based on these results, whole-cell extracts and cell fractions were also prepared from synchronously infected cells.
(ii) VP22 localizes to three distinct subcellular areas during the course of HSV-1 infection. Based on the unique indirect-immunofluorescence staining that was observed, these specific VP22-staining patterns were defined as cytoplasmic, diffuse, and nuclear. The diffuse pattern appears to represent a transition between cytoplasmic and nuclear localization, since VP22 in these cells is present throughout the majority (both cytoplasm and nucleus) of the cell as measured by standard indirect immunofluorescence microscopy.
(iii) VP22 exists in the cytoplasm early in infection and migrates to and accumulates in the nucleus late in infection. To identify the specific phases of viral infection, we compared the localization of VP22 with those of ICP22 and ICP4, representative IE (α) proteins, and gD, a representative L (γ1) protein. VP22 staining in the nucleus was not restricted to replication compartments which accumulated the HSV-1 ICP4 protein (25). Using the synchronized infection system, we monitored VP22 throughout the course of HSV-1 infection. Confocal microscopy confirmed that the major influx of VP22 into the nucleus occurs between 7 and 9 h p.i. VP22 colocalized with gD in the cytoplasm early during infection, while VP22 alone was observed in the nuclei of infected cells after 7 h p.i. Cytoplasmic VP22 colocalized with gD in the cytoplasm early in infection. This area was also the site at which the 58K Golgi marker localized (data not shown), suggesting that both VP22 and gD may associate with the Golgi prior to the entry of VP22 into the nucleus. Although blocking fixed cells with human Ig should prevent the HSV-1 Fc receptor from binding the polyclonal RGST49, we cannot exclude the possibility that the viral Fc receptor is playing a role in the cytoplasmic staining patterns we have observed. Nevertheless, these blocking conditions inhibited the HSV-1 Fc receptor from binding the rabbit polyclonal RGST22 antibody (Fig. 1).
(iv) VP22 extracted from synchronously infected cells migrates as at least three unique electrophoretic species in denaturing gels. In these analyses, the fact that the ICP22 protein was observed earlier in infection than VP22 further supports our conclusion that the infections were synchronized. We defined the three forms of VP22 as VP22a for high mobility, VP22b for intermediate mobility, and VP22c for low mobility. Under our conditions, the lowest-mobility forms were associated with the nuclear fraction. The relative distribution of the forms of VP22 varied during the course of infection. Unexpectedly, a low-mobility species (VP22c) was observed in the nuclear fraction at 5 h p.i., suggesting that very early in infection a subpopulation of VP22 is targeted to the nucleus. It is likely that this population of VP22 is present in such a low abundance that it cannot be detected by indirect immunofluorescence early in infection. No VP22 was detected at 1 h p.i. Therefore, it seems unlikely that the early nuclear form represents VP22 derived from input virus. Since the role played by this early nuclear form of VP22 in infection is not clear, specific studies with VP22 mutants defective in nuclear targeting should help clarify this issue.
(v) Full-length VP22 which partitioned in the nuclear fraction incorporated radiolabel from 32P-orthophosphate, while cytoplasmic VP22 did not. Based on this finding, we conclude that modification of VP22 coincides with its appearance in the nucleus. While the bovine herpesvirus type 1 VP22 homologue was also reported to exhibit a nuclear localization (27), it is unclear whether the bovine protein is modified in a manner similar to that of HSV-1 VP22. The most intriguing finding in this portion of our study was that most of the VP22 protein immunoprecipitated at 13 h p.i. was present in the nuclear matrix while at 9 h p.i. VP22 was cytoplasmic. Since the fractionation buffers used contained 140 mM NaCl, we cannot exclude the possibility that the nuclear form of VP22 which accumulates late in infection is actually less soluble than that produced earlier in infection. In the absence of sonication, it was reported that HSV-1 tegument proteins, in particular VP22, were insoluble in extraction buffers containing Triton X-100, deoxycholate, and SDS (30). An alternative explanation for our results could be the following. One of the initial characterizations of the VP22 protein was that it had the capacity to tightly bind chromatin (26). Since we have routinely been able to extract VP22 from the nuclei of infected cells by using high-salt buffers (4), it is possible that 140 mM NaCl was an insufficient ionic strength to remove VP22 from the other insoluble material in the nuclear matrix. Since all of our fractions were sonicated, our nuclear matrix probably contained insoluble chromatin fragments as well as material previously referred to as matrix (12). If such a scenario turns out to be true, it would mean that the modified form of VP22 which accumulates in the nucleus late in infection has a high affinity for chromatin.
(vi) Our conclusions are based on data from indirect immunofluorescence performed after infected cell monolayers were fixed with formaldehyde and subsequently permeabilized with acetone. Under these conditions, we did not observe any cells in HSV-1(F) plaques that stained for VP22 but not ICP4, consistent with the accepted cascade of synthesis of viral proteins (35). These results are in contrast to those recently described in which COS-1 cells were infected (MOI = 0.1) with a gH-minus derivative of HSV-1 (strain 17) and fixed and permeabilized with 100% methanol (17). In indirect-immunofluorescence experiments with COS-1 cells, the VP22 which was present in cells that also contained β-galactosidase as an internal marker for viral infection had a pattern which we would describe as cytoplasmic. These authors also observed nuclear staining for VP22 in cells devoid of β-galactosidase and concluded that these cells were uninfected. These data are consistent with our results (Fig. 2) obtained when 100% methanol was used for fixing and permeabilization. In plaques fixed and permeabilized only with methanol, VP22 but not ICP4 was observed in the nuclei of some cells.
Previous reports have indicated that the carboxy-terminal half of VP22 contains either a nuclear localization or retention domain since, when this portion of VP22 is fused to the jellyfish green fluorescent protein, it is able to recruit green fluorescept protein to the nuclei of cells during transfection and transient-expression experiments (20). Transfection and transient expression of a copy of VP22 which contains a carboxy-terminal deletion of 34 amino acids also shows only cytoplasmic localization in indirect immunofluorescence studies (17), supporting the theory that this portion of the protein possesses a nuclear targeting or retention activity.
In light of the data presented in Fig. 2, we wish to put forward the following hypothesis to explain these results. We propose that under the conditions of methanol-only fixing and permeabilization, the detection of wild-type VP22 in the nuclei of cells which do not stain for ICP4 (Fig. 2) is due to the high affinity of VP22 for chromatin and matrix at late times during infection (Fig. 8). Possibly, both VP22 and ICP4 are originally present in all cells but in the presence of methanol ICP4 is not retained in the nuclei while VP22 is retained due to its potential chromatin- and matrix-associating property. An alternative model would be that under methanol-only conditions, VP22 may seep out of the nuclei of infected cells and, due to its chromatin- and matrix-associating property, is retained in the nuclei of adjacent cells. Support for this latter theory comes from the finding (17) that exogenous VP22 added to COS-1 cells is observed in the nucleus when cells are fixed and permeabilized with methanol only.
Together, these findings imply that VP22 plays a complex and perhaps tightly regulated role during HSV infection. While VP22 is known to accept a variety of posttranslational modifications, we have investigated only its incorporation of phosphate in this study. The development of appropriate genetic and biochemical system is required to determine the function of modified VP22 during the course of productive infection.
ACKNOWLEDGMENTS
We thank Bernard Roizman (University of Chicago) for the HSV-1(F) virus, Scott Henderson (Mount Sinai School of Medicine) for expert technical advice concerning confocal microscopy, and Tom Moran for advice and Kerryn Mortimer and Helen Park (Mount Sinai School of Medicine) for expert technical help during the development and production of the G49 monoclonal antibody.
These studies were supported in part by grants from the U.S. Public Health Service (AI38873), the American Cancer Society (JFRA 634), and an unrestricted grant from the National Foundation for Infectious Diseases. Confocal laser scanning microscopy was performed at the MSSM-CLSM core facility, with the support of funding from an NIH shared instrumentation grant and an NSF Major Research Instrumentation grant. J.A.B. is a Markey Research Fellow and thanks the Lucille P. Markey Charitable Trust for support. L.E.P. is a U.S. Public Health Service Predoctoral Trainee (GM08553).
REFERENCES
- 1.Avitabile E, Di Gaeta S, Torrisi M R, Ward P L, Roizman B, Campadelli-Fiume G. Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J Virol. 1995;69:7472–7482. doi: 10.1128/jvi.69.12.7472-7482.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker D E, Roizman B. Identification of three genes nonessential for growth in cell culture near the right terminus of the unique sequences of long component of herpes simplex virus 1. Virology. 1990;177:684–691. doi: 10.1016/0042-6822(90)90534-x. [DOI] [PubMed] [Google Scholar]
- 3.Blaho J A, Michael N, Kang V, Aboul-Ela N, Smulson M E, Jacobson M K, Roizman B. Differences in the poly(ADP-ribosyl)ation patterns of ICP4, the herpes simplex virus major regulatory protein, in infected cells and in isolated nuclei. J Virol. 1992;66:6398–6407. doi: 10.1128/jvi.66.11.6398-6407.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaho J A, Mitchell C, Roizman B. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL47, and UL49 gene products. J Biol Chem. 1994;269:17401–17410. [PubMed] [Google Scholar]
- 5.Blaho J A, Roizman B. Analyses of HSV proteins for posttranslational modifications and enzyme functions. In: Brown S M, Maclean A R, editors. Methods in molecular medicine: herpes simplex virus protocols. Vol. 10. Totowa, N.J: Humana Press Inc.; 1998. pp. 237–256. [DOI] [PubMed] [Google Scholar]
- 6.Blaho J A, Zong C S, Mortimer K A. Tyrosine phosphorylation of the herpes simplex virus type 1 regulatory protein ICP22 and a cellular protein which shares antigenic determinants with ICP22. J Virol. 1997;71:9828–9832. doi: 10.1128/jvi.71.12.9828-9832.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boehmer P E, Lehman I R. Herpes simplex virus DNA replication. Annu Rev Biochem. 1997;66:347–384. doi: 10.1146/annurev.biochem.66.1.347. [DOI] [PubMed] [Google Scholar]
- 8.Braun D K, Roizman B, Pereira L. Characterization of posttranslational products of herpes simplex virus gene 35 proteins binding to the surfaces of full capsids but not empty capsids. J Virol. 1984;49:142–153. doi: 10.1128/jvi.49.1.142-153.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Browne H, Bell S, Minson T, Wilson D W. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J Virol. 1996;70:4311–4316. doi: 10.1128/jvi.70.7.4311-4316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burkham J, Coen D M, Weller S K. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J Virol. 1998;72:10100–10100. doi: 10.1128/jvi.72.12.10100-10107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campadelli G, Brandimarti R, Di Lazzaro C, Ward P L, Roizman B, Torrisi M R. Fragmentation and dispersal of Golgi proteins and redistribution of glycoproteins and glycolipids processed through the Golgi apparatus after infection with herpes simplex virus 1. Proc Natl Acad Sci USA. 1993;90:2798–2802. doi: 10.1073/pnas.90.7.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang Y E, Roizman B. The product of the UL31 gene of herpes simplex virus 1 is a nuclear phosphoprotein which partitions with the nuclear matrix. J Virol. 1993;67:6348–6356. doi: 10.1128/jvi.67.11.6348-6356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Bruyn Kops A, Uprichard S L, Chen M, Knipe D M. Comparison of the intranuclear distributions of herpes simplex virus proteins involved in various viral functions. Virology. 1998;252:162–178. doi: 10.1006/viro.1998.9450. [DOI] [PubMed] [Google Scholar]
- 14.Ejercito P M, Kieff E D, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968;2:357–64. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 15.Elliott G, Mouzakitis G, O’Hare P. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J Virol. 1995;69:7932–7941. doi: 10.1128/jvi.69.12.7932-7941.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott G, O’Hare P. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J Virol. 1998;72:6448–6455. doi: 10.1128/jvi.72.8.6448-6455.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott G, O’Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 18.Elliott G, O’Reilly D, O’Hare P. Phosphorylation of the herpes simplex virus type 1 tegument protein VP22. Virology. 1996;226:140–145. doi: 10.1006/viro.1996.0638. [DOI] [PubMed] [Google Scholar]
- 19.Elliott G D, Meredith D M. The herpes simplex virus type 1 tegument protein VP22 is encoded by gene UL49. J Gen Virol. 1992;73:723–726. doi: 10.1099/0022-1317-73-3-723. [DOI] [PubMed] [Google Scholar]
- 20.Fang B, Xu B, Kock P, Roth J A. Intercellular trafficking of VP22-GFP fusion proteins is not observed in cultured mammalian cells. Gene Ther. 1998;5:1420–1424. doi: 10.1038/sj.gt.3300741. [DOI] [PubMed] [Google Scholar]
- 21.Hall L M, Draper K G, Frink R J, Costa R H, Wagner E K. Herpes simplex virus mRNA species mapping in EcoRI fragment I. J Virol. 1982;43:594–607. doi: 10.1128/jvi.43.2.594-607.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herold B C, WuDunn D, Soltys N, Spear P G. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J Virol. 1991;65:1090–1098. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc Natl Acad Sci USA. 1975;72:1276–1280. doi: 10.1073/pnas.72.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knipe D M, Senechek D, Rice S A, Smith J L. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J Virol. 1987;61:276–284. doi: 10.1128/jvi.61.2.276-284.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knopf K W, Kaerner H C. Virus-specific basic phosphoproteins associated with herpes simplex virus type 1 (HSV-1) particles and the chromatin of HSV-1-infected cells. J Gen Virol. 1980;46:405–414. doi: 10.1099/0022-1317-46-2-405. [DOI] [PubMed] [Google Scholar]
- 27.Liang X, Chow B, Li Y, Raggo C, Yoo D, Attah-Poku S, Babiuk L A. Characterization of bovine herpesvirus 1 UL49 homolog gene and product: bovine herpesvirus 1 UL49 homolog is dispensable for virus growth. J Virol. 1995;69:3863–3867. doi: 10.1128/jvi.69.6.3863-3867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liptak L M, Uprichard S L, Knipe D M. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J Virol. 1996;70:1759–1767. doi: 10.1128/jvi.70.3.1759-1767.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukonis C J, Burkham J, Weller S K. Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: only a subset are likely to be precursors to replication compartments. J Virol. 1997;71:4771–4781. doi: 10.1128/jvi.71.6.4771-4781.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison E E, Stevenson A J, Wang Y F, Meredith D M. Differences in the intracellular localization and fate of herpes simplex virus tegument proteins early in the infection of Vero cells. J Gen Virol. 1998;79:2517–2528. doi: 10.1099/0022-1317-79-10-2517. [DOI] [PubMed] [Google Scholar]
- 31.Morrison E E, Wang Y F, Meredith D M. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J Virol. 1998;72:7108–7114. doi: 10.1128/jvi.72.9.7108-7114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newcomb W W, Homa F L, Thomsen D R, Ye Z, Brown J C. Cell-free assembly of the herpes simplex virus capsid. J Virol. 1994;68:6059–6063. doi: 10.1128/jvi.68.9.6059-6063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pomeranz, L. E., and J. A. Blaho. 1999. Unpublished data.
- 33a.Pomeranz, L. E., and J. A. Blaho. 9 February 1999, posting date. Electronic data. [Online.] http://www.mssm.edu/micro/blaho/lpdns/html. [19 May 1999, last date accessed.]
- 34.Quinlan M P, Chen L B, Knipe D M. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell. 1984;36:857–868. doi: 10.1016/0092-8674(84)90035-7. [DOI] [PubMed] [Google Scholar]
- 35.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2231–2295. [Google Scholar]
- 36.Spear P G, Roizman B. Proteins specified by herpes simplex virus. IV. Site of glycosylation and accumulation of viral membrane proteins. Proc Natl Acad Sci USA. 1970;66:730–737. doi: 10.1073/pnas.66.3.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spear P G, Roizman B. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J Virol. 1972;9:143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trus B L, Homa F L, Booy F P, Newcomb W W, Thomsen D R, Cheng N, Brown J C, Steven A C. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J Virol. 1995;69:7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uprichard S L, Knipe D M. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology. 1997;229:113–125. doi: 10.1006/viro.1996.8430. [DOI] [PubMed] [Google Scholar]
- 40.Ward P L, Avitabile E, Campadelli-Fiume G, Roizman B. Conservation of the architecture of the Golgi apparatus related to a differential organization of microtubules in polykaryocytes induced by syn- mutants of herpes simplex virus 1. Virology. 1998;241:189–199. doi: 10.1006/viro.1997.8972. [DOI] [PubMed] [Google Scholar]
- 41.Ward P L, Ogle W O, Roizman B. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J Virol. 1996;70:4623–4631. doi: 10.1128/jvi.70.7.4623-4631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhong L, Hayward G S. Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient cotransfection assays. J Virol. 1997;71:3146–3160. doi: 10.1128/jvi.71.4.3146-3160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]