Abstract
Hepatitis C virus (HCV) glycoproteins E1 and E2, when expressed in eukaryotic cells, are retained in the endoplasmic reticulum (ER). C-terminal truncation of E2 at residue 661 or 715 (position on the polyprotein) leads to secretion, consistent with deletion of a proposed hydrophobic transmembrane anchor sequence. We demonstrate cell surface expression of a chimeric glycoprotein consisting of E2 residues 384 to 661 fused to the transmembrane and cytoplasmic domains of influenza A virus hemagglutinin (HA), termed E2661-HATMCT. The E2661-HATMCT chimeric glycoprotein was able to bind a number of conformation-dependent monoclonal antibodies and a recombinant soluble form of CD81, suggesting that it was folded in a manner comparable to “native” E2. Furthermore, cell surface-expressed E2661-HATMCT demonstrated pH-dependent changes in antigen conformation, consistent with an acid-mediated fusion mechanism. However, E2661-HATMCT was unable to induce cell fusion of CD81-positive HEK cells after neutral- or low-pH treatment. We propose that a stretch of conserved, hydrophobic amino acids within the E1 glycoprotein, displaying similarities to flavivirus and paramyxovirus fusion peptides, may constitute the HCV fusion peptide. We demonstrate that influenza virus can incorporate E2661-HATMCT into particles and discuss experiments to address the relevance of the E2-CD81 interaction for HCV attachment and entry.
Enveloped viruses acquire their lipid membranes by budding through host cellular membranes (reviewed in reference 35). The majority of enveloped viruses bud at the plasma membrane. However, several viruses assemble and bud at internal membranes such as those of the endoplasmic reticulum (ER) (e.g., rotaviruses), ER-Golgi intermediate compartments (e.g., coronaviruses), or the Golgi complex (e.g., bunyaviruses). This behaviour generally reflects the targeting of the viral glycoproteins (gps) within subcompartments of the ER or Golgi complex. In the latter cases, viruses are released from infected cells either by cell lysis or after transport through the cellular secretory pathway to the cell surface.
Hepatitis C virus (HCV), the major cause of non-A, non-B hepatitis, is an enveloped virus classified in the Flaviviridae family (reviewed in references 3 and 39). The genome encodes two putative envelope gps, E1 (polyprotein residues 192 to 383) and E2 (residues 384 to 746), which are released from the viral polyprotein by signal peptidase cleavage(s) (13, 18, 43). Both gps are heavily modified by N-linked glycosylation and are believed to be type I integral transmembrane proteins, with C-terminal hydrophobic anchor domains.
Expression of the E1E2 gps in mammalian cell lines demonstrates their ER retention with no cell surface gp expression detectable (8, 37, 46, 47). Immunoelectron microscopic studies localized the gps to the ER (7, 8). We (10) and others (4) reported the presence of ER retention “signals” within the C-terminal regions of both E1 and E2 gps, explaining these observations. Consistent with these data, truncation of E2 at its C terminus leads to its secretion from expressing cells (26, 29, 30, 45, 47). These observations are consistent with a model of HCV particle morphogenesis occurring by budding into the ER, as reported for other members of the Flaviviridae.
When expressed in tissue culture cells, the E1 and E2 gps interact to form noncovalently linked complexes, whose size is consistent with E1E2 heterodimers (6, 8). In addition to these noncovalently associated E1E2 complexes, a significant proportion of E1 and E2 are present in disulfide-linked aggregates, which are believed to result from a nonproductive folding pathway (1a, 6, 8, 13). Since HCV cannot be propagated efficiently in vitro, it has been difficult to study “native” E1E2 gp forms as they exist on the virus particle. It is critical when studying the biological activity of the HCV gps to distinguish between molecules that undergo productive folding and assembly and those that follow a nonproductive pathway(s) resulting in misfolding and aggregation (7). Recently, Dubuisson and colleagues reported a number of conformation-dependent monoclonal antibodies (MAbs) (H2 and H53) which specifically recognize nondisulfide-bridged E2, both alone and when complexed with E1, allowing the study of gp complexes which may represent “native” prebudding forms of the HCV gp complex (4, 6, 30).
gps exposed on the virus surface mediate entry into target cells. This process requires binding of the virus particle to a receptor(s) present at the surface of the host cell, followed by fusion of the viral and cellular membranes. For viruses such as influenza virus and the flavivirus tick-borne encephalitis virus, particles internalize after receptor binding and fuse with the endosomal membranes. The low pH within the endosomal compartment induces a major structural rearrangement of the gps, resulting in exposure of a fusion peptide which destabilizes membranes, leading to fusion (reviewed in references 11, 17, and 50). The mechanism by which HCV enters target cells is currently unknown; however, the E2 gp is thought to be responsible for initiating virus attachment to a receptor on potential host cells (42). Indeed, a soluble form of a C-terminally truncated E2 gp was used to identify CD81 as a putative receptor for HCV (36). CD81 is a broadly expressed protein and is reported to be involved in a variety of biological responses including adhesion, morphology, proliferation, activation, and differentiation of T-, B-, and other cell types (reviewed in reference 23).
Generation of viral pseudotypes is one of the most widely used methods for assaying functional receptors, allowing attachment, penetration, and uncoating to be studied. Recent reports that vesicular stomatitis virus (VSV) expressing chimeric HCV E2 gps, comprising the putative E2 ectodomain fused to the transmembrane and cytoplasmic domains of VSV G protein, allowed entry into target cells suggested that the ectodomain of E2 was sufficient to confer viral attachment and entry (22, 28). We were interested in studying the antigenic conformation of E2 expressed at the cell surface and whether such a protein was able to induce CD81-dependent cell fusion. Here, we demonstrate cell surface expression of a chimeric gp consisting of E2 residues 384 to 661 fused to the transmembrane and cytoplasmic domains of influenza A virus hemagglutinin (HA) (E2661-HATMCT). These data are consistent with a previous report demonstrating cell surface expression of truncated versions of E2 fused to the transmembrane domain of CD4 or a glycosylphosphatidylinositol anchor (4). The E2661-HATMCT chimeric gp was able to bind a number of conformation-dependent MAbs and a recombinant soluble form of CD81, suggesting that it was folded in a manner comparable to that of native E2. Furthermore, cell surface-expressed E2661-HATMCT demonstrated pH-dependent changes in antigen conformation, consistent with an acid-mediated fusion mechanism. However, E2661-HATMCT was unable to induce cell fusion of CD81-positive HEK cells after neutral- or low-pH treatment. We demonstrate that influenza virus can incorporate E2661-HATMCT into particles and discuss possible experiments to address the relevance of the E2-CD81 interaction for HCV attachment and entry.
MATERIALS AND METHODS
Materials.
MAbs specific for E1 (3/8d and 3/8ow), E2 (1/39, 6/82a, and 6/16), CD81 (5A6 [33]), and glutathione S-transferase (GST) (2/18) were raised by standard procedures. MAbs specific for conformation-dependent epitopes (H2, H31, H33, H44, H50, H53, H60, and H61) were a gift from J. Dubuisson (Institut Pasteur, Lille, France). MAbs specific for influenza A virus NP and fluorescein isothiocyanate (FITC)-, phycoerythrin (PE)- and horseradish peroxidase (HRP)-conjugated antibodies were purchased from Harlan Sera-Labs. The MAb against MHC class I was purchased from Sigma. Enzymes used for cloning were purchased from Gibco-BRL Life Technologies or New England Biolabs. Dulbecco’s minimum essential medium (DMEM), fetal calf serum (FCS), HEPES, and l-glutamine were obtained from Gibco-BRL Life Technologies.
Construction of recombinant cDNA.
A cDNA cassette allowing replacement of the ectodomain or transmembrane and cytoplasmic domains of influenza A virus HA was constructed. Unique restriction sites were introduced into the cDNA of HA by PCR mutagenesis. PCR was carried out with sense (W5506; 5′-TCTGGATACAAAGACTGGGCCCTGTGGATTTCCTTTGCC-3′) and antisense (W5501; 5′-GGGCCCCTGCAGGTCGACTCAAATGCAAATGTTGCA-3′) primers on the plasmid pGEM1+HA template (X-31 strain; kindly supplied by D. Steinhauer, National Institute for Medical Research, London, United Kingdom). The resulting product was used as a primer in a secondary reaction with sense primer (W5502; 5′-GGGCCCGATATCAGCAAAAGCAGGGGATAATTC-3′) with pGEM1+HA as the template. The product of this secondary reaction was digested with EcoRV and PstI and ligated with pBluescript SK(+) (Stratagene) similarly digested. DNA sequencing of the resulting plasmid, designated pBS+HA/CAS, confirmed the introduction of the unique restriction sites. The vector pCDM8 (Invitrogen) was used for expression in eukaryotic cells. The ApaI site within the polyomavirus ori of pCDM8 was destroyed through digestion with ApaI, treatment with T4 DNA polymerase, and self-ligation to form pCDM8(−ApaI). Plasmid pBS+HA/CAS was digested with HindIII and PstI. This fragment was ligated with pCDM8(−ApaI) similarly digested, to form plasmid pCDM8(−ApaI)+HA/CAS(HindIII-PstI). This plasmid was used to generate the fusion protein between the E2 and HA transmembrane and cytoplasmic domain sequences. HCV E2 sequence was amplified by PCR with plasmid pBRTM/HCV1-3011 (kindly supplied by C. M. Rice, Washington University, St. Louis, Mo.) as the template. The sequence encoding HCV residues 364 to 661 was amplified with sense (E2/FWD; 5′-GCGCAAGCTTCCATGGTGGGGAACTGG-3′) and antisense (Y0704; 5′-TATATAGGGCCCCCTCGGACCTGTCCCTGTC-3′) primers, while the sequence encoding HCV residues 364 to 715 was amplified with the same sense primer and the antisense primer Y0705 (5′-TATATAGGGCCCCCTTAATGGCCCAGGACGCG-3′). Both these PCR products were digested with HindIII and ApaI and ligated with pCDM8(−ApaI)+HA/CAS (HindIII-PstI), similarly digested. The resulting plasmids were designated pE2661-HATMCT and pE2715-HATMCT. Corresponding vectors also encoding E1 sequence were generated. Primers Y0699 (5′-GCGAGCAAGCTTCCATGGGTTGCTCTTTCTCTATC-3′) and Y0704 were used in PCR with pBRTM/HCV1-3011 as the template. The product of this reaction was digested with HindIII and ApaI and ligated with pCDM8(−ApaI)+HA/CAS(HindIII-PstI) similarly digested to form the plasmid pE1E2661-HATMCT. To construct pE1E2715-HATMCT, pE1E2661-HATMCT was digested with HindIII and SapI and ligated with pE2715-HATMCT similarly digested. Plasmid pE1E2, encoding the full sequence of E1 and E2, with the endogenous signal peptide, was constructed by PCR amplification with Y0699 and Y5862 (5′-GATATCCTGCAGTCACGCCTCCGCTTGGGATATGAG-3′), using pBRTM/HCV1-3011 as the template. The product of this reaction was digested with HindIII and PstI and ligated with pCDM8(−ApaI) similarly digested. Plasmid pE2 was constructed by PCR of the E2 sequence with E2/FWD primer and Y5862 with pBRTM/HCV1-3011 as the template. The product of this reaction was digested with HindIII and PstI. As a result of the cloning strategy described here, each recombinant chimeric protein possesses an additional Gly-Ala amino acid pair at the junction of the ectodomain (E2 sequence) and transmembrane domain (HA sequence).
Indirect immunofluorescence.
HEK (293) cells were grown in DMEM supplemented with 10% FCS and 2 mM l-glutamine. Subconfluent monolayers grown in 100-mm-diameter dishes were transfected with 10 μg of plasmid by the calcium phosphate coprecipitation method. Precipitates were incubated with cells for 4 h at 37°C before being replaced with DMEM containing 2% FCS. At 48 h posttransfection, the cells were washed once with phosphate-buffered saline (PBS), fixed with 3% paraformaldehyde for 30 min at room temperature, washed with PBS, quenched with 10 mM glycine in PBS for 10 min at room temperature, washed, and permeabilized with 0.1% Triton X-100 in PBS. The permeabilization step was omitted for measurements of surface immunofluorescence. Cells were incubated with PBS containing 1% FCS and 0.05% sodium azide (P/F/A) and then incubated with primary antibodies for 1 h at room temperature. Cocktails of anti-E1 (3/8d and 3/80w) or anti-E2 (1/39, 6/82a, and 6/16) MAbs were used. After incubation with primary antibodies, the cells were washed twice with P/F/A, incubated with FITC-conjugated anti-mouse or anti-rat antibodies (at 1/500 dilution) for 1 h at room temperature, and washed three times with P/F/A. Immunofluorescence was visualized under an Axiovert 135 fluorescence microscope (Zeiss).
Expression and purification of GST-CD81EC2.
The human CD81 EC2 was made from a gel-purified HincII-RsaI fragment, coding for amino acids 116 to 202, of the cDNA clone and ligated to pGEX-2T (Pharmacia) which had been cut with EcoRI and blunted with T4 polymerase. The pGST-CD81EC2 construct was examined by sequencing to confirm the orientation and absence of mutations. SURE Escherichia coli (Stratagene) transformed with the plasmid was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and harvested after 3 h by centrifugation, and the pellet was lysed by sonication. The GST-CD81EC2 fusion protein was recovered by affinity chromatography on glutathione-Sepharose 4B (Pharmacia). The purified fusion protein reacted with the anti-CD81 MAbs 5A6 and 1D6 when unreduced as determined by Western blotting. As noted for cellular CD81, the reduced recombinant fusion protein did not react with the antibodies (1).
Flow-cytometric analysis.
HEK cells were transfected as described above. At 48 h posttransfection, the cells were harvested with PBS containing 0.2 mM EDTA and washed with PBS twice. They were incubated for 30 min at room temperature in PBS containing 1% FCS and 0.05% sodium azide (P/F/A). Viable-cell counts were determined (trypan blue exclusion), and the cells were resuspended at 107/ml in P/F/A. A total of 106 cells were incubated with 100 μl of primary antibodies (anti-E2 linear MAbs; 1/39, 6/82a, and 6/16 equal volumes of tissue-culture supernatant or anti-E2 conformational MAbs; H2, H31, H33, H44, H50, H53, H60, and H61 at 10 μg/ml, kindly supplied by J. Dubuisson, Institut Pasteur de Lille) or with 100 μl of recombinant CD81 protein, diluted in P/F/A for 1 h at room temperature. The cells were washed three times with P/F/A before addition of 100 μl of PE-conjugated secondary antibody (at 1/100 dilution). Experiments assessing the binding of GST fusion proteins to transfected cells included an additional incubation with an anti-GST MAb (100 μl of tissue-culture supernatant). After incubation for 1 h at room temperature, the cells were washed three times with P/F/A and analyzed with a FACScan apparatus. The data were processed with CellQuest software (Becton Dickinson).
Cell-cell fusion assay.
HEK cells were infected with influenza A virus A/WSN/33 at a range of multiplicities of infection in serum-free DMEM for 7 h at 37°C. The virus inoculum was removed, and the cells were incubated for 1 h at 37°C with DMEM containing 1% FCS. The cells were washed free of FCS, incubated for 3 min at room temperature in PBS containing 10 mM morpholineethanesulfonic acid (MES) and 10 mM HEPES at either pH 5.0 or pH 7.0, washed with PBS, and incubated at 37°C in DMEM containing 2% FCS. Transfected cells were treated similarly. Following overnight incubation, the cells were fixed with methanol-acetone and E2 or NP antigen was visualized by indirect immunofluorescence as described above. To visualize nuclear DNA, a mountant containing propidium iodide (Vectashield; Vector Laboratories) was used. The cells were visualized with a confocal microscope (Bio-rad).
Generation and analysis of pseudotyped influenza viruses.
COS-7 cells were electroporated either with empty vector, pCDM8, or with 15 μg of plasmid pE2661-HATMCT as described elsewhere (2). Following electroporation, the cells were resuspended in DMEM containing 10% FCS and 10 mM HEPES (pH 7.4) and allowed to recover at 37°C overnight. They were then infected with influenza A virus A/PR8/34 at a multiplicity of infection of 3. After 24 h, supernatants were collected and clarified of cellular debris by centrifugation at 15,000 rpm in a Beckman SW55 rotor. To confirm that the released virus contained the E2661-HATMCT protein, virus was purified by centrifugation at 45,000 rpm in a Beckman SW55 rotor through a 1-ml cushion of 30% sucrose in NTE (100 mM NaCl, 10 mM Tris-HCl [pH 7.8], 1 mM EDTA). Virus pellets were resuspended in 10 μl of NTE and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting to detect the incorporation of E2661-HATMCT into particles. Cell lysates were generated by lysis of transfected COS-7 cells, or MDCK cells stably expressing the A/WSN/33 M2 protein, grown to confluency in 35-mm-diameter dishes. The cells were washed once in ice-cold PBS and incubated with lysis buffer (100 mM NaCl, 50 mM iodoacetamide, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, 20 mM Tris HCl [pH 7.5]) on ice for 10 to 30 min. After SDS-PAGE, the E2 antigen was detected by Western blotting with rat anti-E2 MAbs followed by an anti-rat HRP-conjugated secondary antibody. The influenza A virus M2 protein was detected by using a mouse MAb, 14C2 (kindly supplied by R. A. Lamb, Northwestern University, Evanston, Ill.), followed by an anti-mouse HRP-conjugated secondary antibody. Proteins were visualized following exposure to enhanced chemiluminescence detection reagents (Amersham Life Sciences) and photographic film.
RESULTS
Truncated E2 with transmembrane and cytoplasmic domains of influenza virus HA is expressed at the cell surface.
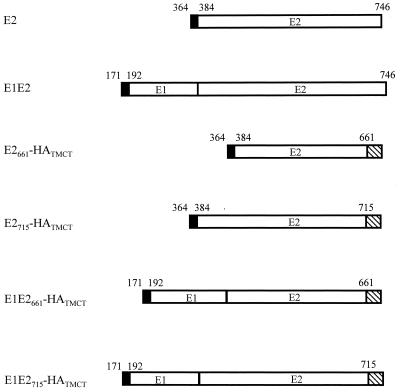
Since C-terminal truncation of E2 results in protein secretion from the cell (29, 30, 45, 47), we reasoned that addition of a transmembrane domain to such a truncated form may result in localization at the plasma membrane. To test this hypothesis, cDNA encoding the chimeric gps was constructed, consisting of the E2 ectodomain (from amino acids 384 to 661 or 715) fused to the transmembrane and cytoplasmic domains of influenza A virus HA (Fig. 1). Since the E2 gp acts as a chaperone for E1 folding (30), we were interested in determining any effects of coexpression of the full-length E1 protein on both E1 and E2 localization. Plasmids encoding both E1 and the chimeric E2 gps were therefore constructed (Fig. 1). All plasmids contained endogenous signal sequences to direct translocation to the ER, including polyprotein residues 364 to 383 for E2 chimeras and 171 to 191 for E1-encoding plasmids.
FIG. 1.
Schematic representation of the proteins expressed in these studies. HCV E1 or E2 sequences were fused to the transmembrane and cytoplasmic domains of influenza A virus HA protein. The amino acid position on the HCV polyprotein is indicated above the bars. Signal sequences are indicated by solid boxes, while the HA sequence is shown by hatching. These chimeric proteins were cloned in the eukaryotic expression vector pCDM8.
HEK (293) cells were transfected with the plasmids shown in Fig. 1, and 48 h posttransfection the cells were fixed, with or without Triton X-100 permeabilization, to monitor internal and cell surface-expressed antigen, respectively. Indirect immunofluorescence was performed with MAbs specific for both E1 and E2 proteins. The results are summarized in Table 1. As expected, full-length E1 and E2 could not be detected at the cell surface whereas E2661-HATMCT could be detected. Coexpression of E1 did not result in E1 expression at the cell surface, nor did it have any detectable effect(s) on E2661-HATMCT cell surface expression. When E2715-HATMCT was expressed, weak fluorescence could be detected both intracellularly and at the cell surface, suggesting that E2715-HATMCT was expressed less efficiently than E2661-HATMCT. Similar results were obtained when E2 expression was quantified by analysis of transfected cells by flow cytometry (data not shown). The reduced expression of the E2715-HATMCT gp is consistent with previous observations about the secretion of E2, truncated at residue 715 relative to 661, and may relate to the folding efficiency of the truncated proteins (4, 26, 30).
TABLE 1.
Summary of indirect immunofluorescence observed for E1 and E2 localization on transiently expressing cells.
| Plasmid | Fluorescence intensity witha:
|
|||
|---|---|---|---|---|
| Anti-E1
|
Anti-E2
|
|||
| Internal | Surface | Internal | Surface | |
| Vector | − | − | ||
| pE2 | +++ | − | ||
| pE1E2 | +++ | − | +++ | − |
| pE2661-HATMCT | +++ | +++ | ||
| pE1E2661-HATMCT | +++ | − | +++ | +++ |
| pE2715-HATMCT | +++ | ++ | ||
| pE1E2715-HATMCT | +++ | − | +++ | ++ |
The intensity of fluorescence across a number of fields is indicated.
Recognition of cell surface E2 by conformation-dependent anti-E2 MAbs: the effect of low-pH treatment.
We were interested in determining if cell surface-expressed E2661-HATMCT was recognized by MAbs reported to specifically interact with correctly folded E2 (4, 6). HEK cells were transfected with pE2661-HATMCT and with control empty vector and at 48 h posttransfection were assayed for their ability to bind a panel of MAbs specific for linear and conformation-dependent epitopes. MAb recognition of transiently expressing cells was analyzed by flow cytometry. All of the conformation-dependent MAbs (H2, H31, H33, H44, H50, H53, H60, and H61) were able to recognize the chimeric gp, with mean fluorescence intensities in the range of 85.3 to 220.3 for E2-expressing cells stained in PBS and with background values for mock-transfected cells of 4.5 to 9.3 (data not shown and Fig. 2). These data suggest that the chimeric E2 gp is expressed in a conformation similar to that suggested for native E2 and may therefore be considered an accurate model of E2 expressed on HCV virions.
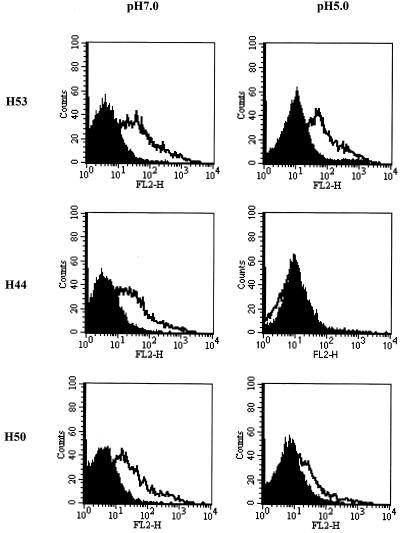
FIG. 2.
Conformation of cell surface E2661-HATMCT. HEK cells were transfected with control empty vector (solid histograms) or with pE2661-HATMCT (empty histograms). At 48 h posttransfection, cells were harvested and treated with pH 5.0 or 7.0 buffer. Cells were washed, resuspended in PBS, and immunostained with MAb H53, H44, or H50. Bound antibody was detected with PE-conjugated rabbit anti-mouse immunoglobulin antibody and flow cytometry.
The entry of flaviviruses into cells is believed to occur by an acid-mediated fusion mechanism, with conformational change(s) being detectable in the envelope gp E after exposure to low pH (14, 16, 20, 40). To determine if a similar mechanism might operate for HCV, E2661-HATMCT-expressing cells were incubated at pH 5 or 7 for 15 min, washed twice, resuspended in PBS, and assayed for their ability to bind the conformation-dependent MAbs. As controls, MHC class I and CD81 expression were monitored and the MAbs were shown to bind equivalently independent of the pH treatment (data not shown). Most of the MAbs bound to the pH 7- or 5-treated cells equivalently, with the exception of MAbs H44 and H50 (data not shown and Fig. 2). MAb H44 failed to recognize pH 5.0-treated E2661-HATMCT, and recognition by H50 was reduced after pH 5.0 treatment (Fig. 2). Since the cells were resuspended in PBS after being treated at pH 5.0 or 7.0, the conformational change(s) that occurred was irreversible. Similar results were observed by enzyme-linked immunosorbent assay for MAb recognition of low-pH-treated soluble E2661 gp (data not shown). These data are consistent with reports regarding the sensitivity of the flavivirus gp E to low pH treatment.
E2661-HATMCT binds recombinant CD81, a putative receptor for HCV.
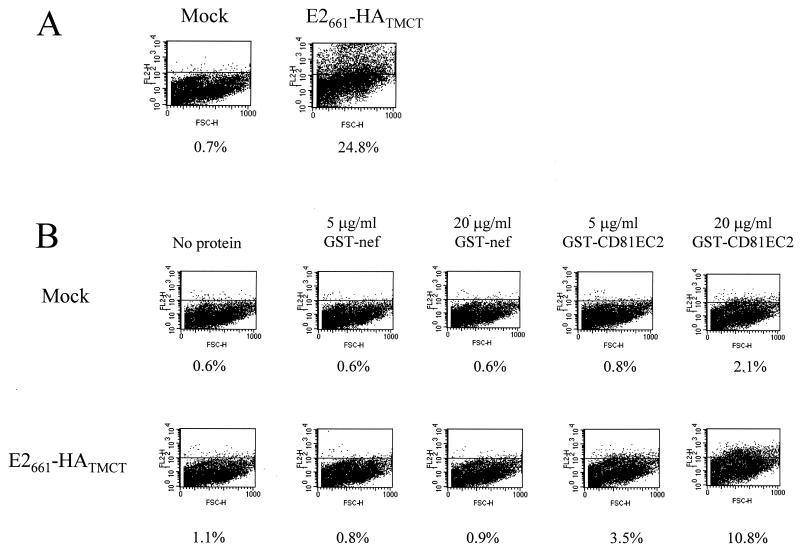
Recently, CD81 has been identified as a putative receptor for HCV (36). Binding of E2 to cells may be blocked by a recombinant fusion protein containing the second extracellular loop (EC2) of CD81 (9, 36). It was of interest, therefore, to determine if cell surface-expressed E2661-HATMCT could bind a recombinant form of CD81, GST-CD81 containing the EC2. This GST-CD81EC2 protein is able to bind a number of conformation-dependent CD81 specific MAbs and to inhibit the interaction of soluble E2 with CD81-positive cells (9). HEK cells were transfected with either pE2661-HATMCT or empty vector and at 48 h posttransfection were monitored for both E2 expression and the ability to bind GST-CD81EC2 and a control protein, GST-nef. FACScan analysis demonstrated that 25% of the cells expressed E2 at their surface (Fig. 3A) and that a percentage of these cells were able to bind GST-CD81EC2. No significant binding of GST-nef to cells transfected with vector or with pE2661-HATMCT was detected. When GST-CD81EC2 (20 μg/ml) was incubated with mock-transfected cells, a low-level binding was observed (2.1% positive cells); however, a higher level of GST-CD81EC2 cell binding was detected for cells expressing E2661-HATMCT, i.e., 10.8% (Fig. 3B). Similar results were observed with a reduced concentration of GST-CD81EC2 (5 μg/ml). These figures are lower than the percentage of cells expressing E2661-HATMCT on their surface, possibly because GST-CD81EC2 binding was not saturated or because the affinity of the anti-GST MAb for the cell-bound GST-CD81EC2 was lower than that of the anti-E2 MAbs for E2. However, we cannot exclude the possibility that the EC1 loop of CD81 (lacking from this recombinant fusion protein) influences the affinity of EC2 for E2. These data confirm that cell surface-expressed chimeric E2 can bind a recombinant form of CD81, with the binding site residing between residues 384 and 661, and that E2661-HATMCT is active in the (putative) receptor-binding function.
FIG. 3.
E2661-HATMCT binds recombinant CD81. (A) Expression of E2661-HATMCT on transfected HEK (293) cells. Cells were transfected with pE2661-HATMCT or with control empty vector. At 48 h posttransfection, the cells were immunostained with rat anti-E2 antibodies followed by PE-conjugated rabbit anti-rat immunoglobulin and were subjected to flow cytometric analysis. The results are presented as dot plots of forward scatter (FSC) against PE fluorescence in the FL2 channel. The percentage of E2-positive cells is indicated. (B) Binding of GST-CD81EC2 to E2661-HATMCT-expressing cells. HEK (293) cells were transfected with control empty vector or with pE2661-HATMCT. At 48 h posttransfection, the cells were incubated with no GST fusion protein, GST-nef, or GST-CD81EC2 at the concentrations indicated. Bound GST fusion protein was detected with a rat anti-GST MAb (2/18) followed by a PE-conjugated rabbit anti-rat immunoglobulin antibody. The percentage of positive cells is indicated below each plot.
E2 at the cell surface does not induce acid-mediated cell-cell fusion.
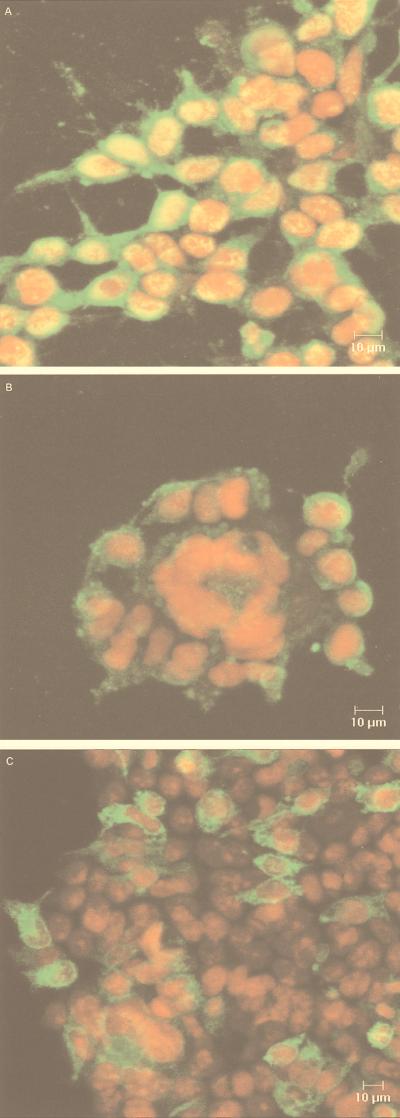
Fusion of cells expressing influenza virus HA protein by acid treatment has been well characterized (reviewed in references 11, 17, and 50). Since flaviviruses have been reported to enter cells via receptor-mediated endocytosis, we were interested in determining if E2661-HATMCT could induce cell-cell fusion in CD81-positive cells after acid treatment. HEK and the glial cell line, U87, were shown to express CD81 with mean fluorescence intensities of 958.4 and 120.3, respectively, by using the CD81-specific MAb 5A6, whereas an irrelevant isotype-matched control MAb gave values of 8.9 and 12.3 (data not shown). HEK cells transiently expressing E2661-HATMCT were treated at pH 5.0 or 7.0 as detailed above and incubated at 37°C overnight. E2-expressing cells were detected by indirect immunofluorescence, using MAbs 1/39, 6/82, and 6/16, and the nuclei were visualized with propidium iodide. After neutral- or low-pH treatment, no cell-cell fusion of cells expressing E2 at their surface was observed (Fig. 4C). As a positive control for the assay, HEK cells were infected with influenza A virus strain A/WSN/33 at different multiplicities of infection. This strain was chosen because cleavage of HA0 to HA1 and HA2, a necessary prelude to fusion, does not require trypsin treatment. Influenza virus-infected cells were identified by using an antibody specific for nucleoprotein (NP). Influenza virus-mediated cell fusion was easily detected, with large syncytia forming around NP-positive cells following low-pH treatment (Fig. 4B) but not after neutral-pH treatment (Fig. 4A). The syncytia were most evident at high multiplicities of infection, in line with previous observations that membrane fusion may be dependent upon the local density of fusion protein (5). However, the U87 glial cell line is a more sensitive indicator cell for studying both influenza virus- and human immunodeficiency virus-mediated cell fusion (24); hence, the experiment detailed above with E2661-HATMCT was repeated in this cell line. Comparable results were obtained, such that no E2661-HATMCT-mediated cell fusion was observed; however, influenza virus-induced fusion was observed at all multiplicities of infection tested (data not shown). These data indicate that under conditions which support cell-cell fusion by the influenza virus HA protein, the chimeric E2661-HATMCT gp does not induce any detectable cell fusion.
FIG. 4.
Cell-cell fusion is not mediated by E2661-HATMCT under conditions which permit HA-mediated fusion. HEK cells were infected with influenza A virus (A and B) or transfected with plasmid pE2661-HATMCT (C). Cell monolayers were treated at pH5.0 (B and C) or pH 7.0 (A) and visualized by indirect immunofluorescence with anti-E2 antibodies for transfected cells, or anti-NP MAb for influenza virus-infected cells, with propidium iodide to visualize nuclei. These micrographs show representative fields from the examined samples.
E2661-HATMCT can be incorporated into influenza virus particles.
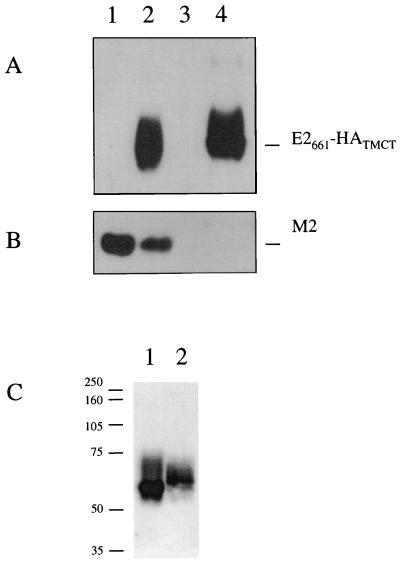
Influenza A viruses do not incorporate significant levels of host cell proteins into their envelopes. However, possession of HA transmembrane and cytoplasmic tail sequences has previously been shown to direct the incorporation of foreign proteins into influenza virus particles (31, 52). Since E2661-HATMCT was expressed at the cell surface and contained the relevant HA sequences, we tested the ability of influenza A virus to incorporate the chimeric gp expressed in COS cells. It is important to note that we (9) and others (36) have previously shown that E2 is unable to bind to COS-expressed CD81, such that high level expression of E2661-HATMCT can be achieved without receptor ligand complex formation. COS-7 cells were electroporated with pE2661-HATMCT (Fig. 5A and B, lanes 2 and 4) or empty vector (lanes 1 and 3). Cells were infected with influenza virus 24 h after transfection (multiplicity of infection, 3; lanes 1 and 2), and the extracellular progeny virus was harvested after a further 24 h. Virions were separated from host cell membrane fragments by ultracentrifugation through a high-density sucrose cushion and characterized for their constituent proteins by Western blotting. The influenza virus protein M2, a minor component of influenza virus, was visible, confirming the presence of influenza A virus particles derived from infected cells (Fig. 5B). E2 antigen was detected in a lysate derived from pE2661-HATMCT-transfected cells (Fig. 5A, lane 4), indicating that expression had occurred in these cells. Furthermore, this protein was incorporated into influenza virus particles, since it was present in progeny virions from cells transfected with pE2661-HATMCT (Fig. 5A, lane 2). The E2661-HATMCT in a lysate of expressing cells and that incorporated into influenza virus virions was compared (Fig. 5C). Since influenza virus virions bud through the plasma membrane, the chimeric molecule would be expected to undergo modification with complex glycans during transport through the secretory transport system. Consistent with this, the E2661-HATMCT present in a lysate from expressing cells migrated more rapidly in SDS-PAGE (Fig. 5C, lane 1) than did that present in influenza virus virions (lane 2).
FIG. 5.
Incorporation of E2661-HATMCT into influenza virus particles. COS-7 cells were either mock transfected or transfected with pE2661-HATMCT and infected at 24 h posttransfection with A/PR/8/34. Virus was harvested and purified 24 h postinfection and analyzed by SDS-PAGE (15% polyacrylamide) and immunoblotting for incorporation of the chimeric protein into particles. (A) Immunoblotting for the presence of E2661-HATMCT with anti-E2 antibodies. Lanes 1 and 2 are purified progeny virus released from mock-transfected cells (lane 1) and cells transfected with pE2661-HATMCT (lane 2). Lanes 3 and 4 are cell lysates prepared from mock-transfected COS-7 cells (lane 3) and COS-7 cells transfected with pE2661-HATMCT (lane 4). (B) To confirm the presence of influenza A virus particles, the same samples were analyzed by SDS-PAGE and immunoblotting to detect M2, using the 14C2 MAb. (C) E2661-HATMCT derived from a lysate of expressing cells (lane 1) and that incorporated into influenza virus virions (lane 2) was compared by SDS-PAGE (10% polyacrylamide) followed by immunoblotting with anti-E2 antibodies.
DISCUSSION
The current understanding of HCV gp function is limited by the lack of a tissue culture system supporting efficient replication of the virus. From studies with transient-expression systems, it is believed that gps E1 and E2 localize to the ER in infected cells (4, 6, 8). By analogy to other flaviviruses, virus morphogenesis may involve budding into the ER and subsequent transport of viral particles through the host cell secretory pathway before release into the extracellular space. Modification of flavivirus E and prM protein glycans by trimming and terminal addition suggests that virions do indeed move through an exocytosis pathway similar to that used for host gps (27, 32).
In this report we describe a truncated form of the HCV E2 gp fused to the transmembrane and cytoplasmic domains of the influenza A virus HA protein. This chimeric protein was expressed at the cell surface, where it was able to bind a number of conformation-dependent MAbs and a recombinant soluble version of the putative HCV receptor, CD81 (Table 1; Fig. 2 and 4). These data suggest that the chimeric gp is folded in a manner comparable to E2 present in native E1E2 complexes and that it is in a form able to bind the putative receptor, CD81. Low-pH treatment of cell surface-expressed E2 resulted in a conformational change(s). However, neutral- or low-pH treatment of CD81-positive cells expressing the chimeric gp at the cell surface did not result in cell fusion.
If HCV virions are indeed transported through the host cell secretory pathway, then E2661-HATMCT should resemble E2 on the surface of virions. Understanding the way in which E2 is glycosylated may help our understanding of E2 conformation and structure. Inhibition of core glycosylation by tunicamycin prevents E2 from folding correctly and being recognized by the conformation-dependent MAbs (1a). Since E2661-HATMCT reacts with such MAbs and since H2 and H53 react with noncovalently associated E1E2 heterodimers (4, 6), this indicates that E2661-HATMCT has a conformation similar to that adopted in E1E2 complexes.
A proteolytic cleavage is a common posttranslational modification of viral membrane proteins (reviewed in reference 21). For example, during virion transit, the prM protein of flaviviruses is cleaved by the host protease furin within a post-Golgi acidic compartment (15, 38, 48). This cleavage is required for the acquisition of virion infectivity. No proteolytic cleavage was detectable in deglycosylated E2661-HATMCT, since no size differences were observed when expressing cells were treated with or without brefeldin A, an inhibitor of the secretory transport system (data not shown). This suggests that, unlike prM, HCV E2 does not undergo proteolytic cleavage as a step in virus maturation.
Previous work has shown that truncation of E2 to residue 661 results in a molecule that is more readily exported from the cell than is a molecule truncated at residue 715 (4, 26, 30). The additional residues could reduce the efficiency of E2 folding and hence of secretory transport. Our data is consistent with this hypothesis, since E2661-HATMCT was detected more readily on the surface of expressing cells than was E2715-HATMCT (Table 1). Given the reported chaperone role of E2 in E1 folding, we were interested in determining whether cell surface expression of E2661-HATMCT would lead to expression of E1 at the cell surface (30). However, E1 was not detected at the cell surface under any circumstances; furthermore, E1 coexpression did not affect the level of E2 transport to the plasma membrane.
It is thought that after receptor-mediated endocytosis, flavivirus entry into target cells proceeds via an acid-mediated fusion event, where the viral envelope fuses with an endosomal membrane. Acid treatment of the flavivirus envelope protein E in mature virions results in a conformational change (14, 16, 20, 40). This conformational change is irreversible and results in the exposure of new antigenic epitopes and the loss of others. Treatment of cell surface-expressed E2661-HATMCT with pH 5.0 buffer caused an irreversible conformational change recognized by MAbs H44 and H50 (Fig. 2). These data suggest that HCV may enter target cells via a mechanism similar to that used by other flaviviruses. However, it should be noted that changes in E2661-HATMCT conformation after pH 5.0 treatment may be unrelated to an acid-mediated mode of entry.
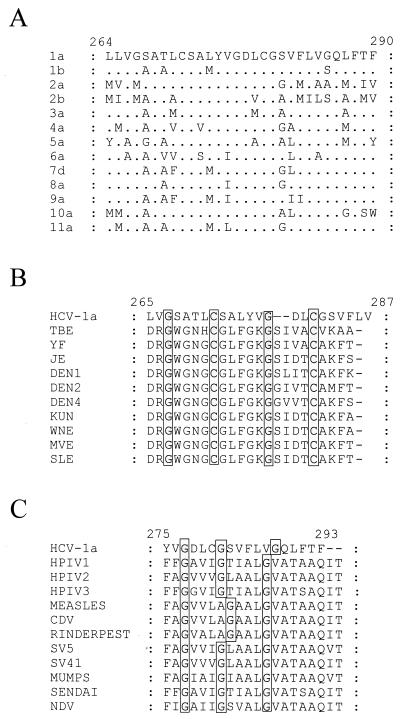
We were unable to demonstrate any E2661-HATMCT-mediated cell-cell fusion (Fig. 4 and data not shown). However, the conditions used in this assay, although compatible with influenza virus HA-mediated fusion, might not support E2-mediated fusion. The production of polykaryotic cells is dependent upon the density of the fusion protein at the cell surface (5), and the expression method used may not result in a sufficient accumulation of cell surface E2661-HATMCT to support fusion. Alternatively, the lipid composition of HEK and U87 cell membranes may not be compatible with E2-mediated fusion (12, 49). In any event, the lack of detectable polykaryons is not definitive evidence that HCV E2 does not have a fusogenic activity, since variant influenza viruses, herpesviruses, and paramyxoviruses exist that do not cause syncytium formation even though they are active in their fusion function (50). Another explanation is that E1 is required, or indeed is responsible, for the fusion event. Since we were unable to demonstrate any fusion activity for E2661-HATMCT, we examined the sequence of E1 for a putative fusion peptide. Interestingly, recombinant E1 protein is secreted when truncated after amino acid 340, only if an internal deletion between residues 262 and 290 is also present (29). This internal deletion spans a hydrophobic domain, possibly containing a fusion peptide, that could act as a transmembrane anchor when E1 is truncated at residue 340, preventing its secretion. Viral fusion peptides may act as transmembrane anchor domains, converting normally soluble proteins into membrane-bound ones (34). Most fusion peptides are composed of 16 to 26 relatively hydrophobic amino acids (50). The sequence of the internal hydrophobic domain in E1 is relatively highly conserved, with changes usually being conservative (Fig. 6A) (25). Alignment of this region of HCV E1 with the putative fusion peptide from flavivirus E proteins (41) revealed several similarities (Fig. 6B). Two Cys residues are completely conserved between all sequences analyzed. The structural implications of this are unclear, but if these residues were involved in disulfide bonds, the putative fusion peptide may be constrained in some fashion. An Asp residue is present in all the representative HCV sequences and some of the flavivirus sequences. The presence of acidic residues in the fusion peptides of some low-pH-activated viral fusion proteins has been noted previously (51). Two Gly residues are conserved within these putative fusion domains. The Gly residues within the E1 sequences have a spacing similar to that observed in the fusion peptides of the paramyxoviruses, at positions 3, 7, and 12 and at positions 3, 7, and 13 for the majority of HCV sequences analyzed to date (Fig. 6C). In the paramyxovirus F proteins, the Gly residues are believed to be important for the structure of the fusion peptide (19). Given the similarities between the internal hydrophobic region of HCV E1, the putative (flavivirus) and known (paramyxovirus) fusion peptides, we propose that this region may comprise the HCV fusion peptide.
FIG. 6.
A putative fusion peptide within the E1 protein. Numbers above the alignments indicate the amino acid position within the HCV-1 polyprotein. (A) Alignment of the internal hydrophobic domain of representative HCV genotypes (25). (B) The HCV putative fusion peptide contains similarities to the predicted fusion peptide from flavivirus E glycoprotein. Shown are alignments of the HCV-1 sequence with representative flavivirus fusion peptides: YF, yellow fever virus; JE, Japanese encephalitis virus; DEN, Dengue virus; KUN, Kunjin virus; WNE, West Nile virus; MVE, Murray Valley encephalitis virus; SLE, St. Louis encephalitis virus (41). Residues completely conserved across these sequences are shaded. (C) Spacing of Gly residues within the HCV putative fusion peptide is similar to that within the fusion peptides of paramyxovirus F proteins. Alignment of the HCV-1 sequence with representative paramyxovirus fusion peptide sequences: HPIV, human parainfluenza virus; SV, simian virus; CDV, canine distemper virus; NDV, Newcastle disease virus (19). Gly residues are shaded.
Some enveloped viruses are promiscuous in regard to the proteins they will incorporate into their membrane, while others appear to use specific signals in the sequences of their envelope proteins to discriminate between viral and cellular proteins present at the site of budding. Influenza virus appears to utilize transmembrane and cytoplasmic tail sequences to select its major envelope protein, HA, during particle formation (31). Hence, it is not surprising that E2 expressing the HATMCT was incorporated efficiently into influenza virus particles (Fig. 5). In contrast, VSV has the capacity to incorporate foreign gps regardless of their amino acid sequence (44) and has been used extensively to study many viral gps. VSV particles expressing either chimeric HCV E1 or E2 gps were recently reported to confer VSV entry, suggesting that the gps could function independently to mediate binding and entry into target cells (22). The entry of these pseudotyped viruses could be inhibited by sera from chimpanzees immunized with the homologous HCV gps; however, the entry was not shown to be CD81 dependent. Clearly, it will be important to demonstrate whether CD81, either alone or with additional factors, can function as the HCV receptor in allowing pseudotyped virus-cell attachment and entry. Since CD81 is so widely expressed, it is unlikely to be the sole factor determining HCV liver tropism. We are now in an ideal position to answer these questions by studying the receptor requirements for attachment, entry, and uncoating of influenza viruses expressing chimeric HCV gps.
ACKNOWLEDGMENTS
We thank D. Steinhauer for the HA clone, J. Dubuisson for conformation-dependent MAbs specific for HCV E2, R. A. Lamb for antibodies specific for influenza virus proteins, and M. Harris for GST-nef. We are also indebted to Yasmin Chaudhry, Barbara Konig, Moy Robson, and Stephen Poutney for excellent technical assistance. We thank Peter Balfe and Jeff Almond for constructive comments on the manuscript.
The work described herein was supported by The Wellcome Trust and The University of Reading Research Endowment Trust and by Public Health Service grant CA 34233 from the National Institutes of Health (to S.L.). J.M.T. received support through a BBSRC special studentship.
REFERENCES
- 1.Andria, M. L., and S. Levy. Unpublished results.
- 1a.Choukhi A, Ung S, Wychowski C, Dubuisson J. Involvement of endoplasmic reticulum chaperones in the folding of hepatitis C virus glycoproteins. J Virol. 1998;72:3851–3858. doi: 10.1128/jvi.72.5.3851-3858.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu G, Hayakawa H, Berg P. Electroporation for the efficient transfection of mammalian cells with DNA. Nucleic Acids Res. 1987;15:1311–1326. doi: 10.1093/nar/15.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clarke B. Molecular virology of hepatitis C virus. J Gen Virol. 1997;78:2397–2410. doi: 10.1099/0022-1317-78-10-2397. [DOI] [PubMed] [Google Scholar]
- 4.Cocquerel L, Meunier J-C, Pillez A, Wychowski C, Dubuisson J. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J Virol. 1998;72:2183–2191. doi: 10.1128/jvi.72.3.2183-2191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danieli T, Pelletier S L, Henis Y I, White J M. Membrane fusion mediated by the influenza hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deleersnyder V, Pillez A, Wychowski C, Blight K, Xu J, Hahn Y S, Rice C M, Dubuisson J. Formation of native hepatitis C virus glycoprotein complexes. J Virol. 1997;71:697–704. doi: 10.1128/jvi.71.1.697-704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubuisson, J. Folding, assembly and subcellular localization of HCV glycoproteins. Curr. Top. Microbiol. Immunol., in press. [DOI] [PubMed]
- 8.Dubuisson J, Hsu H H, Cheung R C, Greenberg H B, Russell D G, Rice C M. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol. 1994;68:6147–6160. doi: 10.1128/jvi.68.10.6147-6160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flint M, Maidens C M, Loomis-Price L, Shotton C, Dubuisson J, Monk P, Higginbottom A, Levy S, McKeating J A. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J Virol. 1999;73:6235–6244. doi: 10.1128/jvi.73.8.6235-6244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flint, M., and J. A. McKeating. The C-terminal region of the hepatitis C virus E1 glycoprotein confers localisation within the endoplasmic reticulum. J. Gen. Virol., in press. [DOI] [PubMed]
- 11.Gaudin Y, Ruigrok R W H, Brunner J. Low-pH induced conformational changes in viral fusion proteins: implications for the fusion mechanism. J Gen Virol. 1995;76:1541–1556. doi: 10.1099/0022-1317-76-7-1541. [DOI] [PubMed] [Google Scholar]
- 12.Gollins S W, Porterfield J S. pH-dependent fusion between the flavivirus West Nile and liposomal model membranes. J Gen Virol. 1986;67:157–166. doi: 10.1099/0022-1317-67-1-157. [DOI] [PubMed] [Google Scholar]
- 13.Grakoui A, Wychowski C, Lin C, Feinstone S M, Rice C M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guirakhoo F, Bolin R A, Roehrig J T. The Murray Valley encephalitis virus prM protein confers acid resistance to virus particles and alters the expression of epitopes within the R2 domain of E glycoprotein. Virology. 1992;191:921–931. doi: 10.1016/0042-6822(92)90267-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guirakhoo F, Heinz F X, Mandl C W, Holzmann H, Kunz C. Fusion activity of flaviviruses: comparison of mature and immature (prM-containing) tick-borne encephalitis virions. J Gen Virol. 1991;72:1323–1329. doi: 10.1099/0022-1317-72-6-1323. [DOI] [PubMed] [Google Scholar]
- 16.Guirakhoo F, Heinz F X, Kunz C. Epitope model of tick-borne encephalitis virus envelope glycoprotein E: analysis of structural properties, role of carbohydrate side chain, and conformational changes occurring at acidic pH. Virology. 1989;169:90–99. doi: 10.1016/0042-6822(89)90044-5. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez L D, Hoffman L R, Wolfsberg T G, White J M. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 18.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci USA. 1991;88:5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horvath C M, Lamb R A. Studies on the fusion peptide of a paramyxovirus fusion glycoprotein: roles of conserved residues in cell fusion. J Virol. 1992;66:2443–2455. doi: 10.1128/jvi.66.4.2443-2455.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura T, Ohyama A. Association between the pH-dependent conformational change of West Nile flavivirus E protein and virus-mediated membrane fusion. J Gen Virol. 1988;69:1247–1254. doi: 10.1099/0022-1317-69-6-1247. [DOI] [PubMed] [Google Scholar]
- 21.Klenk H-D, Garten W. Activation cleavage of viral spike proteins by host proteases. In: Wimmer E, editor. Cellular receptors for animal viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 241–280. [Google Scholar]
- 22.Lagging L M, Meyer K, Owens R J, Ray R. Functional role of hepatitis C virus chimeric glycoproteins in the infectivity of pseudotyped virus. J Virol. 1998;72:3539–3546. doi: 10.1128/jvi.72.5.3539-3546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy S, Todd S C, Maecker H T. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu Rev Immunol. 1998;16:89–109. doi: 10.1146/annurev.immunol.16.1.89. [DOI] [PubMed] [Google Scholar]
- 24.Lewis J, Balfe P, Arnold C, Kaye S, Tedder R S, McKeating J A. Development of a neutralizing antibody response during acute primary human immunodeficiency virus type 1 infection and emergence of antigenic variants. J Virol. 1998;72:8943–8951. doi: 10.1128/jvi.72.11.8943-8951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maertens G, Stuyver L. Genotypes and genetic variation of hepatitis C virus. In: Harrison T J, Zuckerman A J, editors. The molecular medicine of viral hepatitis. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1997. pp. 183–223. [Google Scholar]
- 26.Maidens C, Shotton C, Flint M, Dubuisson J, Almond J W, McKeating J A. Abstracts of the 5th International Meeting of Hepatitis C virus and Related Viruses, Molecular Virology and Pathogenesis 1998. 1998. Full length and truncated forms of the HCV E2 glycoprotein differ in both their antigenic conformation and in their ability to recognise their putative cellular receptors, abstr. O032. [Google Scholar]
- 27.Mason P W. Maturation of Japanese encephalitis virus glycoproteins produced by infected mammalian and mosquito cells. Virology. 1989;169:354–364. doi: 10.1016/0042-6822(89)90161-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuura Y, Ishii K, Aizaki H, Takigawa S, Tani H, Whitt M A, Miyamura T. Abstracts of the 5th International Meeting of Hepatitis C Virus and Related Viruses, Molecular Virology and Pathogenesis 1998. 1998. Characterization of pseudotype VSV possessing HCV envelope proteins, abstr. O006. [Google Scholar]
- 29.Matsuura Y, Suzuki T, Suzuki R, Sato M, Aizaki H, Saito I, Miyamura T. Processing of E1 and E2 glycoproteins of hepatitis C virus expressed in mammalian and insect cells. Virology. 1994;205:141–150. doi: 10.1006/viro.1994.1629. [DOI] [PubMed] [Google Scholar]
- 30.Michalak J-P, Wychowski C, Choukhi A, Meunier J-C, Ung S, Rice C M, Dubuisson J. Characterization of truncated forms of hepatitis C virus glycoproteins. J Gen Virol. 1997;78:2299–2306. doi: 10.1099/0022-1317-78-9-2299. [DOI] [PubMed] [Google Scholar]
- 31.Naim H Y, Roth M G. Basis for selective incorporation of glycoproteins into the influenza virus envelope. J Virol. 1993;67:4831–4841. doi: 10.1128/jvi.67.8.4831-4841.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowak T, Farber P M, Wengler G, Wengler G. Analyses of the terminal sequences of West Nile virus structural proteins and of the in vitro translation of these proteins allow the proposal of a complete scheme of the proteolytic cleavages involved in their synthesis. Virology. 1989;169:365–376. doi: 10.1016/0042-6822(89)90162-1. [DOI] [PubMed] [Google Scholar]
- 33.Oren R, Takahashi S, Doss C, Levy R, Levy S. TAPA-1, the target of an antiproliferative antibody, defines a new family of transmembrane proteins. Mol Cell Biol. 1990;10:4007–4015. doi: 10.1128/mcb.10.8.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paterson R G, Lamb R A. Ability of the hydrophobic fusion-related external domain of a paramyxovirus F protein to act as a membrane anchor. Cell. 1987;48:441–452. doi: 10.1016/0092-8674(87)90195-4. [DOI] [PubMed] [Google Scholar]
- 35.Pettersson R F. Protein localization and virus assembly at intracellular membranes. Curr Top Microbiol Immunol. 1991;170:67–106. doi: 10.1007/978-3-642-76389-2_3. [DOI] [PubMed] [Google Scholar]
- 36.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner A J, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 37.Ralston R, Thudium K, Berger K, Kuo C, Gervase B, Hall J, Selby M, Kuo G, Houghton M, Choo Q-L. Characterization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia viruses. J Virol. 1993;67:6753–6761. doi: 10.1128/jvi.67.11.6753-6761.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Randolph V B, Winkler G, Stollar V. Acidotropic amines inhibit proteolytic processing of flavivirus prM protein. Virology. 1990;174:450–458. doi: 10.1016/0042-6822(90)90099-d. [DOI] [PubMed] [Google Scholar]
- 39.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 931–959. [Google Scholar]
- 40.Roehrig J T, Johnson A J, Hunt A R, Bolin R A, Chu M C. Antibodies to Dengue 2 virus E-glycoprotein synthetic peptides identify antigenic conformation. Virology. 1990;177:668–675. doi: 10.1016/0042-6822(90)90532-v. [DOI] [PubMed] [Google Scholar]
- 41.Roehrig J T, Hunt A R, Johnson A J, Hawkes R A. Synthetic peptides derived from the deduced amino acid sequence of the E-glycoprotein of Murray valley encephalitis virus elicit antiviral antibody. Virology. 1989;171:49–60. doi: 10.1016/0042-6822(89)90509-6. [DOI] [PubMed] [Google Scholar]
- 42.Rosa D, Campagnoli S, Moretto C, Guenzi E, Cousens L, Chin M, Dong C, Weiner A J, Lau J Y N, Choo Q-L, Chien D, Pileri P, Houghton M, Abrignani S. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc Natl Acad Sci USA. 1996;93:1759–1763. doi: 10.1073/pnas.93.5.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santolini E, Migliaccio G, Monica N L. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J Virol. 1994;68:3631–3641. doi: 10.1128/jvi.68.6.3631-3641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schnell M J, Buonocore L, Kretzchmar E, Johnson E, Rose J K. Foreign glycoproteins expressed from recombinant vesicular stomatitis viruses are incorporated efficiently into virus particles. Proc Natl Acad Sci USA. 1996;93:11359–11365. doi: 10.1073/pnas.93.21.11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selby M J, Glazer E, Masiarz F, Houghton M. Complex processing and protein:protein interactions in the E2:NS2 region of HCV. Virology. 1994;204:114–122. doi: 10.1006/viro.1994.1515. [DOI] [PubMed] [Google Scholar]
- 46.Selby M J, Choo Q-L, Berger K, Kuo G, Glazer E, Eckart M, Lee C, Chien D, Kuo C, Houghton M. Expression, identification and subcellular localization of the proteins encoded by the hepatitis C viral genome. J Gen Virol. 1993;74:1103–1113. doi: 10.1099/0022-1317-74-6-1103. [DOI] [PubMed] [Google Scholar]
- 47.Spaete R R, Alexander D A, Rugroden M E, Choo Q-L, Berger K, Crawford K, Kuo C, Leng S, Lee C, Ralston R, Thudium K, Tung J W, Kuo G, Houghton M. Characterization of the hepatitis C virus E2/NS1 gene product expressed in mammalian cells. Virology. 1992;188:819–830. doi: 10.1016/0042-6822(92)90537-y. [DOI] [PubMed] [Google Scholar]
- 48.Stadler K, Allison S L, Schalich J, Heinz F X. Proteolytic activation of tick-borne encephalitis virus by furin. J Virol. 1997;71:8475–8481. doi: 10.1128/jvi.71.11.8475-8481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stegmann T, Nir S, Wilschut J. Membrane fusion activity of influenza virus. Effects of gangliosides and negatively charged phospholipids in target liposomes. Biochemistry. 1989;28:1698–1704. doi: 10.1021/bi00430a041. [DOI] [PubMed] [Google Scholar]
- 50.White J M. Viral and cellular membrane fusion proteins. Annu Rev Physiol. 1990;52:675–697. doi: 10.1146/annurev.ph.52.030190.003331. [DOI] [PubMed] [Google Scholar]
- 51.Zhang L, Ghosh H P. Characterization of the putative fusogenic domain in vesicular stomatitis virus glycoprotein G. J Virol. 1994;68:2186–2193. doi: 10.1128/jvi.68.4.2186-2193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Y, König M, Hobom G, Neumeier E. Membrane-anchored incorporation of a foreign protein in recombinant influenza virions. Virology. 1998;246:83–94. doi: 10.1006/viro.1998.9169. [DOI] [PubMed] [Google Scholar]