Abstract
Serum-free mouse embryo (SFME) cells are a neural stem cell line that is dependent upon epidermal growth factor (EGF) for survival. Removal of EGF results in the G1 arrest and apoptosis of SFME cells. We have shown that the expression of simian virus 40 large T antigen in SFME cells blocks apoptosis and allows cell survival and division in the absence of EGF. Therefore the presence of T antigen abrogates the EGF requirement. The steady-state levels of p53, p21, and mdm-2 do not increase as SFME cells undergo apoptosis upon EGF withdrawal. Furthermore, the amino-terminal 136 amino acids (N136) of T antigen are sufficient to block death and to promote proliferation in the absence of EGF, while the carboxy-terminal fragment (C251–708), which contains the p53 binding site, is unable to block death. Taken together, these data suggest that SFME cells deprived of EGF undergo p53-independent apoptosis. Mutations that disrupt either the J domain or Rb family binding abolish the ability of T antigen to block SFME cell apoptosis and to promote cell growth. We conclude that T antigen must act on one or more members of the Rb family to inhibit SFME cell apoptosis.
Tissue homeostasis is maintained as a balance by the growth and the death of cells. Proliferation, growth arrest, and apoptosis must occur in an ordered, scheduled manner, proceeding at defined times during the development and life span of an organism. Disruption of these events offsets the balance, and homeostasis is destroyed. The disregulation of homeostasis due to the loss of proliferative or apoptotic control contributes to tumorigenesis.
Apoptosis is a tightly regulated process that occurs in response to a cascade of intracellular signals. Required for the normal development of multicellular organisms, apoptosis can proceed in response to numerous and diverse stimuli, for example UV radiation (9, 20, 28) or the removal of a required growth factor such as nerve growth factor (30; reviewed in reference 10). Apoptosis proceeds through a multitude of signal transduction pathways that are the subject of intense investigation. Much effort has gone into distinguishing pathways that require the activation of the tumor suppressor p53 and those that proceed in a p53-independent manner. Apoptosis also serves as a defense mechanism for the cell, occurring in response to foreign invaders such as viruses. In response, viruses have evolved mechanisms to manipulate cellular apoptosis. There are numerous examples of gene products from diverse viral species that act to induce or inhibit apoptosis (reviewed in reference 47). The large T antigen of simian virus 40 (SV40) disrupts homeostasis by modulating cellular proliferation and apoptosis. The expression of SV40 large T antigen in transgenic mice induces, among other things, choroid plexus tumors (45) and intestinal hyperplasia (22).
Mouse embryo cells cultured in basal nutrient medium supplemented with serum eventually undergo growth crisis and senescence. The resulting established cell lines are often genomically altered, immortalized, and/or tumorigenic, displaying gross chromosomal abnormalities (48). Serum-free mouse embryo (SFME) cells are a preastrocyte cell line (38, 41) that was derived and cultured in a basal nutrient medium in which serum is replaced by a defined array of growth factors and other supplements. These procedures have allowed the extended proliferation of SFME cells with no detectable growth crisis or gross genomic alteration. In addition, they are nontumorigenic in syngeneic or athymic mice (26). Interestingly, SFME cells are growth inhibited by the presence of serum and arrest in the G1 phase of the cell cycle (36). Epidermal growth factor (EGF) is required for SFME cell survival and proliferation. The removal of EGF results in G1 arrest and apoptosis (37). The SFME system offers an opportunity to distinguish the signaling pathways that mediate growth arrest and apoptosis.
Established cell lines are typically cultured in basal nutrient medium supplemented with serum. It has been shown that cells transformed by T antigen are able to grow in concentrations of serum that will not support the survival of the parental cell line (16). T antigen also eliminates the growth factor requirement of established cell lines such as C3H10T1/2 and NIH 3T3 cells grown in serum-free media (7, 8). T antigen interacts with a number of cellular factors including the cellular tumor suppressors p53 and pRb, which are involved in G1 arrest and apoptosis. In accordance with its role as a cellular tumor suppressor, p53 is a key component of many cell death pathways (27, 50). It acts as one of the primary elements involved in generating the signal to arrest in G1 (20, 21, 23) and to undergo apoptosis (27, 50). Similarly, pRb plays a major role in G1 checkpoint control (18, 34) and recently has been linked to apoptosis with the discovery of a consensus caspase cleavage site in its C terminus (46). The transforming potential of SV40 correlates with the ability of T antigen to interact with pRb and p53 (12, 42). Therefore, we used T antigen as a tool to identify the cellular factors involved in SFME apoptosis.
MATERIALS AND METHODS
Cell culture and extracts.
Detailed procedures for the initiation and culture of SFME cells have been published (24, 25). The medium was a 1:1 mixture of Dulbecco’s modified Eagle’s medium and Ham’s F12 (17, 29) supplemented with 15 mM HEPES (pH 7.4), 1.2 g of sodium bicarbonate per liter, penicillin (120 mg/ml), streptomycin (200 mg/ml), and ampicillin (25 mg/ml) (F/D). Cells were cultured in medium supplemented with 10 mg of insulin per ml, 10 mg of transferrin per ml, 10 mg of high-density lipoprotein per ml, 50 ng of EGF per ml, and 10−8 M sodium selenite, on dishes or flasks precoated with 10 mg of fibronectin per ml. The cells were maintained at 37°C in a 5% CO2–95% air atmosphere. Growth factors were obtained from Sigma Chemical Co. (St. Louis, Mo.). For the preparation of extracts, the cells were scraped into F/D and centrifuged. The cell pellet was resuspended in lysis buffer (50 mM Tris [pH 8.0], 5 mM EDTA, 150 mM NaCl, 0.5% Nonidet P-40) containing a cocktail of protease inhibitors (1 mg of leupeptin per ml, 0.7 mg of pepstatin per ml, 2.5 mg of E64 per ml, 50 ng of phenylmethylsulfonyl fluoride per ml, 1 mg of aprotinin per ml, 10 mg of soybean trypsin inhibitor per ml, 10 mg of tolylsulfonyl phenylalanyl chloromethyl ketone [TPCK] per ml, 1 mM EDTA, 1 mM dithiothreitol). Samples were incubated on ice for 20 min, vortexed, and centrifuged for 30 min at 20,800 × g at 4°C. The protein concentration was determined by the Bradford assay (39).
Transfection of SFME cells.
SFME cells were transfected by a calcium phosphate method (2) with minor adjustments. Two hours prior to the start of the procedure, F/D (supplemented) plus 10% charcoal-stripped calf serum (Sigma) was added to a 10-cm dish of cells that were at 80 to 90% confluent. The 10% charcoal-stripped calf serum allows the proliferation of SFME cells provided that they are supplemented with all of the required growth factors, and it seems to protect the cells from the CaCl2, which tends to kill them otherwise. Plasmid DNA (10 mg) and 20 mg of carrier DNA (fish sperm DNA [Sigma]) were ethanol precipitated, prepared, and added to the cells as described previously (2). If the plasmid did not contain a G418 resistance gene, the cells were cotransfected with 2 mg of a plasmid containing the G418 resistance gene. The cells were incubated in the precipitate for 4 to 6 h, at which time the medium was replaced with F/D (supplemented) plus 10% charcoal-stripped calf serum. The following morning, the cells were split into three dishes, allowing them to recover. When they appeared healthy, selection for G418 resistance was applied by adding G418 (Sigma) (its concentration varies and is optimized for each lot) or by placing the cells in medium that contained all of the required factors except EGF. The plasmids used were pRSVbneoN136 (42); pSVneoT, which contains the G418 resistance gene and wild-type T antigen, and PKO-NEO, which expresses the G418 resistance gene (both kindly provided by Edward Harlow); C251-708 (4); and pRSVbneo3213, pRSVbneoN136-3213, pRSVb1135, pRSVb1135-1137, and pRSVbneo5110 (42).
Immunoprecipitation.
Equal concentrations of lysates were used for the immunoprecipitation protocol, and the volumes were adjusted to 500 ml with lysis buffer. To each sample, 50 ml of 50% protein A-Sepharose beads (Pharmacia) was added and the samples were rocked at 4°C for 1 h. The samples were centrifuged at 4°C for 20 min at 20,800 × g, and the supernatants were transferred to fresh tubes containing the appropriate antibody (rabbit anti-p53 antibody was a gift from the Tevethia Laboratory, p21 antibody was a gift from the Beach Laboratory, and mdm-2 antibody was a gift from the Levine laboratory). The samples were rocked for 30 min at 4°C. A 50-ml volume of 50% protein A-Sepharose was added, and the incubation was continued for an additional 15 min before the immune complex was pelleted by centrifugation at 20,800 × g for 20 min at 4°C. The pellet was washed three times with 1 ml of SNNTE (5% Sucrose, 1% Nonidet P-40, 0.5 M NaCl, 50 mM Tris [pH 7.5], 5 mM EDTA) and once with 1 ml of NTE (50 mM NaCl, 1 mM Tris [pH 7.5], 5 mM EDTA) by resuspending the pellet in 1 ml of NTE, vortexing the sample for 10 s, and centrifuging it at 20,800 × g for 2 min. The pellet was resuspended in 20 ml of 5× sample buffer (1.54 g of dithiothreitol, 2 g of sodium dodecyl sulfate, 8 ml of 1 M Tris [pH 6.8], 10 ml of glycerol, 0.003% bromphenol blue) boiled for 5 min, and the proteins were separated on a sodium dodecyl sulfate–8 to 12.5% polyacrylamide gel. The proteins were electrophoresed at 80 to 150 V for 1 to 4 h. The results were analyzed by immunoblotting.
Immunoblot analysis.
Immediately following electrophoresis, the proteins were transferred to a polyvinylidene difluoride membrane (Millipore). They were transferred overnight at 18 V in Western transfer buffer (3.03 g of Tris base, 14.4 g of glycine, and 200 ml of methanol per liter) at 4°C. Following transfer, the membrane was placed in blocking buffer (phosphate-buffered saline, 10% powdered low-fat milk [Carnation]) for 1 h on a rocking platform at room temperature. The blocking buffer was decanted, and the membrane was placed in blotting buffer (1 mM EDTA, 10 mM Tris, 100 mM NaCl, 0.1% powdered low-fat milk) containing the appropriate primary antibody. The following concentrations of primary antibodies were used: anti-T antigen PAb 416 ascitic fluid, 1:2,500; anti-T-antigen PAb 419 ascitic fluid, 1:2,500; anti-T antigen 901 hybridoma supernatant, 1:200; rabbit anti-p53, 1:2,500; mouse anti-p21 ascitic fluid (gift from David Beach), 1:1,000; anti-mdm-2 (gift from Arnold Levine) ascitic fluid, 1:1,000. The membrane was incubated in the primary-antibody solution for 1 h on a rocking platform at room temperature. The primary antibody solution was decanted, and the membrane was washed in rinse buffer (1 mM EDTA, 10 mM Tris, 100 mM NaCl, 1% Tween 20) for 30 min, changing the rinse buffer every 5 min. The membrane was transferred to a secondary-antibody solution composed of blotting buffer and a 1:30,000 dilution of the mouse Fc portion of immunoglobulin G conjugated to horseradish peroxidase (Sigma), or rabbit immunoglobulin G conjugated to horseradish peroxidase. The membrane was incubated for 1 h on a rocking platform at room temperature. The secondary-antibody solution was decanted and the membranes were washed as before. The proteins were detected with the Amersham ECL kit for chemiluminescence.
RESULTS
SFME cells are dependent upon EGF for their survival and proliferation.
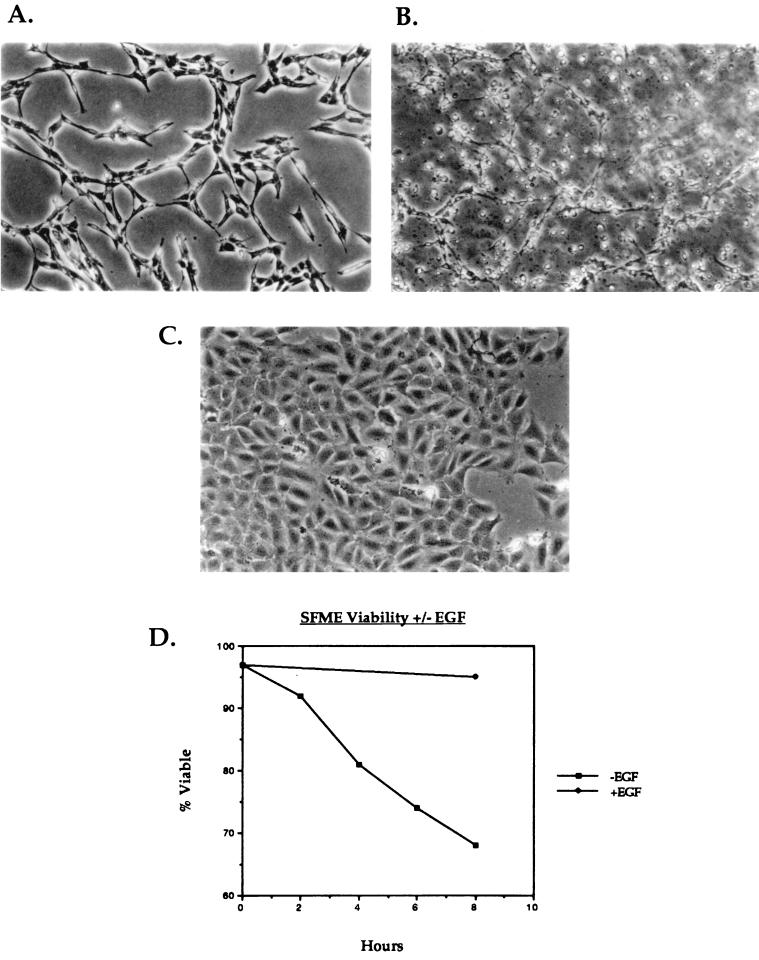
SFME cells are routinely maintained in serum-free medium supplemented with defined growth factors (24–26). Under these conditions, the cells proliferate with no detectable apoptosis and display an extended, fibroblast-like morphology (Fig. 1A and D). Withdrawal of EGF from the medium leads to apoptosis characterized by morphological changes (Fig. 1B and D) as well as biochemical changes in the cells (24). When placed in serum-containing medium, the cells arrest in the G1 phase of the cell cycle and adopt an altered morphology that appears epithelial (Fig. 1C). A trypan blue viability assay (Fig. 1D) reveals that by as early as 8 h after the removal of EGF, >30% of the cells are dead, demonstrating the strict dependence of SFME cells on EGF.
FIG. 1.
SFME cells require EGF for survival and growth. (A to C) Photomicrographs of SFME cells growing in complete serum-free medium (A), after 10 h in serum-free medium that lacks EGF (B), and after 24 h in basal nutrient medium supplemented with 10% FBS (C). (D) Results of a trypan blue viability assay on SFME cells in the presence and absence of EGF.
p53 levels do not increase as SFME cells undergo apoptosis.
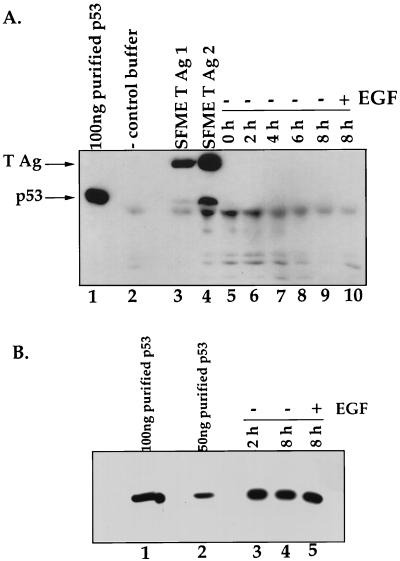
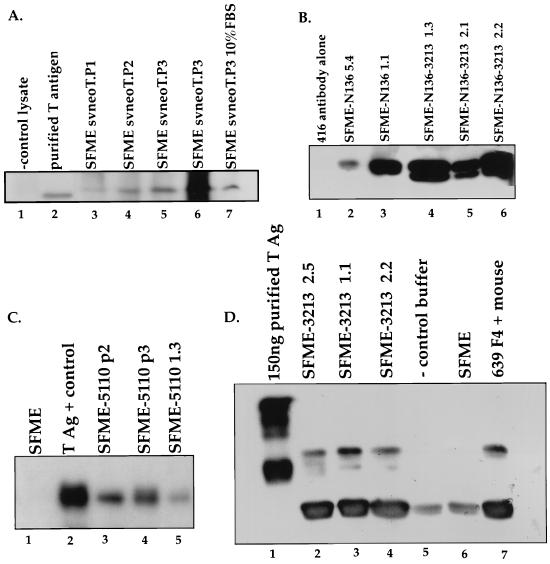
p53-dependent apoptosis pathways are often accompanied by significant increases in the steady-state level of p53 (14, 20). Therefore, we examined the steady-state levels of p53 in SFME cells as they underwent apoptosis. Lysates were generated from cells dividing in the presence of EGF and from cells that were harvested at various times after the removal of EGF. In our first experiment, 800 mg of total protein from each lysate was immunoprecipitated by using an antibody directed against p53 and compared by Western blot analysis probed with antibodies directed against p53 and T antigen (Fig. 2A). We were unable to detect p53 in lysates from cells that were actively growing in the presence of EGF or from cells that were undergoing apoptosis at any of the indicated time points after the removal of EGF, suggesting that under both conditions, p53 levels are relatively low. In contrast, p53 was detectable in lysates from SFME cells that express T antigen, suggesting that the p53 expressed in SFME cells binds and is stabilized by T antigen.
FIG. 2.
(A) Steady-state levels of p53 remain unchanged during SFME apoptosis. Immunoblot analysis of the p53 levels in SFME cells in the presence and absence of EGF is shown. Cells were harvested at 0, 2, 4, 6, and 8 h after the removal of EGF (lanes 5 to 9). Cells that had not been deprived of EGF were harvested 8 h after the medium was replaced (lane 10). SFME T Ag 1 (lane 3) and SFME T Ag 2 (lane 4) are SFME cell lines that stably express T antigen (T Ag) and are grown in the absence of EGF. For all of the above, 800 mg of total protein was analyzed. Lane 1 contains 100 ng of immunopurified p53, and lane 2 contains antibody alone. Samples were immunoprecipitated with a rabbit anti-p53 antibody, and the immunoblot was probed with a p53 antibody and a T-antigen antibody. (B) Immunoblot analysis of p53 levels in lysate from SFME cells that were harvested at 2 and 8 h after the removal of EGF (lanes 3 and 4). Cells that had not been deprived of EGF were harvested 8 h after being fed (lane 5). A 5-mg portion of total protein was analyzed for each sample. Lane 1 contains 100 ng of immunopurified p53, and lane 2 contains 50 ng of immunopurified p53. The Western blot was probed with a rabbit anti-p53 antibody.
When we examined 5 mg of total protein for each sample by immunoblotting (Fig. 2B), we detected p53 in all of the samples. This low level of p53 remains constant as apoptosis proceeds.
p21 and mdm-2 levels do not increase as SFME cells undergo apoptosis.
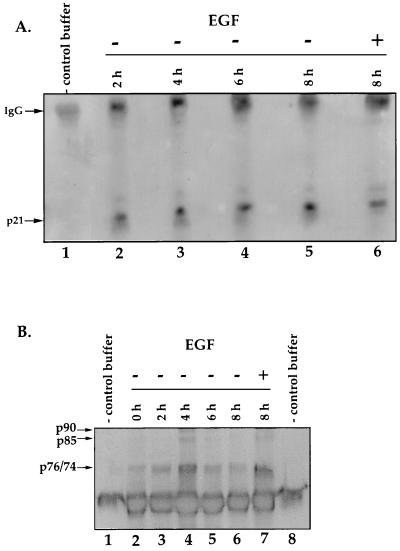
Next, we examined the levels of p21 and mdm-2, two downstream effectors of p53 that are commonly involved in apoptosis. The p21 gene is a p53-responsive gene whose expression levels are known to increase when apoptosis proceeds in a p53-dependent manner (13). Lysates were prepared from cells cultured in medium containing EGF and from cells that were harvested at 2, 4, 6, and 8 h after the removal of EGF. Samples were immunoprecipitated with a monoclonal antibody directed against p21 and analyzed by immunoblotting (Fig. 3A). p21 levels remain unchanged as SFME cells undergo apoptosis.
FIG. 3.
(A) Steady-state levels of p21 and mdm-2 do not increase during SFME apoptosis. An immunoblot of p21 levels from lysates generated from SFME cells that were harvested at 0, 2, 4, 6, and 8 h after the removal of EGF (lanes 2 to 6) is shown. Cells that were growing in the presence of EGF were harvested 8 h after the medium was replaced (lane 6). Lane 1 contains antibody alone. A 5-mg portion of total protein for each sample was immunoprecipitated with a monoclonal p21 antibody. The blot was probed with the same antibody. IgG, immunoglobulin G. (B) Immunoblot analysis of mdm-2 levels from lysates that were generated from SFME cells harvested at 0, 2, 4, 6, and 8 h after the removal of EGF (lanes 2 to 6). Cells growing in the presence of EGF were harvested 8 h after the medium was replaced (lane 7). Lanes 1 and 8 are antibody-alone controls. A 75-mg portion of total protein from each sample was immunoprecipitated with a monoclonal mdm-2 antibody, and the blot was probed with the same antibody.
The mdm-2 gene is a p53-responsive gene that inhibits the transactivation and repression functions of the p53 protein (5, 31, 33, 49). mdm-2 levels often increase during p53-dependent apoptosis (6). We examined the steady-state level of mdm-2 present as cells underwent apoptosis (Fig. 3B) and did not see a steady increase in mdm-2 levels. The small increase seen in the mdm-2 level at 4 h was not reproducible. Therefore, as SFME cells die, p53, p21, and mdm-2 protein levels do not increase. Taken together, these data strongly suggest that p53 is not activated in cells that are deprived of EGF. This suggests that SFME cells undergo p53-independent apoptosis.
SV40 T antigen blocks the apoptosis of SFME cells in the absence of EGF.
T antigen induces and inhibits apoptosis in various systems (1, 3, 19). To determine if SV40 T antigen could alleviate the EGF requirement of SFME cells, a plasmid expressing T antigen was transfected into SFME cells. Transfections were performed in duplicate sets with each containing a mock-transfected dish, a dish that received the G418 resistance plasmid, and a dish that received T antigen cotransfected with the G418 resistance gene. In one set, we removed EGF from the medium and directly selected for the ability to grow in the absence of EGF. Of the 15 colonies that arose (Table 1), 5 were picked and expanded into cell lines. All five cell lines grew in the absence of EGF (see Fig. 5B) as well as in the presence of 10% fetal bovine serum (FBS), and all five cell lines expressed T antigen (see Fig. 6A). The second set of dishes underwent G418 selection in the presence of EGF, and surviving colonies were picked and expanded into cell lines. T-antigen expression was confirmed by immunoblotting. The cell lines were then subjected to selection in the absence of EGF. The results obtained from both rounds of this selection process always correlated. Every cell line expressing T antigen was able to grow in the absence of EGF at a division rate comparable to that of parental cells plated in the presence of EGF. In addition, T-antigen expression allowed cells to overcome the growth-inhibitory effects of serum, supporting proliferation in the presence of 10% FBS.
TABLE 1.
Colony formation when SFME cells are transfected with T antigen or mutants of T antigen and subjected to selection in the absence of EGF and in the presence of G418
| DNAa | No. of colonies obtained
|
No. of colonies expressing proteind | No. of colonies growing without EGFe | |
|---|---|---|---|---|
| Without EGFb | With G418c | |||
| T antigen | 15 | 20 | 5 | 5 |
| N136 | 20 | 31 | 10 | 10 |
| 3213 | 0 | 14 | 8 | 0 |
| N136–3213 | 0 | 26 | 3 | 0 |
| 1135 | 0 | 15 | 6 | 0 |
| 1137–1135 | 0 | 12 | 5 | 0 |
| 5110 | 0 | 18 | 7 | 0 |
| PKO-NEO | 0 | 30 | 0 | |
This column indicates the plasmid that was transfected into the cell. PKO-NEO is a G418 resistance plasmid.
Number of colonies obtained after transfected cells were placed in medium that did not contain EGF.
Number of colonies obtained when transfected cells were placed in medium containing G418.
Subset of colonies from the G418 selection that were examined for T-antigen expression.
Number of T-antigen colonies that were able to grow in the absence of EGF.
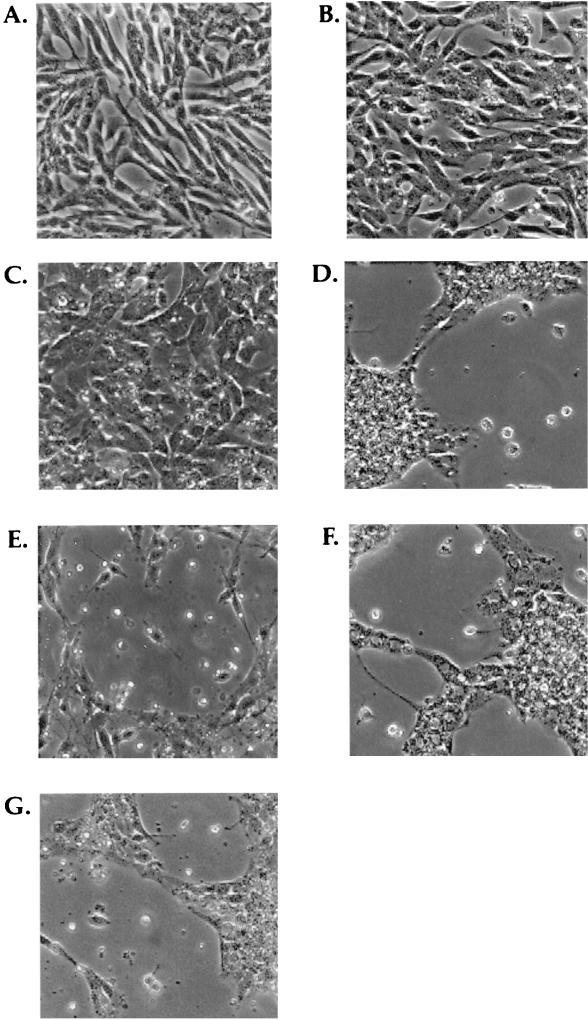
FIG. 5.
Photomicrographs of T-antigen mutants cultured in the absence of EGF. (A) SFME cells growing in complete serum-free medium; (B) SFME T Ag cells growing for 28 h in serum-free medium that lacks EGF; (C) SFME-N136 cells growing for 29 h in serum-free medium that lacks EGF; (D) SFME cells that were mock transfected with carrier DNA cultured for 8 h in serum-free medium that lacks EGF; (E) SFME-1135 cells cultured for 10 h in serum-free medium that lacks EGF; (F) SFME-3213 cells cultured for 8 h in serum-free medium that lacks EGF; (G) SFME-5110 cells cultured for 9 h in serum-free medium that lacks EGF.
FIG. 6.
(A) The ability of T-antigen mutants to block apoptosis is independent of the level of protein expression. Immunoblots of T antigen from SFME-T Ag cell lines are shown. Lanes: 1, 901 antibody alone; 2, 50 ng of immunopurified T antigen; 3 to 6, SFME-T Ag cell lines that grow in the absence of EGF (it is unclear why purified T Ag migrated differently from the T Ag expressed in SFME cells); lane 7, SFME T-antigen cell line that grows in the presence of 10% FBS. Samples were immunoprecipitated, and the blot was probed with 901 antibody. (B) Western blot of N136 and N136–3213 from SFME cell lines. Lanes: 1, 416/419 antibody alone; 2 and 3, SFME-N136 cell lines that grow in the absence of EGF; 4 to 6, SFME-N136–3213 cell lines. The samples were immunoprecipitated, and the blot was probed with 416 and 419 monoclonal T-antigen antibodies. (C) Immunoblot of 5110 from SFME cell lines. Lanes: 1, lysate from SFME parental cells; 2, 100 mg of immunopurified T antigen; 3 to 5, SFME-5110 cell lines. The Western blot was probed with 901 antibody. (D) Immunoblot of 3213 from SFME cell lines. Lanes: 1, 150 ng of purified T antigen; 2 to 4, SFME 3213 cell lines; 5, negative control antibody alone; 6, lysate from SFME parental cells; 7, lysate generated from intestinal cells of a T-antigen-positive transgenic mouse. Samples were immunoprecipitated, and the Western blot was probed with 901 antibody. Again, purified T antigen migrated differently from SFME-expressed T antigen.
In comparison, cells that were transfected with the G418 resistance plasmid alone (PKO-NEO) were subjected to the same selection scheme (Table 1). When selection in the absence of EGF was applied, no colonies were observed. In fact, we have never seen spontaneous colony formation in the mock-infected dishes or in the dishes that received the G418 resistance gene alone under selection in the absence of EGF. Under G418 selection, 30 colonies were obtained. When expanded into cell lines, none were able to grow in 10% FBS or to survive in the absence of EGF.
The J domain and Rb binding motif of T antigen are required to block apoptosis.
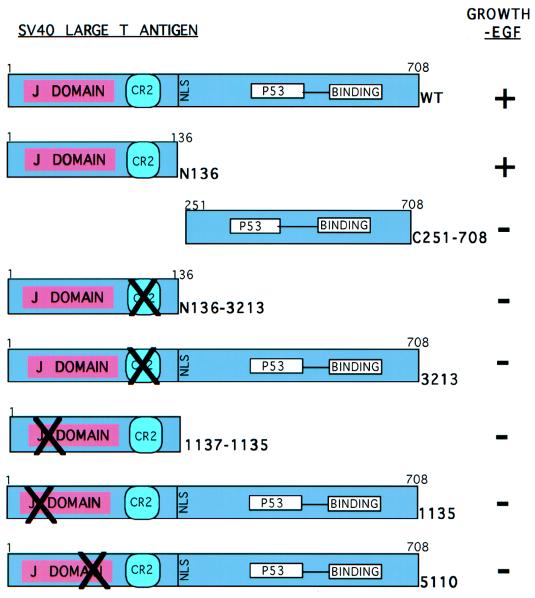
We determined that T antigen alleviates the EGF requirement of SFME cells. We next wanted to determine which activity of T antigen was responsible for this effect. We transfected SFME cells with plasmids that express mutants of T antigen defective in each of the known transforming activities. First, we examined an amino-terminal mutant termed N136, which consists of the first 136 amino acids of T antigen and includes the J domain, conserved region 2 (CR2), and nuclear localization signal (Fig. 4). As shown in Table 1, SFME-N136 cell lines grew in the absence of EGF (Fig. 5C) as well as in the presence of 10% serum. As with the SFME-T-antigen cell lines, SFME-N136 cells displayed a healthy morphology characteristic of the parental line. Protein expression was confirmed by immunoblot analysis (Fig. 6B).
FIG. 4.
SV40 T-antigen mutants transfected into SFME cells. T-antigen mutants were transfected into SFME cells, and the ability of each to confer survival and allow growth in the absence of EGF was determined (shown on the right [data are given in Table 1]). Wt is full-length wild-type T antigen. N136 is the first 136 amino acids of T antigen, defective for the ability to bind p53. C251–708 is composed of amino acids 251 to 708, defective for the J domain and for the ability to interact with the pRb family. N136–3213 is N136 with E107K and E108K. 3213 is full-length T antigen with E107K and E108K, defective for the ability to interact with the pRb family. 1135 (D17–27) and 5110 (D44N) mutations disrupt the J domain.
Next we tested the mutant C251–708, expressing amino acids 251 to 708 of T antigen (Fig. 4). This mutant lacks the J domain, CR2, the nuclear localization signal, and DNA binding activity yet retains the ability to bind p53 (4). When transfected cells were plated in the absence of EGF, no colonies were obtained. When transfected cells were placed under G418 selection, no colonies were obtained and protein expression was confirmed (data not shown), but when these cells were transferred to medium that did not contain EGF, all of the cells died. Thus, N136 allows the survival and growth of SFME cells cultured in the absence of EGF while C251–708 fails to block apoptosis. This supports the conclusion that SFME cells deprived of EGF undergo p53-independent apoptosis.
N136 contains two genetic elements that are required for the transformation of various cell types, the J domain and con served region 2 (CR2) (43). To determine which region of T antigen was required to allow growth in the absence of EGF, we tested mutants with mutations affecting either the J domain or CR2 in the context of a full-length T antigen as well as in the context of N136 (Fig. 4).
CR2 governs the interaction of T antigen with the pRb family of proteins. To determine if an interaction between T antigen and a pRb family member is required to allow SFME cell growth in the absence of EGF, we examined the effect of the 3213 mutation (E107K, E108K), which disrupts that interaction (42). Table 1 shows that SFME cells that expressed 3213 or the double mutant N136–3213 failed to grow in the absence of EGF. Figure 5F shows an SFME-3213 cell line undergoing apoptosis upon removal of EGF. We conclude that T antigen must act on one or more members of the pRb family of proteins to block apoptosis and support growth in the absence of EGF. N136–3213 and 3213 protein expression was confirmed by immunoblot analysis (Fig. 6B and D).
To examine the role of the J domain in the ability of T antigen to abrogate the EGF requirement, mutants 1135 and 5110 were examined (Fig. 4). The 1135 mutation (D17–27) disrupts the J domain. SFME cells were transfected with 1135 and with 1135 carried in an amino-terminal mutant, 1137–1135. When cells transfected with 1135 and 1137–1135 were plated in medium that did not contain EGF, no colonies were obtained (Table 1) and the cells underwent apoptosis (Fig. 5E). Colonies were obtained when transfected cells were placed under G418 selection, cell lines were generated, and protein expression was confirmed by immunoblotting (data not shown). These cells failed to grow in the absence of EGF.
Mutation 5110 (D44N) alters the conserved HPDKGG motif within the J domain. When cells transfected with 5110 were plated in medium that did not contain EGF, no cells survived (Table 1, Fig. 5G). Colonies were obtained when transfected cells were placed under G418 selection, cell lines were generated, and 5110 protein expression was confirmed by immunoblot analysis (Fig. 6C). These cells failed to grow in the absence of EGF. Therefore, mutants of T antigen that contain disruptions in the J domain or the pRb family binding motif are defective in the ability to block apoptosis.
DISCUSSION
Apoptosis is essential for normal development and for maintaining tissue homeostasis. At predetermined points during development as well as throughout the life span of the organism, cells are programmed to die. Failure to do so sometimes contributes to tumorigenesis. In addition to being a required process in development, apoptosis exists as a defense mechanism for the cell. In response to a variety of environmental as well as physiological insults, cells activate their death programs. Consequently, many viruses have evolved mechanisms which allow them to target and manipulate cellular apoptosis. SV40 large T antigen induces apoptosis in a variety of tissues in transgenic mice (1, 3). In this report, we show that T antigen can also block apoptosis.
SFME cells were derived and are maintained in serum-free medium supplemented with EGF and other growth factors. Although they have been passaged in culture for years, SFME cells have not undergone a detectable growth crisis and exhibit karyotypic stability (15). The presence of serum is growth restrictive and results in G1 arrest. When EGF is withdrawn, SFME cells also arrest in the G1 phase of the cell cycle and subsequently undergo apoptosis. By 8 h after the removal of EGF, ∼30% of the cells are dead, and by 48 h after the removal of EGF, ∼90% of the cells are dead (35), demonstrating a strict requirement for EGF and the rapid apoptosis that proceeds in its absence. We have shown that the expression of T antigen abrogates the EGF requirement, allowing the survival and division of SFME cells. T antigen also overcomes the growth-inhibitory effects of serum, since SFME cells that express T antigen grow in the presence of 10% FBS without additional supplements.
We are investigating the mechanism by which T antigen blocks apoptosis. We began by examining which cellular factors are targeted by T antigen in SFME cells. In doing so, we showed that the apoptosis of SFME cells occurs in a p53-independent manner. p53, p21, and mdm-2 levels do not increase as SFME cells undergo apoptosis. Furthermore, N136 is capable of blocking apoptosis whereas C251–708, which binds p53, is not. We conclude that an interaction between T antigen and p53 is not required for the ability of T antigen to block apoptosis.
Our data shows that T antigen must interact with one or more members of the pRb family of proteins to block apoptosis and to allow growth in the absence of a required growth factor. Mutants that have the pRb family binding site disrupted, such as 3213 and N136–3213, are defective for the ability to block apoptosis. The role of the pRb family in SFME cell death is not yet known. However, it is known that SFME cells are arrested in the G1 phase of the cell cycle before dying. The pRb family of proteins are major components involved in G1 checkpoint control and are mediators of G1 arrest (32). It is conceivable that their role is to arrest the cells in G1, allowing apoptosis to proceed.
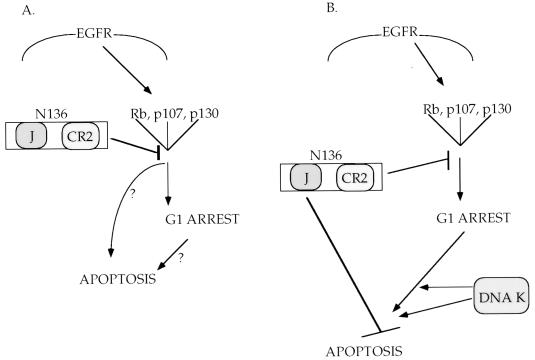
The J domain of T antigen is also required to block SFME cell death. Two possibilities exist: the J domain is targeting an independent cellular factor (Fig. 7B), or J domain function is required for action on the pRb family (Fig. 7A). The J domain of T antigen functionally inactivates the pRb family of proteins (44, 51). We have shown that the J domain must act in cis with the pRb family binding motif to transform cells (42). Similarly, other reports have shown that the J domain and CR2 must function in cis to activate E2F (11, 40). Alternatively, Rb may act as a growth arrest and/or death factor, functioning to promote the G1 arrest and/or apoptosis of SFME cells deprived of EGF. In this instance, it is conceivable that T antigen functions to inactivate Rb, allowing SFME cell survival and division in the absence of EGF. Further work is required to discern the role of the pRb family in SFME cell signaling.
FIG. 7.
Models for the requirement of both the J domain and pRb family binding motif of T antigen to block apoptosis. (A) In the absence of EGF, one or more members of the Rb family function to induce both the G1 arrest and apoptosis of SFME cells. T antigen binds and acts on one or more members of the Rb family, blocking apoptosis and allowing proliferation in the absence of EGF. The J domain and Rb binding motif of T antigen are sufficient to block apoptosis and allow SFME cell proliferation in the absence of EGF. (B) An alternate hypothesis is that while one or members of the Rb family are involved in G1 arrest, it is some other cellular factor (termed DNA K here) that induces apoptosis in the absence of EGF. T antigen must interact with both (sets of) factors, the Rb family through CR2 and DNA K through the J domain, to allow SFME cell survival and proliferation in the absence of EGF. EGFR, EGF receptor.
ACKNOWLEDGMENTS
We thank Arnold Levine, David Beach, and Mary J. Tevethia for providing us with antibodies. We thank Mary J. Tevethia for providing us with plasmid C251–708 and Edward Harlow for providing us with plasmids pSVneoT and PKO-NEO. We thank Angela Helmrich for help with and advice on cell culture. We also thank Jai Vartikar, Kris Sachsenmeier, and Chris Sullivan for comments and helpful discussions in the preparation of the manuscript. We thank Tom Harper for assistance with the figures.
This work was supported by NIH grant CA40586 to James M. Pipas and a grant from the Nihon Kefir Corporation to David Barnes.
REFERENCES
- 1.Allemand I, Grimber G, Kornpost M, Bennoun M, Molina T, Briand P, Joulin V. Compensatory apoptosis in response to SV40 large T antigen expression in the liver. Oncogene. 1995;11:2583–2590. [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994–1997. [Google Scholar]
- 3.Bowman T, Symonds H, Gu L, Yin C, Oren M, Van Dyke T. Tissue-specific inactivation of p53 tumor suppression in the mouse. Genes Dev. 1996;10:826–835. doi: 10.1101/gad.10.7.826. [DOI] [PubMed] [Google Scholar]
- 4.Cavander J F, Conn A, Epler M, Lacko H, Tevethia M J. Simian virus 40 large T antigen contains two independent activities that cooperate with a ras oncogene to transform rat embryo fibroblasts. J Virol. 1995;69:923–934. doi: 10.1128/jvi.69.2.923-934.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, Lin J, Levine A J. Regulation of transcription functions of the p53 tumor suppressor by the mdm-2 oncogene. Mol Med. 1995;1:142–152. [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Wu X, Lin J, Levine A J. mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol Cell Biol. 1996;16:2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiang L C, Silnutzer J, Pipas J M, Barnes D W. Methods for serum-free culture of epithelial and fibroblastic cells. New York, N.Y: Alan R. Liss, Inc.; 1984. pp. 265–276. [Google Scholar]
- 8.Chiang L C, Silnutzer J, Pipas J M, Barnes D W. Selection of transformed cells in serum-free media: a new method for the detection of oncogenes. In Vitro Cell Dev Biol. 1985;21:707–712. doi: 10.1007/BF02620926. [DOI] [PubMed] [Google Scholar]
- 9.Clarke A R, Purdie C A, Harrison D J, Morrison R G, Bird C C, Hooper M L, Wyllie A H. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 10.Collins M K L, Perkins G R, Rodriguez-Tarduchy G, Nieto M A, Lopez-Rivas A. Growth factors as survival factors: regulation of apoptosis. BioEssays. 1994;16:133–138. doi: 10.1002/bies.950160210. [DOI] [PubMed] [Google Scholar]
- 11.DeCaprio, J. A. Personal communication.
- 12.DeCaprio J A, Ludlow W, Figge J, Shew J-Y, Huang C-M, Lee W E, Marsilio E, Paucha E, Livingston D M. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 13.El-Deiry W S, Harper J W, O’Connor P M, Velculescu V E, Canman C E, Jackman J, Pietenpol J A, Burrell M, Hill D E, Yang Y, Wiman K G, Mercer W E, Kastan M B, Kohn K W, Elledge S J, Kinzler K W, Vogelstein B. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 14.El-Deiry W S, Toniko T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 15.Ernst T, Jackson C, Barnes D. Karyotypic stability of serum-free mouse embryo (SFME) cells. Cytotechnology. 1991;5:211–222. doi: 10.1007/BF00556291. [DOI] [PubMed] [Google Scholar]
- 16.Green M. Transformation and oncogenes: DNA viruses. In: Fields B N, editor. Virology. New York, N.Y: Raven Press; 1985. pp. 183–234. [Google Scholar]
- 17.Ham R G, Mckeehan W L. Media and growth requirements. Methods Enzymol. 1979;58:44–93. doi: 10.1016/s0076-6879(79)58126-9. [DOI] [PubMed] [Google Scholar]
- 18.Huang H J, Yee J K, Shew J Y, Chen P L, Bookstein R, Friedmann T, Lee E Y, Lee W H. Suppression of the neoplastic phenotype by replacement of the RB gene in human cancer cells. Science. 1988;242:1563–1566. doi: 10.1126/science.3201247. [DOI] [PubMed] [Google Scholar]
- 19.Jung Y K, Yuan J. Suppression of interleukin-1β converting enzyme (ICE)-induced apoptosis by SV40 large T antigen. Oncogene. 1997;14:1207–1214. doi: 10.1038/sj.onc.1200943. [DOI] [PubMed] [Google Scholar]
- 20.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 21.Kastan M B, Zhan Q, el-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:589–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 22.Kim S H, Roth K A, Coopersmith C M, Pipas J P, Gordon J I. Expression of wild-type and mutant simian virus 40 large tumor antigens in villus-associated enterocytes of transgenic mice. Proc Natl Acad Sci USA. 1994;91:6914–6918. doi: 10.1073/pnas.91.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuerbitz S J, Plunkett B S, Walsh W V, Kastan M B. Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc Natl Acad Sci USA. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loo D, Rawson C, Helmrich A, Barnes D. Serum-free mouse embryo (SFME) cells: growth responses in vitro. J Cell Physiol. 1989;142:210–217. doi: 10.1002/jcp.1041390306. [DOI] [PubMed] [Google Scholar]
- 25.Loo D T, Rawson C L, Ernst T, Shirahata S, Barnes D W. Primary and multipassage culture of mouse embryo cells in serum-containing and serum-free media. In: Baserge R, editor. Cell growth and cell division: a practical approach. Eynsham, England: IRL Press Ltd.; 1989. pp. 17–34. [Google Scholar]
- 26.Loo D T, Fuquay J I, Rawson C R, Barnes D W. Extended culture of mouse embryo cells without senescence: inhibition by serum. Science. 1987;236:200–202. doi: 10.1126/science.3494308. [DOI] [PubMed] [Google Scholar]
- 27.Lowe S, Ruley E. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 28.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 29.Mather J P, Sato G H. The use of hormone-supplemented serum-free media in primary cultures. Exp Cell Res. 1979;124:215–221. doi: 10.1016/0014-4827(79)90271-4. [DOI] [PubMed] [Google Scholar]
- 30.Mesner P W, Winters T R, Green S H. Nerve growth factor withdrawal-induced cell death in neural PC12 cells resembles that in sympathetic neurons. J Cell Biol. 1992;119:1669–1680. doi: 10.1083/jcb.119.6.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Momand J, Zambetti G P, Olson D C, George D, Levine A J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53 mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 32.Nevins J R. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–428. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 33.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of the tumour suppressor p53. Nature (London) 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 34.Qin X Q, Chittenden T, Livingston D M, Kaelin W G. Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 35.Rawson C, Cosola-Smith C, Barnes D. Death of serum-free mouse embryo cells caused by epidermal growth factor deprivation is prevented by cycloheximide, 12-O-tetradecanoylphorbol-13-acetate, or vanadate. Exp Cell Res. 1990;186:177–181. doi: 10.1016/0014-4827(90)90224-x. [DOI] [PubMed] [Google Scholar]
- 36.Rawson C, Loo D, Helmrich A, Ernst T, Natsuno T, Merrill G, Barnes D. Serum inhibition of proliferation of serum-free mouse embryo cells. Exp Cell Res. 1991;192:271–277. doi: 10.1016/0014-4827(91)90186-x. [DOI] [PubMed] [Google Scholar]
- 37.Rawson C L, Loo D T, Duimstra J L, Hedstrom O R, Schmidt E E, Barnes D W. Death of serum-free mouse embryo cells caused by epidermal growth factor deprivation. J Cell Biol. 1991;113:671–680. doi: 10.1083/jcb.113.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakai Y, Rawson C, Lindberg K, Barnes D. Serum and transforming growth factor beta regulate glial fibrillary acidic protein in serum-free-derived mouse embryo cells. Proc Natl Acad Sci USA. 1990;87:8378–8382. doi: 10.1073/pnas.87.21.8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sheng Q, Denis D, Ratnofsky M, Roberts T M, DeCaprio J A, Schaffhausen B. The DnaJ domain of polyomavirus large T antigen is required to regulate the Rb family tumor suppressor function. J Virol. 1997;71:9410–9416. doi: 10.1128/jvi.71.12.9410-9416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solem M, Rawson C, Lindberg K, Barnes D. Transforming growth factor beta regulates cystatin C in serum-free mouse embryo (SFME) cells. Biochem Biophys Res Commun. 1990;172:945–951. doi: 10.1016/0006-291x(90)90767-h. [DOI] [PubMed] [Google Scholar]
- 42.Srinivasan A, McClelland A J, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky J, Pipas J M. The amino-terminal transforming region of simian virus 40 large T and small T antigens functions as a J domain. Mol Cell Biol. 1997;17:4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srinivasan A, Peden K W C, Pipas J M. The large tumor antigen of simian virus 40 encodes at least two distinct transforming functions. J Virol. 1989;63:5459–5463. doi: 10.1128/jvi.63.12.5459-5463.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stubdal H, Zalvide J, Cambell K S, Schweitzer C, Roberts T M, DeCaprio J A. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol Cell Biol. 1997;17:4979–4990. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Symonds H S, McCarthy S A, Chen J, Pipas J M, Van Dyke T. Use of transgenic mice reveals cell-specific transformation by a simian virus 40 T-antigen amino-terminal mutant. Mol Cell Biol. 1993;13:3255–3265. doi: 10.1128/mcb.13.6.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan X, Martin S J, Green D R, Wang J Y J. Degradation of retinoblastoma protein in tumor necrosis factor- and CD95-induced cell death. J Biol Chem. 1997;272:9613–9616. doi: 10.1074/jbc.272.15.9613. [DOI] [PubMed] [Google Scholar]
- 47.Teodoro J, Branton P E. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Todaro G J, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu X, Bayle J H, Olson D, Levine A J. The p53–mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 50.Yonish-Rouach E, Resnitzky E, Lotem J, Sachs L, Kimchi A, Oren M. Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature. 1991;352:345–347. doi: 10.1038/352345a0. [DOI] [PubMed] [Google Scholar]
- 51.Zalvide J, Stubdal H, DeCaprio J A. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol Cell Biol. 1998;18:1408–1415. doi: 10.1128/mcb.18.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]