Abstract
Here we describe, for the first time, recombinants between two highly divergent major groups of human immunodeficiency virus type 1 (HIV-1), M and O, within a Cameroonian woman infected with three different HIV-1 strains, a group O virus, a subtype D virus, and a recently reported IBNG (A/G)-like recombinant virus. Using nested extra-long PCR amplification, we sequenced from the pol region to the env region including accessory genes of the viral genome obtained from the patient’s uncultured peripheral blood mononuclear cells and examined the phylogenetic position of each gene. Compared with sequential blood samples obtained in 1995 and 1996, there were multiple segmental exchanges between three HIV-1 strains (O, D, and IBNG) and all the recombinants appeared to be derived from a common M/O ancestor. Importantly, recombination between groups M and O occurred, even though the homology between these two groups is 69, 76, 68, and 55% in the gag, pol, vif-vpr, and env regions, respectively. Recombination between strains with such distant lineages may contribute substantially to generating new HIV-1 variants.
Accumulated molecular analyses on the genetic diversity of human immunodeficiency virus type 1 (HIV-1) have clarified that this virus family can be classified into group M (major), a rare group O (outlier), and a new group N and that group M can be divided into at least 10 subtypes, designated A through J (19, 21, 28). Recent studies have provided increasing evidence for the importance of recombination in the genetic diversification of HIV-1. It has been shown that 5 to 10% of the HIV-1 strains that have been fully or partially sequenced possess mosaic genomes composed of two different HIV-1 group M subtypes (4, 9, 24). So far, no sequences that are hybrids of group M and group O viruses have been found. In many cases, putative intersubtype recombinants have originated from geographic areas where multiple subtypes are known to be cocirculating, such as Central Africa (1, 4, 8, 27), South America (25), and Southeast Asia (2, 10).
We recently reported the molecular epidemiology of HIV-1 and HIV-2 in Cameroon (29). There is no doubt that a large variety of HIV-1 subtypes and intersubtype recombinants are cocirculating in Cameroon, where almost all the HIV-1 subtypes (A through H), group O, and group N have been characterized (12, 17, 18, 22, 28, 29). In addition, various types of mixed infection, such as between different subtypes of HIV-1 group M, between HIV-1 and HIV-2, and even between HIV-1 groups M and O, were confirmed to occur at a rather high frequency (approximately 10%) in the Cameroonian specimens (29). It seems that the mixed infections with different HIV strains are not as rare as previously thought. Moreover, another in vitro study (16) has shown that mixed infection resulted in biologically active recombinant viruses that spread rapidly. Recombinant viruses appear to have the potential to lead to global pandemics.
We previously reported that one patient, a 22-year-old single Cameroonian woman, was infected with three different HIV-1 strains, a group O virus, a subtype D virus, and an IBNG (A/G)-like recombinant virus (30). Initially, we thought that the third strain was a subtype A virus, but we have found that it was a recently reported IBNG (A/G)-like recombinant virus whose genome contains partial subtype G sequences in a mainly subtype A genome (for details, see Results and Discussion). Here we present the first genetic analysis of HIV-1 “intergroup” (M/O) recombinant viruses from this triply infected patient. We have found multiple segmental exchanges among three HIV-1 strains (O, D, and IBNG) on a single clone in the peripheral blood mononuclear cells (PBMCs) of this individual. Recombinational events such as this case are important because they contribute to the production of new variants of HIV-1.
MATERIALS AND METHODS
Subject.
The patient studied (cm61) was a 22-year-old (1994) single Cameroonian woman diagnosed with AIDS-related complex. She had been living in a suburb near Yaoundé, the capital city of Cameroon, for several years and was occasionally hospitalized with fever, severe weight loss, and chronic diarrhea. As risk factors for infection, she reported only frequent heterosexual contacts. Sera were tested as described previously (29, 30).
Tissue culture.
PBMCs from the patient were separated by Ficoll gradient sedimentation and cocultivated with virus-free human PBMCs from which CD8+ cells were removed or with human T-lymphoid cell lines (molt4#8 [14] or M8166 [3]). All the cultures were maintained for several weeks, and the supernatant was monitored for reverse transcriptase activity as described previously (31) or for the presence of p24/p27 antigen (SIV core antigen assay; Coulter, Miami, Fla.).
PCR and cloning of viral genome segments.
Chromosomal DNA was extracted from PBMCs by using glass milk powder (Prep-A-Gene DNA purification kit; Bio-Rad, Hercules, Calif.). The genome (corresponding to nucleotides [nt] 4075 to 7113 in HIV-1LAI) was amplified by nested extra-long PCR (XL-PCR kit; Perkin-Elmer, Foster City, Calif.) with primers HIV-1pol3 (5′-TAAAAGGAGAAGCCATGCATGGACAAGTAGA-3′) and M10 (5′-CCAATTGTCCCTCATATCTCCTCCTCCAGG-3′) in the first round and unipol1 (5′-AGTGGATTCATAGAAGCAGAAGT-3′) and M8 (5′-TCCTTGGATGGGAGGGGCATACATTGC-3′) in the second round. Cycling conditions included a hot start (1 min at 95°C), then 10 cycles of denaturation (95°C) for 15 s and extension (68°C) for 5 min, and then 20 cycles of denaturation (95°C) for 15 s and extension (65°C) for 15 min, with 15-s increments per cycle. The XL-PCR products (approximately 3,070 bp) were shortened by use of a kilosequence deletion kit (Takara Shuzo Co. Ltd., Otsu, Japan). Smaller, overlapping fragments containing the pol region (corresponding to nt 4075 to 4362 in HIV-1LAI) and the env region (corresponding to nt 6567 to 7113 in HIV-1LAI) of the genome were amplified by conventional PCR and cloned separately with PCR primers as described previously (29). DNA sequencing was carried out by the dideoxy chain termination method with an automated DNA sequencer (373A; Applied Biosystems, Foster City, Calif.). At least 10 plasmid clones were sequenced to obtain the consensus sequence.
Phylogenetic analysis.
DNA sequences were aligned by the ODEN program package of the National Institute of Genetics, Mishima, Japan (13). Nucleotide substitutions of all pairs of the sequences were estimated by the two-parameter (15) and six-parameter (11) methods. The phylogenetic tree was constructed by the neighbor-joining method, and its reliability was estimated by 100 bootstrap replications (5).
Distance plots.
The query sequences were first aligned with a set of reference sequences representing all the established genetic subtypes of HIV-1 (9). The genetic distance (two-parameter and six-parameter) between selected pairs of sequences was determined by moving a window of 300 bp along the genome alignment in 25-bp increments. The distances were plotted at the midpoint of the 300-bp segments.
Bootscanning.
Bootstrap plots (26) were performed on the neighbor-joining trees for a window of 300 bp moving along the alignment in increments of 25 bp. We evaluated 100 replicates generated by the bootstrap resampling for each phylogeny. The percent bootstrap probabilities for each topology were plotted at the midpoint of each window. Breakpoints were assigned to the midpoint of the transitions between segments from different subtypes.
Distribution of informative sites.
To localize intragenic crossover points between regions of DNA sequences, the distribution of phylogenetically informative sites supporting alternative tree topologies was inspected. This was done by surveying the informative sites in a four-sequence alignment including the putative recombinant sequence. There are three possible configurations of the informative sites, two of which support the clustering of the putative recombinant with one parental lineage or the other. The distribution of these two types of sites can be tested by determining whether a break placed at any point along the alignment produces a significant difference in the ratio of the two types of sites on each side of the cut, as assessed by a chi-square value with Yates’s correction for continuity; the optimum position of the breakpoint can be found by maximizing this value (23, 24).
Nucleotide sequence accession numbers.
The nucleotide sequences in this study have been assigned GenBank accession no. U58148 to U58159, AF023085, AF055728 to AF055732, and AF097692 to AF097698.
RESULTS
Serological data and virus isolation.
Blood samples were collected from the patient (cm61) in June 1994, June 1995, and June 1996. All three serum specimens (1994, 1995, and 1996) were dually reactive for HIV-1 and HIV-2 by a particle agglutination test and a Western blotting assay. All sera were then discriminated by using a peptide-based assay and judged to be HIV-1 positive. Virus isolation was performed twice (in 1995 and 1996) by techniques routinely used in our laboratory, but attempts to isolate an infectious HIV-1 strain from subject cm61 were unsuccessful.
Most of the HIV mosaic sequences studied so far have been derived from viruses adapted to grow in immortalized T-cell lines. In the present study, we sought to analyze possible recombinational events in vivo. To exclude the possibility of tissue culture artifacts, we used uncultured PBMC samples instead of cultured cells for sequence analysis. The viral genomes were detected by PCR from the patient in 1994, 1995, and 1996 and designated 94CM61, 95CM61, and 96CM61, respectively. (In this study, CM refers to a HIV strain and cm refers to the patient from whom the strains were obtained.)
Phylogenetic analysis of the env and pol regions.
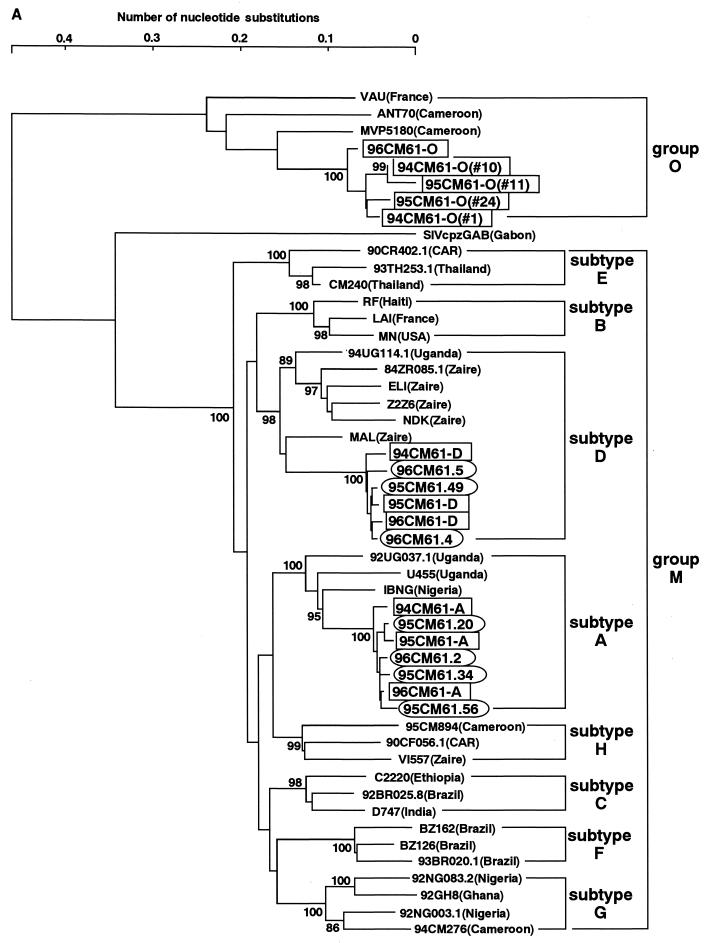
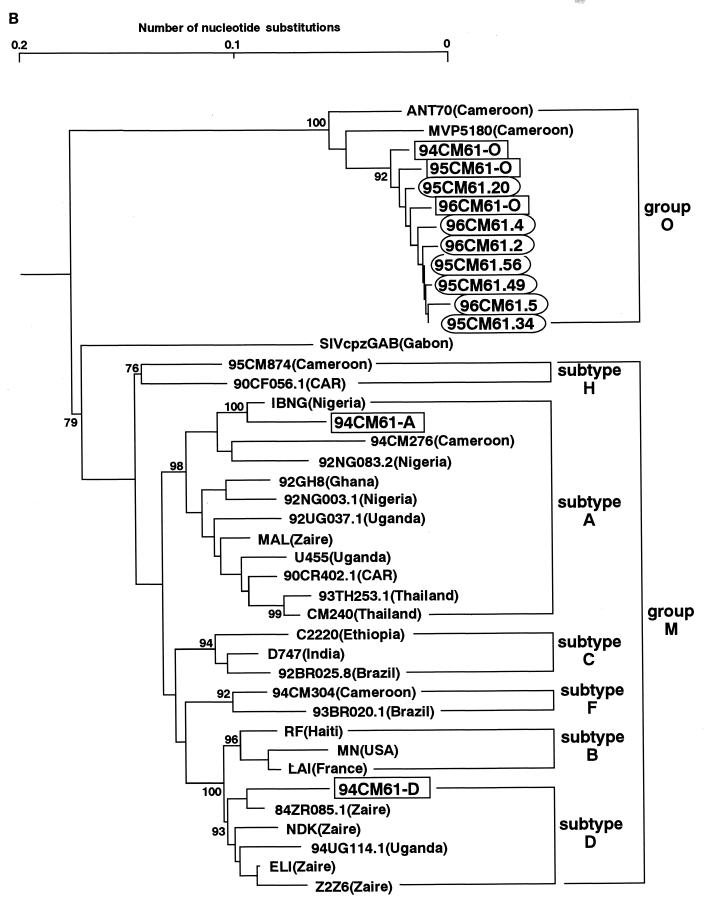
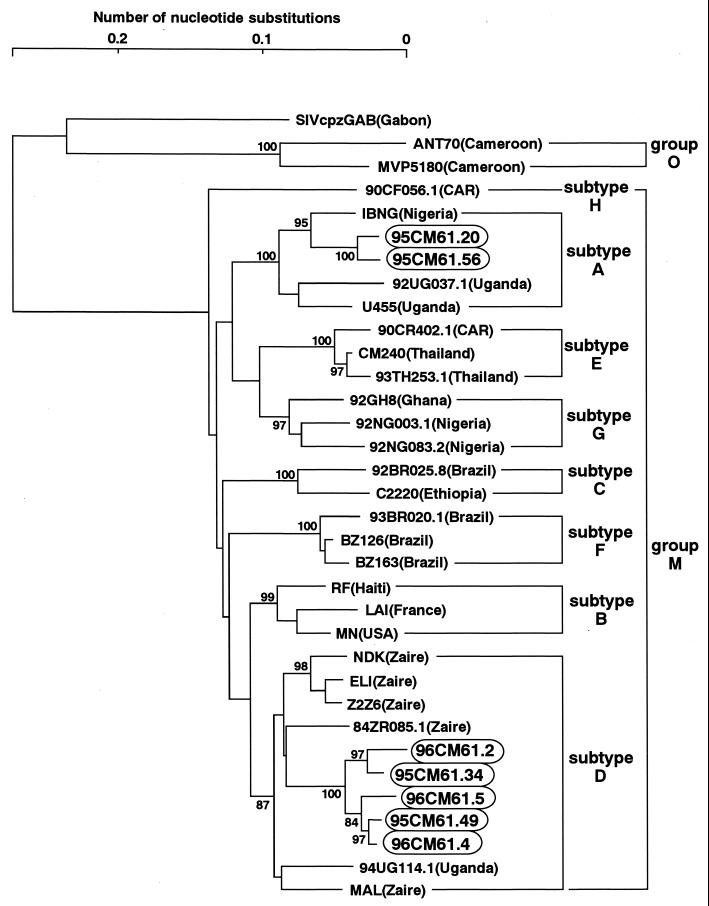
The phylogenetic positions of 94CM61, 95CM61, and 96CM61 were first examined in evolutionary trees based on the env and pol gene sequences. The central portion of gp120 including the V3 region was sequenced, and a phylogenetic tree was constructed (Fig. 1A). Three different types of sequences, subtype A, subtype D, and group O, in the same genomic region were found in each of the three sampling years. The presence of three different sequences in one individual clearly demonstrated that this individual had a triple infection with HIV strains of different origins. It should be noted that the sequences of subtype A were tightly clustered with an IBNG strain. This IBNG strain has been recently reported to be a mosaic virus of subtypes A and G (1). Surveys in Nigeria and Cameroon suggest that this virus is more prevalent than the nonrecombinant subtype A (10, 18).
FIG. 1.
Phylogenetic trees of HIV-1 sequences. The trees were constructed from part of the env sequences including those in the V3 region (approximately 390 bp) (A) and part of the pol sequence that encodes integrase (288 bp) (B). The results obtained by the six-parameter method are shown. Each of the pol and env sequences obtained in 1994, 1995, and 1996 are boxed, and the pol and env sequences from the M/O recombinants are circled. Subtypes are indicated by brackets. Bootstrap values of key nodes are shown.
Figure 1B shows a phylogenetic tree constructed from the pol sequences that encode integrase. The present full-length pol analysis revealed that HIV-1 fell into seven major and clearly defined subtypes: A, B, C, D, F, G, and H (9). In this pol tree, subtypes E and G were included in subtype A. The coexistence of three types of sequences (A, D, and O) in the pol region was again observed in the patient in 1994. It should be noted that the sequence of subtype A was also closely related to the IBNG strain in the pol tree. Taking these results together with the result in the env tree, we presumed that the patient was infected with not a typical subtype A virus but an IBNG (A/G)-like virus other than group O and subtype D viruses. In addition, it is particularly noteworthy that only the group O sequences were found in this patient in 1995 and 1996. The same results were obtained with three different sets of primers for nested PCR in the pol region (29), ruling out the possibility that we missed detecting other sequences (A and D) in the pol region. Based on these results, we consider that the viral population in this patient in 1995 and 1996 might have changed from that in 1994.
Clones and sequences of viral genome segments obtained by XL-PCR amplification.
HIV-1 genomic sequences that were obtained in 1995 and 1996 were repeatedly confirmed by independent XL-PCR amplification and molecular cloning. Twelve clones from the pol region to the env region that were nearly 3,000 bp were obtained from uncultured PBMCs collected in 1995, and seven clones from the same region were obtained from uncultured PBMCs collected in 1996. The purified XL-PCR products were digested with several restriction enzymes (BamHI, EcoRI, HindIII, KpnI, SacI, SalI, SphI, and XbaI). Analysis of these digestions indicated that the former 12 clones were of five types (representative clones were 95CM61.20, 95CM61.34, 95CM61.49, 95CM61.55, and 95CM61.56) and the latter 7 clones were of three types (representative clones were 96CM61.2, 96CM61.4, and 96CM61.5). Complete sequences of each of the above five plus three clones revealed seven potential open reading frames corresponding to the partial pol, vif, vpr, 5′ tat, 5′ rev, vpu, and partial env genes. (After sequencing, the homology between 95CM61.49 and 95CM61.55 turned out to be 98.1%, and these two clones were found to have almost the same mosaic structure.)
Genetic distances and bootscanning.
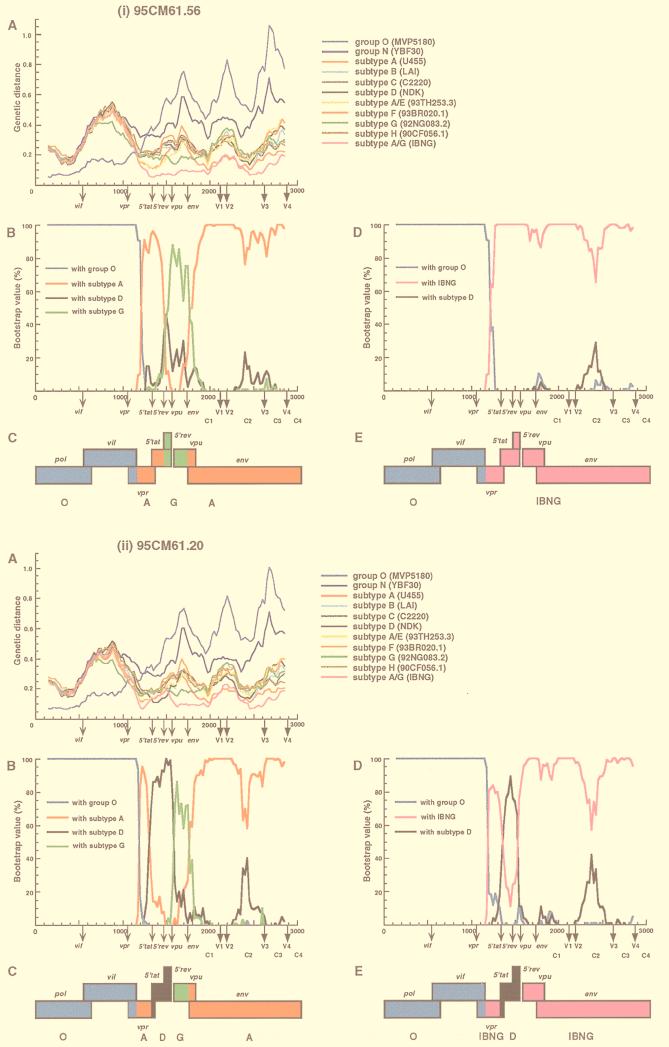
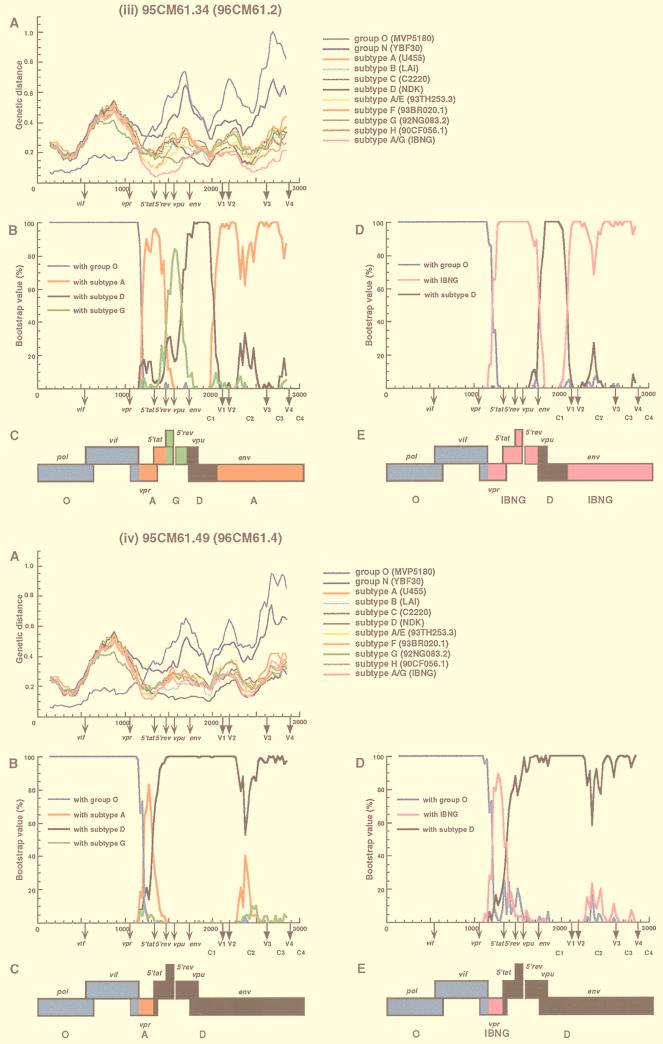
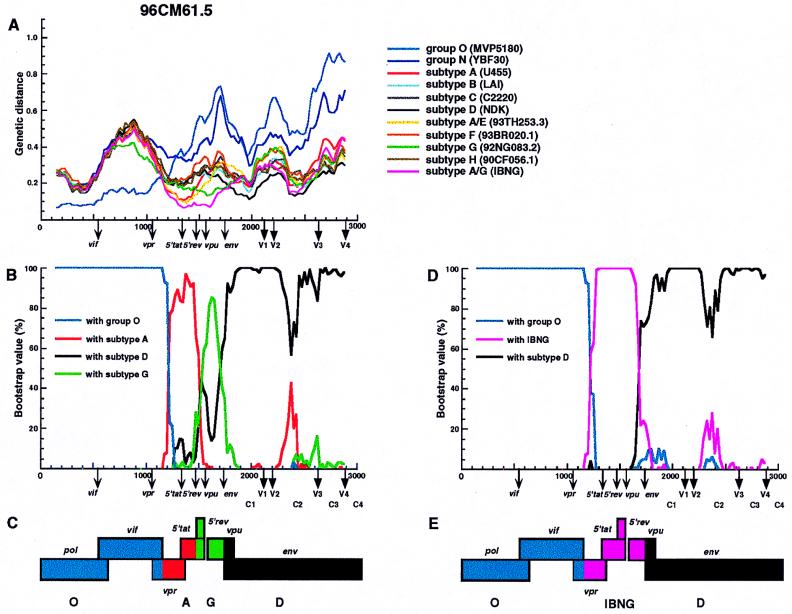
We next determined which segments of the 95CM61 and 96CM61 genomes were derived from the group O, subtype D, or subtype A viruses. Whether a subtype A segment was derived from a typical subtype A virus or from an IBNG (A/G)-like virus, as we presumed above, also needed to be clarified. Distance values were calculated by the two-parameter and six-parameter methods for a window of 300 nt which was moved in steps of 25 nt (2, 10). Since we found no difference between the results obtained by the two methods, only the results obtained by the six-parameter method are shown (Fig. 2A and 3A). These figures show the distance values between 95CM61 (or 96CM61) and 11 other HIV-1 reference strains for each location in the genome. The reference strains are subtypes A (U455), B (LAI), C (C2220), D (NDK), F (93BR020.1), G (92NG083.2), H (90CF056.1), N (YBF30), and O (MVP5180), as well as A/E (93TH253.3) and A/G (IBNG) (9). Note that in the absence of a nonmosaic, full-length subtype G genome (9, 21), we used 92NG083.2, which is known to contain a small A segment in the vif-vpr region, as a subtype G reference, Figure 2A shows the distance plots of four representative clones of 95CM61. The results of 96CM61.2 and 96CM61.4 were quite similar to those of 95CM61.34 and 95CM61.49, respectively. The distance plot profile of the remaining 96CM61.5 was different from those of the preceding four and therefore is shown in Fig. 3A. The genotype that exhibited the parental strain in one segment generally followed the lowest distance in this analysis. Concerning the region from positions 1 to 1175 (the end of the vif region), for example, the distance between 95CM61 (or 96CM61) and MVP5180 was much smaller than the distances between 95CM61 (or 96CM61) and each sequence of the group M subtypes. The same trend was also observed in other clones. This observation indicates that the region from positions 1 to 1175 was derived from a group O-like virus and that a crossover occurred near the end of the vif gene, which is also the 5′ beginning of the vpr gene. As is clearly shown in each plot in Fig. 2A and 3A, 95CM61 and 96CM61 genomes could be related to only group O, subtype D, or IBNG (A/G) viruses and other subtypes were genomically rather distant. After showing the short distances of the respective clones to group O in the pol and vif regions, their distances to IBNG narrowed after the 5′ portion of vpr but the patterns of distances among the seven clones were not uniform, especially after the 5′ tat region. It is emphasized that the distance to IBNG was shorter than to a typical subtype A virus (U455) in the vpr to env regions. These findings suggest that subtype D and IBNG-like viruses were probably parental strains and that various recombinants might have arisen as offspring in this patient in 1995 and thereafter.
FIG. 2.
Analysis of mosaic structures of four types of 95CM61. (A) Each of four respective clones (95CM61.56, 95CM61.20, 95CM61.34, and 95CM61.49) was aligned with 11 full-length HIV-1 isolates (MVP5180, YBF30, U455, LAI, C2220, NDK, 93TH253.3 93BR020.1, 92NG083.2, 90CF056.1, and IBNG) representing groups O and N, subtypes A to H, and A/G recombinant, respectively, and the alignment was sectioned into 300-nt segments, which were moved in steps of 25 nt. The genetic distances between 95CM61 and the respective reference strains were calculated by using the six-parameter method. (B) Breakpoints were fine-mapped by using a five-sequence alignment consisting of 95CM61, U455, NDK, 92NG083.2, and MVP5180. Bootstrap values for the node with either subtype A, subtype D, subtype G, or group O out of 100 bootstrap replications built by the neighbor-joining method were plotted. (D) Because IBNG is known to represent a mosaic of subtypes A and G, breakpoints were then fine-mapped by using a four-sequence alignment consisting of 95CM61 and putative parental strains (IBNG, NDK, and MVP5180), and the magnitude of the bootstrap value supporting the clustering of 95CM61 with IBNG (A/G), subtype D, or group O was plotted, respectively. The distance value (A) and the bootstrap value (B and D) for each segment were plotted at the midpoint of the segment. (C and E) A map of the open reading frames in part of the HIV-1 genome is shown. Segments derived from group O (blue), subtype A (red), subtype D (black), subtype G (green), and an IBNG-like virus (pink) were mapped by the results of diversity and bootstrap plots, as well as phylogenetic tree analyses.
FIG. 3.
Analysis of mosaic structures of 96CM61.5. See the legend to Fig. 2 for details of the method. (The results for 96CM61.2 and 96CM61.4 were quite similar to those for 95CM61.34 and 95CM61.49, respectively.)
To confirm the locations of recombinational breakpoints, we used bootscanning (26) by the neighbor-joining method. We first determined the magnitude of the bootstrap values supporting the clustering of 95CM61 (or 96CM61) with reference subtypes A, D, and G and group O (Fig. 2B and 3B). To determine whether the IBNG-like virus was truly a parental strain, we then performed bootstrap plot analyses with IBNG, subtype D, and group O (Fig. 2D and 3D). The bootstrap value (100 replications) of the node joining 95CM61 (or 96CM61) with each parental strain was plotted for each segment of the genome. These results agreed fairly well with those of the genetic distance analysis described above. In fact, all the clones of 95CM61 and 96CM61 joined MVP5180 with high bootstrap values (100%) until the beginning of the vpr gene, whereas the bootstrap values fell and remained low (0%) from vpr to gp120. This bootstrap profile clearly indicates that the pol and vif regions of all the clones were derived from group O. The plot of bootstrap values revealed detailed structures of the respective clones from the middle portion of vpr to gp120. In the case of clone 95CM61.56 (Fig. 2i panel B), the region from vpr to 5′ tat and the majority of env showed rather high bootstrap values with subtype A, and the 5′ rev region and the 5′ half of vpu alone showed significantly high values with subtype G. Since we knew from a recent report that the corresponding genomic segment of an IBNG strain was subtype G (20), we predicted that the vpr-to-env region might have originated from an IBNG-like virus. As predicted, the high bootstrap values in Fig. 2i panel D clearly show that these regions consistently clustered with IBNG. Figure 2i panel B shows that the IBNG virus was truly mosaic: the 5′ rev-vpu segments were derived from subtype G and the rest of the regions were derived from subtype A as previously reported. In clone 95CM61.20 (Fig. 2ii panel D), however, high bootstrap values with subtype D occurred in the 5′ tat-rev region while the remaining regions were derived from an IBNG-like virus. The vpu sequence of 95CM61.20 clustered with subtype G (Fig. 2ii panel B), whereas the env sequence of 95CM61.20 was that of subtype A. However, this apparently complicated mosaic pattern can be easily explained if we assume that 95CM61.20 was a recombinant between group O and IBNG-like viruses possessing a subtype D segment in the 5′ tat-rev region. The case of clone 95CM61.34 (and also 96CM61.2) is slightly different from that of the previous clone. As shown in Fig. 2iii panel B, very high bootstrap values (>80%) were found in most parts of the 95CM61.34 clone, indicating that the vpr and 5′ tat regions and the V1-to-C4 region of gp120 were derived from subtype A, the sequence from the 5′ rev region to the middle of the vpu region was derived from subtype G, and the signal peptide and the C1 region of gp120 were derived from subtype D. This apparently complicated mosaicism can also be simplified if we assume that clone 95CM61.34 was a recombinant between group O and IBNG-like viruses possessing a subtype D segment in the signal peptide and the C1 region of gp120 (Fig. 2iii panel D). The phylogenetic analysis in the signal peptide and the C1 sequences (Fig. 4) gave similar results. In the tree, both 95CM61.34 and 96CM61.2 belonged to subtype D. By contrast, in clone 95CM61.49 (also 96CM61.4) (Fig. 2iv panel B), the central portion of the vpr gene had a small segment of subtype A and most other regions from 5′ tat to gp120 were of subtype D with high bootstrap values. By making a deduction similar to that mentioned above, this clone can be explained as a recombinant between group O and subtype D viruses possessing an IBNG-like segment in the central portion of vpr. In Fig. 3B, very high bootstrap values supported the clustering of 96CM61.5 with subtype A in the central portion of vpr and 5′ tat and its clustering with subtype G in the 5′ rev-vpu region. By contrast, 96CM61.5 clearly clustered with subtype D in the env region. We can explain this clone as a recombinant between group O and subtype D viruses possessing an IBNG-like segment from the vpr region to the 5′ rev region.
FIG. 4.
Phylogenetic relationships of 95CM61 and 96CM61 in the signal peptide and C1 regions (approximately 400 bp). The tree was rooted by using group O and SIVcpzGAB as outgroups. The sequences of 95CM61 and 96CM61 are circled. Subtypes are indicated by brackets. Bootstrap values of key nodes are shown.
Recombination breakpoint analysis based on informative site distribution analysis.
Tables 1 and 2 show the results of an investigation into the distribution of phylogenetically informative sites around the observed breakpoints. Breakpoints were inserted at each possible point between adjacent informative sites, and a 2 × 2 heterogeneity chi-square value with Yates’s correction for continuity was calculated for the sites to either side of the breakpoint that supports the clustering of the putative recombinant with each of the consensus sequences. Within the precision attainable by this technique, common breakpoints were identified at the end of vif in all the clones (Tables 1 and 2). Other breakpoints in 95CM61.34, 96CM61.2, and 96CM61.5 were also identified by this analysis. As described in the preceding section, it is evident that both 95CM61.49 and 96CM61.4 had a crossover point from an IBNG-like virus to a subtype D-like virus. However, there was no evidence that 95CM61.20 is an “intersubtype” (IBNG-D-IBNG) recombinant. The lengths from the vpr region to the 5′ tat-rev region were probably too short and contained no apparent informative sites within these ambiguous sequences (Table 1).
TABLE 1.
Informative-site analysis of 95CM61 and 96CM61
| Genome | Subtype | Region (nt) | No. of informative sitesa
|
||
|---|---|---|---|---|---|
| IBNG | D | O | |||
| 95CM61.56 | O | 1–1150 | 5 | 6 | 156 |
| IBNG | 1195–3007 | 96 | 15 | 18 | |
| 95CM61.20 | O | 1–1147 | 6 | 5 | 147 |
| IBNG | 1195–3004 | 91 | 16 | 15 | |
| 95CM61.34 | O | 1–1150 | 7 | 6 | 155 |
| IBNG | 1195–1719 | 33 | 0 | 6 | |
| D | 1780–2062 | 3 | 9 | 2 | |
| IBNG | 2107–3016 | 44 | 12 | 12 | |
| 95CM61.49 | O | 1–1150 | 6 | 6 | 156 |
| IBNG | 1195–1343 | 6 | 0 | 2 | |
| D | 1376–3006 | 17 | 55 | 11 | |
| 96CM61.2 | O | 1–1147 | 7 | 6 | 148 |
| IBNG | 1195–1719 | 40 | 0 | 4 | |
| D | 1755–2062 | 3 | 10 | 2 | |
| IBNG | 2107–3019 | 44 | 13 | 13 | |
| 96CM61.4 | O | 1–1147 | 6 | 6 | 151 |
| IBNG | 1195–1343 | 9 | 1 | 0 | |
| D | 1376–3021 | 12 | 56 | 17 | |
| 96CM61.5 | O | 1–1150 | 6 | 6 | 156 |
| IBNG | 1189–1719 | 27 | 1 | 5 | |
| D | 1755–3031 | 7 | 38 | 9 | |
The distribution of informative sites supporting each of three lineages is presented in the columns labeled IBNG, D, and O. In column IBNG, the apparent recombinant is most closely related to a representative of the IBNG (A/G)-like recombinant virus (HIV-1IBNG); in column D, it is most closely related to a representative of subtype D (HIV-1NDK); and in column O, it is most closely related to a strain of group O (HIV-1MVP5180). Crossover points were located so as to maximize statistical significance, assessed by a chi-square with Yates’s correction for continuity of the difference in the distribution of sites supporting phylogenies IBNG plus D and O (1 to 1147 or 1 to 1150) and phylogenies IBNG and D (1195 to 3007, 1195 to 3004, 1195 to 3016, 1195 to 3006, 1195 to 3019, 1195 to 3021 or 1189 to 3031).
TABLE 2.
Informative-site analysis of 95CM61 and 96CM61
| Genome | Subtype | Region (nt) | No. of informative sitesa
|
||
|---|---|---|---|---|---|
| A | D | G | |||
| 95CM61.56 | A | 1208–1454 | 8 | 3 | 2 |
| G | 1509–1686 | 2 | 3 | 7 | |
| A | 1773–3007 | 49 | 15 | 16 | |
| 95CM61.20 | A | 1208–1346 | 6 | 2 | 1 |
| D | 1379–1456 | 1 | 8 | 0 | |
| G | 1509–1688 | 2 | 3 | 7 | |
| A | 1775–3004 | 49 | 17 | 16 | |
| 95CM61.34 | A | 1208–1454 | 8 | 2 | 2 |
| G | 1509–1686 | 1 | 2 | 8 | |
| D | 1760–2002 | 1 | 15 | 3 | |
| A | 2113–3016 | 36 | 14 | 12 | |
| 95CM61.49 | A | 1208–1346 | 6 | 2 | 2 |
| D | 1379–3006 | 23 | 85 | 25 | |
| 96CM61.2 | A | 1208–1454 | 9 | 2 | 2 |
| G | 1509–1686 | 0 | 2 | 8 | |
| D | 1760–2002 | 1 | 16 | 3 | |
| A | 2113–3019 | 43 | 15 | 12 | |
| 96CM61.4 | A | 1208–1346 | 5 | 2 | 1 |
| D | 1379–3021 | 20 | 84 | 24 | |
| 96CM61.5 | A | 1208–1454 | 9 | 2 | 2 |
| G | 1509–1686 | 0 | 2 | 8 | |
| D | 1760–3031 | 12 | 58 | 17 | |
The distribution of informative sites supporting each of three lineages is presented in columns A, D, and G. In column A, the apparent recombinant is most closely related to a representative of subtype A (HIV-1U455); in column D, it is most closely related to a representative of subtype D (HIV-1NDK); and in column G, it is most closely related to a strain of subtype G (HIV-192NG083.2).
Evidence of “intergroup” recombination.
In the present study, to exclude the possibility of PCR-mediated artifacts, the viral clones were verified on two independent XL-PCR amplifications by using sequential blood samples obtained in 1995 and 1996, and essentially the same mosaic patterns for 95CM61.34 and 96CM61.2 and for 95CM61.49 and 96CM61.4 were obtained. Our multiple recombinants are clearly not the products of PCR-mediated artifacts, because it is unlikely for recombinant Tth DNA polymerase to repeatedly generate crossovers at the same positions.
The present analyses provide evidence that the 95CM61 and 96CM61 genomes had multiple breakpoints between genomic segments from different groups or subtypes. Multiple crossover points involving a group O virus, a subtype D virus, and an IBNG-like virus were indicated in the genomes of 95CM61 and 96CM61 (Fig. 2E and 3E). Breakpoints from group O to group M were at the end of vif and were identified by three different methods: the distance plot, bootscanning, and informative site analyses. It is striking that the pol-vif region was uniformly derived from a group O virus and that the central portion of vpr was uniformly derived from an IBNG-like virus, while five mosaic patterns (seven recombinant forms) of the 5′ tat-rev, vpu, and env genes differed from each other. Although the patterns are complicated, they can be easily understood by presuming that all the recombinants originated from group O, subtype D, and IBNG-like viruses. Additional breakpoints have been mapped between vpr and 5′ tat in 95CM61.49, 96CM61.4, and 95CM61.20, at the 5′ beginning of env in 95CM61.34, 96CM61.2, and 96CM61.5, and at the 3′ end of the C1 region in 95CM61.34 and 96CM61.2. (The end of 5′ rev in 95CM61.20 may be another breakpoint, but additional evidence is needed.)
DISCUSSION
To clarify the ongoing process of generation and selection of HIV-1 variants, we have used a set of genetic analyses to identify M/O recombinants in this study. First, the presence of three different sequences in the pol and env regions in patient cm61 in 1994 clearly demonstrated that this patient had a triple infection with HIV strains: a group O virus, a subtype D virus, and an IBNG-like virus (Fig. 1). The existence of three different HIV-1 env sequences was confirmed in sequential samples taken 1 year apart: 95CM61 was obtained in 1995, and 96CM61 was obtained in 1996 (Fig. 1A). Nevertheless, in the phylogenetic analysis based on the pol region, we detected only one type (group O) in cm61 in both 1995 and 1996 (Fig. 1B), although the PCR conditions were the same as those used in 1994. At that point, we thought that recombination might have occurred between groups M and O. Thus, we attempted to amplify 95CM61 and 96CM61 by XL-PCR spanning the region from pol to the env. As a result, four different types of genome were obtained from 95CM61 and three types were obtained from 96CM61. Two of the latter were the same as two of those of 95CM61.
When and how these HIV-1 M/O recombinants arose in this individual are of particular interest. Before answering these question, it is important to distinguish between (i) mixed infection of this patient with three genetically distinct HIV strains and (ii) infection with an “intergroup” recombinant HIV strain. It is evident that three different strains (O, D, and IBNG) were present in this patient in 1994, because these three sequences were demonstrated in both the pol and env regions. Nevertheless, we did not obtain any evidence for the presence of such viruses by XL-PCR in patient cm61 in 1995 (95CM61) and 1996 (96CM61). However, phylogenetic analyses based on the pol and env regions revealed that respective sequences of the pol and env regions from the M/O recombinants were essentially identical to those of the pol and env regions, which were obtained by independent PCRs with the specific primers for each region (Fig. 1). The viral population of the pol sequences converged on a single form (form O) in 1995 and 1996, in contrast to a mixture of forms [forms O, IBNG (A/G), and D] in 1994. Moreover, this patient claimed that she had no history of blood transfusion or intravenous drug use. So-called IBNG-like A/G recombinants were recognized much earlier than 1994 (1) and have been common in Cameroon (29). In addition, we could not find any subtype G sequences in the env analysis (Fig. 1A), and the distance plots indicated that the distances for IBNG were continuously shorter than those for subtypes A and G (Fig. 2A and 3A). Based on these facts, we conclude that the patient initially had a mixed infection consisting of group O, subtype D, and IBNG-like viruses and that M/O recombination presumably occurred in this patient after she acquired the mixed infection, although we cannot exclude the possibility of simultaneous transmission with parental and recombinant strains.
Additional evidence supports the derivation of one M/O recombinant from another in the patient. This is based on the analysis of identified breakpoints in four 95CM61 recombinants and three 96CM61 recombinants that were obtained 12 months later. Figures 2D and 3D show the results of bootscanning around the observed breakpoints, and Tables 1 and 2 show the results of the distribution of phylogenetically informative sites around the observed breakpoints. The breakpoints between genetic segments from group O to M were at the 3′ end of vif and were identically present in all seven of the molecular clones that were obtained in 1995 and 1996, even though the sequences in the vif-to-vpr region are not similar between groups M and O. A possible explanation for the existence of a common breakpoint in all the recombinants is that the recombinants were derived from a common M/O ancestor. Another possibility is that some selection process occurred after recombinational events between two highly divergent HIV-1 groups. In other words, only selected recombinants in the pool of newly generated recombinant viruses may have survived due to their having a certain breakpoint (such as between vif and vpr) that gave them an evolutionary advantage. Coincidentally, we have found the same crossover point in vif of YBF30, the only fully sequenced strain of HIV-1 group N. This strain is known to be a recombinant of divergent viral lineages within the HIV-1/SIVcpz group (7). Other breakpoints appeared mostly near the boundaries of the respective genes. The high frequency of recombination that occurs only near the beginning or end of the respective genes seems to reflect a common adaptive strategy for recombination. In addition, it is noteworthy that respective recombinants rather quickly changed their population within the patient. At least four types of recombinants, varying from a rather simple type (O-IBNG) to more complex types of mosaicisms (O-IBNG-D and O-IBNG-D-IBNG), were present in the patient PBMCs. At least two of the four types existed continuously as revealed in sequential blood samples obtained in 1995 and 1996. These analyses strongly suggest that earlier recombinant forms served as templates for the generation of later ones. However, we were unable to isolate an infectious HIV-1 strain from this patient. Further investigations are needed to clarify what kind(s) of rules determines the survival and subsequent population dynamics of recombinant viruses.
With the spread and mixing of various HIV-1 subtypes, as has been shown to occur in this central African country, the opportunity for coinfection or superinfection has been increasing dramatically in recent years, and subsequent recombinants among these strains are very likely to be generated. It should be emphasized that recombination between groups M and O can occur in vivo, even though the homology between these groups is only 65% on average across the genome. Recombination must be viewed as a viable mechanism not only for increasing HIV-1 variation in infected individuals but also for contributing to the generation of new HIV variants. This fact has also important implications for HIV vaccine strategies that are designed to employ live attenuated viruses (6, 32), which potentially could form recombinants with wild-type strains even if the two viruses are quite divergent.
ACKNOWLEDGMENTS
This work was supported in part by International Scientific Research Program grant 08041173 from Monbusho (Ministry of Education) and by Research Fellowships of Japan Society for the Promotion of Science for Young Scientists.
We are grateful to Beatrice H. Hahn, University of Alabama at Birmingham, for critically reviewing the manuscript and for helpful suggestions.
REFERENCES
- 1.Carr J, Salminen M, Albert J, Sanders-Buell E, Gotte D, Birx D, McCutchan F. Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology. 1998;247:22–31. doi: 10.1006/viro.1998.9211. [DOI] [PubMed] [Google Scholar]
- 2.Carr J, Salminen M, Koch C, Gotte D, Artenstein A, Hegerich P, St. Louis D, Burke D, McCutchan F. Full-length sequence and mosaic structure of a human immunodeficiency virus type 1 isolate from Thailand. J Virol. 1996;70:5935–5943. doi: 10.1128/jvi.70.9.5935-5943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clapham P, Weiss R, Dalgleish A, Exley M, Withby D, Hogg N. Human immunodeficiency virus infection of monocytic and T-lymphocytic cells: receptor modulation and differentiation induced by phorbol ester. Virology. 1987;158:44–51. doi: 10.1016/0042-6822(87)90236-4. [DOI] [PubMed] [Google Scholar]
- 4.Cornelissen M, Kampinga G, Zorgdrager F, Goudsmit J the UNAIDS Network for HIV Isolation and Characterization. Human immunodeficiency virus type 1 subtypes defined by env show high frequency of recombinant gag genes. J Virol. 1996;70:8209–8212. doi: 10.1128/jvi.70.11.8209-8212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 6.Fultz P, Yue L, Wei Q, Girard M. Human immunodeficiency virus type 1 intersubtype (B/E) recombination in a superinfected chimpanzee. J Virol. 1997;71:7990–7995. doi: 10.1128/jvi.71.10.7990-7995.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao F, Bailes E, Robertson D, Chen Y, Rodenburg C, Michael S, Cummins L, Arthur L, Peeters M, Shaw G, Sharp P, Hahn B. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 8.Gao F, Robertson D, Carruthers C, Li Y, Bailes E, Kostrikis L, Salminen M, Bibollet-Ruche F, Peeters M, Ho D, Shaw G, Sharp P, Hahn B. An isolate of human immunodeficiency virus type 1 originally classified as subtype I represents a complex mosaic comprising three different group M subtypes. J Virol. 1998;72:10234–10241. doi: 10.1128/jvi.72.12.10234-10241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao F, Robertson D, Carruthers C, Morrison S, Jian B, Chen Y, Barre-Sinoussi F, Girard M, Srinivasan A, Abimiku A, Shaw G, Sharp P, Hahn B. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J Virol. 1998;72:5680–5698. doi: 10.1128/jvi.72.7.5680-5698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao F, Robertson D, Morrison S, Hui H, Craig S, Decker J, Fultz P, Girard M, Shaw G, Hahn B, Sharp P. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J Virol. 1996;70:7013–7029. doi: 10.1128/jvi.70.10.7013-7029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gojobori T, Ishii K, Nei M. Estimation of average number of nucleotide substitutions when the rate of substitution varies with nucleotide. J Mol Evol. 1982;18:414–423. doi: 10.1007/BF01840889. [DOI] [PubMed] [Google Scholar]
- 12.Gürtler L, Zekeng L, Tsague J, von Brunn A, Afane Ze E, Eberle J, Kaptué L. HIV-1 subtype O: epidemiology, pathogenesis, diagnosis, and perspectives of the evolution of HIV. Arch Virol. 1996;11:S195–S202. doi: 10.1007/978-3-7091-7482-1_17. [DOI] [PubMed] [Google Scholar]
- 13.Ina Y. ODEN: molecular evolutionary analysis system for DNA and amino acid sequences (version 1.1). DNA Data Bank of Japan (DDBJ) Mishima, Japan: National Institute of Genetics; 1991. [Google Scholar]
- 14.Kikukawa R, Koyanagi Y, Harada S, Kobayashi N, Hatanaka M, Yamamoto N. Differential susceptibility to the acquired immunodeficiency syndrome retrovirus in cloned cells of human leukemic T-cell line Molt-4. J Virol. 1986;57:1159–1162. doi: 10.1128/jvi.57.3.1159-1162.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 16.Kuwata T, Miyazaki Y, Igarashi T, Takehisa J, Hayami M. The rapid spread of recombinants during a natural in vitro infection with two human immunodeficiency virus type 1 strains. J Virol. 1997;71:7088–7091. doi: 10.1128/jvi.71.9.7088-7091.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mauclere P, Loussert-Ajaka I, Damond F, Fagot P, Souquieres S, Monny Lobe M, Mbopi Keou F-X, Barré-Sinoussi F, Saragosti S, Brun-Vézinet F, Simon F. Serological and virological characterization of HIV-1 group O infection in Cameroon. AIDS. 1997;11:445–454. doi: 10.1097/00002030-199704000-00007. [DOI] [PubMed] [Google Scholar]
- 18.Mboudjeka, I., L. Zekeng, J. Takehisa, E. Ido, T. Miura, H. Moriyama, M. Yamashita, L. Kaptué, and M. Hayami. HIV-1 genetic variability in the northern part of Cameroon. AIDS Res. Hum. Retroviruses, in press. [DOI] [PubMed]
- 19.McCutchan F, Salminen M, Carr J, Burke D. HIV-1 genetic diversity. AIDS. 1996;10:S13–S20. [PubMed] [Google Scholar]
- 20.Myers G, Korber B, Foley B, Jeang K, Mellors J, Wain-Hobson S. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.M: Los Alamos National Laboratory; 1996. [Google Scholar]
- 21.Myers G, Korber B, Hahn B, Foley B, Mellors J, Leitner T, McCutchan F, Kuiken C. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.M: Los Alamos National Laboratory; 1997. [Google Scholar]
- 22.Nkengasong J, Janssens W, Heyndrickx L, Fransen K, Ndumbe P, Motte J, Leonaers A, Ngolle M, Ayuk J, Piot P, van der Groen G. Genotypic subtypes of HIV-1 in Cameroon. AIDS. 1994;8:1405–1412. doi: 10.1097/00002030-199410000-00006. [DOI] [PubMed] [Google Scholar]
- 23.Robertson D, Hahn B, Sharp P. Recombination in AIDS viruses. J Mol Evol. 1995;40:249–259. doi: 10.1007/BF00163230. [DOI] [PubMed] [Google Scholar]
- 24.Robertson D, Sharp P, McCutchan F, Hahn B. Recombination in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 25.Sabino E, Shpaer E, Morgado M, Korber B, Diaz R, Bongertz V, Cavalcante S, Galvao-Castro B, Mullins J, Mayer A. Identification of human immunodeficiency virus type 1 envelope genes recombinant between subtypes B and F in two epidemiologically linked individuals from Brazil. J Virol. 1994;68:6340–6346. doi: 10.1128/jvi.68.10.6340-6346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salminen M, Carr J, Burke D, McCutchan F. Identification of breakpoints in intergenotypic recombinants of HIV-1 by bootscanning. AIDS Res Hum Retroviruses. 1995;11:1423–1425. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- 27.Salminen M, Carr J, Robertson D, Hegerich P, Gotte D, Koch C, Sanders-Buell E, Gao F, Sharp P, Hahn B, Burke D, McCutchan F. Evolution and probable transmission of intersubtype recombinant human immunodeficiency virus type 1 in a Zambian couple. J Virol. 1997;71:2647–2655. doi: 10.1128/jvi.71.4.2647-2655.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon F, Mauclere P, Roques P, Loussert-Ajaka I, Muller-Trutwin M, Saragosti S, Georges-Courbot M, Barre-Sinoussi F, Brun-Vezinet F. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat Med. 1998;4:1032–1037. doi: 10.1038/2017. [DOI] [PubMed] [Google Scholar]
- 29.Takehisa J, Zekeng L, Ido E, Mboudjeka I, Moriyama H, Miura T, Yamashita M, Gürtler L, Hayami M, Kaptué L. Various types of HIV mixed-infections in Cameroon. Virology. 1998;245:1–10. doi: 10.1006/viro.1998.9141. [DOI] [PubMed] [Google Scholar]
- 30.Takehisa J, Zekeng L, Miura T, Ido E, Yamashita M, Mboudjeka I, Gürtler L, Hayami M, Kaptué L. Triple HIV-1 infection with group O and group M of different clades in a single Cameroonian AIDS patient. J Acquired Immune Defic Syndr. 1997;14:81–82. doi: 10.1097/00042560-199701010-00015. [DOI] [PubMed] [Google Scholar]
- 31.Willey R, Smith D, Laskey L, Theodore T, Earl P, Moss B, Capon D, Martin M. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wooley D, Smith R, Czajak S, Desrosiers R. Direct demonstration of retroviral recombination in a rhesus monkey. J Virol. 1997;71:9650–9653. doi: 10.1128/jvi.71.12.9650-9653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]