Abstract
Splicing and posttranscriptional processing of eukaryotic gene transcripts are linked to their nuclear export and cytoplasmic expression. Unspliced pre-mRNAs and intronless transcripts are thus inherently poorly expressed. Nevertheless, human and animal viruses encode essential genes as single open reading frames or in the intervening sequences of other genes. Many retroviruses have evolved mechanisms to facilitate nuclear export of their unspliced mRNAs. For example, the human immunodeficiency virus RNA-binding protein Rev associates with the soluble cellular export receptor CRM 1 (exportin 1), which mediates nucleocytoplasmic translocation of Rev-HIV RNA complexes through the nuclear pore. The transforming human herpesvirus Epstein-Barr virus (EBV) expresses a nuclear protein, SM, early in its lytic cycle; SM binds RNA and posttranscriptionally activates expression of certain intronless lytic EBV genes. Here we show that both the trans-activation function and cytoplasmic translocation of SM are dependent on association with CRM 1 in vivo. SM is also shown to be associated in vivo with other components of the CRM 1 export pathway, including the small GTPase Ran and the nucleoporin CAN/Nup214. SM is shown to be present in the cytoplasm, nucleoplasm, and nuclear envelope of transfected cells. Mutation of a leucine-rich region (LRR) of SM inhibited CRM 1-mediated cytoplasmic translocation and SM activity, as did leptomycin B, an inhibitor of CRM 1 complex formation. Surprisingly, however, leptomycin B treatment and mutation of the LRR both led to SM becoming more tightly attached to intranuclear structures. These findings suggest a model in which SM is not merely a soluble carrier protein for RNA but rather is bound directly to intranuclear proteins, possibly including the nuclear pore complex.
The Epstein-Barr virus (EBV) protein SM posttranscriptionally activates intronless genes and inhibits expression of intron-containing genes (23, 24, 27, 39). In contrast to the majority of cellular genes, many EBV genes expressed during lytic replication are intronless (2, 26), and SM may therefore be important in enhancing expression of other lytic EBV genes. Activation of intronless genes by SM appears to be exerted both at the level of pre-mRNA stability and nucleocytoplasmic mRNA export (39). SM binds RNA in vitro and is capable of shuttling from nucleus to cytoplasm in a heterokaryon assay (39, 41). It is therefore probable that SM, like the human immunodeficiency virus (HIV) Rev protein, is an RNA transport protein.
It has been shown that several proteins involved in nucleocytoplasmic transport of RNA or protein bind to exportins, such as the recently characterized cellular export receptor CRM 1 (exportin 1) (for a review, see reference 43). Various proteins, including HIV Rev and cyclic AMP-dependent protein kinase inhibitor (PKI), contain leucine-rich regions (LRR) which serve as nuclear export signals (NESs) that are required for nuclear export (12, 45). The mechanism of NES-dependent export has been elucidated by the finding that CRM 1 forms a physical complex with NES-containing peptides and conjugates (14, 15, 33). Complex formation may require the presence of a small GTPase, Ran, associated with GTP (Ran-GTP), resulting in a tripartite complex of CRM 1 with NES-containing proteins and Ran-GTP (14, 38). Current models for NES-dependent nuclear export postulate a gradient of Ran-GTP across the nuclear envelope, with more Ran-GDP present in the cytoplasm. Thus, the CRM 1–Ran-GTP–NES complex is thought to be formed in the nucleus and translocated to the cytoplasm where Ran-GTP is converted to Ran-GDP, accompanied by release of CRM 1 and the NES protein. CRM 1 also associates with the nuclear pore complex (NPC) via CAN/Nup214 and other nucleoporins (13). Thus, CRM 1 is postulated to direct movement of its cargo NES protein to the NPC and dock with components of the NPC. The exact details of these interactions and the mechanism of directional translocation of the CRM 1-NES protein complex through the NPC remain to be delineated.
SM facilitates the expression of intronless mRNAs, as opposed to the unspliced retroviral RNAs transported by Rev-like proteins. Nevertheless, it was considered possible that SM-mediated RNA export occurs via a CRM 1-dependent pathway. We therefore performed immunoprecipitation and immunofluorescence experiments to determine whether SM interacts physically with CRM 1 in vivo. The specificity of CRM 1-mediated effects was confirmed with a specific inhibitor of CRM 1 complex formation, leptomycin B (LMB). The role of CRM 1 in SM function was investigated in reporter assays that measure trans activation of gene expression by SM. These studies revealed an in vivo association of SM with CRM 1 that mediates both nuclear export and functional activity of SM. The subcellular distribution of SM was also analyzed, by cellular fractionation studies. These studies revealed an effect of CRM 1 not only on nuclear export of SM but also on the degree to which SM is bound to structural elements of the nucleus. Finally, the functional role of a putative leucine-rich SM NES was analyzed by site-directed mutagenesis. These experiments demonstrate that the LRR is important in mediating interaction with CRM 1 and suggest a novel mechanism for SM function.
MATERIALS AND METHODS
Immunofluorescence assays.
Cells were grown on glass coverslips prior to washing in phosphate-buffered saline (PBS) and fixation with ice-cold acetone. Fixed cells were incubated in polyclonal rabbit anti-SM antisera at a 1:500 dilution for 1 h at room temperature, washed three times in PBS, and incubated for 1 h with rhodamine-conjugated affinity-purified F(ab′)2 goat anti-rabbit antibodies (Rockland, Gilbertsville, Pa.) at a 1:1,000 dilution. Cells were washed and overlaid with glycerol, and immunofluorescent microscopy was performed with a Nikon Optiphot 2 microscope. Deconvoluted fluoromicrographs were acquired with a DeltaVision deconvolution fluorescent microscope system (Applied Precision, Issaquah, Wash.). Individual optical sections of 200-nm thickness were obtained from slides prepared as described above. Nuclei were counterstained with 0.5 μg of DAPI (4′,6-diamidino-2-phenylindole) per ml, and slides were overlaid with Faramount aqueous mounting medium (DAKO Corporation, Carpinteria, Calif.) prior to microscopy.
Cell lines, plasmids, and antibodies.
SM, antisense control, and CMV-CAT plasmids have been previously described (39). SM mutants were generated by oligonucleotide-directed site-specific mutagenesis (8). CRM 1 cDNA, the influenza virus hemagglutinin (HA)-tagged carboxy-terminal amino acid fragment of CAN/Nup214 (amino acids 1864 to 2090), and polyclonal rabbit anti-CRM 1 (13) were kind gifts of G. Grosveld (St. Jude Children’s Research Hospital, Memphis, Tenn.). CRM 1 cDNA and the HA-CAN/Nup214 fragment were cloned in the pCDNA3 expression vector (Invitrogen Corp.). Polyclonal anti-SM antibodies were generated by injecting rabbits with gel-purified SM–glutathione S-transferase fusion proteins (39). Polyclonal anti-Ran antibodies were purchased from Covance Laboratories (Richmond, Calif.). BJAB, an EBV-negative B lymphoma cell line, and Cos 7 cells have been previously described (16, 31). Anti-FLAG monoclonal antibody was purchased from Sigma (St. Louis, Mo.).
Transfections and reporter assays.
BJAB cells were electroporated with expression constructs, and chloramphenicol acetyltransferase (CAT) assays were performed exactly as previously described (39). Each data point represents pooled results from at least three independent transfections. LMB (kind gift of B. Wolff; Novartis AG) treatment was begun immediately after transfection at a concentration of 10 nM and continued until harvest 16 h later. Cos 7 cells were transfected with LipofectaminePlus, per the manufacturer’s protocol (Gibco Life Sciences).
Immunoprecipitation and immunoblotting.
Cells were lysed 48 h after transfection in immunoprecipitation buffer (Tris-buffered saline [pH 7.4], 1% Triton X-100, 1 mM dithiothreitol, 100 μM GTP-γS, and a mixture of protease inhibitors [Sigma protease inhibitor cocktail no. P2714]). One hundred fifty microliters of each lysate was cleared with preimmune serum, incubated with 1 μl of undiluted polyclonal antibody and 20 μl of protein A-conjugated agarose beads (Sigma) for 90 min at 4°C, and washed four times in immunoprecipitation buffer. Precipitation with anti-HA monoclonal antibodies was performed with 150 μl of 12CA5 hybridoma supernatant (10). Precipitates were boiled in protein loading buffer, electrophoresed, and immunoblotted as previously described (44). Immunoblotting was performed with a 1:400 dilution of polyclonal anti-SM antibody and a 1:7,500 dilution of horseradish peroxidase-linked donkey anti-rabbit immunoglobulin or a 1:4,000 dilution of horseradish peroxidase-linked goat anti-mouse immunoglobulin (Amersham), and immunoreactive proteins were detected by enhanced chemiluminescence.
Cell fractionation and nuclear extractions.
Cos 7 cells were harvested 48 h after transfection by scraping into ice-cold PBS, washed in PBS, and lysed in 0.5% Nonidet P-40, 50 mM Tris–5 mM MgSO4. Under these conditions the nuclei remain intact, as confirmed by light microscopy. Nuclei were separated by centrifugation at 950 × g for 10 min. Nuclei were resuspended in a solution of 250 mM sucrose, 50 mM Tris (pH 7.4), and 5 mM MgSO4 and treated with DNase (250 μg/ml) and RNase A (1 mg/ml) for 2 h at 4°C, followed by washing and resuspension in 50 mM Tris (pH 7.4)–5 mM MgSO4. High-salt extractions were performed by dropwise addition of 2 M NaCl–50 mM Tris (pH 7.4) with constant mixing to a final concentration of 1.6 M NaCl and incubation on ice for 30 min. High-salt extractions with β-mercaptoethanol were performed identically, with the inclusion of β-mercaptoethanol at 1% (vol/vol). The remaining nuclear envelopes were sedimented by centrifugation at 13,000 × g for 30 min. The salt-extracted fraction was desalted and concentrated with a Microcon 10 filter apparatus (Amicon, Beverly, Mass.). Protease inhibitors, as described above, were included at all steps of the isolation process. Equal fractions from each step of the fractionation procedure were prepared for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted as described above. In experiments with LMB, cells were treated with LMB for 16 h after transfection. Treated and untreated cells were harvested at 16 h posttransfection and fractionated exactly as described above.
RESULTS
SM activity is dependent on CRM 1 (exportin 1) function.
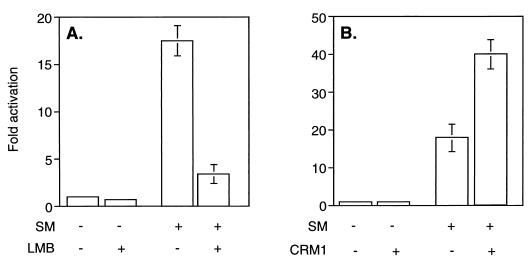
SM-mediated gene activation is correlated with enhanced cytoplasmic accumulation of intronless target gene mRNAs (7, 39). SM contains an LRR resembling an NES found in certain proteins which shuttle from nucleus to cytoplasm (45). Such proteins, notably the HIV Rev protein, bind CRM 1 (exportin 1) and the small GTPase Ran in the nucleus (14). Translocation to the cytoplasm is thought to be followed by hydrolysis of Ran-associated GTP and complex dissociation. Thus, it was considered possible that SM chaperones intronless EBV mRNAs to the cytoplasm via a CRM 1-dependent pathway. We therefore wished to determine whether SM-mediated gene activation is dependent on an interaction with CRM 1. We first examined the effect of a specific inhibitor of CRM 1 complex formation, LMB (47), on SM function. BJAB cells were transfected with an intronless CAT reporter plasmid (CMV-CAT) and an expression vector encoding either SM or antisense SM as a control. Immediately after transfection, cells were incubated in growth medium containing LMB or in control medium without LMB. Cells were harvested after 16 h, at which time viability was not affected by LMB treatment (data not shown) and CAT activity was measured. As previously reported, SM led to activation of CAT expression (approximately 16-fold). LMB itself did not affect baseline expression of CAT activity from the reporter plasmid. However, LMB treatment led to a marked reduction in activation by SM (to four times that of the control) (Fig. 1A).
FIG. 1.
Effects of modulating CRM 1 activity on SM function. (A) Effect of LMB on trans-activation by SM. CAT activity was measured in lysates of BJAB cells transfected with SM or antisense control plasmid and CAT reporter plasmid CMV-CAT. Cells were treated with either LMB (10 nM) or control medium immediately after transfection. Results are means of at least three independent transfections. (B) Effect of CRM 1 overexpression on trans-activation by SM. CAT activity was measured in lysates of BJAB cells transfected with SM or antisense control plasmid, CAT reporter plasmid CMV-CAT, and either control or CRM 1 expression vector.
These experiments indicated that an interaction with CRM 1 was involved in some aspects of gene activation by SM. We therefore attempted to determine whether overexpression of CRM 1 could augment SM-mediated gene activation. A CRM 1 expression plasmid or control vector was cotransfected with CMV-CAT and either SM or antisense SM into BJAB cells, and CAT activity was measured 48 h after transfection. As shown in Fig. 1B, CRM 1 overexpression stimulated SM activation. Activation by SM alone was 17-fold and increased to 40-fold over the control when CRM 1 was cotransfected. CRM 1 overexpression did not increase CAT activity in the absence of SM, demonstrating that CRM 1 does not have a nonspecific stimulatory effect on CAT gene expression.
CRM 1 expression affects intracellular localization of SM.
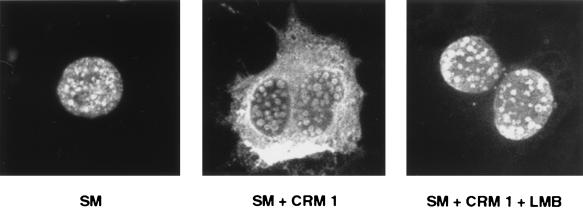
The preceding experiments suggested that CRM 1 may be involved in nucleocytoplasmic translocation of SM. While it has been shown that SM can shuttle from nucleus to cytoplasm in a heterokaryon assay (41), immunofluorescence studies have demonstrated exclusively nuclear localization of SM (6, 48). SM-transfected cells typically display a speckled nuclear fluorescence with nucleolar sparing when stained with anti-SM antibodies (Fig. 2). Such apparently exclusive nuclear localization of other known shuttling proteins has been reported, presumably because the concentration of cytoplasmic proteins is below the limits of detection of conventional indirect immunofluorescence microscopy (34). We reasoned that if cytoplasmic transport of SM is CRM 1 dependent, overexpression of CRM 1 might lead to visible cytoplasmic accumulation of SM. We therefore transfected Cos 7 cells, in which nuclei and cytoplasm are easily differentiated, with SM and either CRM 1 expression vector or control plasmid and examined the transfected cells by indirect immunofluorescence microscopy. As shown in Fig. 2, overexpression of CRM 1 led to dramatic intracellular relocalization of SM. The nuclei of CRM 1 cotransfected cells were relatively depleted of SM, and the cytoplasm became diffusely stained by the anti-SM antibodies.
FIG. 2.
Effects of CRM 1 overexpression and LMB treatment on cellular distribution of SM. Cos 7 cells were grown on coverslips prior to transfection, and immunofluorescence microscopy was performed with anti-SM antibodies. Cells were transfected with SM plasmid alone or with SM and CRM 1 expression plasmids. Cells were also transfected with SM and CRM 1 expression plasmid and treated with LMB immediately after transfection.
To confirm the role of CRM 1 in nucleocytoplasmic export of SM, we studied the effect of LMB on cytoplasmic translocation of SM. Cos 7 cells were transfected with SM and CRM 1 expression vectors, as described above, and incubated in growth medium in the presence or absence of LMB. Sixteen hours after transfection, the cells were fixed and stained with anti-SM antibodies and examined by indirect immunofluorescence microscopy. LMB treatment resulted in exclusively nuclear localization of SM despite overexpression of CRM 1 (Fig. 2). These data thus confirm that cytoplasmic translocation of SM is directly or indirectly dependent on CRM 1 complex formation.
Mutation of a putative NES impairs SM function.
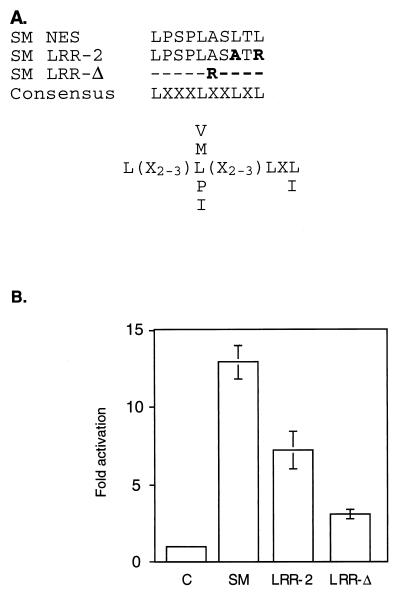
The predicted amino acid sequence of SM contains an LRR which satisfies the consensus requirements (LX2–3LX2–3LXL) for an NES, as found in other proteins known to interact with CRM 1 (4) (Fig. 3A). It should be noted that although there is not strong selection for a particular amino acid at positions denoted by “X”, some amino acids at these positions lead to nonfunctional NESs (4). Thus, not all regions meeting these broad criteria are functional NESs. In order to determine if the putative SM NES (amino acids 227 to 236) is required for SM function, we examined the effect of mutating this region on SM-mediated gene activation. Site-directed mutagenesis was used to generate SM mutants altered in the relevant region. The mutant LRR-2 has leucines 234 and 236 replaced by alanine and arginine, respectively, whereas all 11 amino acids are deleted in the LRR-Δ mutant. We then compared these mutants with wild-type SM in the ability to activate gene expression in the CAT reporter assay (Fig. 3B). Both mutants were impaired in activation function, with LRR-Δ being the least active. We have previously demonstrated that in addition to its putative RNA transport function, SM stabilizes and leads to increased accumulation of target gene RNAs in the nucleus as well as the cytoplasm (39). Therefore, the residual activating function of the SM mutants was not unexpected despite a potential defect in the ability to interact with CRM 1 and hence in the ability to translocate to the cytoplasm.
FIG. 3.
Effect of mutation of a leucine-rich putative SM NES on SM-mediated activation. (A) Amino acids 227 to 236 of SM, with four leucines separated by three, two, and one amino acid, fits the broad consensus sequence described for an NES (3). LRR-2 and LRR-Δ are SM mutants with two leucines altered or the entire LRR deleted and replaced with an arginine, respectively. Amino acid substitutions are shown in bold. “X” represents no selection for a particular amino acid at that site. Preferred amino acids at particular sites are shown by their one-letter codes. (B) BJAB cells were transfected with a CAT reporter plasmid and either SM or a mutant SM plasmid, and CAT activity was measured as described in the text.
Deletion of the LRR abolishes CRM 1-mediated nuclear export of SM.
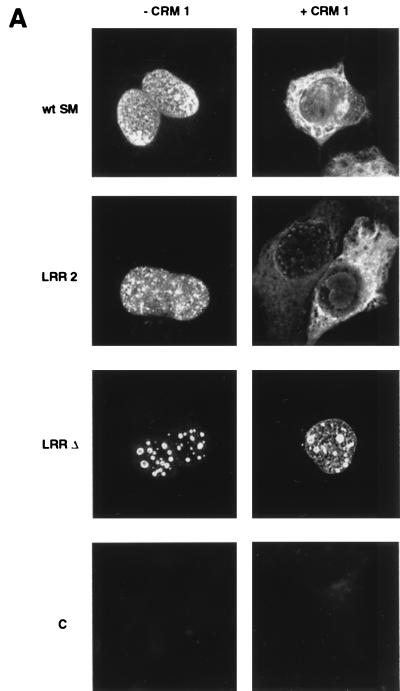
We next wished to determine whether the putative SM NES was important for CRM 1-mediated cytoplasmic localization of SM. Cos cells were transfected with wild-type or LRR mutant plasmids and either CRM 1 plasmid or control vector and examined by immunofluorescence microscopy. In the absence of CRM 1 overexpression, both LRR mutants were detectable only in the nucleus, as expected (Fig. 4). However, both mutants exhibited a more punctate nuclear distribution than wild-type SM. This difference was most obvious with the LRR-Δ mutant, where the fluorescence was most prominent in large nuclear dots. It should be noted that a similar distribution of wild-type SM, albeit not as marked, was observed when SM-transfected cells were treated with LMB (Fig. 2), suggesting that the more diffuse nuclear distribution normally seen with wild-type SM is dependent on interaction with CRM 1.
FIG. 4.
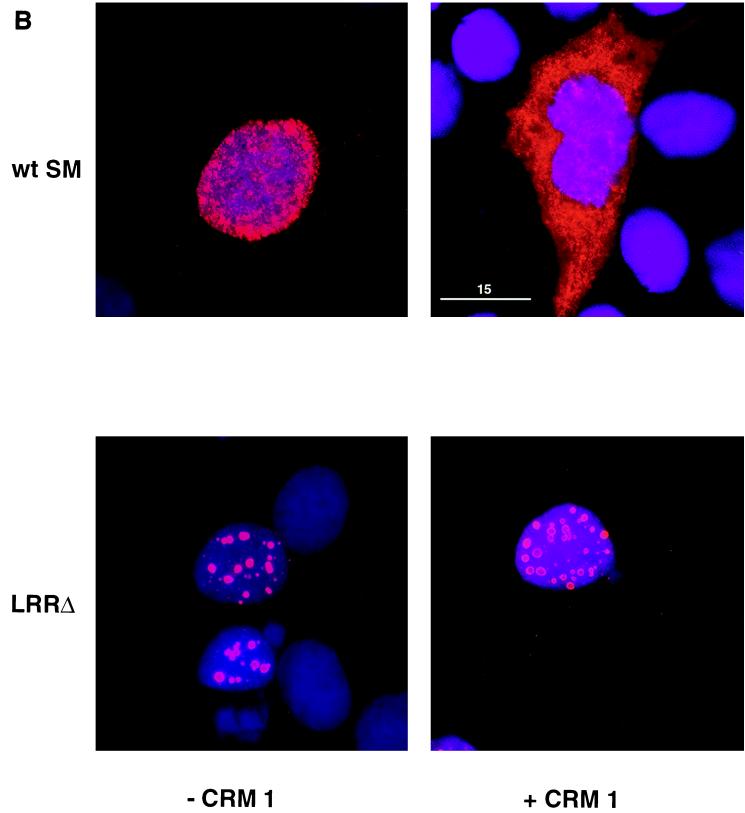
Effects of CRM 1 overexpression on cellular distribution of SM and SM LRR mutants. (A) Immunofluorescence microscopy was performed on SM- or SM mutant-transfected cells with anti-SM antibodies as for Fig. 2. Cos 7 cells were transfected with either wild-type SM (wt SM), SM mutant plasmid LRR-2 or LRR-Δ, or vector plasmid (C), as indicated. Cells were also cotransfected with either control vector (−CRM 1) or CRM 1 expression plasmid (+CRM 1). (B) Cos 7 cells transfected with wt SM or LRR-Δ plasmid and cotransfected with either CRM 1 or control plasmid were stained with anti-SM antibodies. Nuclei were visualized by staining with DAPI. Immunofluorescence images were acquired with a deconvolution fluorescence microscope system. SM staining appears red, and nuclei are purple. Bar, 15 μm.
The effect of CRM 1 overexpression on cytoplasmic translocation of SM in the case of LRR-Δ was also markedly different from wild-type SM or LRR-2. When CRM 1 was overexpressed by cotransfection, the LRR-2 mutant retained the ability to translocate to the cytoplasm (Fig. 4). However, the LRR-Δ mutant displayed a different immunofluorescence pattern, could not be detected in the cytoplasm despite overexpression of CRM 1, and remained confined to large nuclear foci. These data indicate that LRR-2 retains, at least partially, the ability to be exported and interact with CRM 1, whereas deletion of the LRR results in a more severe export defect. These findings thus correlate well with the data on LRR mutant function which revealed that LRR-2 retained more activating capability than LRR-Δ.
In order to confirm these findings and further examine the unusual distribution of the LRR-Δ mutant in the nucleus, SM- and LRR-Δ-transfected cells were examined by immunofluorescence deconvolution microscopy (1). As shown in Fig. 4B, cotransfection of CRM 1 had the expected effect on wild-type SM, causing cytoplasmic translocation, whereas LRR-Δ remained confined to the nucleus. The LRR-Δ mutant was localized to numerous large circular foci (from 10 to 20 per cell) which appeared to be distributed throughout the nucleus and were not confined to the nuclear rim. These foci were less intensely stained in the center and were approximately 1 μm in diameter. The size and distribution of these foci were not affected by overexpression of CRM 1. Whether these foci correspond to areas of pre-mRNA processing or other intranuclear functions remains to be determined.
SM associates with CRM 1 in vivo.
In order to determine whether SM could be found complexed to CRM 1 in vivo, we attempted to immunoprecipitate SM from cell lysates with anti-CRM 1 antibodies. Since CRM 1-NES complexes exist as ternary complexes with the GTP-bound form of Ran (14, 38), immunoprecipitations were performed in the presence of the nonhydrolyzable GTP derivative GTP-γS to maximize the likelihood of detecting SM-CRM 1 complex formation. Proteins immunoprecipitated from SM-transfected Cos 7 cells with anti-CRM 1 antibodies were separated by SDS-PAGE and immunoblotted with anti-SM antibodies. Anti-CRM 1 antibodies precipitated a protein of the appropriate molecular weight (detected by anti-SM antibodies) from SM-transfected cells but not from control-transfected cells (Fig. 5A). Comparison with unprecipitated lysate indicates that approximately 30% of the SM in transfected cells is precipitable by anti-CRM 1 antibodies under these conditions. However, anti-SM antibodies did not precipitate CRM 1 from SM-transfected cells (data not shown), suggesting that SM antibodies may block complex formation or, less likely, that the CRM 1-bound form of SM is not reactive with our antibody. Similar results were obtained with SM-transfected BJAB cells (Fig. 5B).
FIG. 5.
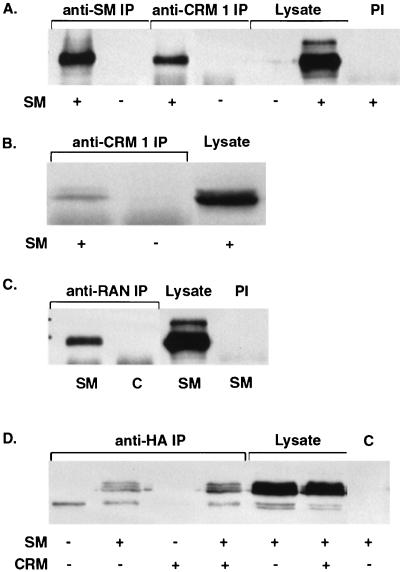
Coimmunoprecipitation of CRM 1 and CRM 1-associated proteins with SM. (A) Lysates of Cos 7 cells transfected with SM or control vector (SM + and −) were immunoprecipitated with anti CRM 1 (anti-CRM 1 IP) or anti-SM (anti-SM IP) antibodies and immunoblotted to detect SM. A control immunoprecipitation of SM-transfected cell lysate with preimmune rabbit serum (PI) is also shown. (B) Lysates of BJAB cells transfected with SM or control vector were immunoprecipitated with anti-CRM 1 antibodies and immunoblotted as in panel A. (C) Lysates of Cos 7 cells transfected with SM and CRM 1 were immunoprecipitated with anti-Ran antibodies (anti-RAN IP) and immunoblotted to detect SM. Control immunoprecipitations with preimmune rabbit serum (PI) are also shown. (D) Cells were transfected with a plasmid expressing an HA-tagged carboxy-terminal fragment of CAN/Nup214 (HA-ΔCAN) and SM or control plasmid. Lysates were immunoprecipitated with anti-HA monoclonal antibody CA125 (anti-HA IP) and immunoblotted with anti-SM antibodies. A control immunoprecipitation (C) performed with an irrelevant monoclonal antibody (anti-FLAG) is also shown. In all panels, lanes containing an equivalent amount of unimmunoprecipitated lysate are indicated.
Since it is known that proteins that are exported by CRM 1 may cooperatively associate with both CRM 1 and the small GTPase Ran (14), we asked whether we could also demonstrate an in vivo association of SM with Ran. Such an association would confirm the functional importance of the CRM 1-SM interaction and suggest that directional export of SM is dependent on the nuclear to cytoplasmic gradient of Ran-GTP. SM-transfected or control-transfected cells were therefore lysed and immunoprecipitated with anti-Ran antibodies. As shown in Fig. 5C, anti-Ran antibodies also immunoprecipitated SM, suggesting that SM forms a tripartite complex with CRM 1 and Ran GTPase in vivo.
A third line of evidence that supports the existence of an SM-CRM 1 interaction was provided by experiments in which the association of SM with a nucleoporin known to interact with CRM 1 was investigated. CRM 1 binds the nucleoporin CAN/Nup214 via a series of FXFG repeats in the carboxy-terminal portion of CAN (13). Expression of a carboxy-terminal fragment of CAN (amino acids 1864 to 2090 [ΔCAN]) containing these repeats has been shown to competitively inhibit CRM 1 function (3, 49). We therefore asked whether a potential indirect association of SM with the carboxy-terminal portion of CAN could be detected by coimmunoprecipitation. Cells were transfected with SM and HA-tagged ΔCAN, lysed, and immunoprecipitated with anti-HA monoclonal antibodies. Immunoprecipitates were analyzed for the presence of SM by SDS-PAGE and immunoblotting. As shown in Fig. 5D, anti-HA antibodies precipitated SM from cells cotransfected with HA-ΔCAN. (Multiple forms of SM, as previously described [7], were visualized in this experiment, as electrophoresis was performed at a higher polyacrylamide concentration). These experiments indicate that SM interacts with at least one component of the NPC, CAN/Nup214, possibly indirectly via CRM 1.
Mutation of the LRR alters intranuclear compartmentalization of SM.
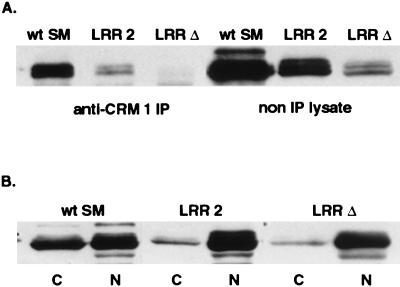
Based on the above findings, we expected that the LRR mutants, particularly LRR-Δ, might not bind to CRM 1 and therefore would not be precipitable with anti-CRM 1 antibodies. Parallel immunoprecipitation experiments were therefore performed with lysates from cells transfected with wild-type SM, LRR-2, and LRR-Δ. As expected, little or no mutant protein was precipitated with anti-CRM 1 antibodies (Fig. 6A, lanes 2 and 3). Surprisingly, however, the total amount of mutant SM protein in unprecipitated cell lysates was also less than that of wild-type SM (Fig. 6, lanes 4, 5, and 6). This was in spite of there being no obvious difference in the amounts of mutant and wild-type SM protein when assessed by immunofluorescence (Fig. 4) and our previous finding of equal amounts of mutant and wild-type SM in immunoblots of whole-cell lysates of similarly transfected cells (data not shown). We therefore further analyzed the amounts and intracellular distribution of mutant and wild-type SM proteins in transfected cells. Cells transfected with mutant or wild-type SM plasmids were lysed in immunoprecipitation buffer containing 1% Triton X-100, and both the lysate and the remaining nuclear pellet were analyzed by immunoblotting. As shown in Fig. 6B, whereas approximately 50% of wild-type SM was found in the soluble lysate, which is expected to contain soluble cytoplasmic and nucleoplasmic SM, only 10 and 5% of LRR-2 and LRR-Δ, respectively, were present in this fraction. Conversely, correspondingly greater amounts of the mutant SM proteins were found in the nuclear pellet fraction.
FIG. 6.
Effect of LRR mutation on intracellular compartmentalization of SM. (A) Detergent-solubilized lysates of cells transfected with SM, LRR-2, or LRR-Δ were immunoprecipitated with anti-CRM 1 antibodies (anti-CRM 1 IP) or electrophoresed directly (non IP lysate) and immunoblotted with anti-SM antibodies. (B) Distribution of SM between detergent-soluble and insoluble fractions. Detergent-soluble (C) and insoluble nuclear pellet (N) fractions of cells transfected with wt SM, LRR-2, or LRR-Δ were analyzed by immunoblotting with anti-SM.
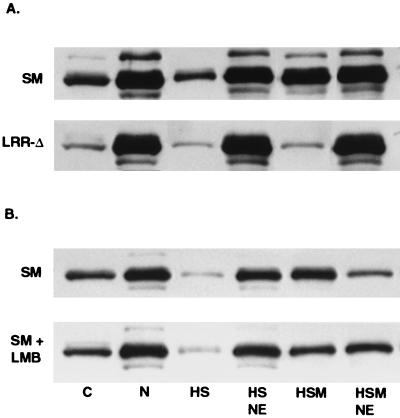
This effect of LRR mutation on intracellular distribution of SM, resulting in a tighter association with nuclear structures, was thus consistent with the immunofluorescence studies described previously which showed that LRR-Δ did not translocate to the cytoplasm. In addition, it was apparent that a large proportion of even wild-type SM remained associated with the nucleus despite detergent treatment. In order to further analyze the nature of the association of SM with nuclear structures, transfected cells were subjected to a further series of fractionation steps. Cos 7 cells transfected with wild-type SM or LRR-Δ plasmid were washed and rapidly lysed in hypotonic buffer containing 0.5% Nonidet P-40. At this detergent concentration, the nuclei remain physically intact and adherent cytoplasm is minimal, as confirmed by light microscopy (data not shown). The lysates were reserved and the nuclei were nuclease treated and extracted with high-salt buffer with or without β-mercaptoethanol. Extraction with β-mercaptoethanol reduces and releases proteins which are oxidatively bound to the nuclear matrix, in addition to the soluble matrix-associated fraction extracted with high salt alone (11, 22). Each fraction and the remaining nuclear envelopes were then subjected to SDS-PAGE and immunoblotted with anti-SM antibodies (Fig. 7A). Approximately 20% of wild-type SM was found in the detergent-soluble fraction, compared to 5% of LRR-Δ. Approximately 10% of the remaining nuclear wild-type SM was extracted with high salt, and an additional 30% was extracted with the inclusion of β-mercaptoethanol, indicating that the latter fraction was also associated with the nuclear matrix. The remaining 60% was associated with the nuclear envelope fraction, which includes the nuclear pore complexes (35). In contrast, the majority of LRR-Δ remained tightly associated with the nuclear envelope fraction and was resistant to the high-salt extraction steps (Fig. 7A). These data indicate that under normal conditions, wild-type SM is found in both a soluble and a tightly nucleus-bound fraction. Further, while SM is also present in an extractable matrix-bound form, a substantial proportion of SM remains tightly associated with the nuclear envelope. In contrast, the majority of LRR-Δ protein is found in the nuclear fractions and particularly in the nuclear envelope fraction.
FIG. 7.
Effects of LRR mutation and LMB treatment on intranuclear compartmentalization of SM. Cells were lysed and separated into soluble (C) and nuclear (N) fractions. Intact nuclei were nuclease treated and extracted with high salt (HS) or high salt plus 2-mercaptoethanol (HSM). Nuclear envelopes remaining after HS extraction (HS NE) or HSM extraction (HSM NE) were collected by centrifugation. Equivalent amounts of each fraction were analyzed by immunoblotting with anti-SM antibodies. (A) Cells were transfected with SM plasmid or LRR-Δ plasmid, as shown, and harvested for fractionation after 48 h. (B) Cells were transfected with SM plasmid and incubated in growth medium alone (SM) or growth medium with LMB (SM + LMB), harvested, and fractionated 16 h posttransfection.
These results were somewhat surprising in the context of the conventional model for CRM 1-mediated export of NES proteins, in which NES proteins are diffusible nucleoplasmic proteins which are bound and transported to the NPC by CRM 1. Further, if CRM 1 mediates docking and interaction with nucleoporins, one might have expected that an inability to interact with CRM 1 would result in less rather than more association with the nuclear envelope. To further test the hypothesis that interaction with CRM 1 modulates the intranuclear attachment of SM, we examined the effect of LMB on the intranuclear compartmentalization of SM. If the increased affinity of LRR-Δ for the nuclear envelope and matrix were indeed due to decreased CRM 1 binding, LMB treatment should result in similar changes in distribution of wild-type SM. SM-transfected cells were treated with LMB and harvested 16 h posttransfection to minimize toxicity. The cells were fractionated, and the distribution of SM in each fraction was compared to those from non-LMB-treated cells by immunoblotting. SM was present in both the soluble and nuclear fractions of untreated cells, as expected, and could be extracted from the nuclei with high salt plus β-mercaptoethanol (Fig. 7B). The proportion of SM that was extractable with high salt and β-mercaptoethanol in non-LMB-treated cells was greater than in the previous experiments and may be a reflection of the shorter time interval between transfection and harvest. Nevertheless, as can be seen by the relative amounts in each fraction, LMB treatment led to a decrease in the amount of SM that was extractable with high salt, particularly in the presence of β-mercaptoethanol, and to a corresponding increase in the amount that remained attached to the nuclear envelope fraction. These data indicate that LMB, by inhibiting the association of SM with CRM 1, alters its intranuclear compartmentalization in a manner that correlates with the effect of LRR mutation. The LRR therefore appears to be required not only for CRM 1-mediated transport to the cytoplasm but also for the proper association of SM with intranuclear structures.
DISCUSSION
In this study, we demonstrate that the SM protein of EBV binds the export receptor CRM 1 and that CRM 1 binding is important for activity and cytoplasmic localization of SM. The role of CRM 1 binding in both aspects of SM function is shown to be specific by the use of an inhibitor of CRM 1 complex formation, LMB. SM is also shown to associate in vivo with the CRM 1-binding GTPase Ran and the carboxy-terminal portion of a nucleoporin, CAN/Nup214, two components of CRM 1-linked nuclear export pathways. Further, we demonstrate that an LRR of SM is required for function and proper intranuclear distribution of SM.
SM is an EBV protein which enhances expression of intronless genes in a gene-dependent manner (23, 30). Although SM clearly has multiple mechanisms of action, including stabilization of RNA and enhancement of posttranscriptional processing (25, 39), it is likely that SM is involved in nucleocytoplasmic export of lytic EBV mRNAs. Several lines of evidence support a role for SM as a carrier protein for RNA. SM has been shown to bind RNA in vitro and to shuttle from cytoplasm to nucleus in a heterokaryon assay (39, 41). SM also enhances cytoplasmic accumulation of target mRNAs (7, 39). Further, SM is homologous to the herpes simplex virus ICP27 protein, which has been shown to shuttle from nucleus to cytoplasm and bind RNA in both locations in vivo (40).
Our finding that there is a functionally important association of SM with CRM 1 in vivo indicates that EBV utilizes a cellular export pathway normally utilized for snRNA and 5S RNA export (21) to facilitate lytic EBV gene expression. The need for such mechanisms to enhance viral RNA expression may be a reflection of the inherent inefficiency with which intronless RNAs are expressed (19). It has been known for some time that addition of exogenous intron sequences to cDNA expression constructs enhances their expression (5). The exact reasons for such a stimulatory effect of intervening sequences on gene expression are poorly understood. The presence of intron sequences facilitates 3′ processing and polyadenylation of pre-mRNA, and a direct interaction between U1 snRNP protein U1A and polyadenylation factors has been shown (29, 32). Engagement of mRNA by the polyadenylation machinery also facilitates nucleocytoplasmic export, possibly due to an interaction of polyadenylation factors with the NPC (9, 20). Lack of assembly of spliceosomes on genes encoded as single open reading frames may thus be a relative barrier to entry of intronless mRNAs into a pathway that culminates in nuclear export. SM may allow direct targeting of intronless EBV mRNAs for signal-mediated export via CRM 1. Utilization of the CRM 1 pathway also provides another potential advantage for EBV gene expression since, in mammalian cells, the pathways for mRNA export and CRM 1-mediated export appear to be independent (3, 14, 47). Thus, inhibition of host cell gene expression during EBV replication could occur without necessarily affecting EBV lytic gene expression. In this context, it is relevant that SM and its homologs in herpes simplex virus and herpesvirus saimiri also inhibit expression of spliced host genes (17, 18, 46).
We have shown that the LRR of SM is required for CRM 1-mediated cytoplasmic translocation of SM and for full SM activity. In the case of HIV Rev, several lines of genetic and in vitro evidence indicate that the major function of the NES is CRM 1 binding (14, 15, 33, 42, 47). Mutational analysis of the Rev NES has demonstrated a good correlation between the ability to bind CRM 1 and Rev function (3). It is likely that the SM LRR performs a similar function for the reasons outlined above. Further, LMB treatment produces the same effects as LRR mutation on cytoplasmic localization and SM activity, indicating that the processes are dependent on CRM 1-NES complex formation. However, the additional effects of LMB treatment and LRR mutation on intranuclear distribution of SM suggest that SM may be mechanistically quite different from HIV Rev.
The results of LMB treatment and deletion of the LRR suggest that in the absence of CRM 1 binding, SM remains more tightly associated with nuclear structures and particularly with the nuclear envelope. Such a finding is somewhat surprising in the context of current models for CRM 1-NES protein export, in which SM plays the role of a soluble transport substrate for CRM 1 (Fig. 8A). According to such a model, SM might become at least transiently tethered to the NPC via CRM 1. Inhibition of CRM complex formation by mutation of the NES or LMB treatment, while leading to nuclear retention of SM, would not be expected to increase the attachment of SM to macromolecular nuclear structures. It is unlikely that the LRR mutations described have merely resulted in an overall decrease in solubility and thus intranuclear aggregation of the mutant proteins for several reasons. First, the SM mutants are properly imported to the nucleus and retain partial trans-activating function. Second, a large fraction of wild-type SM is normally associated with the nuclear envelope. Finally, LMB treatment has effects on the intranuclear distribution and attachment of SM that are similar to mutation of the LRR.
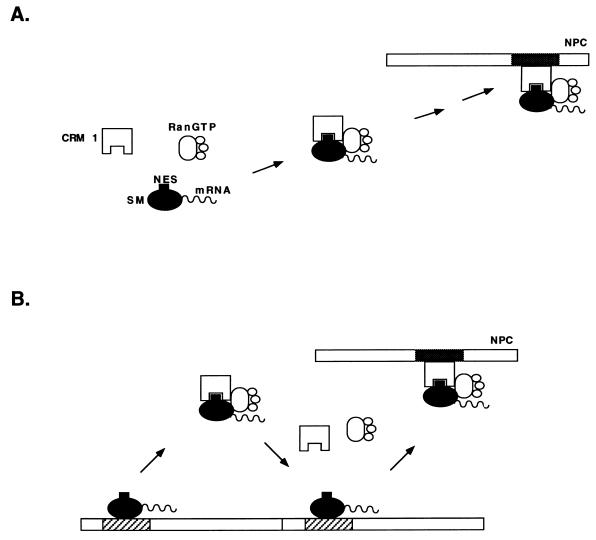
FIG. 8.
Models for intranuclear translocation of SM. (A) Conventional model for CRM 1-mediated export of an NES-containing protein. CRM 1 is shown binding to soluble SM via its NES and transporting it to the NPC. CRM 1 docks at the NPC by binding to an FXFG nucleoporin-binding site (shown in gray). (B) Alternatively, CRM 1 binding detaches SM from its sites on the nuclear matrix or nuclear envelope (diagonal bars). Successive rounds of SM release and binding by CRM 1 constitute a possible mechanism of translocation along the nuclear matrix and through the NPC.
The finding of SM in the nuclear matrix and nuclear envelope fractions suggests an alternative model for SM transport in which SM interacts directly with nuclear matrix and envelope proteins. In such a scenario, SM with its RNA cargo binds directly to one or more nuclear matrix sites. CRM 1 binding to SM in the presence of Ran-GTP would detach SM from its stationary binding site. Binding of SM to another site on the nuclear matrix accompanied by release from CRM 1 could then occur. Successive rounds of CRM 1 binding, release, and matrix binding could thus result in physical translocation of SM to the cytoplasmic face of the NPC. Such interactions would be expected to increase the efficiency of SM complex movement to the NPC, providing a track along the nuclear matrix. It should be noted that this model is similar to one proposed to explain directional translocation of import substrates into the nucleus (36, 37). In that model, the importin α/β-Ran-GDP complex dissociates from its cargo nuclear localization signal (NLS) when it binds NPC components. The import receptor is then released from the NPC on binding Ran-GTP, GTP hydrolysis occurs, NLS cargo is bound, and the cycle is repeated, leading to a “saltatory movement” of the importin-NLS complex across the NPC. Our proposed model is similar in that it postulates a successive series of CRM 1-SM binding and release reactions. However, SM is predicted to be also capable of interacting with nucleoporins or other structural nuclear proteins directly and does not invoke GTP hydrolysis, which does not appear to be required for the export process itself (28). Such a model does not preclude CRM 1-nucleoporin interactions and can therefore involve both SM and CRM 1 as active participants in translocation through the NPC. The components and location of the intranuclear foci of SM accumulation, especially those seen with the LRR-Δ mutant, remain to be determined. Based on the known effects of SM on nuclear mRNA, it is likely that some of these sites are involved in mRNA processing.
In summary, our data demonstrate that SM is a transport protein that functionally interacts with CRM 1 but that it may not be merely a soluble carrier of EBV RNA that acts as a link between the RNA, CRM 1, and the NPC. Rather, it is possible that SM exists in a dynamic structural association with the nuclear matrix and other intranuclear structures. Determination of the exact intranuclear sites of SM accumulation and whether SM associates directly with structural components of the NPC is likely to yield further insights into the mechanism of viral and cellular gene regulation at the level of mRNA transport.
ACKNOWLEDGMENTS
This work was supported by a recruitment grant to S.S. from the John Sealy Memorial Endowment Fund for Biomedical Research.
We express our appreciation to Barbara Wolff of Novartis AG for providing leptomycin B and to Gerard Grosveld for CRM 1 and CAN/Nup214 cDNA and anti-CRM 1 antibodies. We also thank C. Patterson, N. Murray, and A. Fields for many helpful discussions and review of the manuscript.
REFERENCES
- 1.Agard D A, Hiraoka Y, Shaw P, Sedat J W. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. doi: 10.1016/s0091-679x(08)60986-3. [DOI] [PubMed] [Google Scholar]
- 2.Baer R, Bankier A T, Biggin M D. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 3.Bogerd H P, Echarri A, Ross T M, Cullen B R. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J Virol. 1998;72:8627–8635. doi: 10.1128/jvi.72.11.8627-8635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogerd H P, Fridell R A, Benson R E, Hua J, Cullen B R. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callis J, Fromm M, Walbot V. Introns increase gene expression in cultured maize cells. Genes Dev. 1987;1:1183–1200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- 6.Cho M S, Jeang K T, Hayward S D. Localization of the coding region for an Epstein-Barr virus early antigen and inducible expression of this 60-kilodalton nuclear protein in transfected fibroblast cell lines. J Virol. 1985;56:852–859. doi: 10.1128/jvi.56.3.852-859.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook I D, Shanahan F, Farrell P J. Epstein-Barr virus SM protein. Virology. 1994;205:217–227. doi: 10.1006/viro.1994.1637. [DOI] [PubMed] [Google Scholar]
- 8.Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 9.Eckner R, Ellmeier W, Birnstiel M L. Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J. 1991;10:3513–3522. doi: 10.1002/j.1460-2075.1991.tb04915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson I A, Lerner R A, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fields A P, Kaufmann S H, Shaper J H. Analysis of the internal nuclear matrix. Oligomers of a 38 kD nucleolar polypeptide stabilized by disulfide bonds. Exp Cell Res. 1986;164:139–153. doi: 10.1016/0014-4827(86)90461-1. [DOI] [PubMed] [Google Scholar]
- 12.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 13.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fornerod M, Ohno M, Yoshida M. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 15.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 16.Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 17.Hardwicke M A, Sandri-Goldin R M. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J Virol. 1994;68:4797–4810. doi: 10.1128/jvi.68.8.4797-4810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy W R, Sandri-Goldin R M. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang M T F, Gorman C M. Intervening sequences increase efficiency of RNA 3′ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 1990;18:937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y, Carmichael G G. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarmolowski A, Boelens W C, Izaurralde E, Mattaj I W. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaufmann S H, Gibson W, Shaper J H. Characterization of the major polypeptides of the rat liver nuclear envelope. J Biol Chem. 1983;258:2710–2719. [PubMed] [Google Scholar]
- 23.Kenney S, Kamine J, Holley-Guthrie E, Mar E C, Lin J C, Markovitz D, Pagano J. The Epstein-Barr virus immediate-early gene product, BMLF1, acts in trans by a posttranscriptional mechanism which is reporter gene dependent. J Virol. 1989;63:3870–3877. doi: 10.1128/jvi.63.9.3870-3877.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenney S, Kamine J, Markovitz D, Fenrick R, Pagano J. An Epstein-Barr virus immediate-early gene product trans-activates gene expression from the human immunodeficiency virus long terminal repeat. Proc Natl Acad Sci USA. 1988;85:1652–1656. doi: 10.1073/pnas.85.5.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Key S C, Yoshizaki T, Pagano J S. The Epstein-Barr virus (EBV) SM protein enhances pre-mRNA processing of the EBV DNA polymerase transcript. J Virol. 1998;72:8485–8492. doi: 10.1128/jvi.72.11.8485-8492.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2343–2396. [Google Scholar]
- 27.Lieberman P M, O’Hare P, Hayward G S, Hayward S D. Promiscuous trans activation of gene expression by an Epstein-Barr virus-encoded early nuclear protein. J Virol. 1986;60:140–148. doi: 10.1128/jvi.60.1.140-148.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love D C, Sweitzer T D, Hanover J A. Reconstitution of HIV-1 rev nuclear export: independent requirements for nuclear import and export. Proc Natl Acad Sci USA. 1998;95:10608–10613. doi: 10.1073/pnas.95.18.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lutz C S, Murthy K G K, Schek N, O’Conner J P, Manley J L, Alwine J C. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 1996;10:325–337. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- 30.Markovitz D M, Kenney S, Kamine J, Smith M S, Davis M, Huang E-S. Disparate effects of two herpesvirus immediate-early gene trans-activators on the HIV-1 LTR. Virology. 1989;173:750–754. doi: 10.1016/0042-6822(89)90591-6. [DOI] [PubMed] [Google Scholar]
- 31.Menezes J, Liebold W, Klein G, Clements G. Establishment and characterization of an Epstein-Barr virus (EBV)-negative lymphoblastoid B cell line (BJA-B) from an exceptional EBV-genome-negative African Burkitt’s lymphoma. Biomedicine. 1975;22:276–284. [PubMed] [Google Scholar]
- 32.Niwa M, Rose S D, Berget S M. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 33.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 34.Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 35.Radu A, Blobel G, Wozniak R W. Nup155 is a novel nuclear pore complex protein that contains neither repetitive sequence motifs nor reacts with WGA. J Cell Biol. 1993;121:1–9. doi: 10.1083/jcb.121.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radu A, Moore M S, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 37.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 38.Richards S A, Carey K L, Macara I G. Requirement of guanosine triphosphate-bound ran for signal-mediated nuclear protein export. Science. 1997;276:1842–1844. doi: 10.1126/science.276.5320.1842. [DOI] [PubMed] [Google Scholar]
- 39.Ruvolo V, Wang E, Boyle S, Swaminathan S. The Epstein-Barr virus nuclear protein SM is both a post-transcriptional inhibitor and activator of gene expression. Proc Natl Acad Sci USA. 1998;95:8852–8857. doi: 10.1073/pnas.95.15.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandri-Goldin R M. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 1998;12:868–879. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Semmes O J, Chen L, Sarisky R T, Gao Z, Zhong L, Hayward S D. Mta has properties of an RNA export protein and increases cytoplasmic accumulation of Epstein-Barr virus replication gene mRNA. J Virol. 1998;72:9526–9534. doi: 10.1128/jvi.72.12.9526-9534.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 43.Stutz F, Rosbash M. Nuclear RNA export. Genes Dev. 1998;12:3303–3319. doi: 10.1101/gad.12.21.3303. [DOI] [PubMed] [Google Scholar]
- 44.Swaminathan S, Tomkinson B, Kieff E. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc Natl Acad Sci USA. 1991;88:1546–1550. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 46.Whitehouse A, Cooper M, Meredith D M. The immediate-early gene product encoded by open reading frame 57 of herpesvirus saimiri modulates gene expression at a posttranscriptional level. J Virol. 1998;72:857–861. doi: 10.1128/jvi.72.1.857-861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 48.Wong K-M, Levine A J. Identification and mapping of Epstein-Barr virus early antigens and demonstration of a viral gene activator that functions in trans. J Virol. 1986;60:149–156. doi: 10.1128/jvi.60.1.149-156.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zolotukhin A S, Felber B K. Nucleoporins Nup98 and Nup214 participate in nuclear export of human immunodeficiency virus type 1 Rev. J Virol. 1999;73:120–127. doi: 10.1128/jvi.73.1.120-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]