Abstract
Antenatal administration of extracellular vesicles from amniotic fluid stem cells (AFSC-EVs) reverses features of pulmonary hypoplasia in models of congenital diaphragmatic hernia (CDH). However, it remains unknown which lung cellular compartments and biological pathways are affected by AFSC-EV therapy. Herein, we conducted single-nucleus RNA sequencing (snRNA-seq) on rat fetal CDH lungs treated with vehicle or AFSC-EVs. We identified that intra-amniotically injected AFSC-EVs reach the fetal lung in rats with CDH, where they promote lung branching morphogenesis and epithelial cell differentiation. Moreover, snRNA-seq revealed that rat fetal CDH lungs have a multilineage inflammatory signature with macrophage enrichment, which is reversed by AFSC-EV treatment. Macrophage enrichment in CDH fetal rat lungs was confirmed by immunofluorescence, flow cytometry, and inhibition studies with GW2580. Moreover, we validated macrophage enrichment in human fetal CDH lung autopsy samples. Together, this study advances knowledge on the pathogenesis of pulmonary hypoplasia and further evidence on the value of an EV-based therapy for CDH fetuses.
Stem cell extracellular vesicles attenuate multilineage inflammation and macrophage enrichment in fetal CDH hypoplastic lungs.
INTRODUCTION
Pulmonary hypoplasia is characterized by impaired fetal lung development (1). A common cause of pulmonary hypoplasia is congenital diaphragmatic hernia (CDH), a defect due to incomplete closure of the diaphragm and herniation of abdominal organs into the chest (2). Hypoplastic lungs have impaired growth (fewer branches and alveoli), maturation (undifferentiated epithelium and mesenchyme), and vascularization (fewer pulmonary vessels with muscularized wall layers and dysfunctional endothelium) (2). The severity of pulmonary hypoplasia combined with pulmonary hypertension and ventricular dysfunction secondary to CDH are the main determinants of morbidity and mortality (high-income countries, 20 to 30% in the past three decades; low- and middle-income countries, >90%) (2, 3). Because of severe pulmonary hypoplasia, some fetuses die in utero or are electively terminated, some succumb in the first days of life, and many who survive and undergo surgery do not regain normal lung development and have long-term lung morbidity (2).
There is consensus that the antenatal period offers a window of opportunity to reverse pulmonary hypoplasia and attempts have been made to promote fetal lung development antenatally (4, 5). We previously reported that administration of extracellular vesicles derived from amniotic fluid stem cells (AFSC-EVs) promotes branching morphogenesis, rescues tissue homeostasis, and stimulates epithelial cell and fibroblast differentiation in fetal rodent models of pulmonary hypoplasia (6). EVs are lipid-bound nanoparticles that carry small RNA, protein, and lipid cargo that is transferred to target cells to induce biological responses (7). The ability to stimulate lung cell differentiation and rescue dysregulated signaling pathways was observed when AFSC-EVs were administered not only at the pseudoglandular stage but also at the canalicular and saccular stages of lung development, time points that are amenable to human translation (8). Our enzymatic and inhibitory studies proved that the regenerative effects observed in hypoplastic lungs following AFSC-EV treatment were exerted, at least in part, via their RNA cargo (6, 9). AFSC-EV RNA sequencing (RNA-seq) revealed that the cargo contained multiple biomolecules including microRNAs (miRNAs) that regulate the expression of genes involved in lung development, such as the miRNA 17~92 cluster (6). This is relevant as lung developmental processes are partly regulated by multiple miRNAs (10), whose expression is missing or dysregulated in experimental and human CDH lungs (6, 11). Recently, we also demonstrated that antenatal administration of AFSC-EVs improves fetal survival and mechanical ventilation parameters, such as compliance and resistance (12).
It remains undetermined which lung cells are affected by AFSC-EVs and how AFSC-EVs restore the biological pathways required for lung development. Herein, we used single-nucleus RNA-seq (snRNA-seq) to uncover the dysregulated genes and biological pathways in CDH fetal lungs and to determine the effects of in utero AFSC-EV therapy on fetal lung cell populations.
RESULTS
Intra-amniotic administration of AFSC-EVs improves branching morphogenesis and epithelial cell differentiation in fetal rats with CDH
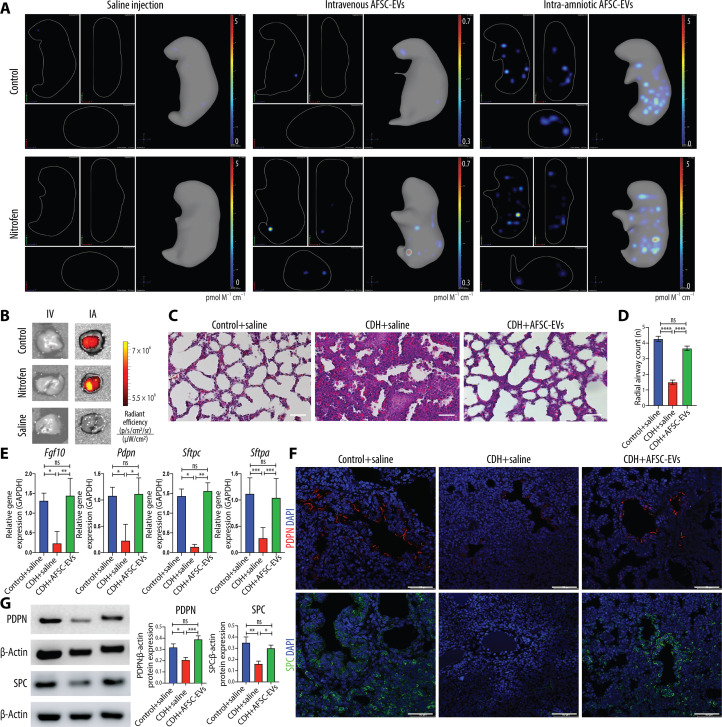
As the canalicular stage of lung development is the earliest time for fetal intervention (13, 14), we selected this stage in rats to trial different routes of AFSC-EV administration. First, we opted for intratracheal instillation of AFSC-EVs. However, given the small size of fetal rats and the technical challenges with this survival surgery, we experienced low survival (20%, n = 10), as also reported by other groups (15, 16), and abandoned this route. We then tested two routes of administration that had high survival: intra-amniotic (IA) (n = 48, survival 84%) and maternal intravenous (IV) (n = 30, survival 100%). All pups that received AFSC-EVs survived the procedure to termination. The concentration of AFSC-EVs administered was based on dose-response experiments that determined a therapeutic dosage of 7.6 × 109 EVs in 100 μl of saline per fetus (6, 9). When we compared the efficiency of AFSC-EV delivery to the fetus, we found that both IA and IV routes successfully delivered AFSC-EVs (ExoGlowVivo labeled) to fetal organs (Fig. 1A, fig. S1, A and B, and movies S1 to S3). However, we detected a positive fluorescent signal in fetal lungs only upon IA injection (Fig. 1B and fig. S1, A and B), which we elected as the optimal route for our experiments. IA injections of saline only or EV-free ExoGlow-Vivo preparations did not yield a fluorescent signal (fig. S1). To validate that IA-injected AFSC-EVs promoted lung growth and maturation in vivo, we assessed lung branching morphogenesis and cell differentiation markers at E21.5. Compared to control, CDH lungs had a reduction in airspace density and lower gene expression levels of fibroblast growth factor 10 (Fgf10; regulator of lung lineage commitment and branching morphogenesis), podoplanin [Pdpn; alveolar type (AT) 1 cell marker], and surfactant proteins C and A (Sftpc and Sftpa, respectively; AT2 cell markers) (Fig. 1, C to E). CDH lungs from fetuses that received an IA injection of AFSC-EVs had restored airspace density and gene expression of Fgf10, Pdpn, Sftpc, and Sftpa back to control levels (Fig. 1, C to E). We validated these findings with immunofluorescence and Western blotting and determined that CDH lungs had reduced levels of PDPN and SPC compared to control (Fig. 1, F and G). Conversely, CDH lungs treated with AFSC-EVs had increased protein expression levels of PDPN and SPC.
Fig. 1. In vivo administration of AFSC-EVs reaches fetal lungs and improves lung development in fetal rats with CDH.
(A) Representative IVIS Spectrum cross-sectional images from three-dimensional (3D) bioluminescence reconstructions of whole fetuses at E21.5. Fetuses randomly received either saline injection (left), IV injection of ExoGlowVivo-stained AFSC-EVs (middle), or IA injection of ExoGlowVivo-stained AFSC-EVs (right) at E18.5 in control fetuses (top row) or fetuses with pulmonary hypoplasia/CDH that received nitrofen (bottom row). Scale bar shows background-corrected fluorescence in pmol M−1 cm−1. Control+saline (n = 3), Control+IV-AFSC-EVs (n = 3), Control+IA-AFSC-EVs (n = 7), Nitrofen+saline (n = 6), Nitrofen+IV-AFSC-EVs (n = 3), and Nitrofen+IA-AFSC-EVs (n = 16). (B) Representative 2D optical images of dissected fetal lungs from the same conditions described in (A), quantified as radiant efficiency [p/s/cm2/sr]/[μW/cm2]. Control+saline (n = 3), Control+IV-AFSC-EVs (n = 3), Control+IA-AFSC-EVs (n = 3), Nitrofen+saline (n = 3), Nitrofen+IV-AFSC-EVs (n = 4), and Nitrofen+IA-AFSC-EVs (n = 9). (C) Representative histology images (hematoxylin and eosin) of fetal lungs from Control+saline, CDH+saline, and CDH+AFSC-EV fetuses. Each condition included fetal lungs from n = 5 experiments. Scale bars, 50 μm. (D) Differences in number of alveoli (RAC) in Control+saline (n = 8), CDH+saline (n = 8), and CDH+AFSC-EVs (n = 9) quantified in at least five fields per fetal lung. ****P < 0.0001; ***P < 0.001; ns, not significant. (E) Gene expression of lung developmental markers Fgf10, Pdpn, and Sftpc and Sftpa. Control+saline (n = 5), CDH+saline (n = 5), and CDH+AFSC-EVs (n = 5). **P < 0.01; *P < 0.05. (F) Representative immunofluorescence images of PDPN (red; top) and SPC (green; bottom) protein expression between Control+saline, CDH+saline, and CDH+AFSC-EV fetuses [4′,6-diamidino-2-phenylindole (DAPI); blue]. Scale bars, 50 μm. (G) Western blot analysis of PDPN and SPC expression in fetal lung quantified by signal intensity normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Control+saline (n = 6), CDH+saline (n = 7), and CDH+AFSC-EVs (n = 6). Groups were compared using Kruskal-Wallis (post hoc Dunn’s nonparametric comparison) for (D) RAC and (E) Fgf10, Pdpn, and Sftpc and one-way ANOVA (Tukey post-test) for (E) Sftpa and (G), according to Shapiro-Wilk normality test.
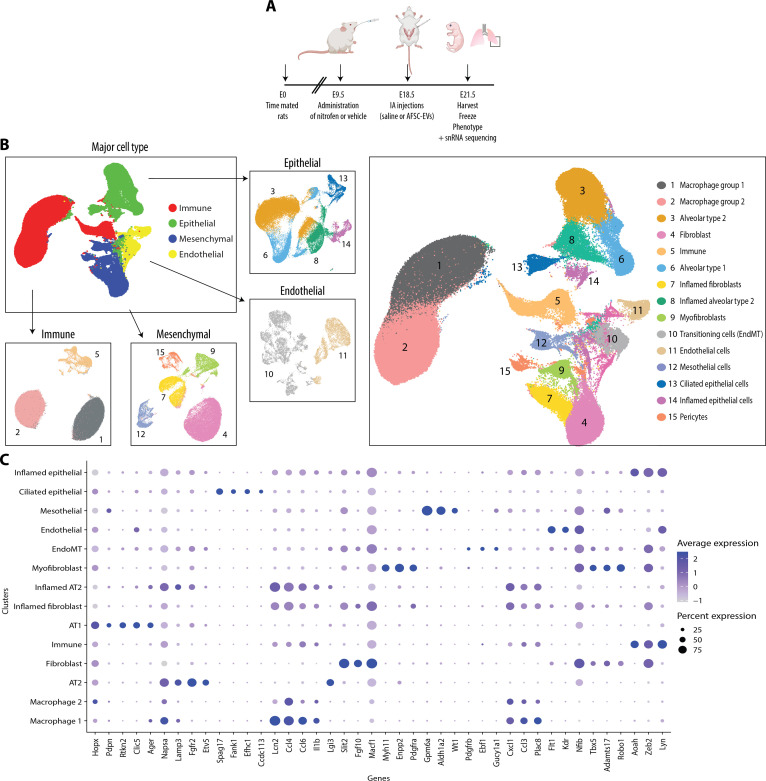
Single-nucleus interrogation of the rat fetal normal and hypoplastic lung identifies four major cell types each with distinct subpopulations
To identify AFSC-EV cell type–specific effects, we conducted snRNA-seq on the left lung harvested at E21.5 from two IA saline-injected controls, three IA saline-injected left-sided CDH fetuses, and three IA AFSC-EV–injected left-sided CDH fetuses (Fig. 2A and fig. S2). We selected fetuses for snRNA-seq studies from a large cohort of pups based on severity of branching morphogenesis [radial alveolar count (RAC) and Fgf10 expression] and expression of key lung differentiation markers (Pdpn, Sftpc, and Sftpa) (fig. S2). We chose these markers in combination with histological changes to represent well-described parameters that are known to be dysregulated in nitrofen-exposed lungs (17). After quality control filtering, we profiled a total of 298,653 nuclei (fig. S3 and table S1). Analysis using bioinformatics tool Seurat (R/4.0.3) revealed 15 distinct clusters representative of the four major cell types that corresponded to epithelial, endothelial, mesenchymal, and immune cells, each containing unique subpopulations (Fig. 2, B and C). We used LungMAP, LungCellMap, Tabula Muris, and Human Protein Atlas annotations from mouse and human lungs to assign cell type identity based on gene expression enrichment of key marker genes (Fig. 2, B and C) (18–22). We identified five distinct epithelial subpopulations, including AT1, AT2, and ciliated epithelial cells (Fig. 2, B and C, and fig. S4). Among these cell types, AT1 cells expressed Hopx, Pdpn, Clic5, and Ager; AT2 cells expressed Napsa, Lamp3, Fgfr2, and Etv5; and ciliated epithelial cells expressed cilia-related genes Dnah12, Hydin, Ak9, and Spag17. In addition, there were two other epithelial cell clusters with an inflammatory signature: Cluster 8 was called “inflamed AT2 cells” as it coexpressed AT2 cell (Lamp3 and Lgi3) and inflammatory markers, whereas cluster 14, broadly called “inflamed epithelial cells” expressed Lcn2, Ccl4, Ccl6, and Il1b. We identified two distinct endothelial clusters: one that had canonical endothelial cell markers Tie2, Flt1, and Kdr and one that coexpressed mesenchymal and endothelial signatures (Nfib, Tbx5, Adamts17, and Robo1) that was termed “EndMT cells” (Fig. 2, B and C, and fig. S4). Four mesenchymal cell subtypes were identified, including fibroblasts, myofibroblasts, mesothelial cells, and pericytes (Fig. 2, B and C, and fig. S4). Among these cell types, fibroblasts expressed Slit2, Fgf10, and Macf1; myofibroblasts expressed Myh11, Enpp2, and Pdgfra; mesothelial cells expressed Gpm6a, Aldh1a2, and Wt1; and pericytes expressed Pdgfrb, Ebf1, and Gucy1a1. Moreover, we identified a mesenchymal cluster that heavily expressed Lcn2, Ccl4, Cxcl1, Ccl3, and Plac8 and was called “inflamed fibroblasts.” Last, three immune cell clusters were detected: two expressing macrophage markers CD68, Adgre-1, CD163, and CD86 and one expressing immune cell markers Aoah, Zeb2, and Lyn (Fig. 2, B and C).
Fig. 2. Single-nucleus interrogation of the rat fetal normal and hypoplastic lung identifies four major cell types each with distinct subpopulations.
(A) Schematic of experimental design and in vivo administration of AFSC-EVs in the rat model of CDH. (B) Global Uniform Manifold Approximation and Projection (UMAP) of all nuclei (n = 298,653) included in our study, further delineated by major cell type and subtype. (C) Expression of known cell type–specific markers used to distinguish cellular subtypes within major cell type clusters. Node size is proportional to the percentage of nuclei within the specified cluster, and node color denotes the average expression across nuclei within the specified cluster.
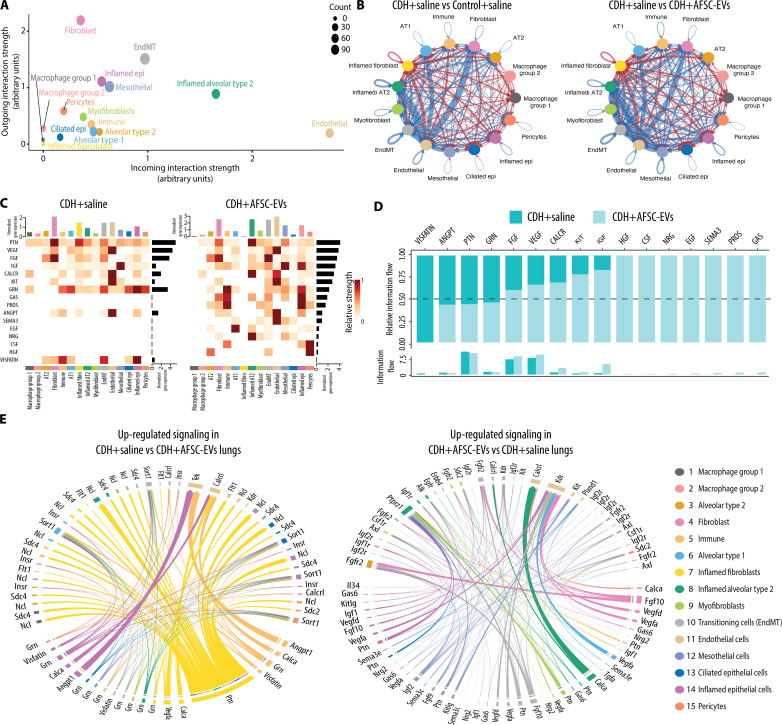
Ligand-receptor analysis of rat fetal lung transcriptomics reveals the biological pathways that are influenced by AFSC-EV administration
To identify signaling pathways activated in CDH lungs treated with saline or AFSC-EVs, we performed ligand-receptor analysis. Of all cell types, lung fibroblasts had the strongest outgoing signals and endothelial cells were the most receptive to incoming ligands (Fig. 3A). Moreover, compared to normal lungs and AFSC-EV–treated CDH lungs, CDH+saline lungs had up-regulated ligand-receptor signaling from fibroblasts to endothelial cells (Fig. 3B). Our ligand-receptor analysis revealed that CDH lungs treated with saline exhibited signaling networks that are involved in inflammation and immune response, such as Visfatin (Fig. 3, C and D). Visfatin is a proinflammatory cytokine that potentiates tumor necrosis factor–α (TNFα) and interleukin-6 (IL-6) production in human peripheral blood mononuclear cells (table S2). The strongest outgoing ligand signal in saline-treated CDH lungs was pleiotrophin (Ptn), a signaling molecule involved in lung development, which, in our experiments, was released from inflamed fibroblast and signaled to its receptors Sdc2 and Ncl on multiple lung cell types (Fig. 3, C to E, and table S2). Moreover, compared to CDH+saline lungs, AFSC-EV–treated CDH lungs had activated signaling networks that control epithelial branching morphogenesis (Fgf10-Fgfr2), surfactant synthesis and alveolarization (Nrg2-Erbb4), distal lung branching and alveologenesis (Igf2-Igf1r), and angiogenesis (Vegfa-Kdr) (Fig. 3, C to E, and table S3).
Fig. 3. Ligand-receptor analysis reveals the biological pathways that are influenced by AFSC-EV treatment of rat fetal hypoplastic lungs.
(A to E) CellChat analysis of signaling pathways in fetal lungs from all three conditions. (A) Comparison of interaction strength of outgoing and incoming signals by specific cluster. Node size represents number of interactions. (B) Statistically significant interactions between clusters (arrows) showing number of interactions that are down-regulated (blue) and up-regulated (red) when comparing Control+saline versus CDH+saline (left) and CDH+saline versus CDH+AFSC-EVs (right). Thickness of arrow indicates interaction strength. (C) Highly expressed ligand-receptor pairs displayed as a heatmap showing outgoing signal strength (top x axis), individual signaling pathways (left y axis), strength of signaling pathway (right y axis), and cell identity (bottom y axis). (D) Shift of signaling pathways related to lung development following AFSC-EV administration to fetal CDH lungs. (E) Chord diagram showing statistically significant up-regulated or down-regulated signaling pathways in each cluster between CDH+saline and CDH+AFSC-EV conditions. Thickness of arrow indicates relative strength of specific pathway.
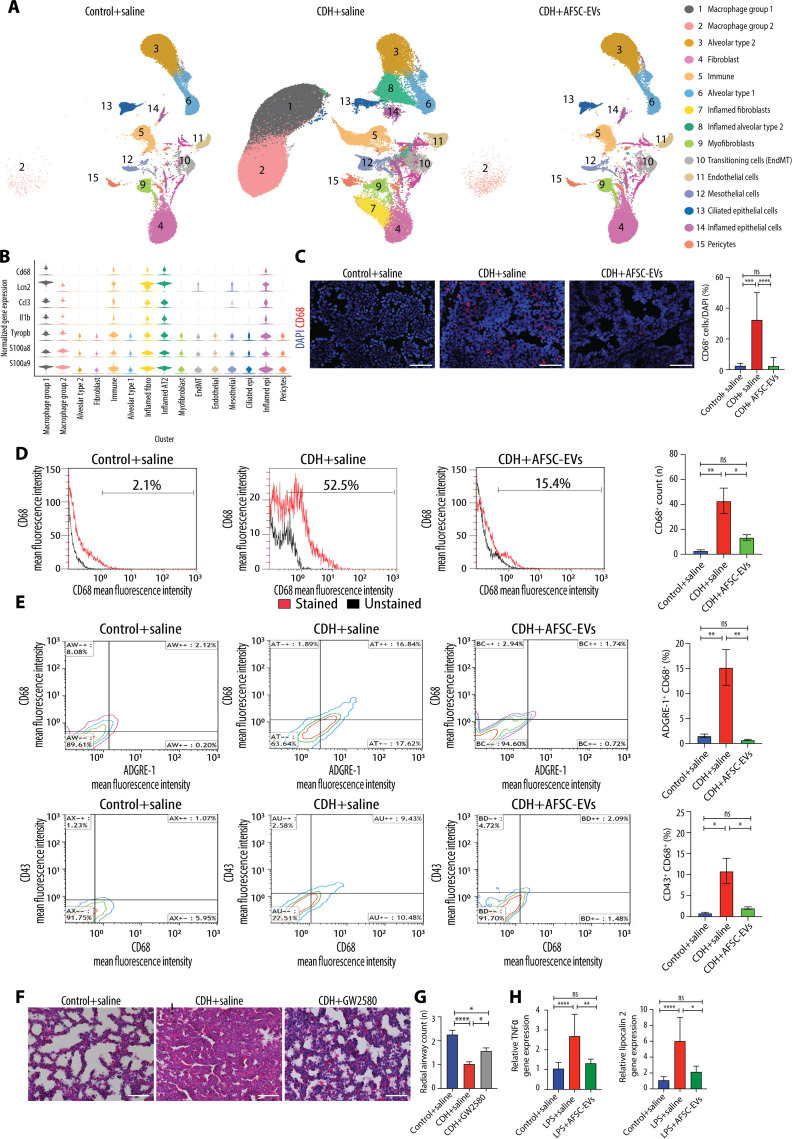
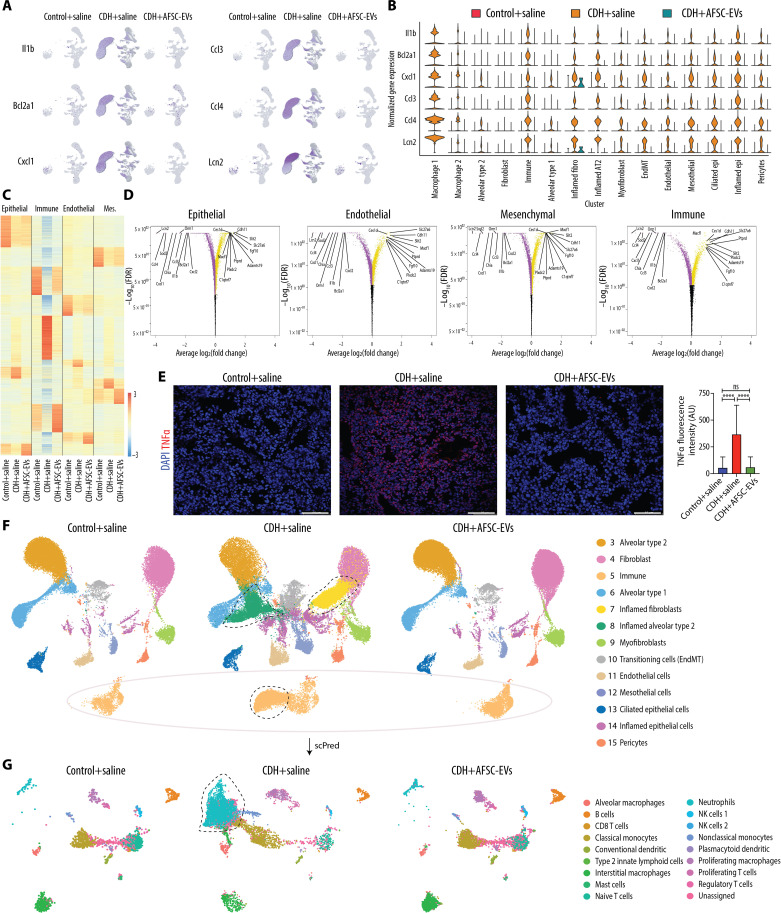
CDH lungs have an inflammatory phenotype with high macrophage density that is reduced to normal levels by AFSC-EV administration
When we analyzed the snRNA-seq data by condition, we found that CDH+saline lungs had notable differences in the pattern and clustering of nuclei (Fig. 4A). Conversely, lungs from Control+saline and CDH+AFSC-EV groups had similar populations and distributions. We found that three clusters were unique to CDH+saline lungs, namely, macrophage group 1 (cluster 1), inflamed fibroblasts (cluster 7), and inflamed AT2 (cluster 8). Moreover, macrophage group 2 was heavily represented in CDH+saline lungs (n = 89,187 nuclei), compared to Control+saline (n = 247 nuclei) and CDH+AFSC-EVs (n = 739 nuclei; table S1). Markers of macrophage identity and function were found in several clusters (Fig. 4B). We further delineated the specific macrophage subtypes contained in clusters 1 and 2 using machine learning and found that most nuclei were alveolar macrophages (n = 140,382, 77%) (fig. S5 and table S4). Using immunofluorescence on an additional cohort of rat fetuses, we confirmed a high density of macrophages in CDH+saline lungs, which was reduced to normal levels in CDH+AFSC-EV lungs (Fig. 4C). Using flow cytometry on additional fetuses not used for snRNA-seq or immunofluorescence studies, we corroborated that CD68+ macrophages were highly expressed in CDH+saline lungs compared to Control+saline and CDH+AFSC-EV lungs (Fig. 4D and data file S3). Moreover, triple staining for CD68, ADGRE-1, and CD43 showed that some macrophages were monocyte-derived (CD68+/CD43high) and some were bone marrow–derived (CD68+/ADGRE-1high) (Fig. 4E and data file S3) (23). To understand if macrophages play a role in pulmonary hypoplasia, we performed inhibition studies, where we blocked macrophage activation using GW2580, a selective inhibitor of colony-stimulating factor 1 receptor (CSF1R) kinase (24). As CSF1R is responsible for macrophage survival, proliferation, and inflammatory responses of lung macrophages (24), GW2580 inhibition effectively modulates macrophage function at relatively low doses that are tolerated in the nitrofen rat model of CDH. CDH fetal lungs that were intra-amniotically injected with GW2580 had a reduction in macrophage density compared to CDH lungs treated with saline (Fig. 4F). GW2580-treated CDH fetal lungs had a less severe degree of pulmonary hypoplasia compared to untreated CDH fetal lungs (Fig. 4, F and G). We then determined if AFSC-EV administration had a direct effect on macrophages by interrogating gene expression of Tnfα and Lcn2 in RAW264.7 cells, an immortalized cell line of macrophages. When we stimulated RAW264.7 cells with lipopolysaccharide (LPS) to mimic activated macrophages, we found that those treated with AFSC-EVs had a reduction in Tnfα and Lcn2 expression compared to stimulated RAW264.7 cells treated with medium alone (Fig. 4H).
Fig. 4. CDH lungs have an inflammatory phenotype with high macrophage density that is reduced to normal levels by AFSC-EV administration.
(A) UMAP of snRNA-seq data split by condition. (B) Violin plots of macrophage and inflammatory marker gene expression across cell types, as measured by snRNA-seq. (C) Representative immunofluorescence images of pan-macrophage marker CD68 in rat fetal lungs from all three conditions, quantified as fluorescence intensity of CD68 per field. Scale bars, 50 μm. Control+saline (n = 8), CDH+saline (n = 6), and CDH+AFSC-EV (n = 8). Groups were compared using Kruskal-Wallis (post hoc Dunn’s nonparametric comparison) for (C), according to Shapiro-Wilk normality test. ***P < 0.001; ****P < 0.0001. (D) Flow cytometry analysis of dissociated lung cells stained for CD68 (red) versus unstained (black) and (E) costained with ADGRE-1 and CD43 (panels are representative of n ≥ 3 pups per group; data file S4). *P < 0.05; **P < 0.01. (F) Representative histology images (hematoxylin and eosin) of fetal lungs from Control+saline, CDH+saline, and CDH+GW2580 fetuses. Scale bars, 50 μm. (G) Differences in number of alveoli (RAC) in Control+saline (n = 7), CDH+saline (n = 8), and CDH+GW2580 (n = 8), quantified in at least seven fields per fetal lung. (H) Gene expression changes in inflammatory markers Tnfα and Lcn2 in RAW264.7 cells stimulated with LPS, relative to Actb housekeeping gene.
CDH fetal lungs have a multilineage inflammatory signature that is dampened by the administration of AFSC-EVs
Differential gene expression analysis of CDH+saline lungs compared to Control+saline showed an extensive inflammatory signature across clusters with up-regulation of Il1b, Bcl2a1, Cxcl1, Ccl3/4, and Lcn2 (Fig. 5, A and B, and table S2). These genes were down-regulated in AFSC-EV–treated lungs (Fig. 5, A and B). Differential gene expression analysis revealed similar patterns between Control+saline and CDH+AFSC-EV lungs regardless of the major cell type (Fig. 5C). Most of the highly differentially expressed genes in CDH+saline lungs were enriched for biological processes related to immune responses and included pathways associated with autophagy, which we also previously showed to be dysregulated in CDH lungs (Fig. 5D and data file S4, late endosomal microautophagy; P = 0.04) (9). We confirmed that the lungs of an additional cohort of CDH+saline rat fetuses were inflamed with up-regulation of TNFα using immunofluorescence, which was restored to normal levels with AFSC-EV treatment (Fig. 5E).
Fig. 5. CDH fetal lungs have a multilineage inflammatory signature that is dampened by the administration of AFSC-EVs.
(A) Featureplot of snRNA-seq data split by condition for six inflammatory genes with high expression in CDH+saline lungs. (B) Violin plot of inflammatory signature genes expression split by condition across cell types, as measured by snRNA-seq. (C) Heatmap displaying differential gene expression by major cell type, showing expression all genes ranked by log2(fold change) and P-adjusted < 0.05 within all conditions. (D) Volcano plots indicating most statistically significant differentially expressed genes by major cell type between CDH+saline-treated and CDH+AFSC-EV–treated groups. FDR, false discovery rate. (E) Representative immunofluorescence images of inflammation marker TNFα in rat fetal lungs from all three conditions, quantified as density per mm2. Scale bars, 50 μm. Control+saline (n = 5), CDH+saline (n = 5), and CDH+AFSC-EV (n = 5). AU, arbitrary units. ****P < 0.0001. (F) UMAP of a subset of data that excludes clusters 1 and 2 (overrepresented in CDH+saline group) split by condition. Outlines indicate nuclei or clusters that are represented in CDH+saline group compared to Control+saline and CDH+AFSC-EV groups. Control+saline (n = 30,064), CDH+saline (n = 45,114), and CDH+AFSC-EV (n = 42,193). (G) UMAP of predicted cell types contained in cluster 5 immune cells from (F), generated by machine learning algorithm (scPred) trained on rat adult lungs. Groups were compared using Kruskal-Wallis (post hoc Dunn’s nonparametric comparison) for (E), according to Shapiro-Wilk normality test.
To investigate the transcriptomic differences across conditions and have a homogeneous comparison with similar number of nuclei within each condition, we created a subset of data by removing macrophage groups 1 and 2 as they were overrepresented in CDH+saline lungs. In this sub-analysis that included 30,064 Control+saline nuclei, 45,114 CDH+saline nuclei, and 42,193 CDH+AFSC-EV nuclei, we again found that all four major cell types were represented (Fig. 5F). Given the inflammatory signature of CDH+saline lungs, we investigated which immune cells were present in cluster 5 using machine learning (see the Supplementary Materials for further details) and found neutrophils, monocytes, T and B cells, and other immune cells (Fig. 5G and table S5). CDH+saline lungs had a higher proportion of neutrophils compared to Control+saline lungs (53 versus 3%, P < 0.0001; Fisher’s exact test), whereas CDH+AFSC-EV lungs had a lower proportion of neutrophils (4%) compared to CDH+saline lungs (P < 0.0001).
Predicted miRNA-mRNA signaling pathways activated by AFSC-EVs
To establish the mechanism behind the effects of AFSC-EVs on CDH lungs, we first reanalyzed the proteomics data of the AFSC-EV cargo (6) and found no proteins with known anti-inflammatory properties. We next investigated the role of the miRNA cargo, which we previously showed to be critical for the effects of AFSC-EVs on branching morphogenesis (6, 9). We used publicly available datasets to generate a network between the miRNAs present in the rat AFSC-EV cargo (6) and the mRNAs identified by snRNA-seq that were down-regulated in CDH+AFSC-EV lungs compared to CDH+saline lungs. Overall, we found 820 predicted miRNA-mRNA targets that regulate several biological processes, including inflammatory/immune responses (fig. S6, A to C). From the 820 predicted miRNA-mRNA pairs, 32 miRNA-mRNA pairs (13 miRNAs and 24 mRNAs) were validated (fig. S6D).
Inflammatory markers are up-regulated in hypoplastic lungs of human fetuses with CDH
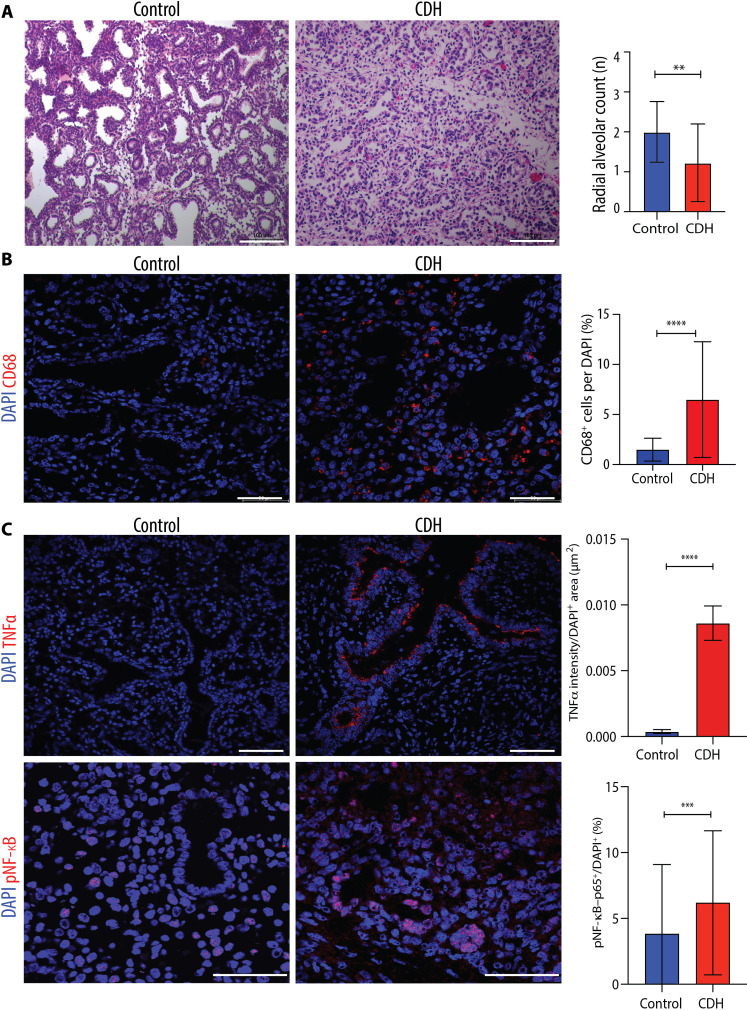
To confirm that the findings observed in fetal rats are relevant to the human CDH condition, we interrogated lung sections from autopsy samples of four human fetuses with CDH that died between gestational weeks 19 and 26 (canalicular stage of lung development) and four controls (no fetal lung pathology or systemic inflammatory conditions; table S6) following ethical approval (nos. 1000074888 and 1000080881, The Hospital for Sick Children, Toronto). We confirmed that compared to controls, the lungs of the CDH fetuses had a lower density of airspaces (Fig. 6A). We then determined that the macrophage density was increased in lungs of CDH fetuses, most predominantly in the parenchyma (Fig. 6B). Moreover, the expression of canonical markers of inflammation such as TNFα and its downstream nuclear factor κB (NF-κB) signaling was up-regulated in lungs of CDH fetuses (Fig. 6C).
Fig. 6. Hypoplastic lungs of human fetuses with CDH have increased macrophage density and up-regulation of inflammatory mediators.
(A) Representative histology images (hematoxylin and eosin) of fetal lungs from autopsy studies of CDH fetuses (n = 4) and controls with no lung pathology or inflammatory condition (n = 4). Scale bars, 100 μm. Quantification of number of alveoli (RAC) in 10 fields per fetal lung. **P < 0.01. (B) Representative immunofluorescence images of pan-macrophage marker CD68 in human fetal lungs autopsy samples from CDH (n = 4) and controls (n = 4), quantified as number per DAPI+ cell (%). Scale bars, 50 μm. ****P < 0.0001. (C) Representative immunofluorescence images of inflammatory mediators TNFα and pNF-κB in the same experimental groups as (B) quantified by fluorescence intensity of TNFα and density of pNF-κB+ cells per field. Scale bars, 50 μm. Groups were compared using two-tailed Mann-Whitney test for (A), (B), and (C) pNF-κB and two-tailed Student’s t test for (C) TNFα, according to Shapiro-Wilk normality test. ***P < 0.001.
DISCUSSION
This study demonstrates that rat CDH fetal hypoplastic lungs have an inflammatory signature with high density of macrophages and up-regulation of biological pathways that are involved in inflammatory and innate immune response. We confirmed that human fetuses with CDH also have an inflammatory status with macrophage enrichment and increased TNFα and phosphorylated NF-κB (pNF-κB) expression. These antenatal findings in fetuses with CDH are in line with similar observations made in postnatally infants with CDH. In human infants with CDH, several studies reported a postnatal up-regulation of pNF-κB in the proximal lung and TNFα in the distal lung as well as high levels of proinflammatory cytokines in the blood (25–34). Similarly, experimental studies using the rat nitrofen model reported high levels of monocyte chemoattractant protein 1, Tnfα, signal transducer and activator of transcription 3 (STAT3), and NF-κB in the lung (35–39), and a study using the lamb model of CDH detected proinflammatory proteins in the tracheal fluid (40). The technology advances offered by snRNA-seq have allowed us to show that all four major lung cell types in CDH rat fetuses had up-regulation of several inflammatory mediators, including Tnfα, Stat3, Lcn2, Il1b, Ccl3/4, and Cxcl1 (table S2). Moreover, ligand-receptor analysis identified increased proinflammatory signaling of visfatin in CDH+saline lungs.
The multilineage inflammatory profile in the CDH lung was accompanied by an increase in macrophage density. Although there is robust literature on the role of macrophages in the adult lung during injury and repair, less is known about macrophage involvement in impaired perinatal lung development. In a healthy state, fetal lung macrophages arise from early developmental embryonic and fetal precursors (41, 42). However, under conditions of sterile inflammation, alveolar spaces can be infiltrated by high numbers of bone marrow hematopoietic cells or monocytes that take up residence in the airways and become alveolar macrophages (42, 43). In the present study, we show that the predominant CD68-expressing lung macrophage population arises from the bone marrow or monocytes and that this population is largely absent in CDH fetal lungs treated with AFSC-EVs. In bronchopulmonary dysplasia (BPD), a condition of premature babies that has some similar features to CDH (44, 45), the inhibition of branching morphogenesis has been shown to be partly caused by activation of fetal lung macrophages, and depletion or targeted macrophage inactivation is protective against impaired branching morphogenesis (46). This is similar to the outcomes of our inhibition studies in CDH lungs. Therefore, we infer that the severity of lung inflammation and macrophage enrichment may contribute to the poor prognosis of babies with CDH and provide an alternative target to stimulate normal lung development.
A promising avenue for targeting multiple molecules and pathways that are dysregulated in CDH lungs is through an EV-based therapy. We showed that antenatal AFSC-EV administration improves dysregulated signaling pathways relevant to lung development and results in enhancement of lung branching morphogenesis and epithelial and mesenchymal maturation during pseudoglandular, canalicular, and saccular stages of lung development (6, 8). In the current study, we provide evidence that AFSC-EVs also have anti-inflammatory effects in a fetal rat model of CDH. The anti-inflammatory effects of stem cell–derived EVs have been recognized within the past decade in numerous experimental and clinical trials under several conditions, including BPD (47, 48). Furthermore, with the advent of SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2)–induced acute respiratory distress syndrome, many research groups have attempted to use EVs from different sources, including mesenchymal stromal cells, as a possible strategy to treat lung inflammation (see the Supplementary Materials) (49–51). Several studies have demonstrated that AFSCs as well as their conditioned medium and EVs have anti-inflammatory effects on different disease models (52–57). In the present study, we have shown that AFSC-EVs reduce the expression of inflammatory mediators in the lung back to control levels. Although immune cells are not predominant in fetal lungs, our snRNA-seq analysis was able to detect gene expression differences in immune cell populations. Moreover, several genes identified by our snRNA-seq analysis to be up-regulated were also observed in a single-cell RNA-seq study of postnatal lungs of mice with BPD (58). This study found that up-regulation of inflammatory cytokine signaling was associated with major structural and cell-to-cell signaling changes in the lung (58).
This and our previous studies show that AFSC-EV administration to rat fetal hypoplastic lungs is associated with anti-inflammatory and regenerative effects likely due to cargo delivery. Our proteomics analysis showed that AFSC-EV cargo contains proteins with molecular function not directly related to modulation of inflammation or lung development (6). Conversely, small RNA-seq of the AFSC-EV cargo identified some miRNAs, such as miR-9, miR-125, and miR-128, that modulate macrophage-mediated inflammatory responses in the lung (59–61). This adds to the previously reported miRNAs that regulate lung developmental processes, such as the miR-17~92 family, which we demonstrated to be key for the AFSC-EV effects on branching morphogenesis with inhibition studies (9). In addition to the analysis of cargo contents, the ligand-receptor analysis of CDH+AFSC-EV lungs allowed us to characterize the cellular cross-talk and dysregulated functional processes in neighboring cells and demonstrated restoration of cellular processes that were dysregulated in CDH+saline lungs, such as up-regulated signaling from fibroblasts to endothelial cells and aberrant endothelial-to-mesenchymal transition. Gene set enrichment analysis also indicated down-regulation of inflammatory pathways in endothelial cells of CDH+AFSC-EV lungs compared to CDH+saline lungs and the improvement of Vegf signaling.
Although we provide evidence that AFSC-EV administration has potential for reversing features of pulmonary hypoplasia in CDH rat fetuses, there are several necessary steps before translating this approach to clinical application. For example, the optimal route of administration will need to be established. Herein, we opted for AFSC-EV IA injection during the saccular stage of lung development when clustered fetal breathing movements occur and intra-amniotically injected products can reach the fetal lung. This route is feasible as access to the amniotic sac is routine during pregnancy for diagnostic procedures and has been used to administer ectodysplasin A to human twins with X-linked hypohidrotic ectodermal dysplasia with profound reversal of disease phenotype (62). We also tested intratracheal administration, a procedure that mimics fetal endoscopic tracheal occlusion in human babies (2, 13). However, given the invasiveness of the procedure in rat fetuses, we had a high rate of fetal demise. Last, we tested maternal IV administration and demonstrated that AFSC-EVs cross the placental barrier, a well-described property of EVs (63). Although this strategy would circumvent the invasiveness of fetal intervention, custom-designed EVs should be constructed to target the fetal lung and avoid off-target effects. Alternatively, fetal circulation can be directly accessed multiple times through the umbilical vein, as recently shown in a fetus with Pompe’s disease (64). Another critical step for translation of EV therapy is the establishment of a stable source of AFSCs from which EVs can be derived. The most feasible solution would be biobanked AFSCs from the amniotic fluid of healthy human pregnancies. AFSC-EVs should be derived in a manner that is scalable and administered as a heterologous therapy.
We acknowledge that our study has limitations. As snRNA-seq transcriptomics requires tissue dissociation, we could not define the location of CD68+ cells within the lung. Although, flow cytometry suggested that the macrophages are either monocyte-derived or bone marrow–derived, in-depth macrophage characterization using spatial transcriptomics and fate mapping are necessary to determine the cellular origin of tissue-resident macrophages. Moreover, using another adult rat lung single-cell RNA dataset, we made predictions on which types of macrophages reside in our snRNA-seq clusters. Although inhibition studies using GW2580 indicated that macrophages play a role in the pathogenesis of pulmonary hypoplasia secondary to CDH, further mechanistic studies using knockout models are necessary to ascertain if macrophages are at the root cause of arrested lung development. Similarly, it remains unclear how AFSC-EVs induce a reduction in macrophage density and which of the other biomolecules present in the AFSC-EV cargo (i.e., proteins, lipids, and other small RNA species) could also play a role. Last, although our study reports a multilineage inflammatory signature in the lung, we did not identify a specific inflamed endothelial cell cluster. Nonetheless, gene set enrichment analysis revealed that CDH+saline lungs had an up-regulation of genes involved in inflammation compared to CDH+AFSC-EVs and Control+saline lungs. Given the evidence that babies with CDH have several organs affected (65), some with an inflammatory response such as the brain (66), the anti-inflammatory and regenerative effects of AFSC-EVs could be beneficial beyond the lung. Further studies are needed to address these important questions before translating these findings to human patients with CDH.
MATERIALS AND METHODS
Extracellular vesicles
CD117+ rat AFSCs were previously characterized as broadly multipotent and nonteratogenic (52), grown in cell culture, and subjected to EV isolation with ultracentrifugation as previously described (6, 8–9, 54). We characterized AFSC-EVs as previously described in accordance with the International Society for Extracellular Vesicles guidelines for proper size, morphology, and expression of canonical EV protein markers (fig. S7, EV-TRACK: EV190001) (6, 8–9, 54, 67). For in vivo tracking experiments, we stained EV-free preparations and AFSC-EVs with ExoGlowVivo according to the manufacturer’s protocol.
Experimental model of CDH
Following ethical approval (no. 49892, The Hospital for Sick Children, Toronto), CDH was induced in rat fetuses with nitrofen administration to dams by oral gavage on E9.5 (6, 16, 68–70). Only 50% of fetal rats develop a diaphragmatic defect, but all develop some degree of pulmonary hypoplasia, which, in part, reflects the variability also observed in human babies with CDH (68–70). For in vivo AFSC-EV administration, three routes were used at E18.5 and described in detail in the Supplementary Materials. At E21.5, only fetuses with a confirmed CDH were used and the left lung was separated for histology, RNA/protein analysis, and snRNA-seq.
Human fetal lung studies
Following ethical approval (nos. 1000074888 and #1000080881, The Hospital for Sick Children, Toronto), we analyzed lung sections from autopsy samples of four human fetuses with CDH that died between gestational weeks 19 and 26 (canalicular stage of lung development) and four controls (no fetal lung pathology or systemic inflammatory conditions; table S7).
Outcome measures
For EV tracking, ExoGlow-Vivo AFSC-EVs were injected through IA or IV, and fetuses and organs were imaged with IVIS. For assessment of lung growth and maturation, lungs were compared for morphometry using radial airspace count and gene and protein expression of SPC and PDPN (6, 8–9). For snRNA-seq studies, a subset of fetal lung samples was chosen (Control+saline, n = 2; CDH+saline, n = 3; and CDH+AFSC-EVs, n = 3), and nuclei were subjected to 10X Genomics protocol (see the Supplementary Materials for further details). For determining miRNA-mRNA regulatory pathways, multiMiR was used with AFSC-EV cargo and down-regulated genes from snRNA-seq dataset. Immunofluorescence assays were used to confirm snRNA-seq findings (i.e., CD68+ macrophages and inflammatory mediators). Flow cytometry was used to sort fetal lung cells for extracellular targets ADGRE-1, CD43, and CD68 (see the Supplementary Materials for further details). To inhibit macrophage activation in rat fetuses with CDH, we intra-amniotically injected GW2580, a selective inhibitor of CSF1R kinase.
Statistical analysis
Data distribution from each group was assessed using Shapiro-Wilk normality test. Groups containing normally distributed data were compared using two-tailed Student’s t test or one-way analysis of variance (ANOVA) (Tukey post-test), while non-normally distributed data were compared using Mann-Whitney or Kruskal-Wallis (post hoc Dunn’s nonparametric comparison) tests. Fisher’s exact test was used to compare proportions of cells in Fig. 5G. A P value of <0.05 was considered statistically significant.
Acknowledgments
We would like to thank G. Raffler, M. S. Gaffi, S. Gandhi, H. Hou, V. Fortuna, and A. Hardy. We are indebted to J. Reyes, D. Chiasson, and G. Somers for selection of autopsy samples included in the manuscript, P. De Coppi for providing rat AFSCs in kind, Lab Animal Service core facility, and A. Darbandi at the Nanoscale Biomedical Imaging Facility at The Hospital for Sick Children.
Funding: This work was supported by the Canadian Institutes of Health Research (CIHR) [CIHR Project Grant (175300; A.Z.) and CIHR FND143309 (M.P.)], American Pediatric Surgical Association Grosfeld Scholarship (A.Z.), SickKids Congenital Diaphragmatic Hernia Fund (R00DH00000; A.Z.), Canada Research Chairs Program (M.D.W.), American Thoracic Society (RP-2020-26; R.L.F.), CIHR Fellowship (176535; K.K.), and German Research Foundation DO (466815475; F.D.). Some of the equipment used in this study was supported by the 3D (Diet, Digestive Tract, and Disease) Centre funded by the Canadian Foundation for Innovation and Ontario Research Fund (project numbers 19442 and 30961).
Author contributions: Conceptualization: L.A., R.L.F., B.K., M.D.W., B.K., and A.Z. Methodology: L.A., R.L.F., B.K., M.L.L., L.M., C.C., M.D.W., and A.Z. Visualization: L.A., R.L.F., B.K., M.L.L., E.Z.-R., K.K., L.M., F.D., M.O., M.B., T.W., C.C., B.K., and A.Z. Funding acquisition: L.A., R.L.F., K.K., F.D., R.W., M.L., M.D.W., M.P., B.K., and A.Z. Project administration: L.A., R.L.F., B.K., M.D.W., and A.Z. Supervision: M.D.W., M.P., B.K., and A.Z. Writing—original draft: L.A., R.L.F., and A.Z. Writing—review and editing: L.A., R.L.F., B.K., R.W., M.L., M.D.W., M.P., B.K., and A.Z.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Relevant information regarding EV isolation and characterization have been submitted to EV-TRACK knowledgebase (EV-TRACK ID: EV190001). Sequencing data generated or analyzed in this study are available in the GEO (Gene Expression Omnibus) database (GSC211914). RNA-seq for AFSC-EV cargo sequencing was accessed from ArrayExpress database (no. E-MTAB-8921) from our previous publication (6).
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S8
Tables S1 to S8
Legends for movies S1 to S3
Legends for data files S1 to S5
References
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S3
Data files S1 to S5
REFERENCES AND NOTES
- 1.Cotten C. M., Pulmonary hypoplasia. Semin. Fetal Neonatal Med. 22, 250–255 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Zani A., Chung W. K., Deprest J., Harting M. T., Jancelewicz T., Kunisaki S. M., Patel N., Antounians L., Puligandla P. S., Keijzer R., Congenital diaphragmatic hernia. Nat. Rev. Dis. Primers. 8, 37 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Global PaedSurg Research Collaboration , Mortality from gastrointestinal congenital anomalies at 264 hospitals in 74 low-income, middle-income, and high-income countries: A multicentre, international, prospective cohort study. Lancet 398, 325–339 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeanty C., Kunisaki S. M., MacKenzie T. C., Novel non-surgical prenatal approaches to treating congenital diaphragmatic hernia. Semin. Fetal Neonatal Med. 19, 349–356 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Figueira R. L., Antounians L., Zani-Ruttenstock E., Khalaj K., Zani A., Fetal lung regeneration using stem cell-derived extracellular vesicles: A new frontier for pulmonary hypoplasia secondary to congenital diaphragmatic hernia. Prenat. Diagn. 42, 364–372 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Antounians L., Catania V. D., Montalva L., Liu B. D., Hou H., Chan C., Matei A. C., Tzanetakis A., Li B., Figueira R. L., da Costa K. M., Wong A. P., Mitchell R., David A. L., Patel K., De Coppi P., Sbragia L., Wilson M. D., Rossant J., Zani A., Fetal lung underdevelopment is rescued by administration of amniotic fluid stem cell extracellular vesicles in rodents. Sci. Transl. Med. 13, eaax5941 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Yáñez-Mó M., Siljander P. R., Andreu Z., Zavec A. B., Borràs F. E., Buzas E. I., Buzas K., Casal E., Cappello F., Carvalho J., Colás E., Cordeiro-da Silva A., Fais S., Falcon-Perez J. M., Ghobrial I. M., Giebel B., Gimona M., Graner M., Gursel I., Gursel M., Heegaard N. H., Hendrix A., Kierulf P., Kokubun K., Kosanovic M., Kralj-Iglic V., Krämer-Albers E.-M., Laitinen S., Lässer C., Lener T., Ligeti E., Linē A., Lipps G., Llorente A., Lötvall J., Manček-Keber M., Marcilla A., Mittelbrunn M., Nazarenko I., Nolte-‘t Hoen E. N. M., Nyman T. A., O'Driscoll L., Olivan M., Oliveira C., Pállinger É., Del Portillo H. A., Reventós J., Rigau M., Rohde E., Sammar M., Sánchez-Madrid F., Santarém N., Schallmoser K., Ostenfeld M. S., Stoorvogel W., Stukelj R., Van der Grein S. G., Vasconcelos M. H., Wauben M. H., De Wever O., Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 4, 27066 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khalaj K., Figueira R. L., Antounians L., Gandhi S., Wales M., Montalva L., Biouss G., Zani A., Treatment with amniotic fluid stem cell extracellular vesicles promotes fetal lung branching and cell differentiation at canalicular and saccular stages in experimental pulmonary hypoplasia secondary to congenital diaphragmatic hernia. Stem Cells Transl. Med. 11, 1089–1102 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khalaj K., Antounians L., Figueira R. L., Post M., Zani A., Autophagy is impaired in fetal hypoplastic lungs and rescued by administration of amniotic fluid stem cell extracellular vesicles. Am. J. Respir. Crit. Care Med. 206, 476–487 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Horowitz A., Simons M., Branching morphogenesis. Circ. Res. 103, 784–795 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Pereira-Terra P., Deprest J. A., Kholdebarin R., Khoshgoo N., DeKoninck P., Munck A. A., Wang J., Zhu F., Rottier R. J., Iwasiow B. M., Correia-Pinto J., Tibboel D., Post M., Keijzer R., Unique tracheal fluid microRNA signature predicts response to FETO in patients with congenital diaphragmatic hernia. Ann. Surg. 262, 1130–1140 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Figueira R. L., Khoshgoo N., Doktor F., Khalaj K., Islam T., Moheimani N., Blundell M., Antounians L., Post M., Zani A., Antenatal administration of extracellular vesicles derived from amniotic fluid stem cells improves lung function in neonatal rats with congenital diaphragmatic hernia. J. Pediatr. Surg. 10.1016/j.jpedsurg.2024.02.029, (2024). [DOI] [PubMed] [Google Scholar]
- 13.Ruano R., Peiro J. L., da Silva M. M., Campos J. A., Carreras E., Tannuri U., Zugaib M., Early fetoscopic tracheal occlusion for extremely severe pulmonary hypoplasia in isolated congenital diaphragmatic hernia: Preliminary results. Ultrasound Obstet. Gynecol. 42, 70–76 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Deprest J. A., Nicolaides K. H., Benachi A., Gratacos E., Ryan G., Persico N., Sago H., Johnson A., Wielgoś M., Berg C., van Calster B., Russo F. M.; TOTAL Trial for Severe Hypoplasia Investigators , Randomized trial of fetal surgery for severe left diaphragmatic hernia. N. Engl. J. Med. 385, 107–118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlon M., Toelen J., Van der Perren A., Vandenberghe L. H., Reumers V., Sbragia L., Gijsbers R., Baekelandt V., Himmelreich U., Wilson J. M., Deprest J., Debyser Z., Efficient gene transfer into the mouse lung by fetal intratracheal injection of rAAV2/6.2. Mol. Ther. 18, 2130–2138 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klaritsch P., Mayer S., Sbragia L., Toelen J., Roubliova X., Lewi P., Deprest J. A., Albumin as an adjunct to tracheal occlusion in fetal rats with congenital diaphragmatic hernia: A placebo-controlled study. Am. J. Obstet. Gynecol. 202, 198.e1–198.e9 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Montalva L., Zani A., Assessment of the nitrofen model of congenital diaphragmatic hernia and of the dysregulated factors involved in pulmonary hypoplasia. Pediatr. Surg. Int. 35, 41–61 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Ardini-Poleske M. E., Clark R. F., Ansong C., Carson J. P., Corley R. A., Deutsch G. H., Hagood J. S., Kaminski N., Mariani T. J., Potter S. S., Pryhuber G. S., Warburton D., Whitsett J. A., Palmer S. M., Ambalavanan N., LungMAP: The molecular atlas of lung development program. Am. J. Physiol. Lung Cell. Mol. Physiol. 313, L733–L740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Travaglini K. J., Nabhan A. N., Penland L., Sinha R., Gillich A., Sit R. V., Chang S., Conley S. D., Mori Y., Seita J., Berry G. J., Shrager J. B., Metzger R. J., Kuo C. S., Neff N., Weissman I. L., Quake S. R., Krasnow M. A., A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 587, 619–625 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karlsson M., Zhang C., Méar L., Zhong W., Digre A., Katona B., Sjöstedt E., Butler L., Odeberg J., Dusart P., Edfors F., Oksvold P., von Feilitzen K., Zwahlen M., Arif M., Altay O., Li X., Ozcan M., Mardinoglu A., Fagerberg L., Mulder J., Luo Y., Ponten F., Uhlén M., Lindskog C., A single-cell type transcriptomics map of human tissues. Sci. Adv. 7, eabh2169 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treutlein B., Brownfield D. G., Wu A. R., Neff N. F., Mantalas G. L., Espinoza F. H., Desai T. J., Krasnow M. A., Quake S. R., Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabula Muris Consortium , Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pridans C., Irvine K. M., Davis G. M., Lefevre L., Bush S. J., Hume D. A., Transcriptomic analysis of rat macrophages. Front. Immunol. 11, 594594 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conway J. G., McDonald B., Parham J., Keith B., Rusnak D. W., Shaw E., Jansen M., Lin P., Payne A., Crosby R. M., Johnson J. H., Frick L., Lin M. H., Depee S., Tadepalli S., Votta B., James I., Fuller K., Chambers T. J., Kull F. C., Chamberlain S. D., Hutchins J. T., Inhibition of colony-stimulating-factor-1 signaling in vivo with the orally bioavailable cFMS kinase inhibitor GW2580. Proc. Natl. Acad. Sci. U.S.A. 102, 16078–16083 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner R., Amonkar G. M., Wang W., Shui J. E., Bankoti K., Hei Tse W., High F. A., Zalieckas J. M., Buchmiller T. L., Zani A., Keijzer R., Donahoe P. K., Lerou P. H., Ai X., A tracheal aspirate-derived airway basal cell model reveals a proinflammatory epithelial defect in congenital diaphragmatic hernia. Am. J. Respir. Crit. Care Med. 207, 1214–1226 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varisco B. M., Nuclear factor-κB keeps basal cells undifferentiated in congenital diaphragmatic hernia. Am. J. Respir. Crit. Care Med. 207, 1122–1123 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohshiro K., Miyazaki E., Taira Y., Puri P., Upregulated tumor necrosis factor-alpha gene expression in the hypoplastic lung in patients with congenital diaphragmatic hernia. Pediatr. Surg. Int. 14, 21–24 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Schaible T., Veit M., Tautz J., Kehl S., Büsing K., Monz D., Gortner L., Tutdibi E., Serum cytokine levels in neonates with congenital diaphragmatic hernia. Klin. Padiatr. 223, 414–418 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Kipfmueller F., Heindel K., Geipel A., Berg C., Bartmann P., Reutter H., Mueller A., Holdenrieder S., Expression of soluble receptor for advanced glycation end products is associated with disease severity in congenital diaphragmatic hernia. Am. J. Physiol. Lung Cell. Mol. Physiol. 316, L1061–L1069 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Perry R., Stein J., Young G., Ramanathan R., Seri I., Klee L., Friedlich P., Antithrombin III administration in neonates with congenital diaphragmatic hernia during the first three days of extracorporeal membrane oxygenation. J. Pediatr. Surg. 48, 1837–1842 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Pavcnik-Arnol M., Bonac B., Groselj-Grenc M., Derganc M., Changes in serum procalcitonin, interleukin 6, interleukin 8 and C-reactive protein in neonates after surgery. Eur. J. Pediatr. Surg. 20, 262–266 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Fleck S., Bautista G., Keating S. M., Lee T. H., Keller R. L., Moon-Grady A. J., Gonzales K., Norris P. J., Busch M. P., Kim C. J., Romero R., Lee H., Miniati D., MacKenzie T. C., Fetal production of growth factors and inflammatory mediators predicts pulmonary hypertension in congenital diaphragmatic hernia. Pediatr. Res. 74, 290–298 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okawada M., Kobayashi H., Tei E., Okazaki T., Lane G. J., Yamataka A., Serum monocyte chemotactic protein-1 levels in congenital diaphragmatic hernia. Pediatr. Surg. Int. 23, 487–491 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Lingappan K., Olutoye O. O. II, Cantu A., Cantu Gutierrez M. E., Cortes-Santiago N., Hammond J. D., Gilley J., Quintero J. R., Li H., Polverino F., Gleghorn J. P.,Keswani S. G., Molecular insights using spatial transcriptomics of the distal lung in congenital diaphragmatic hernia. Am. J. Physiol. Lung Cell. Mol. Physiol. 325, L477–L486 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gosemann J. H., Friedmacher F., Hofmann A., Zimmer J., Kuebler J. F., Rittinghausen S., Suttkus A., Lacher M., Alvarez L., Corcionivoschi N., Puri P., Prenatal treatment with rosiglitazone attenuates vascular remodeling and pulmonary monocyte influx in experimental congenital diaphragmatic hernia. PLOS ONE 13, e0206975 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gosemann J. H., Doi T., Kutasy B., Friedmacher F., Dingemann J., Puri P., Alterations of peroxisome proliferator-activated receptor γ and monocyte chemoattractant protein 1 gene expression in the nitrofen-induced hypoplastic lung. J. Pediatr. Surg. 47, 847–851 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Shima H., Ohshiro K., Taira Y., Miyazaki E., Oue T., Puri P., Antenatal dexamethasone suppresses tumor necrosis factor-alpha expression in hypoplastic lung in nitrofen-induced diaphragmatic hernia in rats. Pediatr. Res. 46, 633–637 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Wagner R., Lieckfeldt P., Piyadasa H., Markel M., Riedel J., Stefanovici C., Peukert N., Patel D., Derraugh G., Min S. L., Gosemann J. H., Deprest J., Pascoe C. D., Tse A., Lacher M., Mookherjee N., Keijzer R., Proteomic profiling of hypoplastic lungs suggests an underlying inflammatory response in the pathogenesis of abnormal lung development in congenital diaphragmatic hernia. Ann. Surg. 278, e411–e421 (2023). [DOI] [PubMed] [Google Scholar]
- 39.Dylong F., Riedel J., Amonkar G. M., Peukert N., Lieckfeldt P., Sturm K., Höxter B., Tse W. H., Miyake Y., Moormann M., Bode L. M., Mayer S., Keijzer R., Lacher M., Ai X., Gosemann J. H., Wagner R., Overactivated epithelial NF-κB disrupts lung development in congenital diaphragmatic hernia. Am. J. Respir. Cell Mol. Biol. 69, 545–555 (2023). [DOI] [PubMed] [Google Scholar]
- 40.Peiro J. L., Oria M., Aydin E., Joshi R., Cabanas N., Schmidt R., Schroeder C., Marotta M., Varisco B. M., Proteomic profiling of tracheal fluid in an ovine model of congenital diaphragmatic hernia and fetal tracheal occlusion. Am. J. Physiol. Lung Cell. Mol. Physiol. 315, L1028–L1041 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guilliams M., De Kleer I., Henri S., Post S., Vanhoutte L., De Prijck S., Deswarte K., Malissen B., Hammad H., Lambrecht B. N., Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med. 23, 1977–1992 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lakhdari O., Yamamura A., Hernandez G. E., Anderson K. K., Lund S. J., Oppong-Nonterah G. O., Hoffman H. M., Prince L. S., Differential immune activation in fetal macrophage populations. Sci. Rep. 9, 7677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mezu-Ndubuisi O. J., Maheshwari A., Role of macrophages in fetal development and perinatal disorders. Pediatr. Res. 90, 513–523 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Varghese N. P., Tillman R. H., Keller R. L., Pulmonary hypertension is an important co-morbidity in developmental lung diseases of infancy: Bronchopulmonary dysplasia and congenital diaphragmatic hernia. Pediatr. Pulmonol. 56, 670–677 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Acker S. N., Mandell E. W., Sims-Lucas S., Gien J., Abman S. H., Galambos C., Histologic identification of prominent intrapulmonary anastomotic vessels in severe congenital diaphragmatic hernia. J. Pediatr. 166, 178–183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalymbetova T. V., Selvakumar B., Rodríguez-Castillo J. A., Gunjak M., Malainou C., Heindl M. R., Moiseenko A., Chao C. M., Vadász I., Mayer K., Lohmeyer J., Bellusci S., Böttcher-Friebertshäuser E., Seeger W., Herold S., Morty R. E., Resident alveolar macrophages are master regulators of arrested alveolarization in experimental bronchopulmonary dysplasia. J. Pathol. 245, 153–159 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Matei A. C., Antounians L., Zani A., Extracellular vesicles as a potential therapy for neonatal conditions: State of the art and challenges in clinical translation. Pharmaceutics 11, 404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delavogia E., Ntentakis D. P., Cortinas J. A., Fernandez-Gonzalez A., Mitsialis S. A., Kourembanas S., Mesenchymal stromal/stem cell extracellular vesicles and perinatal injury: One formula for many diseases. Stem Cells 40, 991–1007 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khalaj K., Figueira R. L., Antounians L., Lauriti G., Zani A., Systematic review of extracellular vesicle-based treatments for lung injury: Are EVs a potential therapy for COVID-19? J. Extracell. Vesicles 9, 1795365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zani-Ruttenstock E., Antounians L., Khalaj K., Figueira R. L., Zani A., The role of exosomes in the treatment, prevention, diagnosis, and pathogenesis of COVID-19. Eur. J. Pediatr. Surg. 31, 326–334 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Sengupta V., Sengupta S., Lazo A., Woods P., Nolan A., Bremer N., Exosomes derived from bone marrow mesenchymal stem cells as treatment for severe COVID-19. Stem Cells Dev. 29, 747–754 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pederiva F., Ghionzoli M., Pierro A., De Coppi P., Tovar J. A., Amniotic fluid stem cells rescue both in vitro and in vivo growth, innervation, and motility in nitrofen-exposed hypoplastic rat lungs through paracrine effects. Cell Transplant. 22, 1683–1694 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Di Bernardo J., Maiden M. M., Hershenson M. B., Kunisaki S. M., Amniotic fluid derived mesenchymal stromal cells augment fetal lung growth in a nitrofen explant model. J. Pediatr. Surg. 49, 859–865 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Antounians L., Tzanetakis A., Pellerito O., Catania V. D., Sulistyo A., Montalva L., McVey M. J., Zani A., The regenerative potential of amniotic fluid stem cell extracellular vesicles: Lessons learned by comparing different isolation techniques. Sci. Rep. 9, 1837 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mezzasoma L., Bellezza I., Orvietani P., Manni G., Gargaro M., Sagini K., Llorente A., Scarpelli P., Pascucci L., Cellini B., Talesa V. N., Fallarino F., Romani R., Amniotic fluid stem cell-derived extracellular vesicles are independent metabolic units capable of modulating inflammasome activation in THP-1 cells. FASEB J. 36, e22218 (2022). [DOI] [PubMed] [Google Scholar]
- 56.Li B., Lee C., O'Connell J. S., Antounians L., Ganji N., Alganabi M., Cadete M., Nascimben F., Koike Y., Hock A., Botts S. R., Wu R. Y., Miyake H., Minich A., Maalouf M. F., Zani-Ruttenstock E., Chen Y., Johnson-Henry K. C., De Coppi P., Eaton S., Maattanen P., Delgado Olguin P., Zani A., Sherman P. M., Pierro A., Activation of Wnt signaling by amniotic fluid stem cell-derived extracellular vesicles attenuates intestinal injury in experimental necrotizing enterocolitis. Cell Death Dis. 11, 750 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O'Connell J. S., Lee C., Farhat N., Antounians L., Zani A., Li B., Pierro A., Administration of extracellular vesicles derived from human amniotic fluid stem cells: A new treatment for necrotizing enterocolitis. Pediatr. Surg. Int. 37, 301–309 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Hurskainen M., Mižíková I., Cook D. P., Andersson N., Cyr-Depauw C., Lesage F., Helle E., Renesme L., Jankov R. P., Heikinheimo M., Vanderhyden B. C., Thébaud B., Single cell transcriptomic analysis of murine lung development on hyperoxia-induced damage. Nat. Commun. 12, 1565 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen S., Wang J., Zhang K., Ma B., Li X., Wei R., Nian H., LncRNA Neat1 targets NonO and miR-128-3p to promote antigen-specific Th17 cell responses and autoimmune inflammation. Cell Death Dis. 14, 610 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cohen T. S., Role of microRNA in the lung’s innate immune response. J. Innate Immun. 9, 243–249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu S., Gao S., Yang Z., Zhang P., miR-128-3p reduced acute lung injury induced by sepsis via targeting PEL12. Open Med. 16, 1109–1120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schneider H., Faschingbauer F., Schuepbach-Mallepell S., Körber I., Wohlfart S., Dick A., Wahlbuhl M., Kowalczyk-Quintas C., Vigolo M., Kirby N., Tannert C., Rompel O., Rascher W., Beckmann M. W., Schneider P., Prenatal correction of X-linked hypohidrotic ectodermal dysplasia. N. Engl. J. Med. 378, 1604–1610 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Sheller-Miller S., Choi K., Choi C., Menon R., Cyclic-recombinase-reporter mouse model to determine exosome communication and function during pregnancy. Am. J. Obstet. Gynecol. 221, 502.e1–502.e12 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Cohen J. L., Chakraborty P., Fung-Kee-Fung K., Schwab M. E., Bali D., Young S. P., Gelb M. H., Khaledi H., DiBattista A., Smallshaw S., Moretti F., Wong D., Lacroix C., El Demellawy D., Strickland K. C., Lougheed J., Moon-Grady A., Lianoglou B. R., Harmatz P., Kishnani P. S., MacKenzie T. C., In utero enzyme-replacement therapy for infantile-onset Pompe’s disease. N. Engl. J. Med. 387, 2150–2158 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Antounians L., Zani A., Beyond the diaphragm and the lung: A multisystem approach to understanding congenital diaphragmatic hernia. Pediatr. Surg. Int. 39, 194 (2023). [DOI] [PubMed] [Google Scholar]
- 66.Blundell M., Doktor F., Figueira R. L., Khalaj K., Biouss G., Antounians L., Zani A., Anti-inflammatory effects of antenatal administration of stem cell derived extracellular vesicles in the brain of rat fetuses with congenital diaphragmatic hernia. Pediatr. Surg. Int. 39, 291 (2023). [DOI] [PubMed] [Google Scholar]
- 67.Théry C., Witwer K. W., Aikawa E., Alcaraz M. J., Anderson J. D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G. K., Ayre D. C., Bach J.-M., Bachurski D., Baharvand H., Balaj L., Baldacchino S., Bauer N. N., Baxter A. A., Bebawy M., Beckham C., Zavec A. B., Benmoussa A., Berardi A. C., Bergese P., Bielska E., Blenkiron C., Bobis-Wozowicz S., Boilard E., Boireau W., Bongiovanni A., Borràs F. E., Bosch S., Boulanger C. M., Breakefield X., Breglio A. M., Brennan M. Á., Brigstock D. R., Brisson A., Broekman M. L., Bromberg J. F., Bryl-Górecka P., Buch S., Buck A. H., Burger D., Busatto S., Buschmann D., Bussolati B., Buzás E. I., Byrd J. B., Camussi G., Carter D. R., Caruso S., Chamley L. W., Chang Y.-T., Chen C., Chen S., Cheng L., Chin A. R., Clayton A., Clerici S. P., Cocks A., Cocucci E., Coffey R. J., Cordeiro-da-Silva A., Couch Y., Coumans F. A., Coyle B., Crescitelli R., Criado M. F., D'Souza-Schorey C., Das S., Chaudhuri A. D., de Candia P., De Santana E. F., Wever O. D., Del Portillo H. A., Demaret T., Deville S., Devitt A., Dhondt B., Vizio D. D., Dieterich L. C., Dolo V., Rubio A. P. D., Dominici M., Dourado M. R., Driedonks T. A., Duarte F. V., Duncan H. M., Eichenberger R. M., Ekström K., Andaloussi S. E., Elie-Caille C., Erdbrügger U., Falcón-Pérez J. M., Fatima F., Fish J. E., Flores-Bellver M., Försönits A., Frelet-Barrand A., Fricke F., Fuhrmann G., Gabrielsson S., Gámez-Valero A., Gardiner C., Gärtner K., Gaudin R., Gho Y. S., Giebel B., Gilbert C., Gimona M., Giusti I., Goberdhan D. C., Görgens A., Gorski S. M., Greening D. W., Gross J. C., Gualerzi A., Gupta G. N., Gustafson D., Handberg A., Haraszti R. A., Harrison P., Hegyesi H., Hendrix A., Hill A. F., Hochberg F. H., Hoffmann K. F., Holder B., Holthofer H., Hosseinkhani B., Hu G., Huang Y., Huber V., Hunt S., Ibrahim A. G.-E., Ikezu T., Inal J. M., Isin M., Ivanova A., Jackson H. K., Jacobsen S., Jay S. M., Jayachandran M., Jenster G., Jiang L., Johnson S. M., Jones J. C., Jong A., Jovanovic-Talisman T., Jung S., Kalluri R., Kano S.-I., Kaur S., Kawamura Y., Keller E. T., Khamari D., Khomyakova E., Khvorova A., Kierulf P., Kim K. P., Kislinger T., Klingeborn M., Klinke D. J. II, Kornek M., Kosanović M. M., Kovács Á. F., Krämer-Albers E.-M., Krasemann S., Krause M., Kurochkin I. V., Kusuma G. D., Kuypers S., Laitinen S., Langevin S. M., Languino L. R., Lannigan J., Lässer C., Laurent L. C., Lavieu G., Lázaro-Ibáñez E., Lay S. L., Lee M.-S., Lee Y. X. F., Lemos D. S., Lenassi M., Leszczynska A., Li I. T., Liao K., Libregts S. F., Ligeti E., Lim R., Lim S. K., Linē A., Linnemannstöns K., Llorente A., Lombard C. A., Lorenowicz M. J., Lörincz Á. M., Lötvall J., Lovett J., Lowry M. C., Loyer X., Lu Q., Lukomska B., Lunavat T. R., Maas S. L., Malhi H., Marcilla A., Mariani J., Mariscal J., Martens-Uzunova E. S., Martin-Jaular L., Martinez M. C., Martins V. R., Mathieu M., Mathivanan S., Maugeri M., McGinnis L. K., McVey M. J., Meckes D. G. Jr., Meehan K. L., Mertens I., Minciacchi V. R., Möller A., Jørgensen M. M., Morales-Kastresana A., Morhayim J., Mullier F., Muraca M., Musante L., Mussack V., Muth D. C., Myburgh K. H., Najrana T., Nawaz M., Nazarenko I., Nejsum P., Neri C., Neri T., Nieuwland R., Nimrichter L., Nolan J. P., Hoen E. N. N., Hooten N. N., O'Driscoll L., O'Grady T., O'Loghlen A., Ochiya T., Olivier M., Ortiz A., Ortiz L. A., Osteikoetxea X., Østergaard O., Ostrowski M., Park J., Pegtel D. M., Peinado H., Perut F., Pfaffl M. W., Phinney D. G., Pieters B. C., Pink R. C., Pisetsky D. S., von Strandmann E. P., Polakovicova I., Poon I. K., Powell B. H., Prada I., Pulliam L., Quesenberry P., Radeghieri A., Raffai R. L., Raimondo S., Rak J., Ramirez M. I., Raposo G., Rayyan M. S., Regev-Rudzki N., Ricklefs F. L., Robbins P. D., Roberts D. D., Rodrigues S. C., Rohde E., Rome S., Rouschop K. M., Rughetti A., Russell A. E., Saá P., Sahoo S., Salas-Huenuleo E., Sánchez C., Saugstad J. A., Saul M. J., Schiffelers R. M., Schneider R., Schøyen T. H., Scott A., Shahaj E., Sharma S., Shatnyeva O., Shekari F., Shelke G. V., Shetty A. K., Shiba K., Siljander P. R.-M., Silva A. M., Skowronek A., Snyder O. L. 2nd, Soares R. P., Sódar B. W., Soekmadji C., Sotillo J., Stahl P. D., Stoorvogel W., Stott S. L., Strasser E. F., Swift S., Tahara H., Tewari M., Timms K., Tiwari S., Tixeira R., Tkach M., Toh W. S., Tomasini R., Torrecilhas A. C., Tosar J. P., Toxavidis V., Urbanelli L., Vader P., van Balkom B. W., van der Grein S. G., Van Deun J., van Herwijnen M. J., Keuren-Jensen K. V., van Niel G., van Royen M. E., van Wijnen A. J., Vasconcelos M. H., Vechetti I. J. Jr., Veit T. D., Vella L. J., Velot É., Verweij F. J., Vestad B., Viñas J. L., Visnovitz T., Vukman K. V., Wahlgren J., Watson D. C., Wauben M. H., Weaver A., Webber J. P., Weber V., Wehman A. M., Weiss D. J., Welsh J. A., Wendt S., Wheelock A. M., Wiener Z., Witte L., Wolfram J., Xagorari A., Xander P., Xu J., Yan X., Yáñez-Mó M., Yin H., Yuana Y., Zappulli V., Zarubova J., Žėkas V., Zhang J.-Y., Zhao Z., Zheng L., Zheutlin A. R., Zickler A. M., Zimmermann P., Zivkovic A. M., Zocco D., Zuba-Surma E. K., Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7, 1535750 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Antounians L., Figueira R. L., Sbragia L., Zani A., Congenital diaphragmatic hernia: State of the art in translating experimental research to the bedside. Eur. J. Pediatr. Surg. 29, 317–327 (2019). [DOI] [PubMed] [Google Scholar]
- 69.Montalva L., Antounians L., Zani A., Pulmonary hypertension secondary to congenital diaphragmatic hernia: Factors and pathways involved in pulmonary vascular remodeling. Pediatr. Res. 85, 754–768 (2019). [DOI] [PubMed] [Google Scholar]
- 70.Iritani I., Experimental study on embryogenesis of congenital diaphragmatic hernia. Anat. Embryol. (Berl.) 169, 133–139 (1984). [DOI] [PubMed] [Google Scholar]
- 71.Kekik C., Besisik S. K., Seyhun Y., Oguz F. S., Sargin D., Carin M. N., Relationship between HLA tissue type, CMV infection, and acute graft-vs-host disease after allogeneic hematopoietic stem cell transplantation: Single-center experience. Transplant. Proc. 41, 3859–3862 (2009). [DOI] [PubMed] [Google Scholar]
- 72.Takayama S., Sakai K., Fumino S., Furukawa T., Kishida T., Mazda O., Tajiri T., An intra-amniotic injection of mesenchymal stem cells promotes lung maturity in a rat congenital diaphragmatic hernia model. Pediatr. Surg. Int. 35, 1353–1361 (2019). [DOI] [PubMed] [Google Scholar]
- 73.Somashekar S. T., Sammour I., Huang J., Dominguez-Bendala J., Pastori R., Alvarez-Cubela S., Torres E., Wu S., Young K. C., Intra-amniotic soluble endoglin impairs lung development in neonatal rats. Am. J. Respir. Cell Mol. Biol. 57, 468–476 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Kobayashi K., Lemke R. P., Greer J. J., Ultrasound measurements of fetal breathing movements in the rat. J. Appl. Physiol. 91, 316–320 (1985). [DOI] [PubMed] [Google Scholar]
- 75.Hsia C., Hyde D. M., Ochs M., Weibel E. R.; ATS/ERS Joint Task Force on Quantitative Assessment of Lung Structure , An official research policy statement of the American Thoracic Society/European Respiratory Society: Standards for quantitative assessment of lung structure. Am. J. Respir. Crit. Care Med. 181, 394–418 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jin S., Guerrero-Juarez C. F., Zhang L., Chang I., Ramos R., Kuan C. H., Myung P., Plikus M. V., Nie Q., Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reimand J., Kull M., Peterson H., Hansen J., Vilo J., g:Profiler—A web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35, W193–W200 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gillespie M., Jassal B., Stephan R., Milacic M., Rothfels K., Senff-Ribeiro A., Griss J., Sevilla C., Matthews L., Gong C., Deng C., Varusai T., Ragueneau E., Haider Y., May B., Shamovsky V., Weiser J., Brunson T., Sanati N., Beckman L., Shao X., Fabregat A., Sidiropoulos K., Murillo J., Viteri G., Cook J., Shorser S., Bader G., Demir E., Sander C., Haw R., Wu G., Stein L., Hermjakob H., D'Eustachio P., The reactome pathway knowledgebase 2022. Nucleic Acids Res. 50, D687–D692 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alquicira-Hernandez J., Sathe A., Ji H. P., Nguyen Q., Powell J. E., scPred: Accurate supervised method for cell-type classification from single-cell RNA-seq data. Genome Biol. 20, 264–281 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hong J., Arneson D., Umar S., Ruffenach G., Cunningham C. M., Ahn I. S., Diamante G., Bhetraratana M., Park J. F., Said E., Huynh C., Le T., Medzikovic L., Humbert M., Soubrier F., Montani D., Girerd B., Trégouët D. A., Channick R., Saggar R., Eghbali M., Yang X., Single-cell study of two rat models of pulmonary arterial hypertension reveals connections to human pathobiology and drug repositioning. Am. J. Respir. Crit. Care Med. 203, 1006–1022 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Herrera-Rivero M., Zhang R., Heilmann-Heimbach S., Mueller A., Bagci S., Dresbach T., Schröder L., Holdenrieder S., Reutter H. M., Kipfmueller F., Circulating microRNAs are associated with pulmonary hypertension and development of chronic lung disease in congenital diaphragmatic hernia. Sci. Rep. 8, 10735 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Piersigilli F., Syed M., Lam T. T., Dotta A., Massoud M., Vernocchi P., Quagliariello A., Putignani L., Auriti C., Salvatori G., Bagolan P., Bhandari V., An omic approach to congenital diaphragmatic hernia: A pilot study of genomic, microRNA, and metabolomic profiling. J. Perinatol. 40, 952–961 (2020). [DOI] [PubMed] [Google Scholar]
- 83.Khoshgoo N., Visser R., Falk L., Day C. A., Ameis D., Iwasiow B. M., Zhu F., Öztürk A., Basu S., Pind M., Fresnosa A., Jackson M., Siragam V. K., Stelmack G., Hicks G. G., Halayko A. J., Keijzer R., MicroRNA-200b regulates distal airway development by maintaining epithelial integrity. Sci. Rep. 7, 6382 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eastwood M. P., Deprest J., Russo F. M., Wang H., Mulhall D., Iwasiow B., Mahood T. H., Keijzer R., MicroRNA 200b is upregulated in the lungs of fetal rabbits with surgically induced diaphragmatic hernia. Prenat. Diagn. 38, 645–653 (2018). [DOI] [PubMed] [Google Scholar]
- 85.Mudri M., Smith S. A., Vanderboor C., Davidson J., Regnault T. R. H., Bütter A., The effects of tracheal occlusion on Wnt signaling in a rabbit model of congenital diaphragmatic hernia. J. Pediatr. Surg. 54, 937–944 (2019). [DOI] [PubMed] [Google Scholar]
- 86.Sanford E. L., Choy K. W., Donahoe P. K., Tracy A. A., Hila R., Loscertales M., Longoni M., MiR-449a affects epithelial proliferation during the pseudoglandular and canalicular phases of avian and mammal lung development. PLOS ONE 11, e0149425 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhu S., He Q., Zhang R., Wang Y., Zhong W., Xia H., Yu J., Decreased expression of miR-33 in fetal lungs of nitrofen-induced congenital diaphragmatic hernia rat model. J. Pediatr. Surg. 51, 1096–1100 (2016). [DOI] [PubMed] [Google Scholar]
- 88.Mulhall D., Khoshgoo N., Visser R., Iwasiow B., Day C., Zhu F., Eastwood P., Keijzer R., miR-200 family expression during normal and abnormal lung development due to congenital diaphragmatic hernia at the later embryonic stage in the nitrofen rat model. Pediatr. Surg. Int. 36, 1429–1436 (2020). [DOI] [PubMed] [Google Scholar]
- 89.Khoshgoo N., Kholdebarin R., Pereira-Terra P., Mahood T. H., Falk L., Day C. A., Iwasiow B. M., Zhu F., Mulhall D., Fraser C., Correia-Pinto J., Keijzer R., Prenatal microRNA miR-200b therapy improves nitrofen-induced pulmonary hypoplasia associated with congenital diaphragmatic hernia. Ann. Surg. 269, 979–987 (2019). [DOI] [PubMed] [Google Scholar]
- 90.Mahood T. H., Johar D. R., Iwasiow B. M., Xu W., Keijzer R., The transcriptome of nitrofen-induced pulmonary hypoplasia in the rat model of congenital diaphragmatic hernia. Pediatr. Res. 79, 766–775 (2016). [DOI] [PubMed] [Google Scholar]
- 91.Li X., Liu H., Lv Y., Yu W., Liu X., Liu C., MiR-130a-5p/Foxa2 axis modulates fetal lung development in congenital diaphragmatic hernia by activating the Shh/Gli1 signaling pathway. Life Sci. 241, 117166 (2020). [DOI] [PubMed] [Google Scholar]
- 92.Ru Y., Kechris K. J., Tabakoff B., Hoffman P., Radcliffe R. A., Bowler R., Mahaffey S., Rossi S., Calin G. A., Bemis L., Theodorescu D., The multiMiR R package and database: Integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res. 42, e133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T., Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li Z., Gong X., Li D., Yang X., Shi Q., Ju X., Intratracheal transplantation of amnion-derived mesenchymal stem cells ameliorates hyperoxia-induced neonatal hyperoxic lung injury via aminoacyl-peptide hydrolase. Int. J. Stem. Cells. 13, 221–236 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Willis G. R., Fernandez-Gonzalez A., Reis M., Yeung V., Liu X., Ericsson M., Andrews N. A., Mitsialis S. A., Kourembanas S., Mesenchymal stromal cell-derived small extracellular vesicles restore lung architecture and improve exercise capacity in a model of neonatal hyperoxia-induced lung injury. J. Extracell. Vesicles 9, 1790874 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Porzionato A., Zaramella P., Dedja A., Guidolin D., Van Wemmel K., Macchi V., Jurga M., Perilongo G., De Caro R., Baraldi E., Muraca M., Intratracheal administration of clinical-grade mesenchymal stem cell-derived extracellular vesicles reduces lung injury in a rat model of bronchopulmonary dysplasia. Am. J. Physiol. Lung Cell. Mol. Physiol. 316, L6–L19 (2019). [DOI] [PubMed] [Google Scholar]
- 97.Braun R. K., Chetty C., Balasubramaniam V., Centanni R., Haraldsdottir K., Hematti P., Eldridge M. W., Intraperitoneal injection of MSC-derived exosomes prevent experimental bronchopulmonary dysplasia. Biochem. Biophys. Res. Commun. 503, 2653–2658 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chaubey S., Thueson S., Ponnalagu D., Alam M. A., Gheorghe C. P., Aghai Z., Singh H., Bhandari V., Early gestational mesenchymal stem cell secretome attenuates experimental bronchopulmonary dysplasia in part via exosome-associated factor TSG-6. Stem Cell Res. Ther. 9, 173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Willis G. R., Fernandez-Gonzalez A., Anastas J., Vitali S. H., Liu X., Ericsson M., Kwong A., Mitsialis S. A., Kourembanas S., Mesenchymal stromal cell exosomes ameliorate experimental bronchopulmonary dysplasia and restore lung function through macrophage immunomodulation. Am. J. Respir. Crit. Care Med. 197, 104–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ahn S. Y., Park W. S., Kim Y. E., Sung D. K., Sung S. I., Ahn J. Y., Chang Y. S., MSK1 functions as a transcriptional coactivator of p53 in the regulation of p21 gene expression. Exp. Mol. Med. 50, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tieu A., Hu K., Gnyra C., Montroy J., Fergusson D. A., Allan D. S., Stewart D. J., Thébaud B., Lalu M. M., Mesenchymal stromal cell extracellular vesicles as therapy for acute and chronic respiratory diseases: A meta-analysis. J. Extracell. Vesicles 10, e12141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Willis G. R., Reis M., Gheinani A. H., Fernandez-Gonzalez A., Taglauer E. S., Yeung V., Liu X., Ericsson M., Haas E., Mitsialis S. A., Kourembanas S., Extracellular vesicles protect the neonatal lung from hyperoxic injury through the epigenetic and transcriptomic reprogramming of myeloid cells. Am. J. Respir. Crit. Care Med. 204, 1418–1432 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ye S. Q., Simon B. A., Maloney J. P., Zambelli-Weiner A., Gao L., Grant A., Easley R. B., McVerry B. J., Tuder R. M., Standiford T., Brower R. G., Barnes K. C., Garcia J. G., Pre-B-cell colony-enhancing factor as a potential novel biomarker in acute lung injury. Am. J. Respir. Crit. Care Med. 171, 361–370 (2005). [DOI] [PubMed] [Google Scholar]
- 104.Lee K. A., Gong M. N., Pre-B-cell colony-enhancing factor and its clinical correlates with acute lung injury and sepsis. Chest 140, 382–390 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Matsuda A., Yang W. L., Jacob A., Aziz M., Matsuo S., Matsutani T., Uchida E., Wang P., FK866, a visfatin inhibitor, protects against acute lung injury after intestinal ischemia-reperfusion in mice via NF-κB pathway. Ann. Surg. 259, 1007–1017 (2014). [DOI] [PubMed] [Google Scholar]
- 106.Ochman M., Maruszewski M., Wojarski J., Żegleń S., Karolak W., Stanjek-Cichoracka A., Przybyłowski P., Zembala M., Kukla M., Serum levels of visfatin, omentin and irisin in patients with end-stage lung disease before and after lung transplantation. Ann. Transplant. 22, 761–768 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Weng T., Liu L., The role of pleiotrophin and beta-catenin in fetal lung development. Respir. Res. 11, 80 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Himburg H. A., Harris J. R., Ito T., Daher P., Russell J. L., Quarmyne M., Doan P. L., Helms K., Nakamura M., Fixsen E., Herradon G., Reya T., Chao N. J., Harroch S., Chute J. P., Pleiotrophin regulates the retention and self-renewal of hematopoietic stem cells in the bone marrow vascular niche. Cell Rep. 2, 964–975 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guardado S., Ojeda-Juárez D., Kaul M., Nordgren T. M., Comprehensive review of lipocalin 2-mediated effects in lung inflammation. Am. J. Physiol. Lung Cell. Mol. Physiol. 321, L726–L733 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bry K., Whitsett J. A., Lappalainen U., IL-1beta disrupts postnatal lung morphogenesis in the mouse. Am. J. Respir. Cell Mol. Biol. 36, 32–42 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stouch A. N., McCoy A. M., Greer R. M., Lakhdari O., Yull F. E., Blackwell T. S., Hoffman H. M., Prince L. S., IL-1β and inflammasome activity link inflammation to abnormal fetal airway development. J. Immunol. 196, 3411–3420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fernandez-Sauze S., Delfino C., Mabrouk K., Dussert C., Chinot O., Martin P. M., Grisoli F., Ouafik L., Boudouresque F., Effects of adrenomedullin on endothelial cells in the multistep process of angiogenesis: Involvement of CRLR/RAMP2 and CRLR/RAMP3 receptors. Int. J. Cancer 108, 797–804 (2004). [DOI] [PubMed] [Google Scholar]
- 113.Zhang S., Patel A., Moorthy B., Shivanna B., Adrenomedullin deficiency potentiates hyperoxic injury in fetal human pulmonary microvascular endothelial cells. Biochem. Biophys. Res. Commun. 464, 1048–1053 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vadivel A., Abozaid S., van Haaften T., Sawicka M., Eaton F., Chen M., Thébaud B., Adrenomedullin promotes lung angiogenesis, alveolar development, and repair. Am. J. Respir. Cell Mol. Biol. 43, 152–160 (2010). [DOI] [PubMed] [Google Scholar]
- 115.Suzuki T., Suzuki S., Fujino N., Ota C., Yamada M., Suzuki T., Yamaya M., Kondo T., Kubo H., c-Kit immunoexpression delineates a putative endothelial progenitor cell population in developing human lungs. Am. J. Physiol. Lung Cell. Mol. Physiol. 306, L855–L865 (2014). [DOI] [PubMed] [Google Scholar]
- 116.Lindsey J. Y., Ganguly K., Brass D. M., Li Z., Potts E. N., Degan S., Chen H., Brockway B., Abraham S. N., Berndt A., Stripp B. R., Foster W. M., Leikauf G. D., Schulz H., Hollingsworth J. W., c-Kit is essential for alveolar maintenance and protection from emphysema-like disease in mice. Am. J. Respir. Crit. Care Med. 183, 1644–1652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 117.Epaud R., Aubey F., Xu J., Chaker Z., Clemessy M., Dautin A., Ahamed K., Bonora M., Hoyeau N., Fléjou J. F., Mailleux A., Clement A., Henrion-Caude A., Holzenberger M., Knockout of insulin-like growth factor-1 receptor impairs distal lung morphogenesis. PLOS ONE 7, e48071 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.He H., Snowball J., Sun F., Na C. L., Whitsett J. A., IGF1R controls mechanosignaling in myofibroblasts required for pulmonary alveologenesis. JCI Insight 6, 144863 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Baker J., Liu J. P., Robertson E. J., Efstratiadis A., Role of insulin-like growth factors in embryonic and postnatal growth. Cell 75, 73–82 (1993). [PubMed] [Google Scholar]
- 120.Liu H., Chang L., Rong Z., Zhu H., Zhang Q., Chen H., Li W., Association of insulin-like growth factors with lung development in neonatal rats. J. Huazhong Univ. Sci. Technol. Med. Sci. 24, 162–165 (2004). [DOI] [PubMed] [Google Scholar]
- 121.Ruttenstock E., Doi T., Dingemann J., Puri P., Insulinlike growth factor receptor type 1 and type 2 are downregulated in the nitrofen-induced hypoplastic lung. J. Pediatr. Surg. 45, 1349–1353 (2010). [DOI] [PubMed] [Google Scholar]
- 122.Jeansson M., Gawlik A., Anderson G., Li C., Kerjaschki D., Henkelman M., Quaggin S. E., Angiopoietin-1 is essential in mouse vasculature during development and in response to injury. J. Clin. Invest. 121, 2278–2289 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ferrara N., Vascular endothelial growth factor: Basic science and clinical progress. Endocr. Rev. 25, 581–611 (2004). [DOI] [PubMed] [Google Scholar]
- 124.Chinoy M. R., Graybill M. M., Miller S. A., Lang C. M., Kauffman G. L., Angiopoietin-1 and VEGF in vascular development and angiogenesis in hypoplastic lungs. Am. J. Physiol. Lung Cell. Mol. Physiol. 283, L60–L66 (2002). [DOI] [PubMed] [Google Scholar]
- 125.Miura da Costa K., Fabro A. T., Becari C., Figueira R. L., Schmidt A. F., Ruano R., Sbragia L., Honeymoon period in newborn rats with CDH is associated with changes in the VEGF signaling pathway. Front. Pediatr. 9, 698217 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Abler L. L., Mansour S. L., Sun X., Conditional gene inactivation reveals roles for Fgf10 and Fgfr2 in establishing a normal pattern of epithelial branching in the mouse lung. Dev. Dyn. 238, 1999–2013 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yin Y., Ornitz D. M., FGF9 and FGF10 activate distinct signaling pathways to direct lung epithelial specification and branching. Sci. Signal. 13, eaay4353 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Danopoulos S., Shiosaki J., Al Alam D., FGF signaling in lung development and disease: Human versus mouse. Front. Genet. 10, 170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Danopoulos S., Thornton M. E., Grubbs B. H., Frey M. R., Warburton D., Bellusci S., Al Alam D., Discordant roles for FGF ligands in lung branching morphogenesis between human and mouse. J. Pathol. 247, 254–265 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Klein J. M., Fritz B. L., McCarthy T. A., Wohlford-Lenane C. L., Snyder J. M., Localization of epidermal growth factor receptor in alveolar epithelium during human fetal lung development in vitro. Exp. Lung Res. 21, 917–939 (1995). [DOI] [PubMed] [Google Scholar]
- 131.Li J., Hu T., Liu W., Xiang B., Jiang X., Effect of epidermal growth factor on pulmonary hypoplasia in experimental diaphragmatic hernia. J. Pediatr. Surg. 39, 37–42 (2004). [DOI] [PubMed] [Google Scholar]
- 132.Peng C. K., Wu C. P., Lin J. Y., Peng S. C., Lee C. H., Huang K. L., Shen C. H., Gas6/Axl signaling attenuates alveolar inflammation in ischemia-reperfusion-induced acute lung injury by up-regulating SOCS3-mediated pathway. PLOS ONE 14, e0219788 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wu Q., Zhou X., Wang Y., Hu Y., LncRNA GAS5 promotes spermidine-induced autophagy through the miRNA-31-5p/NAT8L axis in pulmonary artery endothelial cells of patients with CTEPH. Mol. Med. Rep. 26, 297 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Heeb M. J., Role of the PROS1 gene in thrombosis: Lessons and controversies. Expert Rev. Hematol. 1, 9–12 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Suleiman L., Muataz Y., Négrier C., Boukerche H., Protein S-mediated signal transduction pathway regulates lung cancer cell proliferation, migration and angiogenesis. Hematol. Oncol. Stem Cell Ther. 1, S1658–S3876 (2021). [DOI] [PubMed] [Google Scholar]
- 136.Kagoshima M., Ito T., Diverse gene expression and function of semaphorins in developing lung: Positive and negative regulatory roles of semaphorins in lung branching morphogenesis. Genes Cells 6, 559–571 (2001). [DOI] [PubMed] [Google Scholar]
- 137.Dammann C. E., Nielsen H. C., Carraway K. L. III, Role of neuregulin-1β in the developing lung. Am. J. Respir. Crit. Care Med. 167, 1711–1716 (2003). [DOI] [PubMed] [Google Scholar]
- 138.Liu J., Nethery D., Kern J. A., Neuregulin-1 induces branching morphogenesis in the developing lung through a P13K signal pathway. Exp. Lung Res. 30, 465–478 (2004). [DOI] [PubMed] [Google Scholar]
- 139.Boucherat O., Benachi A., Chailley-Heu B., Franco-Montoya M. L., Elie C., Martinovic J., Bourbon J. R., Surfactant maturation is not delayed in human fetuses with diaphragmatic hernia. PLOS Med. 4, e237 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jones C. V., Alikhan M. A., O'Reilly M., Sozo F., Williams T. M., Harding R., Jenkin G., Ricardo S. D., The effect of CSF-1 administration on lung maturation in a mouse model of neonatal hyperoxia exposure. Respir. Res. 15, 110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S8
Tables S1 to S8
Legends for movies S1 to S3
Legends for data files S1 to S5
References
Movies S1 to S3
Data files S1 to S5