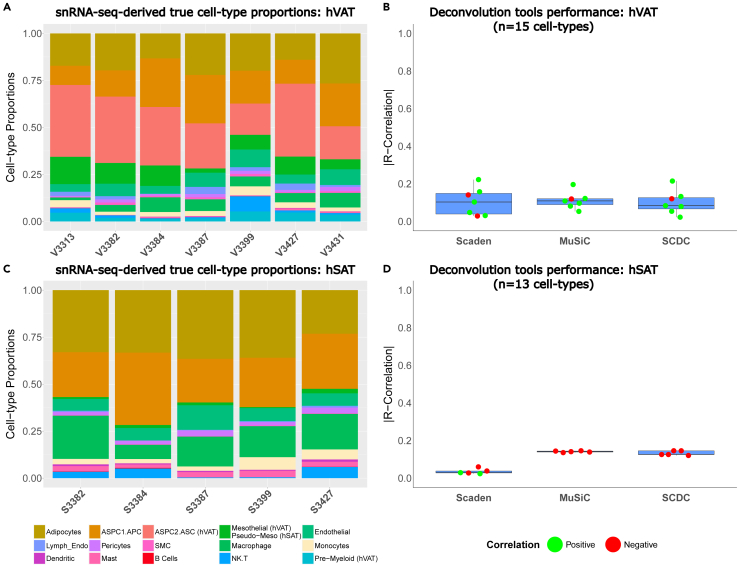
Figure 3.
Performance of cell-type estimation by common deconvolution tools analyzing human visceral and subcutaneous adipose tissue bulk RNA-seq data
(A and C) Cell-type proportions in the initial sample set of 7 hVAT and 5 hSAT samples as assessed by snRNA-seq, respectively. Shown are the proportions of the 15 and 13 cell types, respectively.
(B and D) Accuracy of estimated cell-type proportions achieved by 3 deconvolution tools: MuSiC and SCDC are deconvolution algorithms that utilize cell-type-specific gene expression and Scaden uses a deep neural network ensemble training approach. Boxplot showing each test-case R correlation coefficient between the true proportions (snRNA-seq results) and the tools’ estimated proportions for every sample in the initial sample set (using leave-one-out training methodology, as detailed in STAR Methods).