Abstract
We describe a replication-competent, recombinant vesicular stomatitis virus (VSV) in which the gene encoding the single transmembrane glycoprotein (G) was deleted and replaced by an env-G hybrid gene encoding the extracellular and transmembrane domains of a human immunodeficiency virus type 1 (HIV-1) envelope protein fused to the cytoplasmic domain of VSV G. An additional gene encoding a green fluorescent protein was added to permit rapid detection of infection. This novel surrogate virus infected and propagated on cells expressing the HIV receptor CD4 and coreceptor CXCR4. Infection was blocked by SDF-1, the ligand for CXCR4, by antibody to CD4 and by HIV-neutralizing antibody. This virus, unlike VSV, entered cells by a pH-independent pathway and thus supports a pH-independent pathway of HIV entry. Additional recombinants carrying hybrid env-G genes derived from R5 or X4R5 HIV strains also showed the coreceptor specificities of the HIV strains from which they were derived. These surrogate viruses provide a simple and rapid assay for HIV-neutralizing antibodies as well as a rapid screen for molecules that would interfere with any stage of HIV binding or entry. The viruses might also be useful as HIV vaccines. Our results suggest wide applications of other surrogate viruses based on VSV.
Vesicular stomatitis virus (VSV) is a nonsegmented negative-strand RNA virus (family Rhabdoviridae) that encodes one transmembrane glycoprotein (G). This protein is responsible for the very broad host range and membrane fusion activity of VSV (14, 34) and is the only protein of this virus targeted by neutralizing antibodies (19). VSV readily incorporates foreign viral membrane proteins into its envelope, making it ideal for production of pseudotype viruses carrying the envelope proteins of other viruses (46).
In a recent study, our laboratory described a recombinant VSV with the G gene deleted (ΔG) and expressing instead the human immunodeficiency virus (HIV) receptor CD4 and coreceptor CXCR4 (40). Both CD4 and CXCR4 were incorporated into the VSV envelope and provided a novel targeting specificity to HIV-infected cells expressing the HIV envelope protein (Env). Our group also reported construction of a recombinant VSV expressing an HIV Env-G hybrid protein from an extra gene (18). The Env-G protein was incorporated into VSV virions along with VSV G and was shown to mediate specific infection of CD4+ HeLa cells when infectivity due to VSV G was neutralized.
The purpose of the study described here was to determine if the HIV Env-G gene could completely replace the VSV G gene to generate a viable recombinant virus targeted with the specificity of HIV type 1 (HIV-1). We wanted to examine the pathway of infection by such a surrogate virus, to determine if it might be a useful tool for HIV-neutralization assays and to determine if it could be used to detect specific inhibitors of HIV binding or entry.
Enveloped viruses enter cells either by fusion of their envelope with the host cell plasma membrane or by receptor-mediated endocytosis (reviewed in reference 27). The glycoproteins of enveloped viruses that are endocytosed undergo a low-pH-dependent conformational change in the acidic environment of endosomes. This change results in fusion of the viral membrane with the endosomal membrane and release of viral cores into the cytoplasm. Both influenza virus and VSV are well-studied examples of viruses employing the pH-dependent entry mechanism (27, 44).
The route of HIV-1 entry has been controversial. An early study reported that HIV-1 infection was sensitive to ammonium chloride (25), an inhibitor of endosomal acidification that prevents entry of viruses that use the low-pH pathway. Morphological studies also showed HIV-1 particles in endosomes (32), supporting entry through this route. Subsequent studies contradicted the initial reports: entry of HIV-1 and VSV(HIV) pseudotypes was shown to be resistant to agents that block endosomal acidification, and fusion of HIV-1 with the plasma membrane was observed (29, 42). In addition, mutations altering the rate of CD4 endocytosis were shown to not affect HIV-1 entry (26, 33). It therefore appears that HIV-1 entry can occur in a pH-independent manner at the plasma membrane, although some entry may occur after endocytosis without a requirement for reduced pH.
HIV Env is synthesized as a precursor gp160 molecule that is cleaved to gp120 and gp41 subunits that are noncovalently linked. The glycoprotein apparently forms trimers on the virion envelope with the spikes composed of gp120 bound to the trimeric gp41 glycoprotein oligomer (6, 22). Entry of HIV-1 into cells requires initial binding to CD4 (25, 37), and recent studies have defined chemokine receptor molecules CXCR4 (3, 13) and CCR5 (1, 7, 9–11) as the major coreceptor molecules that are also required. HIV entry occurs in stages: the initial binding to CD4 is followed by conformational changes in the Env protein that allow it to bind coreceptor and subsequently cause fusion of the viral and cellular membranes (23, 43). Efficient incorporation of the gp120-gp41 complex into VSV virions requires replacement of the 150-amino-acid gp41 cytoplasmic tail with the 29-amino-acid cytoplasmic tail of VSV G (18, 31). It was initially thought that the VSV G tail provided a positive incorporation signal for the Env-G protein. However, it has recently been shown that poor incorporation of the HIV Env protein into VSV virions results mainly from a signal within the membrane-proximal 10 amino acids of the gp41 tail which sequesters HIV Env away from VSV budding sites (17).
MATERIALS AND METHODS
Plasmid construction.
The green fluorescent protein (GFP) gene (Green Lantern; Life Technologies) was amplified by PCR by using Vent polymerase (New England Biolabs). The forward primer was 5′GGGCCCCTCGAGGCAAT TGCGCGC TAGCTATGAAAAAAAC TAACAGATATCACCATGAGCAAGGGCGAGGAAC3′. This primer contained the minimally conserved VSV transcription stop and start sequences (underlined) as well as XhoI and NheI sites (bold) for use in further cloning. The reverse primer was 5′GCGGCGCTCTAGATCACTTGTACAGCTCGTCCATG3′. This primer contained an XbaI site (bold). The PCR product was digested with XhoI and XbaI and ligated to pVSV-XN1 (39) that had been digested with XhoI and NheI. The resulting plasmid was called pVSV-GFP.
The HIV gp160G gene, encoding the Env protein of the HIV IIIB strain with the cytoplasmic domain of VSV G replacing its native cytoplasmic domain, has been described previously (31). This gene was excised with MluI and NheI from a plasmid called pVSVΔG-gp160G (16a) and ligated to pVSV-GFP from which the VSV G gene had been removed by digestion with MluI and NheI. The final plasmid was called pVSVΔG-gp160G-GFP.
The 89.6G gene, encoding the Env protein of the HIV 89.6 strain with the cytoplasmic domain of VSV G replacing its native cytoplasmic domain, has also been described. This gene had previously been inserted into a VSV molecular clone, yielding the plasmid pVSV-89.6gp160G (18). The G gene was removed from this plasmid by digestion with MluI and XhoI, and the overhanging cleavage sites were filled in by using T4 DNA polymerase (New England Biolabs). The plasmid was then religated, yielding pVSVΔG-89.6gp160G. This DNA was then digested with XbaI and NheI to remove a fragment containing a portion of the VSV P gene, as well as the VSV M and 89.6G genes. This fragment was then ligated to pVSV-GFP from which the same portion of the VSV P gene, as well as the VSV M and VSV G genes, had been removed at the same restriction sites. The resulting plasmid was called pVSVΔG-89.6G-GFP.
The JRFLG gene, encoding the Env protein of the HIV JRFL strain with the cytoplasmic domain of VSV G replacing its native cytoplasmic domain, was generated by a two-step PCR strategy. First, the sequence coding for the C-terminal 29 amino acid residues of the VSV G protein was amplified by PCR. The forward primer was 5′TCTATAGTGAATAGAGTTAGGATCCATCTTTGCATTAAATTA3′. This primer contained the VSV G cytoplasmic tail sequence, beginning with the codon for the first cytoplasmic residue, fused to the codons for the last four transmembrane and first three cytoplasmic residues of the HIV JRFL env gene (underlined). The reverse primer was 5′ACGTACGTGCTAGCTTACTTTCCAAGTCGGTTC3′. This primer contained an NheI site (bold). The VSV G tail PCR product was purified, and 10% of it was used in a second PCR reaction in which the HIV JRFL env gene was the template (the plasmid was kindly provided by B. Doranz and R. Doms). The forward primer was 5′GGGCCCACGCGTATTATGAGAGTGAAGGGGATCAGG3′. This primer contained an MluI site (bold), followed by the sequence encoding the N-terminal end of the HIV JRFL Env protein. The reverse primer was the same primer used to amplify the VSV G cytoplasmic tail. The PCR product was digested with MluI and NheI, then ligated to pVSV-GFP from which the VSV G gene had been removed by using the same enzymes. The resulting plasmid was called pVSVΔG-JRFLG-GFP.
Virus recoveries.
VSV-GFP virus was recovered from pVSV-GFP by established methods (24, 40). Baby hamster kidney (BHK) cells were plated to approximately 75% confluency on 10-cm-diameter petri dishes. The cells were then infected at a multiplicity of infection (MOI) of 10 with vTF7-3, a recombinant vaccinia virus that expresses T7 RNA polymerase (15). After 1 h, each dish of cells was transfected with 3 μg of pBS-N, 5 μg of pBS-P, 1 μg of pBS-L, and 10 μg of pVSV-GFP by using a cationic liposome reagent (36). Cells were then incubated at 37°C for 48 h. Cell supernatants were passed through a 0.2-μm-pore-size filter to remove vaccinia virus then applied to fresh BHK cells for an additional 48 h at 37°C. Recovery of infectious virus was confirmed by scanning BHK cell monolayers for VSV cytopathic effect and GFP fluorescence. The viral supernatants were then passed through a 0.1-μm-pore-size filter to remove residual vaccinia virus. From the filtered supernatants, individual plaques were isolated and grown on BHK cells. These stocks were then stored at −80°C.
Infectious virus was recovered from pVSVΔG-gp160G-GFP, pVSVΔG-89.6G-GFP, and pVSVΔG-JRFLG-GFP by using the same procedure, with the following modifications. After being infected with vTF7-3, BHK cells were transfected with 3 μg of pBS-N, 5 μg of pBS-P, 1 μg of pBS-L, 4 μg of pBS-G, and 10 μg of full-length VSV plasmid per plate. After 48 h at 37°C, supernatants were filtered and passaged to BHK-G cells (40) that had been induced to express G 12 h previously. After another 48 h incubation at 37°C, the viral supernatants were filtered, and single plaques were picked and propagated on induced BHK-G cells.
Preparation of viral stocks.
VSV-GFP was purified from a single plaque on BHK cells. Virus from this plaque (∼105 PFU) was used to infect ∼107 BHK cells on a 10-cm-diameter petri dish overnight. The medium was clarified by low-speed centrifugation to remove cell debris and virus stocks (∼109 PFU/ml was frozen). The same procedure was used to prepare stocks of ΔG viruses transcomplemented on BHK-G cells, but the titers obtained were ∼100-fold lower. For the preparation of ΔG virus stocks not transcomplemented with VSV G, VSV G-complemented stocks were first expanded by infecting 107 induced BHK-G cells at an MOI of approximately 0.1. The infections were allowed to proceed for approximately 32 h. At this time, nearly all cells showed VSV cytopathic effect, and the viral titer in the supernatants was shown to be at its peak. The entire supernatant from each dish was then transferred to a confluent dish of ∼107 BHK cells. After infection for 3.5 h at 37°C, the viral inocula were withdrawn to remove input, G-complemented virus. Cells were incubated in fresh medium for an additional 11 h, and viral supernatants were then harvested, clarified, and stored at −80°C.
Metabolic labeling and analysis of infected cells.
BHK cells on 6-cm-diameter petri dishes were infected at an MOI of 5 to 10 with wild-type VSV, VSV-GFP, or G-complemented ΔG-gp160G-GFP. After 6 h at 37°C, cells were rinsed three times with methionine-free Dulbecco’s modified Eagle’s medium (DMEM) and then incubated at 37°C with methionine-free DMEM containing 200 μCi of [35S]methionine. After 1 h, cells were rinsed three times with phosphate-buffered saline (PBS) and lysed in 1 ml of detergent solution (1% Nonidet P-40, 0.4% deoxycholate, 50 mM Tris-HCl [pH 8], 62.5 mM EDTA). Labeled lysates were analyzed by electrophoresis on a sodium dodecyl sulfate (SDS)–10% polyacrylamide gel and detected by using a PhosphorImager (Molecular Dynamics).
Immunofluorescence microscopy.
HeLa-CD4 or HeLa cells were plated to near confluency on coverslips and infected at an MOI of 0.75 with VSV-GFP or VSVΔG-gp160G-GFP from BHK cells (not G complemented). To neutralize residual G-complemented virions in the VSVΔG-gp160G-GFP stock, infections with this virus were performed in a 1:1,000 dilution of I1, a monoclonal antibody against VSV G that neutralizes VSV. After 8 to 12 h at 37°C, cells on coverslips were fixed in 3% paraformaldehyde. Coverslips were then washed in PBS containing 10 mM glycine (PBS-glycine) and permeabilized in 1% Triton X-100. After more PBS-glycine washes, coverslips were incubated with a 1:200 dilution of a monoclonal antibody against VSV N protein, followed by a 1:50 dilution of a rhodamine-conjugated donkey anti-mouse antibody (Jackson Research). The coverslips were then mounted on slides, and cells were observed and photographed by using a Nikon Microphot-FX microscope with a 25× objective.
Inhibition of infection by chemokines and receptor antibodies.
Sheep polyclonal antiserum to human CD4, a monoclonal antibody to human CXCR4 (12G5 [12]; AIDS Reagent Program [ARRRP]), and a monoclonal antibody to human CCR5 (2D7 [45]; ARRRP) were each diluted 1:10, 1:50, 1:250, and 1:1,250 in DMEM–10% fetal bovine serum (FBS). Human chemokines SDF-1 (kindly provided by Thomas Turi, Pfizer) and RANTES (regulated upon activation, normal T-cell expressed and secreted) (PeproTech; ARRRP) were each diluted to 0.004, 0.04, 0.4, and 4 μM in DMEM–10% FBS. HeLa-CD4 cells, plated to confluency on 96-well plates, were incubated with 100 μl of each diluted chemokine or antibody for 15 min at room temperature. Noncomplemented VSVΔG-gp160G-GFP was then diluted in DMEM–10% FBS to a final concentration of approximately 100 infectious units per 100 μl. The I1 monoclonal antibody was also included in the virus mixture, at a dilution of 1:1,000 to neutralize infection due to traces of G in the viral stock. VSVΔG-gp160G-GFP (100 μl) was then added to each well of cells. To allow cells to express detectable levels of GFP, infections were allowed to continue for about 10 to 15 h at 37°C. GFP-positive cells were then visualized by fluorescence microscopy, by using a Nikon Microphot-FX microscope with a 10× PlanApo objective.
Inhibition of viral entry by chloroquine and ammonium chloride.
HeLa-CD4 cells plated to confluency in 96-well plates were pretreated for 1 h at 37°C with varying concentrations of chloroquine or ammonium chloride in DMEM–10% FBS. Cells were then infected with approximately 100 infectious units/well of either VSV-GFP or noncomplemented VSVΔG-gp160G-GFP for 90 min at 37°C, in the presence of the compounds. Cells were washed twice in DMEM–10% FBS to remove input virus, and DMEM–10% FBS containing drug was then added to the wells. Following an additional incubation at 37°C for either 2 h (chloroquine) or 5 h (ammonium chloride), the medium was replaced with DMEM, and the samples were incubated again at 37°C. At 10 h postinfection, GFP-positive cells were counted by fluorescence microscopy.
Neutralization of VSVΔG-gp160G-GFP by antiviral sera.
HeLa-CD4 cells were plated to confluency on 96-well plates. Noncomplemented VSVΔG-gp160G-GFP was diluted in DMEM–10% FBS to a final concentration of approximately 100 infectious units per 100 μl. The I1 monoclonal antibody was included in this mixture, at a dilution of 1:1,000 to completely neutralize a low level of infectivity due to residual G. HIV serum immunoglobulin (HIVIg) (ARRRP) and normal human serum were each diluted 1:10, 1:50, 1:250, and 1:1,250 in DMEM–10% FBS. Equal volumes of diluted virus and diluted antibody were then mixed and incubated for 15 min at 37°C, and 200 μl of this mixture was used to infect each well of HeLa-CD4 cells. At 10 to 15 h postinfection, GFP-positive cells were counted by fluorescence microscopy.
Virus titrations.
HeLa-CD4 cells, or derivatives of the CV-1 cell line expressing human CD4, human CD4 and human CXCR4, or human CD4 and human CCR5 (kindly provided by David Kabat) were plated to confluency on 96-well dishes. Noncomplemented stocks of VSVΔG-gp160G-GFP, VSVΔG-89.6G-GFP, and VSVΔG-JRFLG-GFP were diluted serially and used to infect these cells, again in the presence of I1 at 1:1,000 dilution. At 10 to 15 h postinfection, GFP-positive cells were counted by fluorescence microscopy, and viral titer was calculated.
RESULTS
Recovery of recombinant VSVs expressing GFP and HIV-gp160G.
To generate a recombinant VSV expressing GFP for rapid monitoring of VSV infection, we inserted the GFP gene between the G and L genes in a VSV vector DNA plasmid (39). This plasmid expresses the recombinant VSV antigenome RNA under T7 promoter control and allows recovery of infectious VSVs from DNA (Fig. 1). We then recovered virus from this construct in cells expressing the recombinant VSV antigenome RNA as well as the VSV N, P, and L proteins. The recovered virus grew to titers of >109/ml (equivalent to wild-type VSV) and expressed abundant GFP. To generate a recombinant VSV surrogate virus with the cellular targeting specificity of HIV-1, we replaced the G gene in pVSV-GFP with the HIV-1 gp160G gene (18) (Fig. 1). This gene encodes the extracellular and transmembrane domains of the Env protein from the HIV IIIB strain. The cytoplasmic domain was replaced with the cytoplasmic domain of VSV G to facilitate incorporation of the protein into VSV particles. Virus was recovered from this construct in cells expressing the RNA antigenome and the VSV G, N, P, and L proteins (24, 40).
FIG. 1.
Diagrams of recombinant VSV genomes. To generate VSV-GFP, the GFP gene was inserted into the VSV genome sequence preceded by the appropriate VSV transcription start and stop sequences (SS). The ΔG-gp160G-GFP clone was constructed by inserting the gp160G gene, again under the control of VSV transcriptional start and stop sequences, upstream of the GFP gene in the VSV-GFP clone. Diagrams represent the negative-sense RNA viral genome, extending 3′ to 5′ from left to right. XhoI, NheI, and MluI cleavage sites are indicated.
Because VSVΔG-gp160G-GFP does not encode G, we were able to propagate it most easily on the complementing BHK-G cell line (40) where titers of ∼106 PFU/ml were obtained on BHK-G cells. Producing viral stocks specifically targeted to CD4+ cells, however, required that we propagate VSVΔG-gp160G-GFP without G complementation. To do this, we first expanded G-complemented virus on BHK-G cells. We next transferred the cell supernatants to BHK cells and then harvested the supernatants after one round of infection on these cells. Stocks prepared in this way still contained traces of G-complemented virus that would infect BHK cells, but this titer was less than 1% of that seen on Hela-CD4+ cells and was easily neutralized by antibody to VSV G.
Recombinant VSVs express the GFP and HIV-gp160G genes.
To examine the proteins produced by the recombinant viruses, we analyzed whole lysates of infected cells by SDS-polyacrylamide gel electrophoresis (PAGE). Because VSV shuts off host cell protein synthesis within 2 to 3 h of infection, proteins encoded by VSV recombinants can be visualized without immunoprecipitation. Figure 2 shows [35S]methionine labeled lysates of BHK cells infected with VSV, VSV-GFP, or VSVΔG-gp160G-GFP as indicated. As expected, VSV-GFP expressed all five VSV proteins. It also expressed a new protein migrating just ahead of VSV M. The size of this protein (approximately 22 kDa) is that expected for GFP. The VSVΔG-gp160G-GFP virus also expressed the 22-kDa protein as well as a larger protein with a size consistent with that of gp160G. Cells infected with VSVΔG-gp160G-GFP did not express VSV G protein.
FIG. 2.
Protein expression by recombinant VSVs. BHK cells were infected with VSV, VSV-GFP, or VSVΔG-gp160G-GFP at an MOI of 5. Infected cells were incubated for 6 h at 37°C and labeled with [35S]methionine for 1 h. Cell lysates were then analyzed directly by SDS-PAGE. The positions of wild-type VSV proteins, L, G, N, P, and M, are indicated along with the GFP and gp160G proteins encoded by the recombinants.
To examine GFP fluorescence in infected cells, VSV-GFP and VSVΔG-gp160G-GFP were used to infect HeLa-CD4 cells. Cells were then stained with a monoclonal antibody to VSV N, followed by a rhodamine-conjugated secondary antibody. In Fig. 3A and C, immunofluorescence staining shows that both viruses readily infected HeLa-CD4 cells and expressed VSV N. Furthermore, Fig. 3B and D illustrate that the same cells staining positive for N also showed the green fluorescence characteristic of GFP.
FIG. 3.
GFP expression in infected cells. HeLa-CD4 cells were infected for 10 h with VSV-GFP or VSVΔG-gp160G-GFP at an MOI of approximately 0.75. All infections with VSVΔG-gp160G-GFP were performed in the presence of excess I1. The antibody eliminates a low level of infection resulting from carryover of traces of VSV G protein. A and B show the same field of cells infected with VSV-GFP. (A) Rhodamine immunofluorescence detecting VSV N. (B) GFP fluorescence. C and D show the same field of cells infected with VSVΔG-gp160G-GFP. (C) Rhodamine immunofluorescence detecting VSV N protein. (D) GFP fluorescence.
The VSVΔG-gp160G-GFP virus specifically infects CD4+ cells.
Because VSVΔG-gp160G-GFP particles should have only HIV gp160G protein on their surface, we expected noncomplemented virus particles to infect only CD4+ cells. To test this, we infected HeLa or HeLa-CD4 cells with either VSV-GFP or VSVΔG-gp160G-GFP and then stained infected cells for VSV N. Figure 4A and B show that VSV-GFP was able to infect both HeLa and HeLa-CD4 cells. In contrast, VSVΔG-gp160G-GFP infected HeLa-CD4 cells (Fig. 4D) but not HeLa cells (Fig. 4C), indicating a requirement for CD4 in the absence of VSV G protein.
FIG. 4.
CD4-dependent infectivity of VSVΔG-gp160G-GFP. HeLa or HeLa-CD4 cells were infected with VSV-GFP or VSVΔG-gp160G-GFP at an MOI of 0.75 for 10 h. Infection was measured by detection of VSV N protein by using indirect immunofluorescence. (A) HeLa cells infected with VSV-GFP. (B) HeLa-CD4 cells infected with VSV-GFP. (C) HeLa cells infected with VSVΔG-gp160G-GFP. (D) HeLa-CD4 cells infected with ΔG-gp160G-GFP.
Because VSVΔG-gp160G-GFP infected HeLa-CD4 cells, we expected that it would be possible to grow stocks of this virus on these cells. To determine the titers attainable on HeLa-CD4 cells, we inoculated cells with noncomplemented VSVΔG-gp160G-GFP and allowed the infection to spread throughout the culture. Cells showed cytopathic effect within 8 h, but virus harvested from the cells after 24 h, when nearly all cells had been infected, had titers of only 104 PFU/ml. Although we were able to propagate virus indefinitely in HeLa-CD4 cells (as determined by spread of GFP fluorescence and cytopathic effect), the titers of virus grown on these cells were approximately 10- to 100-fold lower than stocks grown on BHK cells. It should be noted that even wild-type VSV produces approximately 100-fold lower titers on HeLa cells than on BHK cells. We therefore prepared all further stocks by using VSV G-complemented virus grown for one round on BHK cells in the absence of VSV G.
Infection by VSVΔG-gp160G-GFP is blocked by antibodies to CD4 or coreceptor.
Besides requiring CD4, HIV-1 infection requires one of a group of chemokine receptor proteins that function as coreceptors. Viral strains are assigned (according to coreceptor use) to one of three groups: R5, for strains that use the CCR5 coreceptor; X4, for strains that use the CXCR4 coreceptor; or R5X4, for strains that use either coreceptor (2). The R5 group includes most virus strains that are transmitted sexually and isolated from newly-infected patients, while the X4 and R5X4 strains are typically isolated from patients in late stages of infection or from isolates adapted for growth on T cell lines (e.g., IIIB).
Because VSVΔG-gp160G-GFP has a gp160 gene derived from the IIIB strain and displayed the same CD4 dependence as HIV-1, we expected that it, like HIV-1 IIIB, would require the CXCR4 cofactor. To test this, we attempted to inhibit VSVΔG-gp160G-GFP infection with antibodies to CD4, CXCR4, or CCR5. We infected HeLa-CD4 cells with approximately 100 infectious units of VSVΔG-gp160G-GFP in the presence of varying concentrations of these antibodies and subsequently determined infectivity by counting GFP-positive cells.
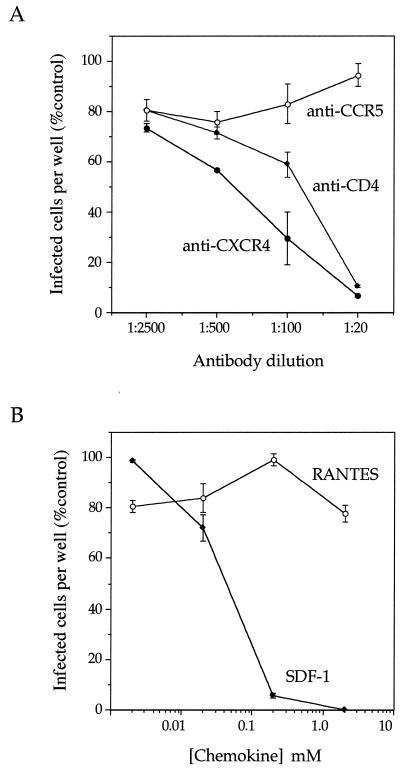
Figure 5A shows the results of this experiment. Increasing concentrations of an anti-CD4 serum resulted in increasing inhibition of VSVΔG-gp160G-GFP infection, consistent with the requirement for CD4 expression. A monoclonal antibody to CXCR4 (12), previously shown to inhibit HIV IIIB infection, also blocked VSVΔG-gp160G-GFP infection. By contrast, a monoclonal antibody to CCR5 (45), previously shown to inhibit infection by macrophage-tropic R5 HIV strains, did not reduce VSVΔG-gp160G-GFP infection.
FIG. 5.
Inhibition of VSVΔG-gp160G-GFP infection through a block of receptor or coreceptor. (A) HeLa-CD4 cells were infected with approximately 100 infectious units of VSVΔG-gp160G-GFP in the presence of indicated dilutions of polyclonal sheep antiserum to human CD4, a monoclonal antibody to human CXCR4, or a monoclonal antibody to human CCR5. (B) HeLa-CD4 cells were infected with approximately 100 infectious units of ΔG-gp160G-GFP in the presence of the indicated concentrations of purified human chemokines SDF-1 or RANTES. After 12 h, GFP-positive cells were counted. Viral infectivity at each concentration of inhibitor is expressed as (number of infected cells per well with inhibitor/number of infected cells per well without inhibitor) × 100. Each dilution of antibody was tested in duplicate; error bars represent the range of results between duplicates.
Infection is blocked by the ligand to CXCR4.
The results described above indicated that VSVΔG-gp160G-GFP retained the cofactor specificity of the HIV strain from which its env gene was taken. To further examine coreceptor usage, we examined the effects of the chemokines SDF-1 and RANTES on infectivity (Fig. 5B). SDF-1 is the ligand for CXCR4 (4, 30) and has previously been shown to inhibit infection by X4 HIV strains such as HIV-1 IIIB. RANTES is a ligand for CCR5 and is known to inhibit infection by macrophage-tropic (R5) HIV strains (8). SDF-1 strongly inhibited ΔG-gp160G-GFP infection at concentrations as low as 0.2 mM. By contrast, RANTES had no effect on infection. We therefore conclude that, like HIV IIIB infection, VSVΔG-gp160G-GFP infection requires CD4 and CXCR4.
Entry by a pH-independent pathway.
Because our data showed that VSVΔG-gp160G-GFP required the same receptor and cofactor as HIV IIIB, we wanted to determine if its entry pathway was pH-dependent or -independent. As described above, the pathway of HIV entry has been controversial, although recent studies favor pH-independent entry by fusion at the cell surface. In contrast, VSV enters cells through an endocytic pathway and requires the mildly acidic pH of the endosome to trigger the membrane fusion activity of G (14, 27, 34).
The weak bases chloroquine and ammonium chloride have previously been used to distinguish between the pH-dependent and -independent pathways. Both compounds inhibit acidification of endosomes, thereby inhibiting VSV entry but not affecting entry of viruses that fuse with the plasma membrane. To examine the pathway of VSVΔG-gp160G-GFP entry, we examined the effects of both compounds on infection. Figure 6 shows that increasing concentrations of either drug increasingly inhibited VSV-GFP infection. In contrast, neither drug had any inhibitory effect on infection by VSVΔG-gp160G-GFP. In fact, there appeared to be a significant increase in infection in the presence of increasing ammonium chloride concentrations. This effect was apparently unrelated to effects on endosomal pH because a similar effect was not observed with chloroquine. We therefore conclude that VSVΔG-gp160G-GFP enters cells through a pH-independent pathway presumably involving fusion with the cell surface.
FIG. 6.
Effect of chloroquine and ammonium chloride on the infectivity of VSV-GFP and ΔG-gp160G-GFP. HeLa-CD4 cells on 96-well plates were pretreated with either drug for 1 h then infected with either virus for 90 min in the presence of the drug. Cells were then incubated for an additional 5 h with chloroquine (A) or for an additional 2 h with ammonium chloride (B). At 10 h postinfection, GFP-positive cells were counted. Each drug concentration was tested in triplicate; error bars represent one standard deviation.
Neutralization by anti-HIV serum.
Because VSVΔG-gp160G-GFP uses the HIV entry pathway and its infection can be monitored readily, we wanted to test its utility in a neutralizing assay for HIV-1. To do this, samples of 100 infectious units of virus were incubated with dilutions of either normal human serum or pooled serum HIV-1 immunoglobulin (HIVIg) from infected donors prior to infection of HeLa cells in 96-well microtiter plates. GFP-positive cells were then counted after 10 to 15 h as a measure of infection.
Figure 7 shows the results of a representative experiment. Normal human serum had no effect on viral infectivity even at the lowest dilution. By contrast, higher concentrations of HIVIg exhibited increased neutralization of infection. Greater than 50% neutralization was seen at a 1:500 dilution, 95% neutralization was achieved at a 1:100 dilution, and complete neutralization was observed at a 1:20 dilution. Quantitatively, these results are similar to those we had observed previously using the same HIVIg sample in an HIV-1 IIIB neutralization assay based on inhibition of syncytia formation in MT-2 cells. In that assay we observed approximately 60% reduction in syncytia at a 1:500 dilution and 95% reduction at a 1:100 dilution (17).
FIG. 7.
Neutralization of ΔG-gp160G-GFP by HIVIg. Approximately 100 infectious units of ΔG-gp160G-GFP were incubated with HIVIg or normal human serum (NHS) at the indicated dilutions for 15 min at 37°C. Virus was then applied to HeLa-CD4 cells in 96-well plates. After 10 h, GFP-positive cells were counted. Viral infectivity at each antibody dilution is expressed as (number of infected cells per well with antibody/number of infected cells per well without antibody) × 100. Each dilution of antibody was tested in duplicate; error bars represent the range of results between duplicates.
Recombinants expressing Env from other HIV strains retain the cofactor specificities of those strains.
A potential application of VSVΔG-gp160G-GFP is in screening for compounds that might inhibit any step of HIV entry, such as receptor or coreceptor binding. Because HIV strains vary in coreceptor usage and sensitivity to neutralization, it would be desirable to have VSV/HIV surrogate viruses displaying alternative Env proteins from primary isolates that use different coreceptors. Env genes were therefore taken from two primary isolates, HIV 89.6, an X4R5 strain (10), and HIV JRFL, an R5 strain (41). As with the initial construct, the env genes from 89.6 and JRFL were engineered to encode the VSV G cytoplasmic domain in place of the normal gp41 cytoplasmic domains, and the GFP gene was included to simplify the assay of infection. Recombinant VSVΔG-gp160G-GFPs were then recovered and characterized. Immunofluorescence showed that both viruses (termed VSVΔG-89.6G-GFP and VSVΔG-JRFLG-GFP) expressed GFP and their respective chimeric EnvG proteins on the cell surface (not shown).
To compare coreceptor requirements, viruses were used to infect derivatives of a CV-1 cell line constitutively expressing CD4, CD4 and CXCR4, or CD4 and CCR5. Table 1 shows the results of such an experiment. The VSVΔG-gp160G-GFP virus did not infect CD4+ or CD4+/CCR5+ cells but did infect CD4+/CXCR4+ cells. The VSVΔG-JRFLG-GFP virus did not infect CD4+ or CD4+/CXCR4+ cells but did infect CD4+/CCR5+ cells. Finally, VSVΔG-89.6G-GFP failed to infect CD4+ cells but did infect CD4+/CXCR4+ and CD4+/CCR5+ cells. All three recombinant VSVs thus have the same X4, X4R5, and R5 specificities as the HIV strains from which their env genes are derived.
TABLE 1.
Infection of CV-1 cells encoding HIV-1 receptor and coreceptors
| Virus | Viral titers (infectious units/ml) on CV-1 cell lines expressinga:
|
||
|---|---|---|---|
| CD4 | CD4 + CXCR4 | CD4 + CCR5 | |
| ΔG-gp160G-GFP | 0 | 6.5 × 103 | 0 |
| ΔG-89.6G-GFP | 0 | 3.1 × 105 | 8.3 × 104 |
| ΔG-JRFLG-GFP | 0 | 0 | 1.7 × 105 |
Viral stocks were grown first on BHK-G cells and then on BHK cells. They were diluted serially and used to infect cells expressing the indicated human proteins. After 15 h, GFP-positive cells were counted. Viral titers are expressed as number of infectious particles per milliliter as determined by counting individual green fluorescent cells or small clusters of two to six cells representing single initial infections.
DISCUSSION
We constructed a recombinant, VSVΔG-gp160G-GFP, for use in assays of HIV-1 entry and neutralization. This surrogate virus expresses a chimeric HIV-gp160G protein in place of VSV G protein. It is thus able to specifically infect cells that express CD4 and CXCR4, the coreceptor for the HIV-IIIB strain from which the env gene was derived. Infection was blocked by polyclonal antibody to CD4, by a monoclonal antibody to CXCR4, by the natural ligand for CXCR4, and by antibodies that neutralize HIV-1. We have also prepared two additional VSVΔG recombinants carrying the hybrid env-G genes derived from R5 or X4R5 HIV strains, and these show the coreceptor specificities of the HIV strains from which they were derived.
Infection of cells by VSV G-gp160G-GFP was insensitive to compounds that block endosomal acidification and block VSV infection, thus supporting a mechanism of HIV entry through pH-independent fusion with the plasma membrane. These results also suggest that in a normal VSV infection exposure to the acidic environment of endosomes is not required except to induce the required conformational change in G. This situation contrasts with that of influenza virus, where the acidic environment of the endosome also has the important role of promoting subsequent dissociation of the matrix protein from the ribonucleocapsid (5, 28).
Assays commonly used to detect HIV neutralization involve mixing of various dilutions of sera with HIV, followed by infection of T cell lines or peripheral blood lymphocytes and measurement of HIV p24 or reverse transcriptase production with time. Counting of syncytia formed by infected cells can be used as a measure of infection for those strains that cause cell-cell fusion. These assays require working with infectious HIV and typically require 4 to 14 days before neutralizing titers can be calculated. In addition, T cell lines employed in many of these assays can be sensitive to cytotoxic components in serum, making assay at low serum dilution very difficult. More recently developed assays employ cell lines expressing indicator proteins such as β-galactosidase under HIV long terminal repeat control and permit detection of infection and determination of neutralization titers in 2 to 3 days (20). The surrogate viruses expressing GFP described here provide a simple alternative for assay of HIV-neutralizing antibodies. The assay can be performed in as little as 10 h, and it does not require working with HIV-1. In addition, the cell lines employed are adherent and insensitive to serum components.
Current HIV therapy employs cocktails of inhibitors of HIV reverse transcripts and protease (16). These compounds effectively block new infections by preventing production of reverse-transcribed DNA for integration, or by preventing cleavage of the HIV polyprotein encoded by gag and blocking assembly of infectious HIV virions. Compounds able to block the initial steps of HIV binding and entry could be included in combination antiviral cocktails and might be identified by using the surrogate viruses we describe here.
HIV-1 infection continues to spread in much of the world at an increasing and alarming rate, and a safe and effective vaccine is urgently needed. It is possible that the surrogate viruses described, or variants expressing additional HIV proteins such as the proteins encoded by gag, would also be useful as HIV vaccines. VSV recombinants expressing an influenza virus hemagglutinin protein (as well as VSV G) have already been shown to have potent mucosal-immunizing activity protecting against influenza virus infection (35).
Because the surrogate viruses described here should specifically target CD4+ cells, including professional antigen-presenting cells such as macrophages and dendritic cells, they might be good inducers of both cellular and humoral immunity to HIV. However, the titers of viruses grown in culture are at best only 104 to 105 infectious units/ml, 104-fold lower than wild-type VSV. The low titers probably result from inefficient particle assembly in the absence of VSV G protein, as well as from the relatively low levels of EnvG incorporation (40). Our experience in vaccine studies of other poorly replicating surrogate viruses in mice (35a) suggests that this level of viral replication would not induce vigorous immune responses. However, the level of replication that might be achieved in vivo in primates expressing the appropriate receptor and coreceptor is unknown, and specific targeting of cells of the immune system might enhance immune responses greatly. Therefore, testing in appropriate animal models is warranted. It also might be possible to select or engineer viruses with higher replication capacity in vitro and in vivo.
We also note that VSVΔG-based surrogate viruses could have applications in the study of many other enveloped viruses. Engineering of the genes encoding the appropriate surface glycoproteins into the VSVΔG-GFP background could allow study of entry of viruses such as hepatitis C that are difficult to propagate in culture. As long as the foreign viral glycoproteins can be expressed well on the cell surface, it is likely that sufficient quantities would be incorporated into budding virions, since substantial amounts of most proteins tested to date are incorporated into the VSV envelope with or without VSV G (21, 38, 40).
ACKNOWLEDGMENTS
We thank Linda Buonocore for helpful advice and assistance and other members of the Rose laboratory for encouragement throughout the course of this work. We are grateful to David Kabat for generously providing the CV-1 cell lines expressing receptor and coreceptors.
This study was supported by NIH grants RO1AI 40357 and RO1AI 24345.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Berger E A, Doms R W, Fenyo E M, Korber B T, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 3.Berson J F, Long D, Doranz B J, Rucker J, Jirik F R, Doms R W. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 5.Bui M, Whittaker G, Helenius A. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J Virol. 1996;70:8391–8401. doi: 10.1128/jvi.70.12.8391-8401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 7.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 8.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 9.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 10.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 11.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 12.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 13.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 14.Florkiewicz R Z, Rose J K. A cell line expressing vesicular stomatitis virus glycoprotein fuses at low pH. Science. 1984;225:721–723. doi: 10.1126/science.6087454. [DOI] [PubMed] [Google Scholar]
- 15.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho D D. Toward HIV eradication or remission: the tasks ahead. Science. 1998;280:1866–1867. doi: 10.1126/science.280.5371.1866. [DOI] [PubMed] [Google Scholar]
- 16a.Johnson, E. Unpublished data.
- 17.Johnson, E., J. Forman, and J. Rose. Unpublished data.
- 17a.Johnson J E, Rodgers W, Rose J K. A plasma membrane localization signal in the HIV-1 envelope cytoplasmic domain prevents localization at sites of VSV budding and incorporation into VSV virions. Virology. 1998;251:244–252. doi: 10.1006/viro.1998.9429. [DOI] [PubMed] [Google Scholar]
- 18.Johnson J E, Schnell M J, Buonocore L, Rose J K. Specific targeting to CD4+ cells of recombinant vesicular stomatitis viruses encoding human immunodeficiency virus envelope proteins. J Virol. 1997;71:5060–5068. doi: 10.1128/jvi.71.7.5060-5068.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelley J M, Emerson S U, Wagner R R. The glycoprotein of vesicular stomatitis virus is the antigen that gives rise to and reacts with neutralizing antibody. J Virol. 1972;10:1231–1235. doi: 10.1128/jvi.10.6.1231-1235.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kretzschmar E, Buonocore L, Schnell M J, Rose J K. High-efficiency incorporation of functional influenza virus glycoproteins into recombinant vesicular stomatitis viruses. J Virol. 1997;71:5982–5989. doi: 10.1128/jvi.71.8.5982-5989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Evidence for cell-surface association between fusin and the CD4-gp120 complex in human cell lines. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 24.Lawson N D, Stillman E A, Whitt M A, Rose J K. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci USA. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 26.Maddon P J, McDougal J S, Clapham P R, Dalgleish A G, Jamal S, Weiss R A, Axel R. HIV infection does not require endocytosis of its receptor, CD4. Cell. 1988;54:865–874. doi: 10.1016/s0092-8674(88)91241-x. [DOI] [PubMed] [Google Scholar]
- 27.Marsh M. The entry of enveloped viruses into cells by endocytosis. Biochem J. 1984;218:1–10. doi: 10.1042/bj2180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 29.McClure M O, Marsh M, Weiss R A. Human immunodeficiency virus infection of CD4-bearing cells occurs by a pH-independent mechanism. EMBO J. 1988;7:513–518. doi: 10.1002/j.1460-2075.1988.tb02839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 31.Owens R J, Rose J K. Cytoplasmic domain requirement for incorporation of a foreign envelope protein into vesicular stomatitis virus. J Virol. 1993;67:360–365. doi: 10.1128/jvi.67.1.360-365.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pauza C D, Price T M. Human immunodeficiency virus infection of T cells and monocytes proceeds via receptor-mediated endocytosis. J Cell Biol. 1988;107:959–968. doi: 10.1083/jcb.107.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelchen-Matthews A, Clapham P, Marsh M. Role of CD4 endocytosis in human immunodeficiency virus infection. J Virol. 1995;69:8164–8168. doi: 10.1128/jvi.69.12.8164-8168.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riedel H, Kondor-Koch C, Garoff H. Cell surface expression of fusogenic vesicular stomatitis virus G protein from cloned cDNA. EMBO J. 1984;3:1477–1483. doi: 10.1002/j.1460-2075.1984.tb01999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts A, Kretzschmar E, Perkins A S, Forman J, Price R, Buonocore L, Kawaoka Y, Rose J K. Vaccination with a recombinant vesicular stomatitis virus expressing an influenza virus hemagglutinin provides complete protection from influenza virus challenge. J Virol. 1998;72:4704–4711. doi: 10.1128/jvi.72.6.4704-4711.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Roberts, A., and J. Rose. Unpublished results.
- 36.Rose J K, Buonocore L, Whitt M A. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques. 1991;10:520–525. [PubMed] [Google Scholar]
- 37.Sattentau Q J, Weiss R A. The CD4 antigen: physiological ligand and HIV receptor. Cell. 1988;52:631–633. doi: 10.1016/0092-8674(88)90397-2. [DOI] [PubMed] [Google Scholar]
- 38.Schnell M J, Buonocore L, Kretzschmar E, Johnson E, Rose J K. Foreign glycoproteins expressed from recombinant vesicular stomatitis viruses are incorporated efficiently into virus particles. Proc Natl Acad Sci USA. 1996;93:11359–11365. doi: 10.1073/pnas.93.21.11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schnell M J, Buonocore L, Whitt M A, Rose J K. The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J Virol. 1996;70:2318–2323. doi: 10.1128/jvi.70.4.2318-2323.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schnell M J, Johnson J E, Buonocore L, Rose J K. Construction of a novel virus that targets HIV-1-infected cells and controls HIV-1 infection. Cell. 1997;90:849–857. doi: 10.1016/s0092-8674(00)80350-5. [DOI] [PubMed] [Google Scholar]
- 41.Smyth R J, Yi Y, Singh A, Collman R G. Determinants of entry cofactor utilization and tropism in a dualtropic human immunodeficiency virus type 1 primary isolate. J Virol. 1998;72:4478–4484. doi: 10.1128/jvi.72.5.4478-4484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stein B S, Gowda S D, Lifson J D, Penhallow R C, Bensch K G, Engleman E G. pH-independent HIV entry into CD4-positive T cells via virus envelope fusion to the plasma membrane. Cell. 1987;49:659–668. doi: 10.1016/0092-8674(87)90542-3. [DOI] [PubMed] [Google Scholar]
- 43.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 44.White J M. Membrane fusion: the influenza paradigm. Cold Spring Harbor Symp Quant Biol. 1995;60:581–588. doi: 10.1101/sqb.1995.060.01.062. [DOI] [PubMed] [Google Scholar]
- 45.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zavada J. Viral pseudotypes and phenotypic mixing. Arch Virol. 1976;50:1–15. doi: 10.1007/BF01317996. [DOI] [PubMed] [Google Scholar]