Abstract
Bamboo is an economically important crop that has gained prominence as an alternative to wood to reduce deforestation and ecosystem destruction. Diseases of bamboo that typically occur on leaves and stems can cause significant loss, reducing the quality and yield of the bamboo. However, there are few reports identifying the fungal species diversity and potential pathogens of bamboo. Here, we describe four new species of plant fungi from the leaves of bamboo within Fujian provinces, China. Fungi were isolated from diseased leaves collected within Fujian province and identified based on their morphological characteristics and multilocus phylogenies using nucleotide sequences derived from combined datasets of the intervening 5.8S nrRNA gene (ITS), the 28S large subunit of nuclear ribosomal RNA gene (LSU), the large subunit of RNA polymerase I (rpb1), the translation elongation factor 1-α gene (tef1-α), and the partial beta-tubulin gene (tub2). These analyses helped reveal and clarify taxonomic relationships in the family Magnaporthaceae. The new species of bambusicolous fungi identified include two species of Bifusisporella, described as B. fujianensis sp. nov. and B. bambooensis sp. nov., and two species of Apiospora, described as A. fujianensis sp. nov. and A. fuzhouensis sp. nov. This study further expands the characterization and distribution of fungi associated with bamboo.
Keywords: Apiospora, Bifusisporella, bambusicolous fungi, molecular phylogeny, morphology, new species
1. Introduction
The bamboo plant (Poales, Bambusoideae) family includes over 1400 different species of monocotyledon, mostly evergreen perennials. Bamboo encompasses the largest members of the grass family, occur naturally in a wide range of different ecosystems, and are cultivated as a highly versatile crop [1]. Bamboos are well known to confer a number of beneficial ecological effects including carbon sequestration and erosion control, include some of the fastest growing plants known, have widespread ornamental use, and represent an important economic crop in regions where they are cultivated. Commercial applications of bamboo as a material for use in building/construction and fabrication of furniture, fabric, paper, cookware, cooking utensils, and many other items stems from its high strength-to-weight ratio and ease of cultivation that includes rapid plant growth. In addition, the plant is a food source for humans and other animals, notably giant and red pandas, as well as bamboo lemurs [2]. Currently, approximately 80% of the world’s bamboo species are found in the eastern and/or southern areas of Asia, with China having the richest bamboo resources in terms of highest diversity and overall cultivated area, accounting for more than 50% of the worldwide bamboo species [3,4]. Bamboos plants show high resistance to microbial diseases, with fungal ascomycetes as the major microorganisms that limit the health and productivity of bamboo forests. Different bamboo fungi can infect various parts of the plant, resulting in nevus (gall-like “tumors”), spotted wilt, leaf damage/necrosis, and other symptoms and diseases that can lead to reduced quality and yield of the bamboo. Deleterious effects of Bambusicolous Ascomycota can impact economic development, and methods for the biological control of some bamboo fungi can reduce losses in bamboo forests and the cultivation industry, helping to maintain diversity, plant populations, and the varied beneficial ecological functions of bamboo forests [5]. Over 1150 different species of ascomycetes may have some association (including pathogens, mutualists, and commensals) with bamboo, of which 350 asexual morphs, 240 hyphomycelia, and 110 coelomycetes have been tentatively identified [6]. These fungi are mainly distributed in the Sordariomycetes, Dothideomycetes, and Eurotiomycetes, with the more representative families of bamboo fungi found in the Magnaporthaceae and Apiosporaceae families within the Sordariomycetes.
Magnaporthaceae classification was proposed by Cannon [7] and included the genus Magnaporthe and its related genera Buergenerula, Clasterosphaeria, Gaeumannomyces, Herbampulla, and Omnidemptus. More recently, additional taxa classified within Magnaporthaceae include Magnaporthiopsis [8], Bussabanomyces, Kohlmeyeriopsis and Slopeiomyces [9], Pseudophhialophora [10], Falciphora [11], Neogaeumanyces [12], Budhanggurabania [13], Falciphoriella and Gaemannomycella [14], and Bifusisporella. Currently, Magnaporthaceae consists of 25 genera and more than 100 species. The genus Bifusisporella was erected by Rejane [15], with B. sorghi designated as the type species. The morphology of Bifusisporella is characterized by septate, branched mycelium with a smooth, hyaline to light brown surface, conidiophores reduced to conidiogenous cells, which can be solitary or aggregated, curved and elongated cylindrical or clavate, and are typically light brown. Conidia are described as dimorphic, with macroconidia slightly more curved than microconidia, and both sickle-shaped, hyaline, and smooth [15].
The Apiosporaceae fungal family belongs to the Ascomycota (Sordariomycetes Amphisphaeriales), with the type genus being Apiospora Sacc. introduced by Saccardo [16] and the type species being A. montagnei Sacc. The sexual morphology of Apiospora is characterized by hyaline ascospores surrounded by thick gelatinous sheaths [17,18,19]. The asexual form of Apiospora is characterized by lenticular conidia that are spherical or subglobose and usually light brown to brown in color [20,21]. Most Apiospora species are associated with plants as endophytes, with some species being economically important plant pathogens [22,23].
In this study, four fungal species, two of which represent new species, found growing on bamboo plants were identified and placed within the Magnaporthaceae, with their taxonomic placement determined based on morphological characteristics and molecular identification. The latter involved multilocus phylogenetic reconstructions using a combined dataset of the intervening 5.8S nrRNA gene (ITS), the 28S large subunit of nuclear ribosomal RNA gene (LSU), the large subunit of RNA polymerase I (rpb1), and the translation elongation factor 1-α gene (tef1-α) nucleotide sequences. Similarly, four additional fungal isolates (again, with two representing new species) were identified and placed within the Apiosporaceae based upon morphological characteristics and molecular taxonomic and phylogenetic analyses using the combined marker loci sequence dataset (ITS + LSU + tef1-α + tub2). Our results identify two new species of Bifusisporella, Bifusisporella fujianensis sp. nov. and Bifusisporella bambooensis sp. nov. (Magnaporthaceae), and two new species of Apiospora, Apiospora fujianensis sp. nov., Apiospora fuzhouensis sp. nov. (Apiosporaceae), which are illustrated and described. This study expands the diversity of fungi infecting the economically and environmentally important bamboo plant.
2. Materials and Methods
2.1. Fungal Isolates and Morphology
Specimens were collected from diseased bamboo leaves in groves located in Fujian province, China. Tissue fragments with a total area of about 25 mm2 were removed from the edges of the bamboo leaves in which disease spots, e.g., necrosis, wilting, and/or discoloration/blackening, were apparent. Samples were soaked in 75% ethanol for 45–60 s, then soaked in sterile deionized water for 45 s and washed with sterile water. Tissue fragments were then transferred to a 5% sodium hypochlorite solution for 30 s, followed by three washes in sterile deionized water for 60 s. The fragments were dried with sterilized filter paper and then transferred to the PDA plates which were incubated at 25 °C for 5–7 days following previously established procedures [24]. Growing edges of fungal mycelia were transferred to new PDA plates and plates were incubated for 5–7 d. The procedure was continued until the fungal culture was pure (typically 2–4 times). To promote sporulation and observe the colony morphology, purified isolates were inoculated in the center of PDA and synthetic low-nutrient agar (SNA) plates and cultured at 25 °C under alternating conditions of 12 h near-ultraviolet light and 12 h dark [25]. At 7 and 14 d of growth on PDA, photos of the colonies were taken with a digital camera, and the morphology of conidiomata, conidiophores, and conidiogenous cells was observed using a stereomicroscope (Nikon SMZ74, Tokyo, Japan). Samples were also prepared for analyses by scanning electron microscope (SEM, Nikon Ni-U; HITACHI SU3500) as described [26]. Fungal micromorphology and structure were measured by Digimizer 5.4.4 software. Single colony purified cultures were cut and stored in 10% sterilized glycerin and sterile water at 4 °C for future detailed study.
2.2. DNA Extraction, PCR Amplification, and Sequencing
Genomic DNA was isolated from fresh mycelia using a fungal DNA extraction mini ki, from cells cultured at 25 °C on PDA for 15–30 days as described [27]. Primers ITS5/ITS4 [28], LROR/LR5 [29], RPB1-Ac/RPB1-Cr [30,31], EF1-983F/EF1-2218R [32], EF1-728F/EF-2 [33], and Bt2a/Bt2b [34] were used for amplification of the intervening 5.8S nrRNA gene (ITS), the 28S large subunit of nuclear ribosomal RNA gene (LSU), the large subunit of RNA polymerase I (rpb1), the translation elongation factor 1-α gene (tef1-α),and the partial beta-tubulin gene (tub2) by polymerase chain reactions (PCR) as described [27]. Primer sequences are given in Table 1.
Table 1.
The primer sequences and programs in this study.
| Locus | Primers | Sequence (5′–3′) | PCR Cycles | References |
|---|---|---|---|---|
| ITS | ITS5 | GGA AGT AAA AGT CGT AAC AAG G | (95 °C: 30 s, 55 °C: 30 s, 72 °C: 1 min) × 35 cycles | [28] |
| ITS4 | TCCTCCGCTTATTGATATGC | |||
| LSU | LROR | GTACCC GCTGAACTTAAGC | (95 °C: 30 s, 52 °C: 30 s, 72 °C: 1 min) × 35 cycles | [29] |
| LR5 | TCCTGAGGGAAACTTCG | |||
| rpb1 | fRPB1-Ac | GAR TGY CCD GGD CAY TTY GG | (95 °C: 30 s, From 57 °C to 72 °C at 0.2 °C/s:30 s, 72 °C: 1 min) × 35 cycles | [30,31] |
| fRPB1-Cr | CCNGCDATNTCRTTRTCCATRTA | |||
| tef1-α | EF1-983F | GCYCCYGGHCAYCGTGAYTT | (95 °C: 30 s, 57/52 °C: 30 s, 72 °C: 1 min) × 35 cycles | [32] |
| EF1-2218R | ATGACACCRACRGCRACRGTYTGYAT | |||
| EF1-728F | CATCGAGAAGTTCGAGAAGG | (95°C: 30 s, 51 °C: 30 s, 72 °C: 1 min) × 35 cycles | [33] | |
| EF-2 | GGARGTACCAGTSATCATGTT | |||
| tub2 | Bt2a | GGTAACCAAATCGGT GCTGCT TTC | (95 °C: 30 s, 56 °C: 30 s, 72 °C: 1 min) × 35 cycles | [34] |
| Bt2b | ACCCTCAGTGTAGTGACCCTTGGC |
PCR amplification of target loci was performed using a Bio-Rad thermal cycler (Hercules, CA, USA) with a 25 μL reaction volume of 12.5 μL 2×Rapid Taq Master Mix (Vazyme, Nanjing, China), with 1 μL (10 μM) for the forward and reverse primers (Sangon, Shanghai, China) and 1 μL for the template genomic DNA in the amplifier, and adjusted with distilled deionized water to a total volume of 25 μL. PCR products were visualized on 1% agarose gel electrophoresis. Bidirectional (both strand) sequencing of PCR products was conducted by the Tsingke Company Limited (Fuzhou, China). Consensus sequences were assembled using MEGA 7.0 [35]. New sequences generated in this study were uploaded to GenBank (https://www.ncbi.nlm.nih.gov, accessed on 19 March 2024, Table 2).
Table 2.
Species names, voucher or culture codes, hosts or substrate, locations, and corresponding GenBank accession numbers of DNA sequences used in this study.
| Species | Culture/Voucher | Host/Substrate | Country | GenBank Accession Number | ||||
|---|---|---|---|---|---|---|---|---|
| ITS | LSU | tef1-a | tub2 | rpb1 | ||||
| Barretomyces calatheae | CBS 129274 = CPC 18464 | Calathea longifolia | Brazil | KM484831 | KM484950 | - | - | KM485045 |
| Bambusicularia brunnea | CBS 133599 | Sasa sp. | Japan | KM484830 | KM484948 | - | - | KM485043 |
| Bambusicularia brunnea | CBS 133600 | Phyllostachys bambusoides | Japan | AB274436 | KM484949 | - | - | KM485044 |
| Bifusisporella bambooensis | CGMCC3.25653 | Bambusoideae sp. | China | PP159031 | PP159039 | PP488459 | - | PP488463 |
| Bifusisporella bambooensis | CGMCC3.27207 | Bambusoideae sp. | China | PP477445 | PP477439 | PP488461 | - | PP488465 |
| Bifusisporella fujianensis | CGMCC3.25651 | Bambusoideae sp. | China | PP159030 | PP159038 | PP488458 | - | PP488462 |
| Bifusisporella fujianensis | CGMCC3.27206 | Bambusoideae sp. | China | PP477444 | PP477438 | PP488460 | - | PP488464 |
| Bifusisporella sorghi | URM 7442 | Sorghum bicolor endophyte | Brazil | MK060155 | MK060153 | MK060157 | - | MK060159 |
| Bifusisporella sorghi | URM 7864 | Sorghum bicolorendophyte | Brazil | MK060156 | MK060154 | MK060158 | - | MK060160 |
| Bifusisporella sichuanensis | SICAUCC 22-0073 | Phyllostachys edulis | China | ON227097 | ON227101 | ON244427 | - | ON244428 |
| Bussabanomyces longisporus | CBS 125232 | Amomum siamense, leaves | Thailand | KM484832 | KM484951 | KM009202 | - | KM485046 |
| Buergenerula spartinae | ATCC 22848 | Spartina alterniflora, leaves | USA | JX134666 | DQ341492 | JX134692 | - | JX134720 |
| Falciphora oryzae | CBS 125863= R5-6-l | Oryza sativa, root, endophyte | China | EU636699 | KJ026705 | JN857963 | - | KJ026706 |
| Falciphoriella solaniterrestris | CBS 117.83 | Soil in potato field | Netherlands | KM484842 | KM484959 | - | - | KM485058 |
| Gaeumannomycella graminis | CPC 26020 = CBS 141384 | Cynodon dactylon × C. transvaalensis | USA | KX306498 | KX306568 | KX306701 | - | KX306633 |
| Gaeumannomycella graminicola | CPC 26025 = CBS 141381 | Stenotaphrum secundatum | USA | KX306495 | KX306565 | KX306698 | - | KX306630 |
| Gaeumannomycella caricis | CPC 26262 = CBS 141374 | Carex rostrata | UK | KX306478 | KX306548 | KX306675 | - | KX306671 |
| Gaeumannomycella caricis | CBS 388.81 | Carex rostrata | UK | KM484843 | KM484960 | KX306674 | - | - |
| Gaeumannomyces floridanus | CPC 26037 | Stenotaphrum secundatum | USA | KX306491 | KX306561 | KX306693 | - | KX306626 |
| Gaeumannomyces fusiformis | CPC 26068 | Oryza sativa | USA | KX306492 | KX306562 | KX306694 | - | KX306627 |
| Gaeumannomyces glycinicola | CPC 26266 | Glycine max | USA | KX306494 | KX306564 | KX306696 | - | KX306629 |
| Gaeumannomyces glycinicola | CPC 26057 | Glycine max | USA | KX306493 | KX306563 | KX306695 | - | KX306628 |
| Gaeumannomyces graminicola | CBS 352.93 | - | - | KM484834 | DQ341496 | KX306697 | - | KM485050 |
| Gaeumannomyces graminis | CPC 26045 | Cynodon dactylon × C. transvaalensis | - | KX306505 | KX306575 | KX306708 | - | KX306640 |
| Gaeumannomyces graminis var. graminis | M33 | - | - | JF710374 | JF414896 | JF710411 | - | JF710442 |
| Gaeumannomyces graminis var. graminis | M54 | - | - | JF414848 | JF414898 | JF710419 | - | JF710444 |
| Gaeumannomyces hyphopodioides | CBS 350.77 | Zea mays, root | UK | KX306506 | KX306576 | - | - | - |
| Gaeumannomyces hyphopodioides | CBS 541.86 | Triticum aestivum, seedling | Germany | KX306507 | KX306577 | KX306709 | - | - |
| Gaeumannomyces oryzicola | CPC 26063 | Oryza sativa | USA | KX306516 | KX306586 | KX306717 | - | KX306646 |
| Gaeumannomyces oryzinus | CPC 26030 | Cynodon dactylon × C. transvaalensis | Bahamas | KX306517 | KX306587 | KX306718 | - | KX306647 |
| Gaeumannomyces radicicola | CBS 296.53 | - | Canada | KM009170 | KM009158 | KM009206 | - | KM009194 |
| Gaeumannomyces setariicola | CPC 26059 | Setaria italica | South Africa | KX306524 | KX306594 | KX306725 | - | KX306654 |
| Gaeumannomyces tritici | CBS 273.36 | Triticum aestivum | Argentina | KX306525 | KX306595 | KX306729 | - | KX306655 |
| Gaeumannomyces walkeri | CPC 26028 | Stenotaphrum secundatum | USA | KX306543 | KX306613 | KX306746 | - | KX306670 |
| Gaeumannomyces wongoonoo | BRIP:60376 | Stenotaphrum secundatum | Australia | KP162137 | KP162146 | - | - | - |
| Kohlmeyeriopsis medullaris | CBS 117849 = JK5528S | Juncus roemerianus | USA | KM484852 | KM484968 | - | - | KM485068 |
| Macgarvieomyces borealis | CBS 461.65 | Juncus effiisus, leaf spots | UK | MH858669 | DQ341511 | KM009198 | - | KM485070 |
| Macgarvieomyces juncicola | CBS 610.82 | Juncus effiisus, stem base | The Netherlands | KM484855 | KM484970 | KM009201 | - | KM485071 |
| Magnaporthaceae, incertaesedis | CPC 26284 = GP57 | Triticum aestivum | UK | KX306546 | KX306616 | KX306677 | - | - |
| Magnaporthiopsissp. | CPC 26038 | Cynodon dactylon × C. transvaalensis | USA | KX306545 | - | KX306676 | - | KX306672 |
| Magnaporthiopsis incrustans | M35 | - | - | JF414843 | JF414892 | - | - | JF710437 |
| Magnaporthiopsis maydis | CBS 133165 = ATCC MYA-3356 | Zeamays | Israel | KX306544 | KX306614 | - | - | - |
| Magnaporthiopsis maydis | CBS 662.82A | Zeamays | Egypt | KM484856 | KM484971 | - | - | KM485072 |
| Magnaporthiopsis cynodontis | RS7-2 = CBS 141700 | ultradwarf bermudagrass roots | USA | KJ855508 | KM401648 | KP282714 | - | KP268930 |
| Magnaporthiopsis cynodontis | RS5-5 | roots | USA | KJ855506 | KM401646 | KP282712 | - | KP268928 |
| Magnaporthiopsis cynodontis | RS3-1 | roots | USA | KJ855505 | KM401645 | KP282711 | - | KP268927 |
| Magnaporthiopsis meyeri-festucae | FF2 | - | - | MF178146 | MF178151 | MF178167 | - | MF178162 |
| Magnaporthiopsis meyeri-festucae | SCR11 | - | - | MF178150 | MF178155 | MF178171 | - | MF178166 |
| Magnaporthiopsis panicorum | CM2S8 | - | - | KF689643 | KF689633 | KF689623 | - | KF689613 |
| Magnaporthiopsis panicorum | CM10s2 | - | - | KF689644 | KF689634 | KF689624 | - | KF689614 |
| Magnaporthiopsis rhizophila | M22 | - | - | JF414833 | JF414882 | JF710407 | - | JF710431 |
| Nakataea sp. | CBS 332.53 | Oryza sativa | USA | KM484867 | KM484981 | - | - | KM485083 |
| Nakataea oryzae | CBS 252.34 | Oryza sativa | Burma | KM484862 | KM484976 | - | - | KM485078 |
| Nakataea oryzae | CBS 288.52 | Oryza sativa, stem | Japan | KM484864 | KM484978 | - | - | KM485080 |
| Neogaeumannomyces bambusicola | MFLUCC11-0390 | Dead culm of bamboo (Bambusae) | Thailand | KP744449 | KP744492 | - | - | - |
| Neopyricularia commelinicola | CBS 128307 = KACC 44083 | Commelina communis, leaves | Korea | FJ850125 | KM484984 | KM009199 | - | KM485086 |
| Neopyricularia commelinicola | CBS 128308 | Commelina communis, leaves | Korea | FJ850122 | KM484985 | - | - | KM485087 |
| Omnidemptus affinis | ATCC 200212 | Panicum effiisum var. effiisum grass leaves | Australia | JX134674 | KX134686 | JX134700 | - | JX134728 |
| Ophioceras dolichostomum | CBS 114926 = HKUCC 3936 = KM 8 | Wood | China | JX134677 | JX134689 | JX134703 | - | JX134731 |
| Ophioceras leptosporum | CBS 894.70 = ATCC 24161 = HME 2955 | Dead stem of dicot plant (probably Urtica dioicd) | UK | JX134678 | JX134690 | JX134704 | - | JX134732 |
| Proxipyricularia zingiberis | CBS 132355 | Zingiber mioga | Japan | AB274433 | KM484987 | - | - | KM485090 |
| Proxipyricularia zingiberis | CBS 133594 | Zingiber mioga | Japan | AB274434 | KM484988 | - | - | KM485091 |
| Pseudoph ialophora eragrostis | CM12m9 | Eragrostis sp. | USA | KF689648 | KF689638 | KF689628 | - | KF689618 |
| Pseudopyricularia cyperi | CBS 133595 | Cyperus iria | Japan | KM484872 | KM484990 | - | - | AB818013 |
| Pseudopyricularia kyllingae | CBS 133597 | Kyllinga brevifolia | Japan | KM484876 | KM484992 | KT950880 | - | KM485096 |
| Pyricularia grisea | BR0029 | Digitaria sanguinalis | Brazil | KM484880 | KM484995 | - | - | KM485100 |
| Pyricularia grisea | CR0024 | Lolium perenne | Korea | KM484882 | KM484997 | - | - | KM485102 |
| Pyricularia ctenantheicola | GR0001 = Ct-4 = ATCC 200218 | Ctenanthe oppenheimiana | Greece | KM484878 | KM484994 | - | KM485098 | |
| Pyricularia oryzae | CBS 365.52 = MUCL 9451 | - | Japan | KM484890 | KM485000 | - | - | KM485110 |
| Slopeiomyces cylindrosporus | CBS 609.75 | Grass root, associated with Phialophora graminicola | UK | KM484944 | KM485040 | JX134693 | - | KM485158 |
| Utrechtiana cibiessia | CBS 128780 = CPC 18916 | Phragmites australis, leaves | Netherlands | JF951153 | JF951176 | - | - | KM485047 |
| Xenopyricularia zizaniicola | CBS 132356 | Zizania latifolia | Japan | KM484946 | KM485042 | KM009203 | - | KM485160 |
| Apiospora acutiapica | KUMCC 20-0209 | - | - | MT946342 | MT946338 | MT947359 | MT947365 | - |
| Apiospora acutiapica | KUMCC 20-0210 | Bambusa bambos | China | - | MT946339 | MT947360 | MT947366 | - |
| Apiospora agari | KUC 21333 | Agarum cribrosum | Korea | - | MH498440 | MH544663 | MH498478 | - |
| Apiospora aquatica | MFLU 18-1628 | Submerged wood | China | MK828608 | MK835806 | - | - | - |
| Apiospora arctoscopi | KUC 21331 | Egg of Arctoscopus japonicus | Korea | - | MH498449 | MN868918 | MH498487 | - |
| Apiospora arctoscopi | KUC 21344 | - | - | MH498528 | - | MN868919 | MH498486 | - |
| Apiospora arctoscopi | KUC 21347 | - | - | MH498525 | - | MN868922 | MH498483 | - |
| Apiospora arundinis | CBS 114316 | Hordeum vulgare | Iran | KF144884 | KF144928 | KF145016 | KF144974 | - |
| Apiospora arundinis | CBS 106.12 | - | - | KF144883 | KF144927 | KF145015 | KF144973 | - |
| Apiospora arundinis | CBS 732.71 | - | - | KF144889 | KF144934 | KF145022 | KF144980 | - |
| Apiospora aurea | CBS 244.83 | Air | Spain | AB220251 | KF144935 | KF145023 | KF144981 | - |
| Apiospora balearica | CBS 145129 | Poaceae | Spain | MK014869 | MK014836 | MK017946 | MK017975 | - |
| Apiospora neobambusae | HMAS LC7106 | - | - | KY494718 | KY494794 | KY806204 | KY705186 | - |
| Apiospora bambusicola | MFLUCC20-0144 | Schizostachyum brachycladum | Thailand | MW173030 | MW173087 | MW183262 | - | - |
| Apiospora biserialis | CGMCC 3.20135 | Bambusoideae sp. | China | MW481708 | MW478885 | MW522938 | MW522955 | - |
| Apiospora camelliae-sinensis | LC5007 | Camellia sinensis | China | KY494704 | KY494780 | KY705103 | KY705173 | - |
| Apiospora camelliae-sinensis | LC8181 | - | - | KY494761 | KY494837 | KY705157 | KY705229 | - |
| Apiospora chiangraiense | MFLU 21-0046 | - | - | MZ542520 | MZ542524 | - | MZ546409 | - |
| Apiospora chromolaenae | MFLUCC 17-1505 | Chromolaena odorata | Thailand | MT214342 | MT214436 | MT235802 | - | - |
| Apiospora cordylines | GUCC 10027 | - | - | MT040106 | - | MT040127 | MT040148 | - |
| Apiospora cyclobalanopsidis | CGMCC 3.20136 | Cyclobalanopsis glauca | China | MW481713 | MW478892 | MW522945 | MW522962 | - |
| Apiospora cyclobalanopsidis | GZCC:20-0103 | - | - | MW481714 | - | MW522946 | MW522963 | - |
| Apiospora descalsii | CBS 145130 | Ampelodesmos mauritanicus | Spain | MK014870 | MK014837 | MK017947 | MK017976 | - |
| Apiospora dichotomanthi | CGMCC 3.18332 | Dichotomanthes tristaniiaecarpa | China | KY494697 | KY494773 | KY705096 | KY705167 | - |
| Apiospora esporlensis | CBS 145136 | Phyllostachys aurea | Spain | MK014878 | MK014845 | MK017954 | MK017983 | - |
| Apiospora esporlensis | 18TJAM004 | - | - | MT856406 | - | MT881953 | MT881991 | - |
| Apiospora euphorbiae | IMI 285638b | Bambusoideae sp. | Bangladesh | AB220241 | AB220335 | - | AB220288 | - |
| Apiospora euphorbiae | ZHKUCC 22-0001 | - | - | OM728647 | OM486971 | OM543543 | OM543544 | - |
| Apiospora fermenti | KUC 21289 | - | - | MF615226 | - | MH544667 | MF615231 | - |
| Apiospora fermenti | KUC 21288 | - | - | MF615230 | - | MH544668 | MF615235 | - |
| Apiospora fujianensis | CGMCC3.25647 | Bambusoideae sp. | China | PP159026 | PP159034 | PP488454 | PP488470 | - |
| Apiospora fujianensis | CGMCC3.25648 | Bambusoideae sp. | China | PP159027 | PP159035 | PP488455 | PP488471 | - |
| Apiospora fuzhouensis | CGMCC3.25649 | Bambusoideae sp. | China | PP159028 | PP159036 | PP488456 | PP488468 | - |
| Apiospora fuzhouensis | CGMCC3.25650 | Bambusoideae sp. | China | PP159029 | PP159037 | PP488457 | PP488469 | - |
| Apiospora gaoyouensis | CFCC52301 | Phragmites australis | China | MH197124 | - | MH236793 | MH236789 | - |
| Apiospora garethjonesii | JHB004 | Culms of dead bamboo | China | KY356086 | KY356091 | - | - | - |
| Apiospora garethjonesii | SICAUCC 22-0028 | - | - | ON228606 | ON228662 | - | ON237654 | - |
| Apiospora garethjonesii | SICAUCC 22-0027 | - | - | ON228603 | ON228659 | - | ON237651 | - |
| Apiospora gelatinosa | HKAS 111962 | Culms of dead bamboo | China | MW481706 | MW478888 | MW522941 | MW522958 | - |
| Apiospora guiyangensis | HKAS 102403 | Dead culms of Poaceae | China | MW240647 | MW240577 | MW759535 | MW775604 | - |
| Apiospora guizhouensis | CGMCC 3.18334 | Air in karst cave | China | KY494709 | KY494785 | KY705108 | KY705178 | - |
| Apiospora guizhouensis | KUMCC 20-0206 | - | - | MT946347 | MT946341 | MT947364 | MT947370 | - |
| Apiospora hainanensis | SAUCC 1681 | Leaf of bamboo | China | OP563373 | OP572422 | OP573262 | OP573268 | - |
| Apiospora hispanica | IMI 326877 | Maritime sand | Spain | AB220242 | AB220336 | - | AB220289 | - |
| Apiospora hydei | CBS 114990 | Bambusoideae sp. | China | KF144890 | KF144936 | KF145024 | KF144982 | - |
| Apiospora hydei | LC 7103 | - | - | KY494715 | KY494791 | KY705114 | KY705183 | - |
| Apiospora hyphopodii | SICAUCC 22-0034 | - | - | ON228605 | ON228661 | - | ON237653 | - |
| Apiospora hysterina | ICMP 6889 | Bambusoideae sp. | New Zealand | MK014874 | MK014841 | MK017951 | MK017980 | - |
| Apiospora hysterina | KUC21438 | - | - | ON764019 | ON787758 | ON806623 | ON806633 | - |
| Apiospora iberica | CBS 145137 | Arundo donax | Portugal | MK014879 | MK014846 | MK017955 | MK017984 | - |
| Apiospora intestini | CBS 135835 | Gut of grasshopper | India | KR011352 | KR149063 | KR011351 | KR011350 | - |
| Apiospora intestini | MFLU:21-0045 | - | - | MZ542521 | MZ542525 | MZ546406 | MZ546410 | - |
| Apiospora italica | AP29118 | - | - | MK014881 | MK014848 | MK017957 | MK017986 | - |
| Apiospora jatrophae | CBS 134262 | Jatropha podagrica | India | JQ246355 | - | - | - | - |
| Apiospora jiangxiensis | CGMCC 3.18381 | Maesa sp. | China | KY494693 | KY494769 | KY705092 | KY705163 | - |
| Apiospora jiangxiensis | SICAU 22-0070 | - | - | ON227094 | ON227098 | ON244431 | ON244432 | - |
| Apiospora kogelbergensis | CBS 113332 | Cannomois virgata | South Africa | KF144891 | KF144937 | KF145025 | KF144983 | - |
| Apiospora kogelbergensis | CBS 117206 | - | - | KF144895 | KF144941 | KF145029 | KF144987 | - |
| Apiospora koreana | KUC 21332 | Egg of Arctoscopus japonicus | Korea | MH498524 | - | MH544664 | MH498482 | - |
| Apiospora koreana | KUC21350 | - | - | MH498521 | - | MN868929 | MH498479 | - |
| Apiospora locuta-pollinis | LC11683 | Brassica campestris | China | MF939595 | - | MF939616 | MF939622 | - |
| Apiospora locuta-pollinis | KUNCC:22-12409 | - | - | OP377737 | OP377744 | OP381091 | - | - |
| Apiospora longistroma | MFLU 15-1184 | Culms of decaying bamboo | Thailand | KU940141 | KU863129 | - | - | - |
| Apiospora malaysiana | CBS 102053 | Macaranga hullettii | Malaysia | KF144896 | KF144942 | KF145030 | KF144988 | - |
| Apiospora malaysiana | CBS:251.29 | - | - | KF144897 | KF144943 | KF145031 | KF144989 | - |
| Apiospora marianiae | AP18219 | Dead stems of Phleum pratense | Spain | ON692406 | ON692422 | ON677180 | ON677186 | - |
| Apiospora marii | CBS 497.90 | Air | Spain | MH873913 | KF144947 | KF145035 | KF144993 | - |
| Apiospora marii | CBS 200.57 | - | - | KF144900 | KF144946 | KF145034 | KF144992 | - |
| Apiospora marina | KUC 21328 | Seaweed | Korea | MH498538 | MH498458 | MH544669 | MH498496 | - |
| Apiospora mediterranea | IMI 326875 | Air | Spain | AB220243 | AB220337 | - | AB220290 | - |
| Apiospora minutispora | 17E 042 | Soil | Korea | LC517882 | - | LC518889 | LC518888 | - |
| Apiospora montagnei | LSU0093 | - | - | MT000394 | MT000490 | - | - | - |
| Apiospora mori | MFLU 18-2514 | Dead leaves of Morus australis | China | MW114313 | MW114393 | - | - | - |
| Apiospora mori | NCYU 19-0364 | - | - | MW114314 | MW114394 | - | - | - |
| Apiospora mukdahanensis | MFLUCC 22-0056 | - | - | OP377735 | OP377742 | OP381089 | - | - |
| Apiospora multiloculata | MFLUCC 21-0023 | Dead culms of Bambusae | Thailand | OL873137 | OL873138 | - | OL874718 | - |
| Apiospora mytilomorpha | DAOM 214595 | Dead blades of Andropogon sp. | India | KY494685 | - | - | - | - |
| Apiospora neobambusae | LC7124 | - | - | KY494727 | KY494803 | KY806206 | KY705195 | - |
| Apiospora neochinense | CFCC 53036 | Fargesia qinlingensis | China | MK819291 | - | MK818545 | MK818547 | - |
| Apiospora neogarethjonesii | HKAS 102408 | Dead culms of Bambusae | China | MK070897 | MK070898 | - | - | - |
| Apiospora neosubglobosa | SICAUCC 22-0039 | - | - | ON228614 | ON228670 | - | ON237662 | - |
| Apiospora obovata | CGMCC 3.18331 | Lithocarpus sp. | China | KY494696 | KY494772 | KY705095 | KY705166 | - |
| Apiospora obovata | LC8177 | - | - | KY494757 | KY494833 | KY705153 | KY705225 | - |
| Apiospora ovata | CBS 115042 | Arundinaria hindsii | China | KF144903 | KF144950 | KF145037 | KF144995 | - |
| Apiospora paraphaeosperma | MFLUCC13-0644 | Dead clumps of Bambusa sp. | Thailand | KX822128 | KX822124 | - | - | - |
| Apiospora phragmitis | CBS 135458 | Phragmites australis | Italy | KF144909 | KF144956 | KF145043 | KF145001 | - |
| Apiospora phragmitis | AP29717A | - | - | MK014892 | MK014859 | MK017968 | MK017997 | - |
| Apiospora phyllostachydis | MFLUCC 18-1101 | Phyllostachys heteroclada | China | MK351842 | MH368077 | MK340918 | MK291949 | - |
| Apiospora piptatheri | CBS 145149 | Piptatherum miliaceum | Spain | MK014893 | MK014860 | - | - | - |
| Apiospora piptatheri | KUC21279 | - | - | MF615229 | - | MH544671 | MF615234 | - |
| Apiospora pseudoparenchymatica | CGMCC 3.18336 | Bambusoideae sp. | China | KY494743 | - | KY705139 | KY705211 | - |
| Apiospora pseudorasikravindrae | KUMCC 20-0208 | - | - | MT946344 | - | MT947361 | MT947367 | - |
| Apiospora pseudorasikravindrae | KUMCC 20-0211 | - | - | MT946345 | - | MT947362 | MT947368 | - |
| Apiospora pseudosinensis | CPC 21546 | Leaf of bamboo | The Netherlands | KF144910 | KF144957 | KF145044 | MN868936 | - |
| Apiospora pseudospegazzinii | CBS 102052 | Macaranga hullettii | Malaysia | KF144911 | KF144958 | KF145045 | KF145002 | - |
| Apiospora pterosperma | CBS 134000 | Machaerina sinclairii | Australia | KF144913 | KF144960 | KF145046 | KF145004 | - |
| Apiospora pusillisperma | KUC 21321 | Seaweed | Korea | MH498533 | MH498453 | MN868930 | MH498491 | - |
| Apiospora qinlingensis | CFCC 52303 | Fargesia qinlingensis | China | MH197120 | - | MH236795 | MH236791 | - |
| Apiospora rasikravindrae | LC5449 | Soil in karst cave | China | KY494713 | KY494789 | KY705112 | KY705182 | - |
| Apiospora rasikravindrae | AP10418 | - | - | MK014896 | MK014863 | - | MK017999 | - |
| Apiospora sacchari | CBS 212.30 | Phragmites australis | UK | KF144916 | KF144962 | KF145047 | KF145005 | - |
| Apiospora sacchari | CBS:664.74 | - | - | KF144919 | KF144965 | KF145050 | KF145008 | - |
| Apiospora saccharicola | CBS 191.73 | Air | The Netherlands | KF144920 | KF144966 | KF145051 | KF145009 | - |
| Apiospora sargassi | KUC 21228 | Sargassum fulvellum | Korea | KT207746 | - | MH544677 | KT207644 | - |
| Apiospora sasae | CBS 146808 | Dead culms of Sasa veitchii | The Netherlands | MW883402 | MW883797 | MW890104 | MW890120 | - |
| Apiospora septata | CGMCC 3.20134 | Bambusoideae sp. | China | MW481711 | MW478890 | MW522943 | MW522960 | - |
| Apiospora serenensis | IMI 326869 | - | Spain | AB220250 | AB220344 | - | AB220297 | - |
| Apiospora serenensis | ATCC 76309 | - | - | AB220240 | AB220334 | - | AB220287 | - |
| Apiospora setariae | CFCC 54041 | Decaying culms of Setaria viridis | China | MT492004 | - | - | - | - |
| Apiospora setostroma | KUMCC 19-0217 | Dead branches of bamboo | China | MN528012 | MN528011 | MN527357 | - | - |
| Apiospora sichuanensis | HKAS 107008 | Dead culms of Poaceae | China | MW240648 | MW240578 | MW759536 | MW775605 | - |
| Apiospora sorghi | URMBRA 9300 | Sorghum bicolor | Brazil | MK371706 | - | - | MK348526 | - |
| Apiospora sphaerosperma | CBS114314 | Leaf of Hordeum vulgare | Iran | KF144904 | KF144951 | KF145038 | KF144996 | - |
| Apiospora sphaerosperma | CBS 142.55 | - | - | KF144908 | KF144955 | KF145042 | AB220303 | - |
| Apiospora stipae | CBS 146804 | Stipa gigantea | Spain | MW883403.1 | MW883798.1 | - | MW890121.1 | - |
| Apiospora subglobosa | MFLUCC 11-0397 | Dead culms of bamboo | Thailand | KR069112 | KR069113 | - | - | - |
| Apiospora subrosea | CGMCC 3.18337 | Bambusoideae sp. | China | KY494752 | KY494828 | KY705148 | KY705220 | - |
| Apiospora subrosea | LC 7291 | - | - | KY494751 | KY494827 | KY705147 | KY705219 | - |
| Apiospora taeanensis | KUC 21322 | Seaweed | Korea | MH498515 | - | MH544662 | MH498473 | - |
| Apiospora taeanensis | KUC 21359 | - | - | MH498513 | - | MN868935 | MH498471 | - |
| Apiospora thailandica | MFLUCC 15-0202 | - | - | KU940145 | KU863133 | - | - | - |
| Apiospora thailandica | MFLUCC 15-0199 | - | - | KU940146 | KU863134 | - | - | - |
| Apiospora tropica | MFLUCC 21–0056 | - | - | OK491657 | OK491653 | - | OK560922 | - |
| Apiospora vietnamensis | IMI 99670 | Citrus sinensis | Vietnam | KX986096 | KX986111 | - | KY019466 | - |
| Apiospora xenocordella | CBS 478.86 | Soil from roadway | Zimbabwe | KF144925 | KF144970 | KF145055 | KF145013 | - |
| Apiospora xenocordella | LC3486 | - | - | KY494687 | KY494763 | KY705086 | KY705158 | - |
| Apiospora yunnana | MFLUCC 150002 | Culms of Decaying bamboo | China | KU940147 | KU863135 | - | - | - |
| Apiospora yunnana | SICAU 22-0072 | - | - | ON227096 | ON227100 | ON244425 | ON244426 | - |
Notes: newly generated sequences are in bold.
2.3. Phylogenetic Analyses
Based on maximum likelihood (ML) and Bayesian inference (BI) analyses, phylogenetic trees were constructed to explore the phylogeny relationships of the fungal strains, grouping them into either the Magnaporthaceae or Apiosporaceae families. Corresponding gene loci of the reference sequences were downloaded from GenBank. Ophioceras dolichostomum (CBS 114926) was selected as an outgroup taxonomic unit for the phylogeny of Magnaporthaceae, and Sporocadus trimorphus (CBS 114203) was selected as an outgroup taxonomic unit for the phylogeny of Apiosporaceae. All sequences were aligned using the MAFFT v. 7 online service (http://mafft.cbrc.jp/alignment/server/, accessed on 2 February 2024) [36] and manually adjusted in BioEdit v.7.2.6.1 [37] and MEGA 7.0 [35].
In addition, four simultaneous Markov Chain Monte Carlo (MCMC) chains, starting with 2,000,000 generations of random trees, were sampled every 100th generation, resulting in a total of 20,000 trees. The first 25% of trees were discarded as burn-in of each analysis. Branches with significant Bayesian Posterior Probabilities (BYPP > 0.90) were estimated in the remaining 15,000 trees [38]. Phylogenetic trees were plotted with FigTree v.1.4.4 [39] and embellished with Adobe Illustrator CS6. New sequences generated in this study have been deposited in GenBank (https://www.ncbi.nlm.nih.gov, accessed on 19 March 2024).
3. Results
3.1. Phylogenetic Analyses
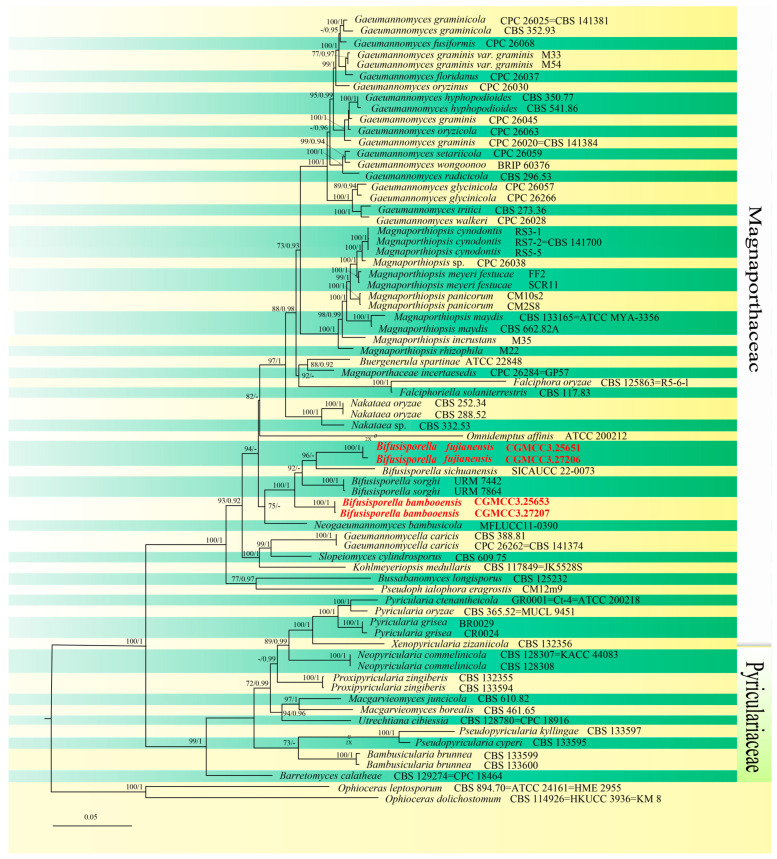
Samples of bamboo plants showing obvious fungal growth were collected from the Baizhu Garden of Fujian Agriculture and Forestry University and West Lake Park of Fuzhou City, Fujian Province, China. A total of eight fungal isolates with different morphological appearances were single-colony purified. For each fungal isolate, ~2637 bp of nucleotide sequences corresponding to portions of the ITS, LSU, rpb1, and tef1-αloci (ITS: 1–369; LSU: 370–1146; rpb1: 1147–1893; tef1-α: 1894–2775) were isolated. Based upon initial BLAST results, four of these sequences were isolated, combined with sequences from 68 closely related species as determined by BLAST searches, as well as homologous regions from Ophioceras dolichostomum (CBS 114926) and Ophioceras leptosporum (CBS 894.70) (Ophioceraceae, Magnaporthales) used as the outgroup for phylogenetic analyses. These analyses showed 1297 distinct patterns, with 1349 bp identical, 614 variable, including gaps, and 812 bp which were parsimony-informative. Maximum likelihood phylogenies were inferred using IQ-TREE under the TIM2 + R4 + F model for 5000 ultrafast bootstraps, as well as the Shimodaira–Hasegawa-like approximate likelihood-ratio test [nst = 6, rates = invgamma], with an average standard deviation of split frequencies = 0.005812. The topological results obtained from the ML analysis were consistent with the results of the BI analysis connecting the combined datasets. As a result, the ML tree is shown, and the BI posterior probabilities are placed on it (Figure 1). Based on phylogenetic resolution and morphological analysis (given below), we report two of the four isolates as new species of Magnaporthaceae: Bifusisporella fujianensis and Bifusisporella bambooensis. The new species B. fujianensis was most closely related to B. sichuanensis (SICAUCC 22-0073) (ML-BS: 96%, BYPP: 0.76), and B. bambooensis to B. sorghi (URM 7442, URM 7864) (ML-BS: 100%, BYPP: 1).
Figure 1.
ML tree generated from combined ITS, LSU, rpb1, and tef1-α sequence data of Magnaporthaceae and Pyriculariaceae. The maximum likelihood (ML) bootstrap support values and Bayesian posterior probabilities (BYPP) bootstrap support values above 70% and 0.90 are shown at the first and second position. Species with sequences obtained in this study are in boldface and newly generated sequences were indicated in red. Ophioceras dolichostomum (CBS 114926) and O.leptosporum (CBS894.70) (Ophioceraceae) were used as the outgroup. Yellow-green strips represent different neighboring species.
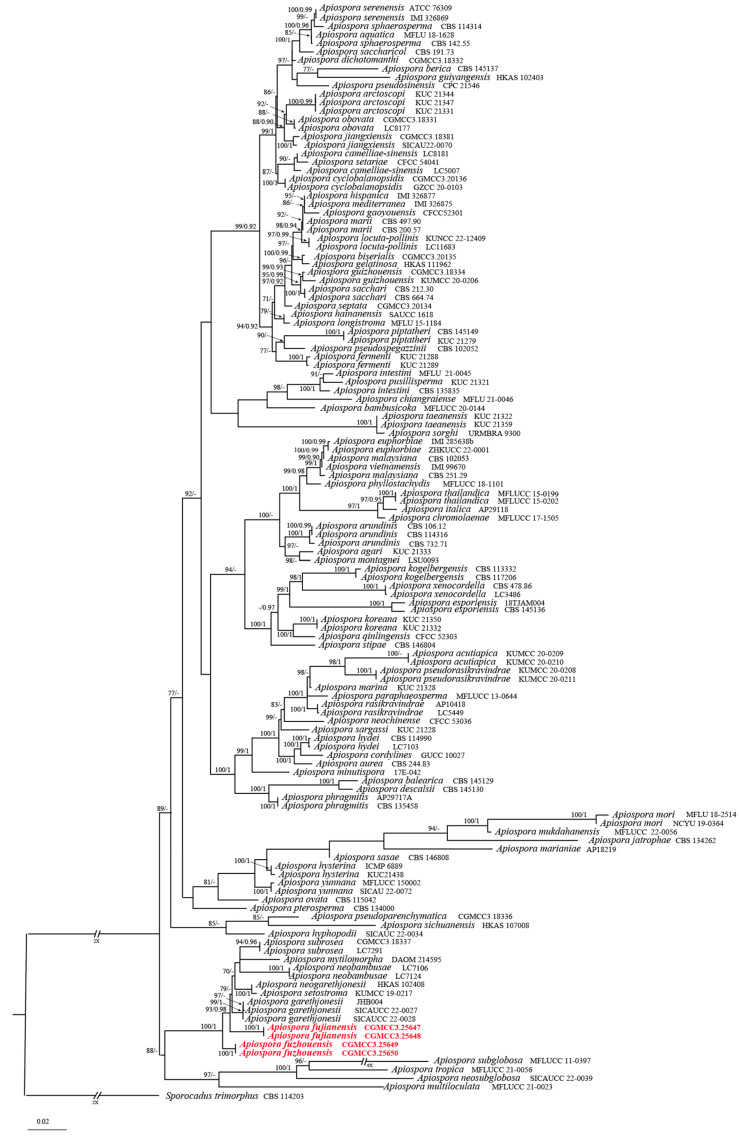
Initial BLAST results of sequences derived from the remaining four isolates indicated placement of these within the Apiosporaceae family. Analyses using sequences derived from the four genetic loci examined, namely the ITS + LSU + tef1-α + tub2 concatenated sequence dataset which had an aligned length of 1941 total characters (ITS: 1–507, LSU: 508–1341, tub2: 1342–1830, tef1-α: 1831–1941), supported the classification of two of the isolates as new species. Based on these and morphological data (below), a new species, Apiospora fujianensis, was identified, related to A. garethjonesii (SICAUCC 22-0028), with good support (ML-BS: 93% and BYPP: 0.98). Designation of the other new species, A. fuzhouensis, was similarly strongly supported (100% ML/1 PP), with the species forming a separate branch within Apiospora. For A. fuzhouensis loci analyses, 924 distinct patterns were identified, with 1186 bp constant, 157 variable and included gaps, and 598 bp which were parsimony-informative. Maximum likelihood phylogenies were inferred using IQ-TREE [40], under the GTR + R3 + F model for 5000 ultrafast bootstraps [41], as well as the Shimodaira–Hasegawa-like approximate likelihood-ratio test [nst = 1, rates = invgamma] (Figure 2).
Figure 2.
Phylogram of Apiospora based on combined ITS, LSU, tef1-α, and tub2 genes. ML bootstrap support values (ML-BS ≥ 70%) and Bayesian posterior probability (BYPP ≥ 0.90) are shown as first and second position above nodes, respectively. Strains from this study are shown in red. Some branches were shortened according to the indicated multipliers.
3.2. Taxonomy
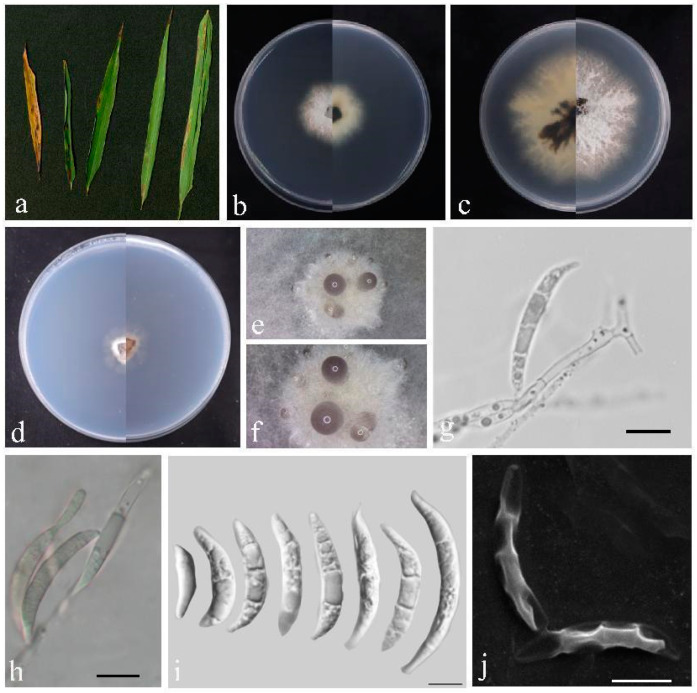
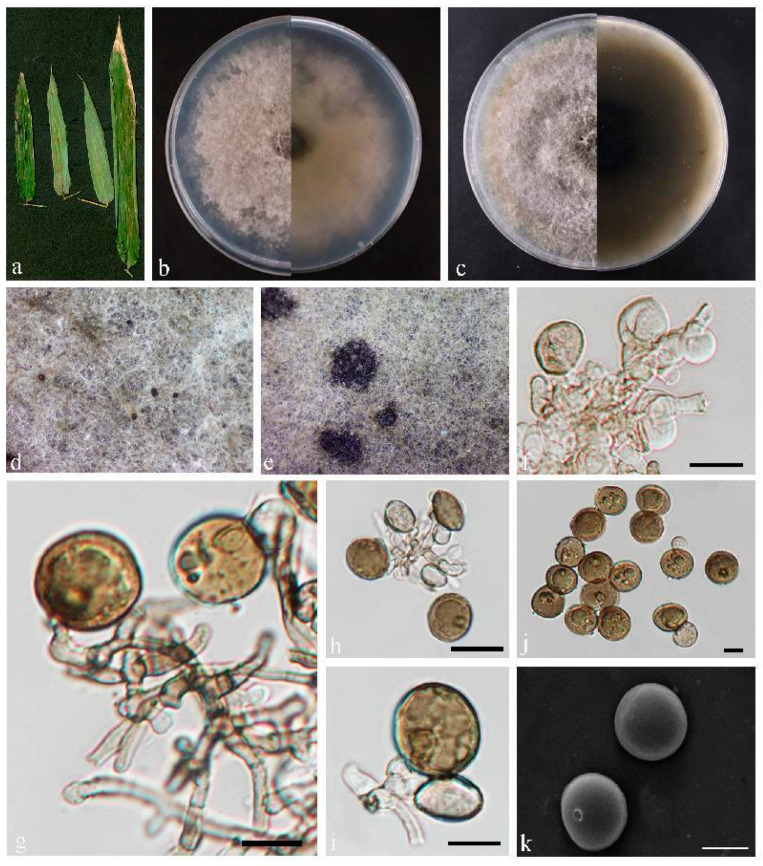
Bifusisporella fujianensis sp. nov. Z.Y. Zhao and J.Z. Qiu, (Figure 3).
Figure 3.
Bifusisporella fujianensis (HMAS 352712). (a) Leaves of host plant. (b,c) Upper and reverse view of colony after incubation for 7 days on PDA and 14 days. (d) Upper and reverse view of colony after incubation for 14 days on SNA (containing pine needle). (e,f) Conidiomata sporulating on PDA. (g,h) Conidiogenous cells and conidia. (i–k) Conidia. Scalebar = 10 µm (g–k).
MycoBank: MB852815.
Etymology: Named after Fujian Province where the fungus was collected.
Holotype: China, Fujian Province, Fujian University of Agriculture and Forestry (119°14′35.14″ E, 26°5′2.55″ N), from diseased leaves of bamboo in China, March 2023, Z. Y. Zhao (holotype HMAS352712; ex-type living culture CGMCC3.25651).
Description: Leaf spots irregularly shaped, sunken in the center, brown or tan in color. Conidiomata elevated on agar, solitary, spherical, gradually transitioning from white hyaline to black, conidiophores reduced to conidiophores cells. Conidiogenous cells were phialidic, solitary or aggregated, curved, elongated, cylindrical or rod-shaped, light brown, 8.9–14.3 × 5.8–8.1 µm. Conidia were dimorphic, falcate or curved moon-shaped, smooth or cracked surface, transparent in color, 0–3 septa, 37.3–56.3 × 3.6–5.7 µm, mean = 45.1 × 4.4 µm. No sexual morphology was observed.
Culture characteristics: Colonies flattened on PDA with feathery margins, white, on SNA surface and reverse, white. The calculated growth rate was 0.6 cm/day at 25 °C.
Material examined: China, Fujian Province, Fujian University of Agriculture and Forestry (119°14′35.14″ E, 26°5′2.55″ N), from diseased leaves of bamboo in China, March 2023, Z. Y. Zhao (paratype HMAS352713; ex-paratype living culture CGMCC3.27206).
Notes: The strain of the genus Bifusisporella was identified as a new species; nucleotide comparison of ITS, LSU, tef1-α, and rpb1 (CGMCC3.25651) showed differences with the sequences of B. sichuanensis (SICAUCC 22-0071), similarities are 12.3% (64/522), 4.1% (33/797), 4.1% (36/884), and 8.5% (58/684). In addition, the asexual morph of B. sichuanensis was not observed and the sexual morph of B. fujianensis was not observed.
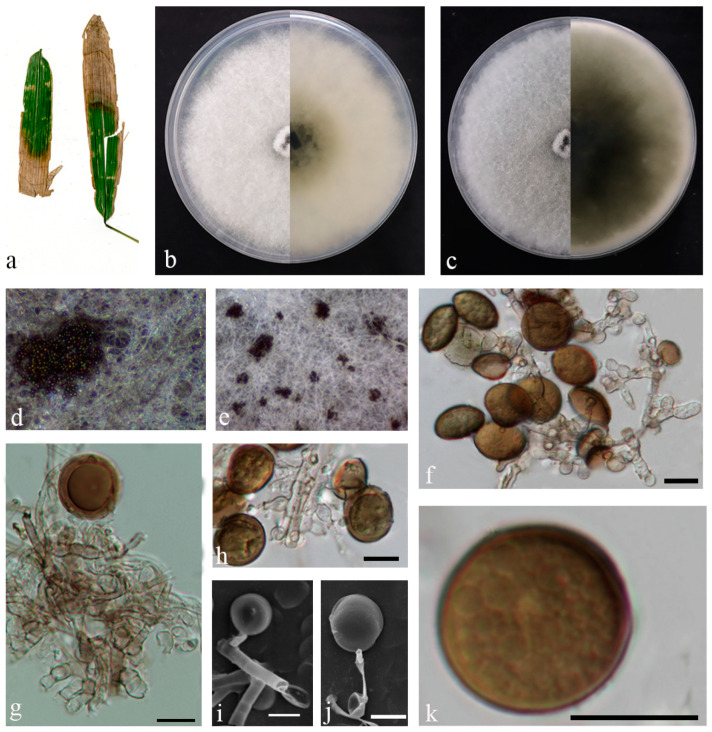
Bifusisporella bambooensis sp. nov. Z.Y. Zhao and J.Z. Qiu, (Figure 4).
Figure 4.
Bifusisporella bambooensis (HMAS 352714). (a) Leaves of host plant. (b,c) Upper and reverse view of colony after incubation for 7 days on PDA and 14 days. (d) Upper and reverse view of colony after incubation for 14 days on SNA (containing pine needle). (e,f) Conidiomata sporulating on PDA. (g,h) Conidiogenous cells and conidia.(i,j) Conidia. Scalebar = 10 µm (g–j).
MycoBank: MB852816.
Etymology: The epithet “bambooensis” refers to the host, which is bamboo.
Holotype: China, Fujian Province, Fujian University of Agriculture and Forestry, (119°14′35.14″ E, 26°5′2.55″ N), from diseased leaves of bamboo. March 2023, Z.Y. Zhao (holotype HMAS352714; ex-type living culture CGMCC3.25653).
Description: Leaf spots were pike-shaped, color gradually changing from blackish brown to white from outside to inside, Conidiomata bulging on agar, black to hyaline, aggregated and spherical, conidiophores reduced to conidiophores cells. Conidiogenous cells were phialidic, singly or in groups, curved, elongated, cylindrical, 7.2–21.0 × 4.2–6.4 µm, conidia were dimorphic, falcate or curved moon-shaped, smooth or cracked surface, hyaline, 0–3 septa, 10.8–45.0 × 2.8–4.9 µm, mean = 25.0 × 3.7 µm. No sexual morphology was observed.
Culture characteristics: Colonies flattened on PDA, irregular black center, fading to white with white feathery margins, on SNA surface and reverse, white. Calculated growth rate was 1.0–1.4 cm/day at 25 °C. The growth rate was 0.5 cm/day.
Material examined: China, Fujian Province, Fujian University of Agriculture and Forestry (119°14′35.14″ E, 26°5′2.55″ N), from diseased leaves of bamboo in China, March 2023, Z.Y. Zhao (paratype HMAS352715; ex-paratype living culture CGMCC3.27207).
Notes: Bifusisporella bambooensis is phylogenetically close (100% ML and 1BYPP), but distinct from B. sorghi (URM 7442). Compared to Bifusisporella sorghi, Bifusisporella bambooensis sp. nov. has larger conidiogenous cells and conidia (7.2–21.0 × 4.2–6.4 vs. 5.0–19.5 × 3.0–4.0 μm; 10.8–45.0 × 2.8–4.9 vs. 19.0–34.0 × 3.0–4.0 μm); nucleotide comparison of ITS, tef1-α and rpb1 (CGMCC3.25653) showed separation from B. sorghi (URM 7442), with differences of 6% (29/487), 5.3% (23/437), and 9.4% (65/693), respectively.
Apiospora fujianensis sp. nov. Z.Y. Zhao and J.Z. Qiu, (Figure 5).
Figure 5.
Apiospora fujianensis (HMAS 352716). (a) Leaves of host plant. (b,c) Upper and reverse view of colony after incubation for 7 days on PDA and 14 days. (d,e) Conidiomata sporulating on PDA. (f–j) Conidiogenous cells and conidia. (k) Conidia. Scale bars = 10 µm (f–k).
MycoBank: MB852818.
Etymology: Named after Fujian Province where the fungus was collected.
Holotype: China, Fujian Province, West Lake Park,119°17′47.09″ E,26°5′57.90″ N, from diseased leaves of bamboo in China, October 2022, J.H. Chen (holotype HMAS352716; ex-type living culture CGMCC3.25647).
Description: Leaf spots irregularly shaped, brown or tan in color. Conidiomata on agar were elevated, solitary or aggregated, spherical, black, conidiophores cells were solitary or aggregated, hyaline rounded, 3.5–5.8 × 3.5–5.2 µm. Conidia were rounded or ellipsoidal, contained globular contents, brown, 7.5–17.0 × 5.6–18.2 µm, mean = 14.0 × 11.6 µm. No sexual morphology was observed.
Culture characteristics: Colonies flattened on PDA, fluffy mycelium, black center with white margins over time; calculated growth rate of 1.3 cm/day at 25 °C.
Material examined: China, Fujian Province, West Lake Park, 119°17′47.09″ E, 26°5′57.90″ N, from diseased leaves of bamboo in China, October 2022, J.H. Chen (paratype HMAS352717; ex-paratype living culture CGMCC3.25648).
Notes: In the present study, two strains were obtained from diseased leaves of bamboo and differed from each other with a high degree of statistical support (BYPP = 0.98 and ML-BS = 93%), although overall analyses indicated that both isolates represented different strains of the same species.
Apiospora fuzhouensis sp. nov. Z.Y. Zhao and J.Z. Qiu, (Figure 6).
Figure 6.
Apiospora fuzhouensis (HMAS 352718). (a) Leaves of host plant. (b,c) Upper and reverse view of colony after incubation for 7 days on PDA and 14 days. (d,e) Conidiomata sporulating on PDA. (f–i) Conidiogenous cells and conidia. (j,k) Conidia. Scale bars = 10 µm (f–k).
MycoBank: MB852820.
Etymology: Named after Fuzhou, Fujian Province, where the fungus was collected.
Holotype: China, Fujian Province, Fujian University of Agriculture and Forestry, (119°14′35.14″ E, 26°5′2.55″ N), from diseased leaves of bamboo. March 2023, Z.Y. Zhao (holotype HMAS352718; ex-type living culture CGMCC3.25649).
Description: Leaf spots irregular in shape, brown or tan in color. Conidiomata on agar are elevated, solitary or aggregated, spherical, black, Conidiophores hyaline to light brown, smooth, fusiform, subcylindrical, conidiophore cells were solitary or aggregated, hyaline rounded, 1.5–9.1 × 2.4–7.2 µm. Conidia were rounded or ellipsoidal, brownish, 11.3–19.3 × 8.7–19.5 µm, mean = 14.8 × 14.4 µm. No sexual forms were observed.
Culture characteristics: Colonies flattened on PDA; mycelium fluffy, black; calculated growth rate 1.2 cm/day.
Material examined: China, Fujian Province, Fujian University of Agriculture and Forestry, (119°14′35.14″ E, 26°5′2.55″ N), from diseased leaves of bamboo. March 2023, Z.Y. Zhao (paratype HMAS352719; ex-paratype living culture CGMCC3.25650).
Notes: Two strains were obtained from diseased leaves of bamboo and differed from each other with a high degree of statistical support (100% ML/1 PP, Figure 2), although overall analyses indicated that both isolates represented different strains of the same species. The nucleotide comparison of ITS sequences of A. garethjonesii (SICAUCC 22-0028) revealed 39 bp (39/542 bp, 7.2%) nucleotide differences. The nucleotide comparison of tub2 sequences of A. garethjonesii (SICAUCC 22-0028) revealed 20 bp (20/524 bp, 3.8%) nucleotide differences. Morphologically, the conidia of A. fuzhouensis were slightly smaller than those of A. garethjonesii (SICAUCC 22-0028). Therefore, the two strains are proposed as a new species.
4. Discussion
As interest in bamboo has intensified due to its wide range of beneficial environmental effects as well as agricultural, industrial, and even foodstuff uses, identification of pathogens, that can decrease quality and/or yield of the plant has also gained interest. Here, we have identified four new species of fungi from diseased bamboo leaves found in Fujian Province, China. Identification was conducted using morphological and molecular phylogenetic analyses, with the former, i.e., characterization of the conidiomata, conidiophores, and conidiogenous cells used as important lines of evidence supporting species identification [42] and the latter (molecular approaches) allowing for phylogenetic placement and confirmation of new species designations.
Two of new species were identified as belonging to the Bifusisporella genus. Previously, Silva et al. isolated an endophyte, Bifusisporella sorgh, from healthy sorghum leaves in Brazil [15], and another endophyte, B. sichuanensis, has been reported from leaves of Sichuan poplar [43]. Most Bifusisporella species have sickle-shaped ditype conidia and are commonly found in Poaceae. The newly described species in this report, B. fujianensis, grouped with B. sichuanensis, but was distinct from the latter in both morphology and multilocus sequence analyses, whereas B. bambooensis potentially represents a separate clade. Morphological differences between the species were evident, particularly concerning the conidia (Table 3).
Table 3.
The location, hosts or substrate, and main morphological characters of Bifusisporella.
| Species | Location | Host/Substrate | Conidiogenous Cells | Size of Conidiophore Cells (µm) |
Conidia | Size of Conidia (µm) |
References |
|---|---|---|---|---|---|---|---|
| Bifusisporella bambooensis sp. nov. | China | Bambusoideae sp. | Cylindrical | 7.2–21.0 × 4.2–6.4 | falcate or curved moon-shaped | 10.8–45.0 × 2.8–4.9 | In this study |
| Bifusisporella sorghi | Brazil | Sorghum bicolor | Cylindrical orclavate | 5.0–19.5 × 3.0–4.0 | falcate | Macroconidia 19.0–34.0 × 3.0–4.0 Microconidia 7.0–14.5 × 1.0–2.0 |
[15] |
| Bifusisporella fujianensis sp. nov. | China | Bambusoideae sp. | Cylindricalor rod-shaped | 8.9–14.3 × 5.8–8.1 | falcate or curved moon-shaped | 37.3–56.3 × 3.6–5.7 | In this study |
| Bifusisporella sichuanensis | China | Phyllostachys edulis | - | - | - | - | [43] |
The remaining two new species identified in this report were found to belong to the Apiospora genus. Crous and Groenewald synonymized Apiospora with Arthrinium [44]; however, with additional genetic data from the Arthrinium type species, A. caricicola, Apiospora and Arthrinium were separated into two distinct genera [19]. Biogeographically, most specimen of Arthrinium have been found in temperate and boreal zones, whereas those of Apiospora have been mainly collected in tropical and subtropical regions, with the latter genus displaying a relatively wider distribution area. Currently, therefore, based on the molecular phylogenetic analysis of multigene loci (ITS, LSU, and exon sequences of tef1-α and tub2), Arthrinium and Apiospora are considered to represent independent lineages within the Apiosporaceae [19], confirming that the overall genetic, morphological, and ecological differences between Apiospora and Arthrinium are sufficient to support the taxonomic separation of the two genera. Apiospora are characterized by round/lenticular conidia, which are mainly found in Poaceae. Based on morphology and molecular analyses, Apiospora fujianensis sp. nov. and Apiospora fuzhouensis sp. nov. were described as two new species within Apiospora.
As a neo-tropical region, fungal species diversity in Fujian and surrounding areas appears particularly robust [26,27]. However, thus far, only a few species of fungi have been found in bamboo leaves, with our understanding of the diversity of fungal parasitism on bamboo incomplete. This is likely due to bamboo being particularly hardy and resistant to many microorganisms combined with a lack of specimen and data support [45]. These factors necessitate the collection of diverse specimens [5], as well as exploring the reaction/defense by the plant. Here, we provide candidate fungi, with further genomic and physiological studies aimed towards understanding the nature of these fungi on bamboo warranted.
Acknowledgments
We would like to thank Sen Liu, Chengjie Xiong, Longbin Lin, Weibin Zhang, Jinhui Chen, Pengyu Lai, Zhiang Heng, Ziyi Wu, Ruiya Chen, Chenjie Yang, and Mengjia Zhu for their help with sample collection.
Author Contributions
Conceptualization, Z.Z., X.G. and J.Q.; methodology, Z.Z. and T.M.; software, H.P. and Y.L.; validation, Z.L., J.X. and X.C.; formal analysis, X.Z.; investigation, H.L.; resources, P.J.; data curation, Z.Z., T.M., N.O.K., H.P., Y.L., Z.L., J.X., X.C., X.Z., H.L., M.Y.J.-S., P.J. and S.H.; writing—original draft preparation, Z.Z., T.M. and M.Y.J.-S.; writing—review and editing, Z.Z., N.O.K. and J.Q.; visualization, S.H.; supervision, X.G. and J.Q.; project administration, X.G. and J.Q.; funding acquisition, J.W., X.G. and J.Q. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All newly generated sequences were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 19 March 2024). All new taxa were linked with MycoBank (https://www.mycobank.org/ (accessed on 13 March 2024)).
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was funded by the National Natural Science Foundation of China (No. 32270029, U1803232, 31670026), the National Key R & D Program of China (No. 2017YFE0122000), a Social Service Team Support Program Project (No. 11899170165), Science and Technology Innovation Special Fund (Nos. KFB23084, CXZX2019059S, CXZX2019060G) of Fujian Agriculture and Forestry University, a Fujian Provincial Major Science and Technology Project (No. 2022NZ029017), an Investigation and evaluation of biodiversity in the Jiulong River Basin (No. 082·23259-15), and Macrofungal and microbial resource investigation project in Longqishan Nature Reserve (No. SMLH2024(TP)-JL003#).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Manandhar R., Kim J.H., Kim J.T. Environmental, social and economic sustainability of bamboo and bamboo-based construction materials in buildings. J. Asian Archit. Build. Eng. 2019;18:49–59. doi: 10.1080/13467581.2019.1595629. [DOI] [Google Scholar]
- 2.Binfield L., Britton T.L., Dai C., Innes J. Evidence on the social, economic, and environmental impact of interventions that facilitate bamboo industry development for sustainable livelihoods: A systematic map protocol. Environ. Evid. 2022;11:33. doi: 10.1186/s13750-022-00286-8. [DOI] [Google Scholar]
- 3.Liu W.Y., Hui C.M., Wang F., Wang M., Liu G.L. Review of the Resources and Utilization of Bamboo in China. Bamboo Curr. Future Prospect. 2018;8:133–142. doi: 10.5772/intechopen.76485. [DOI] [Google Scholar]
- 4.Dlamini L.C., Fakudze S., Makombe G.G., Muse S., Zhu J. Bamboo as a valuable resource and its utilization in historical and modern-day China. BioResources. 2022;17:1926–1938. doi: 10.15376/biores.17.1.Dlamini. [DOI] [Google Scholar]
- 5.Jiang H.B., Phookamsak R., Hongsanan S., Bhat D.J., Mortimer P.E., Suwannarach N., Kakumyan P., Xu J. A review of bambusicolous Ascomycota in China with an emphasis on species richness in southwest China. Stud. Fungi. 2022;7:20. doi: 10.48130/SIF-2022-0020. [DOI] [Google Scholar]
- 6.Yu X.D., Zhang S.N., Liu J.K. Additions to Bambusicolous Fungi of Savoryellaceae from Southwest China. J. Fungi. 2023;9:571. doi: 10.3390/jof9050571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thongkantha S., Jeewon R., Dhanasekaran V., Lumyong S., McKenzie E., Hyde K. Molecular phylogeny of Magnaporthaceae (Sordariomycetes) with a new species, Ophioceras chiangdaoense from Dracaena loureiroi in Thailand. Fungal Divers. 2009;34:157–173. [Google Scholar]
- 8.Luo J., Zhang N. Magnaporthiopsis, a new genus in Magnaporthaceae (Ascomycota) Mycologia. 2013;105:1019–1029. doi: 10.3852/12-359. [DOI] [PubMed] [Google Scholar]
- 9.Klaubauf S., Tharreau D., Fournier E., Groenewald J.Z., Crous P.W., Vries R.P., Lebrun M. Resolving the polyphyletic nature of Pyricularia (Pyriculariaceae) Stud. Mycol. 2014;79:85–120. doi: 10.1016/j.simyco.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo J., Walsh E., Zhang N. Four new species in Magnaporthaceae from grass roots in New Jersey Pine Barrens. Mycologia. 2014;106:580–588. doi: 10.3852/13-306. [DOI] [PubMed] [Google Scholar]
- 11.Luo J., Walsh E., Zhang N. Toward monophyletic generic concepts in Magnaporthales: Species with Harpophora asexual states. Mycologia. 2015;107:641–646. doi: 10.3852/14-302. [DOI] [PubMed] [Google Scholar]
- 12.Liu J.K., Hyde K.D., Jones E.B.G., Ariyawansa H.A., Bhat D.J., Boonmee S., Maharachchikumbura S.S.N., McKenzie E.H.C., Phookamsak R., Phukhamsakda C., et al. Fungal diversity notes 1–110: Taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 2015;72:1–197. doi: 10.1007/s13225-015-0324-y. [DOI] [Google Scholar]
- 13.Crous P.W., Wingfield M., Guarro J., Hernández-Restrepo M., Sutton D., Acharya K., Barber P., Boekhout T., Dimitrov R., Dueñas M., et al. Fungal Planet description sheets: 320–370. Persoonia. 2015;34:167–266. doi: 10.3767/003158515X688433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernández-Restrepo M., Groenewald M., Elliott M.L., Canning G., McMillan V., Crous P.W. Take-all or nothing. Stud. Mycol. 2016;83:19–48. doi: 10.1016/j.simyco.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silva R.M.F., Oliveira R.J.V., Bezerra J.D.P., Bezerra J.L., Souza-Motta C.M., Silva G.A. Bifusisporella sorghi gen. et sp. nov. (Magnaporthaceae) to accommodate an endophytic fungus from Brazil. Mycol. Prog. 2019;18:847–854. doi: 10.1007/s11557-019-01494-2. [DOI] [Google Scholar]
- 16.Pintos Á., Alvarado P. New studies on Apiospora (Amphisphaeriales, Apiosporaceae): Epitypification of Sphaeria apiospora, proposal of Ap. marianiae sp. nov. and description of the asexual morph of Ap. sichuanensis. MycoKeys. 2022;92:63–78. doi: 10.3897/mycokeys.92.87593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao C.F., Senanayake I.C., Dong W., Thilini Chethana K.W., Tangtrakulwanich K., Zhang Y.X., Doilom M.K. Taxonomic and Phylogenetic Updates on Apiospora: Introducing Four New Species from Wurfbainia villosa and Grasses in China. J. Fungi. 2023;9:1087. doi: 10.3390/jof9111087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu R.Y., Li D.H., Zhang Z.X., Liu S.B., Liu X.Y., Wang Y.X., Zhao H., Liu X.Y., Zhang X.G., Xia J.W., et al. Morphological and phylogenetic analyses reveal two new species and a new record of Apiospora (Amphisphaeriales, Apiosporaceae) in China. MycoKeys. 2023;95:27–45. doi: 10.3897/mycokeys.95.96400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pintos Á., Alvarado P. Phylogenetic delimitation of Apiospora and Arthrinium. Fungal Syst. Evol. 2021;7:197–221. doi: 10.3114/fuse.2021.07.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon S.L., Cho M., Lee Y.M., Lee H., Kim C., Kim G.H., Kim J.J. Diversity of the Bambusicolous Fungus Apiospora in Korea: Discovery of New Apiospora Species. Mycobiology. 2022;50:302–316. doi: 10.1080/12298093.2022.2133808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyde K.D., Frohlich J., Taylor J.E. Fungi from palms. XXXVI. Reflections on unitunicate ascomycetes with apiospores. Sydowia. 1998;50:21–80. [Google Scholar]
- 22.Zhang J.Y., Chen M.L., Boonmee S., Wang Y.X., Lu Y.Z. Four New Endophytic Apiospora Species Isolated from Three Dicranopteris Species in Guizhou, China. Fungal Divers. 2023;9:1096. doi: 10.3390/jof9111096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian X.G., Karunarathna S.C., Mapook A., Promputtha I., Xu J.C., Bao D.F., Tibpromma S. One New Species and Two New Host Records of Apiospora from Bamboo and Maize in Northern Thailand with Thirteen New Combinations. Life. 2021;11:1071. doi: 10.3390/life11101071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su D., Zhang W.H., Sun R., Zhang Z.T., Lyu G. First Report of Botryosphaeria dothidea Causing Leaf Spot on Kadsura coccinea in China. Plant Dis. 2021;105:2714. doi: 10.1094/PDIS-01-21-0150-PDN. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z.X., Liu X.Y., Tao M.F., Liu X.Y., Xia J.W., Zhang X.G., Meng Z. Taxonomy, Phylogeny, Divergence Time Estimation, and Biogeography of the Family Pseudoplagiostomataceae (Ascomycota, Diaporthales) J. Fungi. 2023;9:82. doi: 10.3390/jof9010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu S., Zhu M., Keyhani N.O., Wu Z., Lv H., Heng Z., Chen R., Dang Y., Yang C., Chen J., et al. Three New Species of Russulaceae (Russulales, Basidiomycota) from Southern China. J. Fungi. 2024;10:70. doi: 10.3390/jof10010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mu T., Chen J., Zhao Z., Zhang W., Stephenson S.L., Yang C., Zhu M., Su H., Liu P., Guan X., et al. Morphological and phylogenetic analyzes reveal two new species of Melanconiella from Fujian Province, China. Front. Microbiol. 2023;14:1229705. doi: 10.3389/fmicb.2023.1229705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White T.J., Bruns T.D., Lee S.B., Taylor J.W. Amplification and direct sequencing offungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; New York, NY, USA: 1990. pp. 315–322. [DOI] [Google Scholar]
- 29.Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matheny P.B., Liu Y.J., Ammirati J.F., Hall B.D. Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales) Am. J. Bot. 2002;89:688–698. doi: 10.3732/ajb.89.4.688. [DOI] [PubMed] [Google Scholar]
- 31.Castlebury L., Rossman A., Sung G., Hyten A., Spatafora J. Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycol. Res. 2004;108:864–872. doi: 10.1017/S0953756204000607. [DOI] [PubMed] [Google Scholar]
- 32.Rehner S.A., Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98. doi: 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- 33.O’Donnell K., Kistler H.C., Cigelnik E., Ploetz R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He J., Han X., Luo Z.L., Xian L., Tang S.M., Luo H.M., Niu K.Y., Su X.J., Li S.H. Species diversity of Ganoderma (Ganodermataceae, Polyporales) with three new species and a key to Ganoderma in Yunnan Province, China. Front. Microbiol. 2022;13:1035434. doi: 10.3389/fmicb.2022.1035434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. doi: 10.1021/bk-1999-0734.ch008. [DOI] [Google Scholar]
- 39.Rani A.K., Hina S., Iqbal H., Shabbir Z., Izhar A., Usman M., Khalid A.N. Identification and taxonomic position of a new Pseudoomphalina species from Pakistan based on light, scanning electron microscopy, and molecular analysis. Microsc. Res. Tech. 2023;86:1144–1153. doi: 10.1002/jemt.24387. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen L.T., Schmidt H., von Haeseler A., Minh B. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minh B.Q., Nguyen M.A., von Haeseler A. Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hughes S.J. Conidiophores, conidia, and classification. Can. J. Bot. 2011;31:577–659. doi: 10.1139/b53-046. [DOI] [Google Scholar]
- 43.Zeng Q., Lv Y.C., Xu X.L., Deng Y., Wang F.H., Liu S.Y., Liu L.J., Yang C.J., Liu Y.G. Morpho-Molecular Characterization of Microfungi Associated with Phyllostachys (Poaceae) in Sichuan, China. J. Fungi. 2022;8:702. doi: 10.3390/jof8070702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crous P.W., Groenewald J.Z. A phylogenetic re-evaluation of Athrinium. IMA Fungus. 2013;4:133–154. doi: 10.5598/imafungus.2013.04.01.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alves-Silva G., Drechsler-Santos E.R., da Silveira R.M. Bambusicolous Fomitiporia revisited: Multilocus phylogeny reveals a clade of host-exclusive species. Mycologia. 2020;112:633–648. doi: 10.1080/00275514.2020.1741316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All newly generated sequences were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 19 March 2024). All new taxa were linked with MycoBank (https://www.mycobank.org/ (accessed on 13 March 2024)).