Summary
Male infertility is a major concern affecting reproductive health. Biallelic deleterious variants of most DNAH gene family members have been linked to male infertility, with intracytoplasmic sperm injection (ICSI) being an efficacious way to achieve offspring. However, the association between DNAH12 and male infertility is still limited. Here, we identified one homozygous variant and two compound heterozygous variants in DNAH12 from three infertile Chinese men. Semen analysis revealed severe asthenozoospermia, abnormal morphology, and structure of sperm flagella. Furthermore, the Dnah12 knock-out mouse revealed severe spermatogenesis failure and validated the same male infertility phenotype. Favorable fertility outcomes were achieved through ICSI in three human individuals and Dnah12 knock-out mice. Collectively, our study indicated that biallelic variants of DNAH12 can induce male infertility in both human beings and mice. Notably, evidence from DNAH12 enhanced that ICSI was an optimal intervention to achieve favorable fertility outcomes for infertile males with DNAH gene family variants.
Subject areas: Reproductive medicine, Genetics, Genomics, Human Genetics, Molecular biology
Graphical abstract
Highlights
-
•
DNAH12 deleterious variants were found in infertile Chinese males with asthenozoospermia
-
•
Dnah12-KO mice validated the male infertility phenotype observed in affected males
-
•
Infertile males caused by DNAH family gene variants can have favorable ICSI outcomes
Reproductive medicine; Genetics; Genomics; Human Genetics; Molecular biology
Introduction
Infertility has a global incidence of approximately 8–12%, and male factors can be responsible for up to 50% of infertility cases. It is a multifaceted clinical condition that can arise due to various factors that impede sperm production, maturation, secretion, motility, or fertilization.1,2 The causes of male infertility are numerous, and genetic and environmental factors may play a vital role.3 The advancements in genetic testing technologies and methods have recently identified a series of monogenic pathogenic factors contributing to male infertility. It has been reported that at least 2,000 genes are involved in the process of spermatogenesis.4,5,6 Various genes are responsible for performing distinct functions at different stages of spermatogenesis, including Deleted in Azoospermia (DAZ) gene family,7,8 cilia and flagella associated protein (CFAP) gene family,9,10 Dynein Axonemal Heavy Chain (DNAH) family,11,12 and so forth.
DNAH gene family plays a crucial role in the structure and function of cilia and flagella, which are mainly involved in cell motility and ATP hydrolysis. There are currently 13 identified members in the DNAH gene family, which encodes for axonemal dynein heavy chain proteins. Several members of this gene family have been identified to generate an inner dynein arm heavy chain in cilia and flagella, including DNAH1 (OMIM: 603332), DNAH2 (OMIM: 603333), DNAH7 (OMIM: 610061) and DNAH10 (OMIM: 605884). While a portion of members compose an outer dynein arm heavy chain, such as DNAH8 (OMIM: 603337), DNAH11 (OMIM: 603339), DNAH17 (OMIM: 610063),13,14,15 and so forth. The study of the DNAH gene family and its related proteins is crucial for understanding the molecular mechanisms underlying cilia and flagella function, as well as the pathogenesis of cilia/flagella-related disorders. Variations in the DNAH gene family have been associated with various genetic disorders, such as primary ciliary dyskinesia (PCD), characterized by defects in respiratory cilia and/or multiple morphological abnormalities of the flagella of sperm (MMAF).16,17 It is worth noting that DNAH1 is the first gene determined to be responsible for MMAF and male infertility in humans.11 Subsequently, some other genes in the DNAH gene family have been identified to be associated with male infertility in humans and mice, such as DNAH2, DNAH6 (OMIM: 603336), DNAH7, DNAH8, DNAH9 (OMIM: 603330), DNAH10, DNAH17 (OMIM: 610063), and so forth. with various phenotypes of male infertility.18,19,20,21,22,23,24 Intracytoplasmic sperm injection (ICSI) is a highly effective technique that has been proven to assist infertile couples in achieving successful pregnancy, particularly in cases of severe male factor infertility, including those with variants of DNAH gene family members. However, there remains a lack of robust evidence supporting the effect of ICSI on infertile cases led by variants in the rest of the DNAH gene family members.
Similar to previously reported genes, variants in the DNAH12 (OMIM: 603340) gene have also been described as a potential contributing factor in male infertility.25,26 However, there remains to be more strong evidence supported by the animal models and replication of clinical cases to validate the precise role of DNAH12 in spermatogenesis. Furthermore, no studies have hitherto elucidated an optimal intervention for these cases to achieve favorable fertility outcomes. In this study, we have identified biallelic deleterious variants of the DNAH12 gene in three unrelated male patients with infertility. The generation of the Danh12 gene knock-out animal model also confirmed the male infertility phenotype in mice. Furthermore, ICSI was performed on human individuals carrying DNAH12 gene variants and the Dnah12−/− mouse model, resulting in favorable fertility outcomes in all cases. Our findings bolstered the strong evidence that ICSI is an optimal intervention for achieving favorable fertility outcomes in cases carrying variants of another DNAH gene family member-DNAH12, which serves as a novel pathogenic gene of male infertility.
Results
Overview of DNAH gene family and clinical outcomes of cases carrying variants of this gene family members
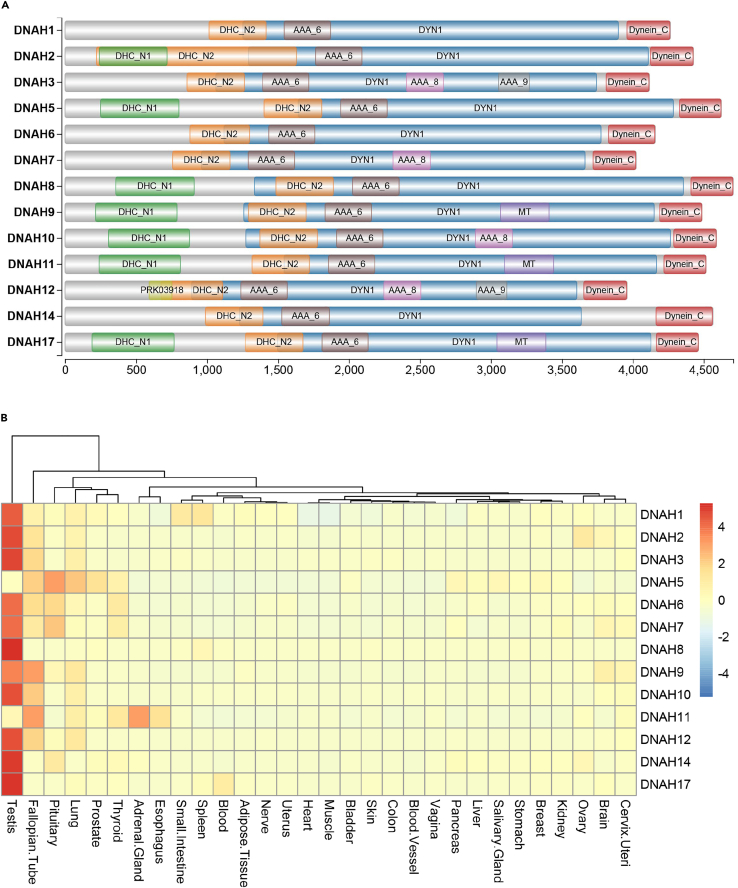
Currently, 13 members of DNAH gene family have been identified, with four same structural domains: DHC_N2, AAA_6, DYN1, and Dynein_C. Several members share an additional domain of DHC_N1, such as DNAH2, DNAH5, DNAH8, DNAH9, DNAH10, DNAH11 and DNAH17. The detailed information was gathered from the National Center for Biotechnology Information (NCBI, www.ncbi.nlm.nih.gov/) and summarized in Figure 1A. Additionally, the expression pattern of all 13 members was presented in Figure 1B based on the database of the Genotype-Tissue Expression Portal (GTEx, www.gtexportal.org/home/). It is worth noting that 11 genes have been found to be linked with human diseases, such as PCD and/or male infertility. These genes display significantly elevated expression levels in the testis, except for DNAH5 and DNAH11. And no male infertility phenotype has yet been observed in patients affected by these two genes. We further presented a comprehensive overview of fertility outcomes reported in prior studies concerning genetic variants within the DNAH gene family (as shown in Table 1). This compilation suggested that most infertile cases linked to DNAH gene family variants can achieve successful pregnancy outcomes through standard ICSI, including DNAH1, DNAH2, DNAH6, DNAH7, DNAH8, DNAH9, and DNAH10. However, it should be noted that patients with DNAH17 variants may require additional artificial oocyte activation (AOA) during the ICSI procedure.
Figure 1.
Overview of DNAH gene family members
(A) Schematic representation of the structural domains of DNAH family members.
(B) Expression pattern of DNAH family proteins. DHC_N1: dynein heavy chain, N-terminal region 1; DHC_N2: dynein heavy chain, N-terminal region 2; MT: microtubule-binding stalk of dynein motor; AAA_6: Hydrolytic ATP binding site of dynein motor region; AAA_8: ATP-binding dynein motor region D4; AAA_9: ATP-binding dynein motor region D5; DYN1:Dynein heavy chain 1; Dynein_C: Dynein heavy chain C-terminal domain.
Table 1.
Summary of clinical characteristics of DNAH family members
| Gene | Protein localisation | Phenotype (human) | Phenotype (KO mice) | Human |
KO mice |
Reference | ||
|---|---|---|---|---|---|---|---|---|
| Fertility strategy | Pregnancy | Fertility strategy | Pregnancy | |||||
| DNAH1 | IDA | Asthenozoospermia, MMAF, PCD | Reduced ciliary beating, Asthenozoospermia, |
ICSI | Yes | IVF | NA∗ | Ben Khelifa et al11, Neesen et al27, Wambergue et al28, Khan et al29, Guan et al30 |
| DNAH2 | IDA | Asthenozoospermia, MMAF, sperm head anomaly | Asthenozoospermia, MMAF, sperm head anomaly | ICSI | Yes | / | / | Li et al25, Hwang et al31, Li et al32 |
| DNAH5 | ODA | PCD | PCD | / | / | / | / | Hornef et al33, Nothe-Menchen et al34, Olbrich et al35 |
| DNAH6 | IDA | Asthenozoospermia, MMAF, sperm head anomaly | / | ICSI | Yes | / | / | Tu et al23, Li et al36 |
| DNAH7 | IDA | Asthenozoospermia, MMAF, PCD | / | ICSI | Yes | / | / | Gao et al18, Pereira et al37, Wei et al38 |
| DNAH8 | ODA | Asthenozoospermia, MMAF, | Asthenozoospermia, MMAF | ICSI | Yes | ICSI | Yes | Liu et al20, Ferreux et al39, Yang et al40 |
| DNAH9 | ODA | Asthenozoospermia, PCD | PCD | ICSI | Yes | / | / | Tang et al21, Loges et al41, Zheng et al42 |
| DNAH10 | IDA | Asthenozoospermia, MMAF, PCD | Asthenozoospermia, MMAF, PCD | ICSI | Yes | / | / | Tu et al22, Li et al43, Wang et al44 |
| DNAH11 | ODA | PCD | PCD, asthenozoospermia | / | / | / | / | Dougherty et al45, Lucas et al, 46 |
| DNAH14 | / | PCD | / | / | / | / | / | Guan et al.30 |
| DNAH17 | ODA | Asthenozoospermia, MMAF | Asthenozoospermia, MMAF | ICSI | No | / | / | Whitfield et al.24, Zhang et al47, Song et al48, Song et al49, Xue et al50 |
| ICSI+AOA | Yes | |||||||
KO: knock out; IVF: in vitro fertilization; ICSI: intracytoplasmic sperm injection; MMAF: multiple morphological abnormalities of the sperm flagella; PCD: primary ciliary dyskinesia; AOA: artificial oocyte activation; IDA: inner arm dynein; ODA: out arm dynein; NDD: neurodevelopmental disorders; NA: not available; ∗ The number of 8-cell stage embryos was reduced in Dnah1 KO mice, the pregnancy outcome was not available.
Out of the 13 members of the DNAH gene family, the association of DNAH3 and DNAH12 with human genetic diseases is still not well-established, which attracts our attention to investigate their roles in male infertility. Furthermore, we also aim to determine whether ICSI can overcome male infertility caused by these potential pathogenic gene variants.
Identification of biallelic DNAH12 variants in three unrelated infertile Chinese men
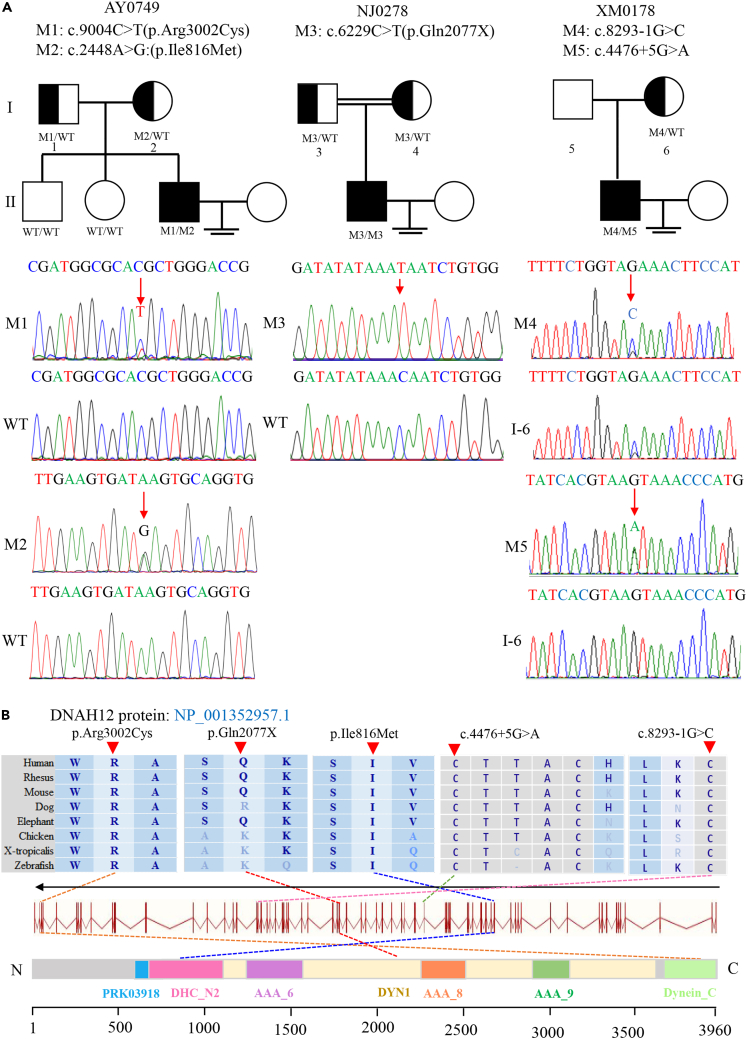
Biallelic variants of DNAH12 were identified in three unrelated Chinese men by WES in our study cohort, accounting for 0.2%(3/1532). Demographic details of three patients were recorded in Table S1. Proband NJ0278 carried a homozygous stop-gain variant (c.6229C>T[p.Gln2077X]), while proband AY0749 and XM0178 harbored compound heterozygous missense and splicing variants (c.9004C>T[p.Arg3002Cys]/c.2448A>G[p.Ile816Met]; c.8293-1G>C/c.4476 + 5G>A, respectively). These variants were validated and confirmed for segregation analysis through Sanger sequencing (Figure 2). No other potential variants of known genes inherited in a correct pattern (autosomal recessive or X-link) were discovered (Table S2).
Figure 2.
Identification of bi-allelic DNAH12 variants in three Chinese men with male infertility
(A) Pedigree analysis of three families affected by bi-allelic DNAH12 variants identified by WES. Black-filled squares indicate infertile men in these families. Sanger sequencing results are shownunder the pedigrees. The mutated positions are indicated by red arrows.
(B) Schematic representation of the domains of DNAH12 protein. Dotted lines with different colors indicate the positions of different DNAH12 variants identified in the present study. Sequence alignment shows the conservation of mutated residues among different species. M, mutation type; WT, wild type.
The homozygous stop-gain variant of DNAH12 identified in NJ0278 was rare in the public databases, with a variant frequency of less than 0.001. Furthermore, bioinformatics software predicts this novel variant as pathogenic and will lead to a truncated DNAH12 protein (Table 2). The two heterozygous missense variants (c.9004C>T and c.2448A>G) of DNAH12 identified in AY0749 were highly conserved across species, and various bioinformatics prediction tools suggest that they have damaging effects (Figure 2; Table 2). Biallelic variants of DNAH12 identified in XM0178 included a canonical splicing variant, which could result in the erroneous splicing of DNAH12-mRNA, and a non-canonical splicing variant predicted to be detrimental by the SPCards platform (www.genemed.tech/spcards/home/), a manually curated database and analytic platform for splicing variants in the human genome. Collectively, these findings indicated that all variants of DNAH12 identified in this study were potentially pathogenic.
Table 2.
Identified biallelic variants of DNAH12 and ICSI outcomes in three infertile Chinese men
| Subject | AY0749 | NJ0278 | XM0178 | ||||
|---|---|---|---|---|---|---|---|
| cDNA Mutation | c.9004C>T | c.2448A>G | c.6229C>T | c.8293-1G>C | c.4476 + 5G>A | ||
| Protein Alteration | p.Arg3002Cys | p.Ile816Met | p.Gln2077X | – | – | ||
| Mutation Type | Missense | Missense | Stop-gain | Splicing | Splicing | ||
| Affected Allele | Het | Het | Hom | Het | Het | ||
| ACMG classification | VUS | VUS | VUS | VUS | VUS | ||
| Allele Frequency in Population | |||||||
| 1KGP | 0 | 2.20E-03 | 0 | 0 | 0 | ||
| gnomAD | 1.28E-05 | 5.00E-04 | 2.71E-05 | 0 | 0 | ||
| gnomAD-EAS | 9.18E-05 | 6.30E-03 | 4.00E-04 | 0 | 0 | ||
| Function prediction | |||||||
| SIFT | B | D | NA | NA | NA | ||
| PolyPhen-2 | D | D | NA | NA | NA | ||
| Mutation Taster | D | B | D | D | NA | ||
| CADD | 29.4 | 24.6 | 38 | 25 | 23.6 | ||
| Splicing prediction | Score (threshold) | ||||||
| spliceAI | – | – | – | 0.9579(>0.1) | 0.8813(>0.1) | ||
| dbscSNV ADA | – | – | – | 1.0(>0.6) | 0.9998(>0.6) | ||
| dbscSNV RF | – | – | – | 0.9440(>0.6) | 0.9260(>0.6) | ||
| SPiCE | – | – | – | 1.0(>0.115) | 0.9999(>0.115) | ||
| ICSI outcomes | |||||||
| Male age (years) | 31 | 41 | 33 | ||||
| Female age (years) | 28 | 37 | 31 | ||||
| No. of ICSI cycles | 1 | 1 | 1 | ||||
| No. of oocytes retrieved | 17 | 14 | 16 | ||||
| No. of oocytes injected | 11 | 14 | 16 | ||||
| Fertilization rate (%) | 100(11/11) | 100(14/14) | 93.8(15/16) | ||||
| Cleavage rate (%) | 90.9(10/11) | 100(14/14) | 93.3(14/15) | ||||
| 8-Cell formation rate (%) | 63.6(7/11) | 50.0(7/14) | 42.9(6/14) | ||||
| Blastocyst formation rate (%) | 54.5(6/11) | 21.4(3/14) | 58.3(7/12) | ||||
| High quality blastocyst rate (%) | 36.4(4/11) | 21.4(3/14) | 33.3(4/12) | ||||
| No. of transfer cycles | 1 | 1 | – | ||||
| Number of embryos transferred per cycle | 1 | 1 | – | ||||
| Implantation rate (%) | 100 | 100 | – | ||||
| Clinical pregnancy rate per transfer cycle (%) | 100 | 100 | – | ||||
| Miscarriage rate (%) | 0 | 100 | – | ||||
| Delivery | 1 | 0 | – | ||||
RefSeq accession number of DNAH12 is ENST00000351747; Variants with CADD values greater than 20 are considered to be deleterious; Abbreviations: 1KGP, 1000 Genomes Project; gnomAD, the Genome Aggregation Database; VUS: variant of uncertain significance; NA, not available; B, benign; D, damaging; ICSI: intracytoplasmic sperm injection.
Clinical features and sperm phenotypes of the probands carrying DNAH12 variants
All three probands were diagnosed with primary male infertility. Proband NJ0278 was from a consanguineous family. Medical history-taking, physical examination, laboratory, and imaging testing revealed no abnormal findings. No typical PCD-related symptoms (such as wet cough, recurrent chest infections, perennial rhinosinusitis, otitis media with effusion, bronchiectasis, and hearing impairment) were described, and no signs of pulmonary infection and situs inversus were detected in the imaging testing. Routine analysis of semen samples demonstrated severe asthenozoospermia in all three probands, with normal sperm concentration and total count. Detailed information can be found in Table S3.
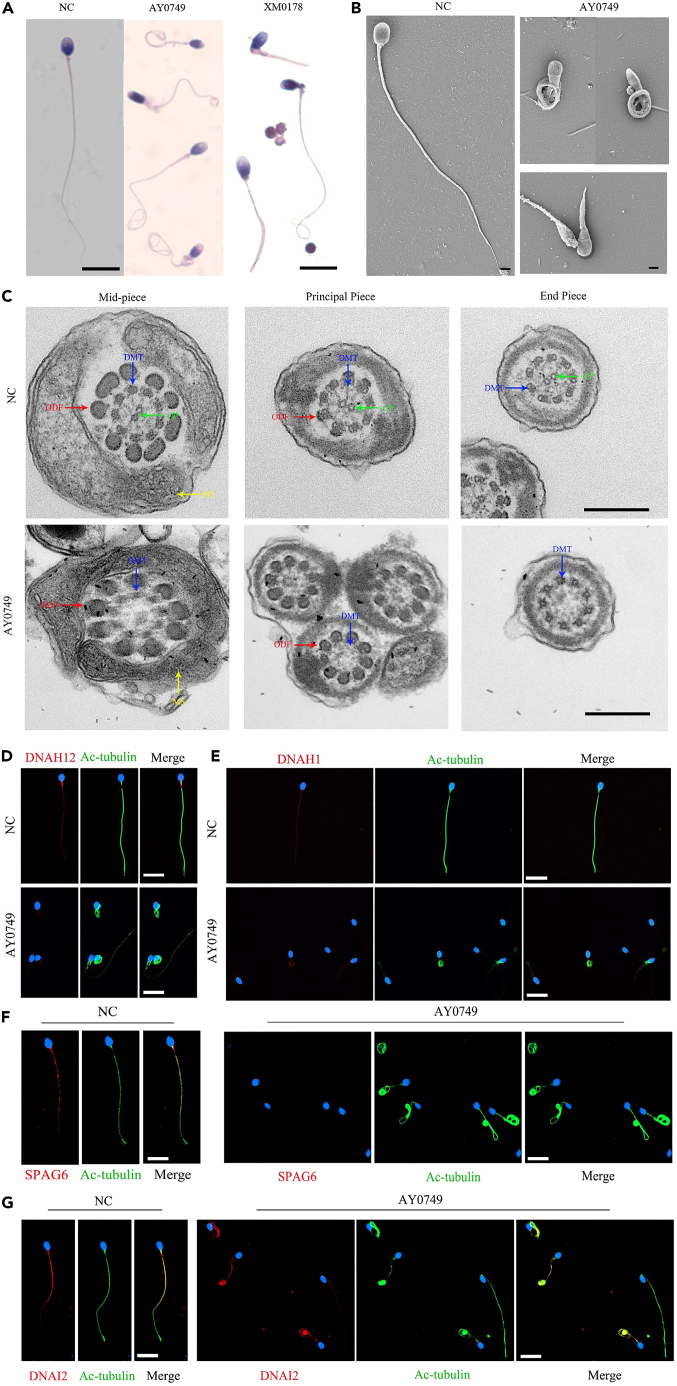
Spermatozoa from AY0749 and XM0178 were collected for further morphology and ultrastructure analysis. Morphology results indicated that most spermatozoa of proband AY0749 and XM0178 presented with typical MMAF phenotypes, further SEM examination confirmed the abnormal sperm phenotypes observed in proband AY0749, both indicating the multiple morphological abnormalities of the sperm flagella (Figures 3A and 3B). A detailed description of sperm flagella morphology was presented in Table S3. Subsequent TEM examination revealed a defective "9 + 2" flagella structure in most spermatozoa from proband AY0749, primarily characterized by the absence of central microtubules (Figure 3C).
Figure 3.
Sperm morphology, ultrastructural deficiency, and immunofluorescence staining in individuals carrying biallelic DNAH12 variants
(A and B) The morphology of spermatozoa from proband AY0749 and XM0178 under light microscopy and scanning electron microscopy (SEM). A: Scale bar, 10μm; B: Scale bar, 2μm.
(C) Abnormal sperm flagella ultrastructure was observed in proband AY0749 compared to normal spermatozoa by transmission electron microscopy (TEM), including the absence of central pair apparatus. Scale bar, 500nm.
(D) Spermatozoa from normal male control and AY0749 harboring bi-allelic DNAH12 variants were stained with anti-DNAN12 (red) and anti-Ac-tubulin (green) antibodies. A notable absence of the DNAH12 signal was revealed in the flagella of AY0749 spermatozoa. Scale bar, 10μm.
(E) DNAH1 signal (red) was present in both control and AY0749. Scale bar, 10μm. Scale bar, 10μm.
(F) SPAG6 signal (red) was present normally in the spermatozoa from control, while absent in proband AY0749. Scale bar, 10μm.
(G) DNAI2 signal (red) was present in both control and AY0749. Scale bar, 10μm. NC, normal control; CP, central pair apparatus (green arrows); DMT, peripheral microtubule doublet (blue arrows); ODF, outer dense fiber (red arrows); MS, mitochondrial sheath (yellow arrows).
Expression analysis of DNAH12 and some sperm flagella-associated genes in spermatozoa with DNAH12 gene defects
Firstly, we employed spermatozoa obtained from AY0749 and XM0178 to investigate the quantitative and localized expression of DNAH12 using RT-PCR and IF analysis. The RT-PCR experiment revealed a significant decrease in the quantitative expression of DNAH12-mRNA compared to the control group in proband AY0749 and XM0178 (Figure S1A). Furthermore, the immunofluorescence experiment indicated a notable absence of the DNAH12 signal in the flagella of spermatozoa from AY0749 (Figure 3D). These results suggested that these cases experienced defects of the localization and quantitative expression of DNAH12 due to biallelic variants of DNAH12. We further selected several markers representing different structures of sperm flagella, including DNAH1, DNAI2, and SAPG6 for IF analysis. Our results revealed that the expression levels of DNAH1 and DNAI2 were predominantly normal, whereas the expression of SAPG6 was mostly absent (Figures 3E–3G). Similar results were presented in Figure S1B, considering the protein expression levels of DNAI2 and SPAG6. Furthermore, combined with the observation of largely missing central microtubules in spermatozoa from AY0749, these findings potentially supported an association between biallelic variants of DNAH12 and central microtubule defects.
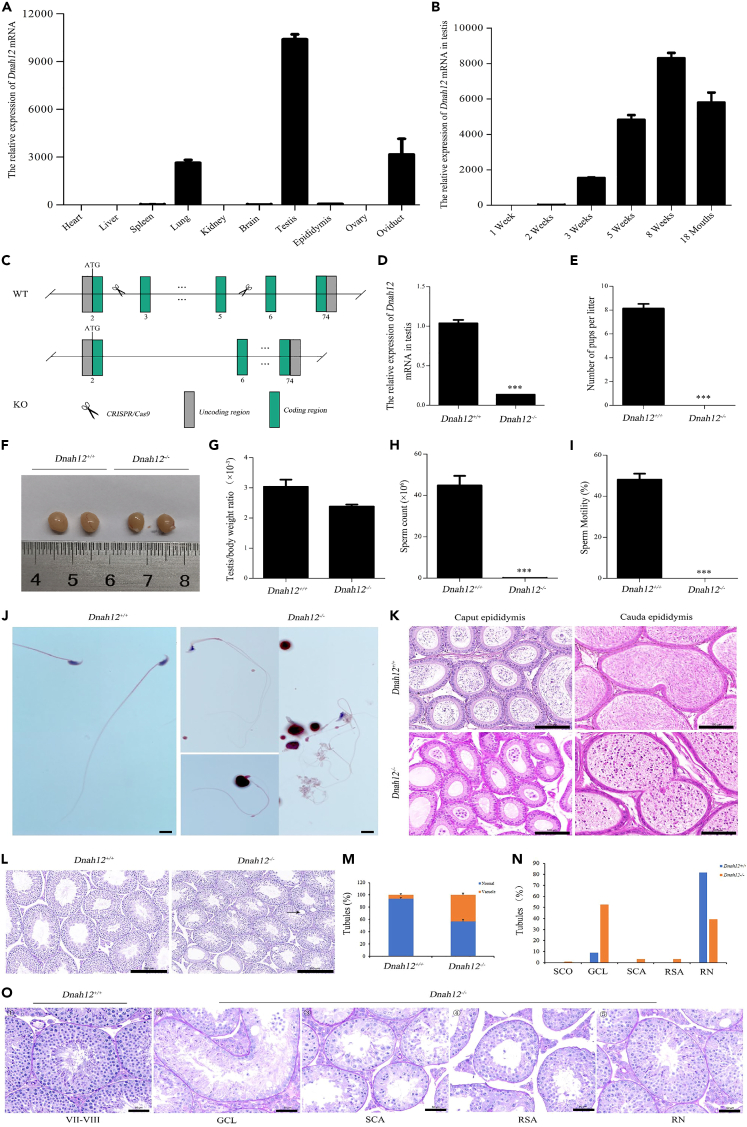
Phenotypical characterization of Dnah12−/− male mice
Firstly, we evaluated the mRNA expression levels of Dnah12 in various organs of wild-type (WT) adult mice. The results showed that dnah12 exhibited higher expression in the testis, oviduct, and lung (Figure 4A). Subsequently, we detected the Dnah12 mRNA expression levels in the testis at various ages, revealing a correlation with sexual maturation, with the peak at eight weeks of age (Figure 4B).
Figure 4.
Generation, reproductive, and pathogenic phenotypes of Dnah12−/− male mice
(A) The mRNA expression levels of Dnah12 in various organs of WT adult mice.
(B) Dnah12 mRNA expression levels in the testis correlate with sexual maturation, peaking at eight weeks of age.
(C) Dnah12−/− mice were generated by CRISPR-Cas9 technology through the targeted excision of three exons from exon 3 to exon 5 of the Dnah12 gene.
(D) RT-PCR analysis revealed a significant decrease of Dnah12 mRNA expression in the testis of Dnah12−/− male mice compared to WT. Statistics: Student’s t test, t = 15.80, ∗∗∗p < 0.001.
(E) Dnah12−/− male mice were completely infertile. Statistics: Student’s t test, t = 13.86, ∗∗∗p < 0.001.
(F) No difference was found in the testis size from WT and Dnah12−/− male mice.
(G) No significant difference was found in the testis/body weight ratio between WT and Dnah12−/− male mice. Statistics: Student’s t test, t = 2.00, p = 0.116.
(H)Dnah12−/− male mice demonstrated a significantly decreased sperm count compared to WT. Statistics: Student’s t test, t = 52.42, ∗∗∗p < 0.001.
(I) Dnah12−/− male mice presented sperm without motility. Statistics: Student’s t test, t = 13.47, ∗∗∗p < 0.001.
(J) The morphological assessment revealed severe abnormalities in the Dnah12−/− spermatozoa, particularly characterized by anomalous sperm heads and coiled flagella. Scale bar: 10μm.
(K) Compared with WT, a severely decreased number of spermatozoa in the epididymis of Dnah12−/− male mice, with numerous exfoliating cells. Scale bar: 100μm.
(L) Significant vacuolization was presented in the seminiferous tubules in the testis of Dnah12−/− male mice. Scale bar: 200μm.
(M) A significantly higher proportion of vacuolized seminiferous tubules in the testis from Dnah12−/− male mice. Statistics: Chi-square test, χ2 = 13.47, ∗∗∗p < 0.001.
(N) Quantifying seminiferous tubules at different spermatogenic stages in adult WT and Dnah12−/− mice. Spermatogenic stages were divided into five groups based on the presence and arrangement of germ cells under PAS staining of testicular sections. It showed an increase in the GCL, SCA, and RSA ratios in the testis from Dnah12−/− mice.
(O) Representative seminiferous tubules of PAS stained testicular sections of adult WT and Dnah12−/− mice. ① showed stage Ⅶ-Ⅷ seminiferous tubule of WT mice with the well-organized distribution of germ cells. ②, ③, ④respectively exhibited different types degenerated germ cells of Dnah12−/− mice, including GCL, SCA, and RSA, ⑤ exhibited RN testicular tubule from adult Dnah12−/− mice. SCO: Sertoli cell-only tubules; GCL: massive germ cell lost tubules; SCA: spermatocyte cells arrest tubules; RSA: round spermatids as most advanced spermatogenic cells in tubules; RN: relatively normal tubules; WT: wild type. Scale bar: 50μm.
To investigate the influence of DNAH12 variants on spermatogenesis and flagellogenesis, we performed CRISPR-Cas9 technology to generate Dnah12−/− mice, which was achieved through the targeted excision of three exons from exon 3 to exon 5 of Dnah12 gene (Figure 4C). Subsequent RT-PCR analysis revealed a significant decrease of Dnah12 mRNA expression in the testis of Dnah12−/− male mice compared to WT male mice (Figure 4D). Strikingly, our experiments demonstrated that Dnah12.
−/− male mice were completely infertile (Figure 4E). No significant differences were also discerned in the testis weights, sizes, and the testis/body weight ratio between WT and Dnah12−/− male mice. (Figures 4F and 4G).
To further elucidate the cause of male infertility in Dnah12−/− mice, we examined semen characteristics by analyzing sperm retrieved from the cauda epididymis of Dnah12−/− mice. In contrast to WT, Dnah12−/− mice demonstrated a significant decline of sperm concentration and a complete absence of motility (Figures 4H and 4I). The morphological assessment revealed severe spermatozoa abnormalities, particularly characterized by anomalous sperm heads and flagella (Figure 4J). Given the substantially reduced sperm count in the cauda epididymis of Dnah12−/− male mice, we conducted a histopathological analysis of the testes and epididymides in WT and Dnah12−/− male mice. The results indicated a severe decrease in sperm within the epididymis and testis of Dnah12−/− mice compared with WT. Specifically, we observed an almost absence of mature sperm in the cauda of the epididymis in Dnah12−/− male mice, along with significant vacuolization in the seminiferous tubules (Figures 4K–4M). Notably, it was shown that the testis of Dnah12−/− male mice exhibited widespread impairment of spermatogenesis across multiple tubules, with a significant increase in the number of massive germ cell lost tubules at various stages of spermatogenesis (Figures 4N and 4O).
Clinical outcomes of ICSI in humans and mice carrying biallelic DNAH12 variants
All three probands with biallelic DNAH12 variants accepted ICSI treatment. Notably, proband AY0749 and NJ0278 achieved successful clinical pregnancy outcomes. For proband AY0749, his wife underwent a single ICSI cycle, retrieving 11 mature oocytes and four high-quality blastocysts. After embryo transfer, successful live birth was achieved. Similarly, a positive clinical pregnancy outcome was observed in proband NJ0278 and his wife. Additionally, the partner of XM0178 retrieved 16 mature oocytes, obtained four high-quality blastocysts and was waiting for embryo transfer (Table 2). We also performed ICSI using spermatozoa from WT and Dnah12−/− male mice. A comparable successful blastocyst generation was achieved following ICSI in both Dnah12−/− and WT male mice (Figure S2). In conclusion, our data strongly substantiated the proposition that ICSI may serve as a promising therapeutic option for infertile cases carrying DNAH12 variants.
Discussion
This study first presented a comprehensive overview of clinical fertility outcomes reported concerning variants within the DNAH gene family. ICSI has been proven to be an optimal treatment for male infertility caused by variants of most members in this gene family. Our focus is directed toward two members, DNAH12 and DNAH3, that have not yet been well studied concerning human genetic disorders. We have effectively identified homozygous and compound heterozygous variants of the DNAH12 gene in three unrelated individuals with male infertility. The potential deleterious effects of these variants have been substantiated through rigorous bioinformatics analysis and in vivo experiments. Coupled with the phenotypical characterization of Dnah12−/− male mice, these findings strongly suggest that DNAH12 is likely to play a pivotal role in male fertility, and biallelic deleterious variants of DNAH12 may lead to severe sperm abnormalities and male infertility. Moreover, the evidence from DNAH12 in humans and mice indicated enhanced the conclusion that ICSI was an optimal intervention for achieving favorable fertility outcomes for male infertile cases carrying variants of the DNAH gene family.
DNAH12, a member of the DNAH gene family, is located at chromosome 3p14.3, and consists of 78 exons. It encodes a predicted 3960-amino acid protein. This gene exhibits high expression in ciliated cells of the testis, lung, brain, and fallopian tube tissues, according to NCBI, GTEx, and Human Protein Atlas (HPA) databases. The role of the DNAH gene family in sperm axoneme assembly and male fertility has been well-established.18,19,20,21,22,23,24,51 However, there is still limited comprehension regarding the significance of DNAH12 in male fertility and spermatogenesis defects. The first insufficient evidence of establishing a link between DNAH12 variants and male infertility was gathered by Oud et al., who identified two heterozygous variants (c.5393T>C[p.Phe1798Ser]/c.7438C>T[p.Pro2480Ser]) of DNAH12 in an Argentinian patient with MMAF. However, their status as compound heterozygous variants has not be confirmed.26 Subsequently, Li et al. investigated 67 unrelated Chinese patients diagnosed with MMAF and found one patient harboring compound heterozygous variants (c.6533C>G[p.Ser2178∗]/c.4166C>T[p.Pro1389Leu]) in DNAH12, which were confirmed to be inherited from his parents, respectively.25 These two cases offered solely reliable evidence for an association between DNAH12 variants and male infertility.
This present study identified three male infertility cases carrying damaging biallelic variants of DNAH12 without other known asthenoteratospermia-related gene. Proband NJ0278 carried one stop-gain variant of DNAH12 gene, resulting in a truncated protein and seriously affecting its function. Concerning proband AY0749, who harbored compound heterozygous variants in DNAH12, RT-PCR experiment confirmed a significant decrease in the expression levels of DNAH12 mRNA in the sperm, indicating the damaging effects of this compound heterozygous variants. The morphological examination and SEM both suggested the presence of MMAF phenotype in the sperm. Furthermore, TEM revealed the absence of central microtubules. Additionally, for proband XM0178 with MMAF, the compound heterozygous splicing variants were predicted to be deleterious by multiple bioinformatics tools. And importantly, the RT-PCR experiment also confirmed a significant decrease expression of DNAH12 mRNA. Therefore, based on these evidences, we proposed that biallelic deleterious variants of DNAH12 could be responsible for male infertility in humans, potentially associated with asthenoteratospermia characterized by MMAF. Unfortunately, we encountered difficulties in obtaining enough spermatozoa from the cases of NJ0278 and XM0178, which prevented us from conducting further morphological evaluation, SEM, TEM, and other functional examinations.
To provide additional evidence for the significant role of the DNAH12 gene in male reproductive function, we generated Dnah12−/− mice. As anticipated, male Dnah12−/− mice demonstrated complete infertility, mirroring the reproductive defects observed in humans with DNAH12 gene variants. Subsequent investigations further revealed a substantial decrease in sperm count in male Dnah12−/− mice. The exceedingly low number of sperms hindered our ability to definitively establish a correlation between DNAH12 and MMAF phenotype. Nevertheless, the utilization of male Dnah12−/− has successfully confirmed that defects in the DNAH12 gene can result in male infertility phenotype.
The application of ICSI contributes to favorable fertility outcomes in two cases. A similar outcome was also observed in male Dnah12−/− mice, where a successful generation of blastocysts was obtained. These findings strongly indicate that ICSI is an effective intervention for achieving positive clinical outcomes for infertile cases carrying DNAH12 defects. The summarized results for the DNAH gene family were described in Table 1. However, it should be noted that for individuals with variants of DNAH17, the combination of AOA and ICSI is necessary. The evidence from this study enhanced the efficacy of ICSI in achieving favorable pregnancy outcomes for male infertile cases caused by variants in the DNAH gene family.
In conclusion, our genetic and functional analyses in human subjects and mice model strongly suggest that bi-allelic variants of DANH12 are a crucial genetic cause of asthenoteratospermia-associated male infertility. Moreover, the evidence from DNAH12 in both humans and mice enhanced the conclusion that ICSI was an optimal intervention for achieving favorable fertility outcomes for male infertile cases carrying variants of the DNAH gene family. This study provides further perspectives to comprehend and provide guidance to patients afflicted by DNAH-related male infertility.
Limitations of the study
There are certain limitations associated with this study. Firstly, we only recruited 3 cases carrying biallelic variants of DNAH12. To validate the causality of DNAH12 gene variants and male infertility in humans, it is necessary to replicate the study using a larger sample size of male infertility cases. Secondly, we only obtained one patient’s semen sample for in-depth analysis. Furthermore, although Dnah12−/− mice showed male infertility phenotype, the precise mechanism responsible for the notable reduction in sperm count and impaired spermatogenesis requires further investigation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Anti-β actin mAb | Zsgb-bio | Cat#TA-09; RRID: AB_2636897 |
| Anti-SPAG6 antibody (rabbit) | Sigma Aldrich | Cat#HPA038440; RRID: AB_10671742 |
| Anti-DNAH1 antibody (rabbit) | Sigma Aldrich | Cat#HPA036806; RRID: AB_10670849 |
| Anti-DNAI2 antibody (rabbit) | Proteintech | Cat#17533-1-AP; RRID: AB_2096670 |
| Anti-DNAH12 antibody (rabbit) | Thermo Fisher Scientific | Cat#PA5-63952; RRID: AB_2640661 |
| Anti-Acetylated Tubulin (mouse) | Sigma Aldrich | Cat#T6793; RRID: AB_609894 |
| Donkey Anti-Mouse IgG(H/L), Alexa Fluor 488-AffiniPure | Jackson ImmunoResearch | Cat#715-545-150; RRID: AB_2340846 |
| Donkey Anti-Rabbit IgG(H/L), Alexa Fluor 594-AffiniPure | Jackson ImmunoResearch | Cat#711-585-152; RRID: AB_2340621 |
| Goat Anti-Rabbit IgG(H + L)(peroxidase/ HRP conjugated) |
Elabscience | Cat#E-AB-1003; RRID: AB_2921220 |
| Goat Anti-Mouse IgG(H + L)(peroxidase/ HRP conjugated) |
Elabscience | Cat# E-AB-1001; RRID: AB_2715613 |
| Biological samples | ||
| Frozen human peripheral blood sample | This study | N/A |
| Frozen human semen samples | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RIPA lysis buffer | Beyotime | Cat#P0013B |
| 4% paraformaldehyde | Biosharp | Cat#BL539A |
| Gluta fixative solution | Biosharp | Cat#BL911A |
| Osmium(VIII) oxide | Sigma Aldrich | Cat#1.24505 |
| TRIzol reagent | Invitrogen | Cat#AM9738 |
| PMSF Protease Inhibitor | Beyotime | Cat#P1046 |
| Hoechst 33342 | Thermo Fisher Scientific | Cat#62249 |
| Triton X-100 | Biosharp | Cat#BS084 |
| Antifade Mounting Medium | Beyotime | Cat#P0126 |
| Critical commercial assays | ||
| Sure Select XT Human All Exon Kit | Agilent | Cat#5191-4028 |
| PrimeScript RT reagent Kit | TaKaRa | Cat#RR037A |
| PAS stain Kit | Beyotime | Cat#C0142S |
| Hematoxylin and Eosin Staining Kit | Beyotime | Cat#C0105S |
| SYBR GreenⅠMaster | Roche | Cat#04887352001 |
| Experimental models: Organisms/strains | ||
| Mouse:C57BL/6JGpt | Jiangsu GemPharmatech Co., Ltd | N/A |
| Oligonucleotides | ||
| Primers used in this study are listed In Tables S3–S6 | This study | N/A |
| Software and algorithms | ||
| Sorting Intolerant From Tolerant | SIFT | RRID:SCR_012813 http://sift-dna.org/ |
| PolyPhen-2 | PolyPhen-2 | RRID:SCR_013189 http://genetics.bwh.harvard.edu/pph2/ |
| Mutation Taster | Mutation Taster | RRID:SCR_010777 http://www.mutationtaster.org/ |
| UCSC | UCSC | RRID:SCR_005780 http://genome.ucsc.edu/ |
| Uniprot | Uniprot | RRID:SCR_002380 https://www.uniprot.org/ |
| Genome Aggregation Database | GnomAD | RRID:SCR_014964 http://gnomad.broadinstitute.org/ |
| GeneCards | GeneCards | RRID:SCR_002773 http://genecards.org |
| NCBI | NCBI | RRID:SCR_006472 http://www.ncbi.nlm.nih.gov |
| Mouse Genome Informatics | Mouse Genome Informatics | RRID:SCR_006460 http://www.informatics.jax.org/ |
| Burrows-Wheeler Aligner | Burrows-Wheeler Aligner | RRID:SCR_010910 http://bio-bwa.sourceforge.net/ |
| Picard | Picard | RRID:SCR_006525 http://broadinstitute.github.io/picard/ |
| ANNOVAR | ANNOVAR | RRID:SCR_012821 http://www.openbioinformatics.org/annovar/ |
| GraphPad Prism | GraphPad Prism | RRID:SCR_002798 http://www.graphpad.com/ |
| ImageJ | Wayne Rasband, NIH | RRID:SCR_003070 http://imagej.net/ |
| SlideViewer | SlideViewer | RRID:SCR_024885 http://www.3dhistech.com/research/software-downloads/ |
| Others | ||
| Whole-exome sequencing data for the infertile males |
This study | https://doi.org/10.57760/sciencedb.08464 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Pro. Xiaojin He (xiaojinhe@sjtu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Data: Whole-exome sequencing data have been deposited at Science DataBank and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
Code: This paper does not report original code.
-
•
Additional information: Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Human participants and clinical samples
In this study, a research cohort study involving 1532 patients diagnosed with male infertility has been undertaken to investigate the genomic etiology from several reproductive centers in China since 2018. All participants had a normal somatic chromosome karyotype of 46, XY, and were free from Y chromosome microdeletions. Individuals with other risk factors associated with infertility, including cryptorchidism, radiotherapy, and chemotherapy, epididymitis, epididymal-orchitis, undescended testis, hypogonadism, varicocele as well as sexually transmitted infections were also excluded. All participants signed the written informed consent. Whole blood samples were collected from patients and their parents for sequencing and genetic analysis. Semen samples were collected and evaluated form all patients according to the fifth edition of WHO guideline. This study was approved by the ethics committees of the First Affiliated Hospital of Anhui Medical University, the First Affiliated Hospital of Nanjing Medical University, and Xiamen Maternity and Child Care Hospital (PJ2020-12-36).
Mice
The Dnah12 gene was edited using CRISPR/Cas9 technology to create the Dnah12−/− male mouse model. Specifically, the region containing exons 3 to 5 of the Dnah12-201 transcript (ENSMUST00000022433.11) was knocked out. Homozygous Dnah12 deficiency were identified using two PCR primers (see Table S4 for details). The animal ethics committee of Anhui Medical University approved this study (LLSC20230107). Adult male mice, aged 8–10 weeks were used for further analysis.
Method details
Genetic sequencing and bioinformatic analysis
Genomic DNA was extracted from the peripheral blood sample of each individual for whole exome sequencing (WES). The Sure Select XT Human All Exon Kit (Agilent) was used to amplify the exons, which were then sequenced on the Illumina HiSeq X-TEN platform. The generated clean reads were aligned to the human genome assembly GRCh38/hg38 using the Burrows-Wheeler Aligner (BWA) software. Duplicated reads were removed, and the quality of genomic variants was assessed using the Picard software. Pathogenic gene variants were annotated and analyzed using ANNOVAR software and further investigated in various databases and bioinformatics tools. The detailed procedures used to identify candidate pathogenic variants were described as follows21,43,52: (1)variant frequency below 1% in the 1KGP, GnomAD, and GnomAD-EAS databases; (2)loss-of-function variants, including start-gain, stop-loss, frameshift indel, and splice site variants; (3)deleterious missense variants predicted by SIFT, PolyPhen-2, Mutation Taster, and CADD; (4) pathogenic variants of testis-specific expressed genes(testis-specific expression was defined as an average expression value of≥3 reads per kilobase per million map reads in the human testis and 1-folds higher than the average expression value in other tissues based on the GTEx); (5) bi-allelic or X-linked variants following inherited model(Figure S3). Sanger sequencing was utilized to validate the candidate variants with the primers presented in Table S5.
Semen parameters and sperm morphological evaluation
The standard seminal parameters were evaluated in the laboratory by using fresh semen samples obtained through masturbation from individuals who abstained from sexual activity for 2–7 days, in accordance with the fifth edition of WHO guideline.53 The remaining fresh sperm samples were then washed twice with PBS buffer and fixed with 4% paraformaldehyde before being dehydrated and stained with H&E or Papanicolaou. Over 200 spermatozoa were counted and subjected to morphological analysis using Hematoxylin and Eosin (H&E) and Papanicolaou staining. Each participant provided semen samples for parameter assessment at least two occasions.
Scanning electron microscopy (sem) and transmission electron microscopy (tem) analyses
The spermatozoa were initially fixed in 2.5% glutaraldehyde buffer at 4°C overnight, respectively, followed by being washed with PBS buffer. For SEM, the fixed spermatozoa were subjected to dehydration using a gradient of ethanol solutions (30%, 50%, 70%, 80%, 90%, 100%, 100%), dried using a K850 CO2 critical point dryer (Quorum Technologies, Lewes, UK), and coated with a conductive material using a Cressington 108 Auto carbon coater (Cressington Scientific Instruments Ltd, Watford, UK). Finally, they were observed using a GeminiSEM 300 scanning electron microscope (ZEISS, Oberkochen, Germany) to obtain an ultrastructural view. To facilitate TEM analysis, the spermatozoa were fixed and rinsed four times with PBS buffer every hour before being post-fixed in a 1% osmic acid solution. Following a previously described protocol, the samples were further processed and subjected to examination using a Talos L120C G2 transmission electron microscope (Thermo Fisher Scientific, Waltham, MA, USA) for a detailed ultrastructural analysis of the spermatozoa.
Real-time PCR (RT-PCR) analysis
Total RNA was extracted from spermatozoa and testis tissues using TRIzol reagent (Invitrogen, Waltham, MA, USA) and reverse transcribed into cDNA using SuperScript III Reverse Transcriptase and oligo (dT) primers (TaKaRa Bio, Kusatsu, Shiga, Japan). GAPDH was used as internal control, and the primers used for RT-qPCR are listed in Table S6. Data analysis was performed using the 2−ΔΔCt method.
Western blot (WB) analysis
Spermatozoa or tissues from humans or mice were homogenized and lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime Biotechnology) containing phosphatase and protease inhibitors (Beyotime Biotechnology). The lysates were heated at 100°C for 10 min and stored at −20°C for subsequent WB assays. The following antibodies were used: rabbit polyclonal anti-SPAG6 (Sigma-Aldrich, HPA038440, 1:1000), rabbit polyclonal anti-DNAI2 (Proteintech, 17533-1-AP, 1:1000), mouse polyclonal anti-β-actin (ZSGB-Bio, TA-09, 1:5000), and HRP-conjugated secondary antibody (1:10000).
Immunofluorescence (IF) analysis
Fixed human spermatozoa were subjected to IF analysis as previously described, including smear preparation, antigen blocking, and antibody incubation. The following antibodies were used: rabbit polyclonal anti-SPAG6 (Sigma-Aldrich, HPA038440, 1:100), rabbit polyclonal anti-DNAH1 (Sigma-Aldrich, HPA036806, 1:100), rabbit polyclonal anti-DNAI2 (Proteintech, 17533-1-AP, 1:200), rabbit polyclonal anti-DNAH12(Thermo, PA5-63952,1:100), mouse polyclonal anti-ac-tubulin (Sigma, T6793, 1:500), and secondary anti-mouse Alexa Fluor 488 (Yeasen Biotechnology, 34106ES60, 1:800) and anti-rabbit Alexa Fluor 594 antibodies (Jackson ImmunoResearch, 111–585-003, 1:800). Hoechst nuclear stain (Thermo, 62249, 1:800) was used to visualize the nuclei.
Ovarian stimulation, ICSI, and embryo culture
Controlled ovarian stimulation was conducted using either a standard gonadotropin-releasing hormone antagonist or agonist protocol, depending on the ovarian reserve of the female partners. Follicular growth was monitored via serial ultrasonography. Ovulation was triggered by administering human chorionic gonadotropin (hCG) when either at least three follicles with a diameter greater than 17 mm or two follicles with a diameter greater than 18 mm were observed in the ovaries. Cumulus-oocyte complexes were retrieved 34–38 h after the hCG trigger through ultrasound-guided transvaginal aspiration. Following retrieval, cumulus-oocyte complexes were isolated from the follicular fluid and cultured in IVF fertilization medium (Cook, Sydney, Australia) at 37°C in a 6% CO2 environment for 4–6 h. Each oocyte was assessed for maturity under an inverted microscope, and only metaphase II stage oocytes were selected for subsequent ICSI insemination. The ICSI procedure adhered to the guidelines set by the European Society of Human Reproduction and Embryology, as described previously.54 After injection, the inseminated oocytes were transferred to cleavage culture medium (COOK, Australia Pty. Ltd) supplemented with 10% serum substitute supplement (SSS) (IrvineScientiffc, Santa Ana, USA). The culture was maintained at 37°C in a 6% CO2 environment for early embryo development. After 16–18 h, fertilization was assessed, and the fertilized oocytes were further cultured in the same medium for an additional 2 days. On day 3 post-ICSI, the embryos were transferred to blastocyst medium (COOK, Australia Pty. Ltd) and cultured for another 2-3 days to reach the blastocyst stage. On day 5 or day 6, the blastocysts were evaluated according to the Gardner grading criterion and cryopreserved by vitrification for future thawed embryo transfer (T-ET). Following embryo transfer, serum HCG levels were measured on the 14th day, and the presence of a fetal heartbeat at the 8th week confirmed a clinical pregnancy.
Mating test
To evaluate the fertility of Dnah12+/+ and Dnah12−/− mice, at least three adult male mice, aged 8–10 weeks, were utilized for a mating test that spanned six months. Female mice were examined for the presence of a vaginal plug and, if found, were transferred to a separate cage. In cases where no plug was detected or if the female mouse failed to conceive after two weeks, a replacement was utilized. Documentation of pregnancy outcomes and the count of offspring was maintained.
Periodic acid-schiff (PAS) staining
To assess the morphology of seminiferous tubules in the testes of Dnah12+/+ and Dnah12−/− male mice, paraffin sections were stained using the PAS staining assay kit (Beyotime Biotechnology, C0142S). The procedure included dewaxing via a xylene solution, hydration with gradient ethanol solutions (100%-90%-80%-70%-ddH2O), an oxidation process, Schiff staining, and hematoxylin staining. Images were captured using the Pannoramic MIDI Ⅱ digital slicing scanner (3D HISTECH), and tube morphology was assessed using the SlideViewer software (version 2.6.0).
Quantification and statistical analysis
Statistical analyses were conducted using SPSS.20.0(Chicago, USA). Data are presented as mean ± SD or percentage at least three independent experiments. Student’s t test, ANOVA, and chi-square test were used to assess statistical significance. Statistical significance was defined as p < 0.05. All tests and p values are described in the corresponding figure legends and/or results.
Acknowledgments
We would like to thank all the individuals and their family members for participating in this study. And this study was supported by the National Key R&D Program of China (2021YFC2700901), National Natural Science Foundation of China (81971441, 82101681 and 82071697), University Natural Foundation of Anhui Educational Committee (2022AH010072), Key Project of Natural Science Research of Anhui Educational Committee (2023AH053287), and Basic and Clinical Cooperation Research Promotion Plan of Anhui Medical University (2023xkjT036).
Author contributions
H.G., K.W., X.Y., and X.H were involved in the study design and execution, article writing, editing, and review. H.Y., K.L., Q.S., Z.W., and C.X. were involved in clinical sample collection and analysis. D.T., D.L., P.Z., and M.L. performed the mouse experiments. Y.G., H.W., and X.N. were involved in whole-exome sequencing and screening for the mutations. X.H., Y.C., X.Y., and Y.S. were involved in the project supervision, article editing, and review, and critical discussion. All authors have agreed with the submission.
Declaration of interests
The authors declare no competing interests.
Published: June 24, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110366.
Contributor Information
Yunxia Cao, Email: caoyunxia5927@ahmu.edu.cn.
Yanwei Sha, Email: shayanwei928@126.com.
Xiaoyu Yang, Email: yxy1921@163.com.
Xiaojin He, Email: xiaojinhe@sjtu.edu.cn.
Supplemental information
References
- 1.Agarwal A., Baskaran S., Parekh N., Cho C.L., Henkel R., Vij S., Arafa M., Panner Selvam M.K., Shah R. Male infertility. Lancet. 2021;397:319–333. doi: 10.1016/S0140-6736(20)32667-2. [DOI] [PubMed] [Google Scholar]
- 2.Minhas S., Bettocchi C., Boeri L., Capogrosso P., Carvalho J., Cilesiz N.C., Cocci A., Corona G., Dimitropoulos K., Gül M., et al. European Association of Urology Guidelines on Male Sexual and Reproductive Health: 2021 Update on Male Infertility. Eur. Urol. 2021;80:603–620. doi: 10.1016/j.eururo.2021.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Agarwal A., Mulgund A., Hamada A., Chyatte M.R. A unique view on male infertility around the globe. Reprod. Biol. Endocrinol. 2015;13:37. doi: 10.1186/s12958-015-0032-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krausz C., Riera-Escamilla A. Genetics of male infertility. Nat. Rev. Urol. 2018;15:369–384. doi: 10.1038/s41585-018-0003-3. [DOI] [PubMed] [Google Scholar]
- 5.Krausz C., Cioppi F., Riera-Escamilla A. Testing for genetic contributions to infertility: potential clinical impact. Expert Rev. Mol. Diagn. 2018;18:331–346. doi: 10.1080/14737159.2018.1453358. [DOI] [PubMed] [Google Scholar]
- 6.Jiao S.Y., Yang Y.H., Chen S.R. Molecular genetics of infertility: loss-of-function mutations in humans and corresponding knockout/mutated mice. Hum. Reprod. Update. 2021;27:154–189. doi: 10.1093/humupd/dmaa034. [DOI] [PubMed] [Google Scholar]
- 7.Fu X.F., Cheng S.F., Wang L.Q., Yin S., De Felici M., Shen W. DAZ Family Proteins, Key Players for Germ Cell Development. Int. J. Biol. Sci. 2015;11:1226–1235. doi: 10.7150/ijbs.11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y., Xiao C., Zhang S., Zhoucun A., Li X., Zhang S. Preliminary study of the relationship between DAZ gene copy deletions and spermatogenic impairment in Chinese men. Fertil. Steril. 2006;85:1061–1063. doi: 10.1016/j.fertnstert.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 9.Tang S., Wang X., Li W., Yang X., Li Z., Liu W., Li C., Zhu Z., Wang L., Wang J., et al. Biallelic Mutations in CFAP43 and CFAP44 Cause Male Infertility with Multiple Morphological Abnormalities of the Sperm Flagella. Am. J. Hum. Genet. 2017;100:854–864. doi: 10.1016/j.ajhg.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu H., Li W., He X., Liu C., Fang Y., Zhu F., Jiang H., Liu W., Song B., Wang X., et al. NovelCFAP43 andCFAP44 mutations cause male infertility with multiple morphological abnormalities of the sperm flagella (MMAF) Reprod. Biomed. Online. 2019;38:769–778. doi: 10.1016/j.rbmo.2018.12.037. [DOI] [PubMed] [Google Scholar]
- 11.Ben Khelifa M., Coutton C., Zouari R., Karaouzène T., Rendu J., Bidart M., Yassine S., Pierre V., Delaroche J., Hennebicq S., et al. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am. J. Hum. Genet. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu W., An M., Xu Y., Gao Q., Lu M., Li Y., Zhang L., Wang H., Xu Z. Mutational landscape of DNAH1 in Chinese patients with multiple morphological abnormalities of the sperm flagella: cohort study and literature review. J. Assist. Reprod. Genet. 2021;38:2031–2038. doi: 10.1007/s10815-021-02201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canty J.T., Tan R., Kusakci E., Fernandes J., Yildiz A. Structure and Mechanics of Dynein Motors. Annu. Rev. Biophys. 2021;50:549–574. doi: 10.1146/annurev-biophys-111020-101511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inaba K. Sperm flagella: comparative and phylogenetic perspectives of protein components. Mol. Hum. Reprod. 2011;17:524–538. doi: 10.1093/molehr/gar034. [DOI] [PubMed] [Google Scholar]
- 15.Satir P., Christensen S.T. Overview of structure and function of mammalian cilia. Basic Clin. Androl. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- 16.Nsota Mbango J.F., Coutton C., Arnoult C., Ray P.F., Touré A. Genetic causes of male infertility: snapshot on morphological abnormalities of the sperm flagellum. Basic Clin Androl. 2019;29:2. doi: 10.1186/s12610-019-0083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sironen A., Shoemark A., Patel M., Loebinger M.R., Mitchison H.M. Sperm defects in primary ciliary dyskinesia and related causes of male infertility. Cell. Mol. Life Sci. 2020;77:2029–2048. doi: 10.1007/s00018-019-03389-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Y., Liu L., Shen Q., Fu F., Xu C., Geng H., Lv M., Li K., Tang D., Song B., et al. Loss of function mutation in DNAH7 induces male infertility associated with abnormalities of the sperm flagella and mitochondria in human. Clin. Genet. 2022;102:130–135. doi: 10.1111/cge.14146. [DOI] [PubMed] [Google Scholar]
- 19.Gao Y., Tian S., Sha Y., Zha X., Cheng H., Wang A., Liu C., Lv M., Ni X., Li Q., et al. Novel bi-allelic variants in DNAH2 cause severe asthenoteratozoospermia with multiple morphological abnormalities of the flagella. Reprod. Biomed. Online. 2021;42:963–972. doi: 10.1016/j.rbmo.2021.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Liu C., Miyata H., Gao Y., Sha Y., Tang S., Xu Z., Whitfield M., Patrat C., Wu H., Dulioust E., et al. Bi-allelic DNAH8 Variants Lead to Multiple Morphological Abnormalities of the Sperm Flagella and Primary Male Infertility. Am. J. Hum. Genet. 2020;107:330–341. doi: 10.1016/j.ajhg.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang D., Sha Y., Gao Y., Zhang J., Cheng H., Zhang J., Ni X., Wang C., Xu C., Geng H., et al. Novel variants in DNAH9 lead to nonsyndromic severe asthenozoospermia. Reprod. Biol. Endocrinol. 2021;19:27. doi: 10.1186/s12958-021-00709-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tu C., Cong J., Zhang Q., He X., Zheng R., Yang X., Gao Y., Wu H., Lv M., Gu Y., et al. Bi-allelic mutations of DNAH10 cause primary male infertility with asthenoteratozoospermia in humans and mice. Am. J. Hum. Genet. 2021;108:1466–1477. doi: 10.1016/j.ajhg.2021.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tu C., Nie H., Meng L., Yuan S., He W., Luo A., Li H., Li W., Du J., Lu G., et al. Identification of DNAH6 mutations in infertile men with multiple morphological abnormalities of the sperm flagella. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-52436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitfield M., Thomas L., Bequignon E., Schmitt A., Stouvenel L., Montantin G., Tissier S., Duquesnoy P., Copin B., Chantot S., et al. Mutations in DNAH17, Encoding a Sperm-Specific Axonemal Outer Dynein Arm Heavy Chain, Cause Isolated Male Infertility Due to Asthenozoospermia. Am. J. Hum. Genet. 2019;105:198–212. doi: 10.1016/j.ajhg.2019.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Wang Y., Wen Y., Zhang T., Wang X., Jiang C., Zheng R., Zhou F., Chen D., Yang Y., Shen Y. Whole-exome sequencing of a cohort of infertile men reveals novel causative genes in teratozoospermia that are chiefly related to sperm head defects. Hum. Reprod. 2021;37:152–177. doi: 10.1093/humrep/deab229. [DOI] [PubMed] [Google Scholar]
- 26.Oud M.S., Houston B.J., Volozonoka L., Mastrorosa F.K., Holt G.S., Alobaidi B.K.S., deVries P.F., Astuti G., Ramos L., McLachlan R.I., et al. Exome sequencing reveals variants in known and novel candidate genes for severe sperm motility disorders. Hum. Reprod. 2021;36:2597–2611. doi: 10.1093/humrep/deab099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neesen J., Kirschner R., Ochs M., Schmiedl A., Habermann B., Mueller C., Holstein A.F., Nuesslein T., Adham I., Engel W. Disruption of an inner arm dynein heavy chain gene results in asthenozoospermia and reduced ciliary beat frequency. Hum. Mol. Genet. 2001;10:1117–1128. doi: 10.1093/hmg/10.11.1117. [DOI] [PubMed] [Google Scholar]
- 28.Wambergue C., Zouari R., Fourati Ben Mustapha S., Martinez G., Devillard F., Hennebicq S., Satre V., Brouillet S., Halouani L., Marrakchi O., et al. Patients with multiple morphological abnormalities of the sperm flagella due to DNAH1 mutations have a good prognosis following intracytoplasmic sperm injection. Hum. Reprod. 2016;31:1164–1172. doi: 10.1093/humrep/dew083. [DOI] [PubMed] [Google Scholar]
- 29.Khan R., Zaman Q., Chen J., Khan M., Ma A., Zhou J., Zhang B., Ali A., Naeem M., Zubair M., et al. Novel Loss-of-Function Mutations in DNAH1 Displayed Different Phenotypic Spectrum in Humans and Mice. Front. Endocrinol. 2021;12 doi: 10.3389/fendo.2021.765639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan Y., Yang H., Yao X., Xu H., Liu H., Tang X., Hao C., Zhang X., Zhao S., Ge W., Ni X. Clinical and Genetic Spectrum of Children With Primary Ciliary Dyskinesia in China. Chest. 2021;159:1768–1781. doi: 10.1016/j.chest.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang J.Y., Nawaz S., Choi J., Wang H., Hussain S., Nawaz M., Lopez-Giraldez F., Jeong K., Dong W., Oh J.N., et al. Genetic Defects in DNAH2 Underlie Male Infertility With Multiple Morphological Abnormalities of the Sperm Flagella in Humans and Mice. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.662903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Sha Y., Wang X., Ding L., Liu W., Ji Z., Mei L., Huang X., Lin S., Kong S., et al. DNAH2 is a novel candidate gene associated with multiple morphological abnormalities of the sperm flagella. Clin. Genet. 2019;95:590–600. doi: 10.1111/cge.13525. [DOI] [PubMed] [Google Scholar]
- 33.Hornef N., Olbrich H., Horvath J., Zariwala M.A., Fliegauf M., Loges N.T., Wildhaber J., Noone P.G., Kennedy M., Antonarakis S.E., et al. DNAH5 mutations are a common cause of primary ciliary dyskinesia with outer dynein arm defects. Am. J. Respir. Crit. Care Med. 2006;174:120–126. doi: 10.1164/rccm.200601-084OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nothe-Menchen T., Wallmeier J., Pennekamp P., Hoben I.M., Olbrich H., Loges N.T., Raidt J., Dougherty G.W., Hjeij R., Dworniczak B., et al. Randomization of Left-right Asymmetry and Congenital Heart Defects: The Role of DNAH5 in Humans and Mice. Circ Genom Precis Med. 2019;12 doi: 10.1161/CIRCGEN.119.002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olbrich H., Häffner K., Kispert A., Völkel A., Volz A., Sasmaz G., Reinhardt R., Hennig S., Lehrach H., Konietzko N., et al. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat. Genet. 2002;30:143–144. doi: 10.1038/ng817. [DOI] [PubMed] [Google Scholar]
- 36.Li L., Sha Y.W., Xu X., Mei L.B., Qiu P.P., Ji Z.Y., Lin S.B., Su Z.Y., Wang C., Yin C., Li P. DNAH6 is a novel candidate gene associated with sperm head anomaly. Andrologia. 2018;50 doi: 10.1111/and.12953. [DOI] [PubMed] [Google Scholar]
- 37.Pereira R., Barbosa T., Gales L., Oliveira E., Santos R., Oliveira J., Sousa M. Clinical and Genetic Analysis of Children with Kartagener Syndrome. Cells. 2019;8 doi: 10.3390/cells8080900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei X., Sha Y., Wei Z., Zhu X., He F., Zhang X., Liu W., Wang Y., Lu Z. Bi-allelic mutations in DNAH7 cause asthenozoospermia by impairing the integrality of axoneme structure. Acta Biochim. Biophys. Sin. 2021;53:1300–1309. doi: 10.1093/abbs/gmab113. [DOI] [PubMed] [Google Scholar]
- 39.Ferreux L., Bourdon M., Chargui A., Schmitt A., Stouvenel L., Lorès P., Ray P., Lousqui J., Pocate-Cheriet K., Santulli P., et al. Genetic diagnosis, sperm phenotype and ICSI outcome in case of severe asthenozoospermia with multiple morphological abnormalities of the flagellum. Hum. Reprod. 2021;36:2848–2860. doi: 10.1093/humrep/deab200. [DOI] [PubMed] [Google Scholar]
- 40.Yang Y., Jiang C., Zhang X., Liu X., Li J., Qiao X., Liu H., Shen Y. Loss-of-function mutation in DNAH8 induces asthenoteratospermia associated with multiple morphological abnormalities of the sperm flagella. Clin. Genet. 2020;98:396–401. doi: 10.1111/cge.13815. [DOI] [PubMed] [Google Scholar]
- 41.Loges N.T., Antony D., Maver A., Deardorff M.A., Güleç E.Y., Gezdirici A., Nöthe-Menchen T., Höben I.M., Jelten L., Frank D., et al. Recessive DNAH9 Loss-of-Function Mutations Cause Laterality Defects and Subtle Respiratory Ciliary-Beating Defects. Am. J. Hum. Genet. 2018;103:995–1008. doi: 10.1016/j.ajhg.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng R., Yang W., Wen Y., Xie L., Shi F., Lu D., Luo J., Li Y., Zhang R., Chen T., et al. Dnah9 mutant mice and organoid models recapitulate the clinical features of patients with PCD and provide an excellent platform for drug screening. Cell Death Dis. 2022;13:559. doi: 10.1038/s41419-022-05010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li K., Wang G., Lv M., Wang J., Gao Y., Tang F., Xu C., Yang W., Yu H., Shao Z., et al. Bi-allelic variants in DNAH10 cause asthenoteratozoospermia and male infertility. J. Assist. Reprod. Genet. 2022;39:251–259. doi: 10.1007/s10815-021-02306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang R., Yang D., Tu C., Lei C., Ding S., Guo T., Wang L., Liu Y., Lu C., Yang B., et al. Dynein axonemal heavy chain 10 deficiency causes primary ciliary dyskinesia in humans and mice. Front. Med. 2023;17:957–971. doi: 10.1007/s11684-023-0988-8. [DOI] [PubMed] [Google Scholar]
- 45.Dougherty G.W., Loges N.T., Klinkenbusch J.A., Olbrich H., Pennekamp P., Menchen T., Raidt J., Wallmeier J., Werner C., Westermann C., et al. DNAH11 Localization in the Proximal Region of Respiratory Cilia Defines Distinct Outer Dynein Arm Complexes. Am. J. Respir. Cell Mol. Biol. 2016;55:213–224. doi: 10.1165/rcmb.2015-0353OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lucas J.S., Adam E.C., Goggin P.M., Jackson C.L., Powles-Glover N., Patel S.H., Humphreys J., Fray M.D., Falconnet E., Blouin J.L., et al. Static respiratory cilia associated with mutations in Dnahc11/DNAH11: a mouse model of PCD. Hum. Mutat. 2012;33:495–503. doi: 10.1002/humu.22001. [DOI] [PubMed] [Google Scholar]
- 47.Zhang B., Khan I., Liu C., Ma A., Khan A., Zhang Y., Zhang H., Kakakhel M.B.S., Zhou J., Zhang W., et al. Novel loss-of-function variants in DNAH17 cause multiple morphological abnormalities of the sperm flagella in humans and mice. Clin. Genet. 2021;99:176–186. doi: 10.1111/cge.13866. [DOI] [PubMed] [Google Scholar]
- 48.Song B., Liu C., Gao Y., Marley J.L., Li W., Ni X., Liu W., Chen Y., Wang J., Wang C., et al. Novel compound heterozygous variants in dynein axonemal heavy chain 17 cause asthenoteratospermia with sperm flagellar defects. J Genet Genomics. 2020;47:713–717. doi: 10.1016/j.jgg.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 49.Song B., Yang T., Shen Q., Liu Y., Wang C., Li G., Gao Y., Cao Y., He X. Novel mutations in DNAH17 cause sperm flagellum defects and their influence on ICSI outcome. J. Assist. Reprod. Genet. 2023;40:2485–2492. doi: 10.1007/s10815-023-02897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xue Y., Cheng X., Xiong Y., Li K. Gene mutations associated with fertilization failure after in vitro fertilization/intracytoplasmic sperm injection. Front. Endocrinol. 2022;13 doi: 10.3389/fendo.2022.1086883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Touré A., Martinez G., Kherraf Z.E., Cazin C., Beurois J., Arnoult C., Ray P.F., Coutton C. The genetic architecture of morphological abnormalities of the sperm tail. Hum. Genet. 2021;140:21–42. doi: 10.1007/s00439-020-02113-x. [DOI] [PubMed] [Google Scholar]
- 52.Tan C., Meng L., Lv M., He X., Sha Y., Tang D., Tan Y., Hu T., He W., Tu C., et al. Bi-allelic variants in DNHD1 cause flagellar axoneme defects and asthenoteratozoospermia in humans and mice. Am. J. Hum. Genet. 2022;109:157–171. doi: 10.1016/j.ajhg.2021.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooper T.G., Noonan E., von Eckardstein S., Auger J., Baker H.W.G., Behre H.M., Haugen T.B., Kruger T., Wang C., Mbizvo M.T., Vogelsong K.M. World Health Organization reference values for human semen characteristics. Hum. Reprod. Update. 2010;16:231–245. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- 54.Van Landuyt L., De Vos A., Joris H., Verheyen G., Devroey P., Van Steirteghem A. Blastocyst formation in in vitro fertilization versus intracytoplasmic sperm injection cycles: influence of the fertilization procedure. Fertil. Steril. 2005;83:1397–1403. doi: 10.1016/j.fertnstert.2004.10.054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data: Whole-exome sequencing data have been deposited at Science DataBank and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
Code: This paper does not report original code.
-
•
Additional information: Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.