Abstract
A cDNA corresponding to the coding region of VP1, the putative RNA-dependent RNA polymerase, of infectious bursal disease virus (IBDV) was cloned and inserted into the genome of a vaccinia virus inducible expression vector. The molecular mass and antigenic reactivity of VP1 expressed in mammalian cells are identical to those of its counterpart expressed in IBDV-infected cells. The results presented here demonstrate that VP1 is efficiently incorporated into IBDV virus-like particles (VLPs) produced in mammalian cells coexpressing the IBDV polyprotein and VP1. Incorporation of VP1 into VLPs requires neither the presence of IBDV RNAs nor that of the nonstructural polypeptide VP5. Immunofluorescence, confocal laser scanning microscopy, and immunoprecipitation analyses conclusively showed that VP1 forms complexes with the structural polypeptide VP3. Formation of VP1-VP3 complexes is likely to be a key step for the morphogenesis of IBDV particles.
Infectious bursal disease virus (IBDV), a member of the Birnaviridae family (3), infects young chickens, causing a disease characterized by the destruction of the bursa of Fabricious that inflicts major losses on the poultry industry worldwide (for reviews see references 25, 29, and 34).
The genome of IBDV is formed by two segments of double-stranded RNA (dsRNA) of 3.2 kb (segment A) and 2.8 kb (segment B) (16). Segment A contains two partially overlapping open reading frames (ORFs). The first one encodes a nonstructural polypeptide of 17 kDa, VP5, which is dispensable for replication in vitro but important for virus-induced pathogenicity (26, 28), and the second one encodes a 109-kDa polyprotein that is autoproteolytically cleaved, rendering three polypeptides, VPX, VP3, and VP4. VPX is further processed to produce a polypeptide known as VP2 (23). VPX, VP2, and VP3 form the virus capsid while VP4 appears to be responsible for the proteolytic maturation of the polyprotein (6, 16, 18). Segment B encodes VP1, a 95-kDa protein that shares a number of primary sequence features with RNA polymerases from diverse origins (2). Although direct evidence demonstrating its RNA polymerase activity is lacking, VP1 is considered to be the RNA-dependent RNA polymerase (RdRp) responsible for the replication of the genome and the synthesis of mRNAs.
IBDV particles are nonenveloped icosahedrons with a diameter of 60 to 70 nm (11, 15, 31). Cryoelectron microscopy and image processing analysis showed that the capsid is formed by a single shell with a thickness of approximately 9 nm with a T=13 symmetry (1). It has been suggested that the external surface might be built of trimeric subunits formed by VP2 and that the inner surface might be built of trimeric subunits formed by VP3. The positively charged C terminus of the latter might interact with the genomic dsRNA molecules (1, 16). VP1 is the third, less abundant, structural polypeptide. In virus particles, VP1 is found as a “free” protein as well as covalently linked to both ends of the dsRNA genome segments (17, 22, 24).
Despite recent advances arising from the application of new approaches, such as the use of reverse genetics for the generation of IBDV mutants (27, 28), many aspects of IBDV’s molecular biology remain poorly understood.
The use of heterologous expression systems has been instrumental for the understanding of morphogenesis and structure of viruses of different families, and specifically the members of the Reoviridae, the larger family of dsRNA viruses (reviewed in reference 38).
It has recently been demonstrated that expression of the IBDV polyprotein in either mammalian or insect cells by using recombinant viral vectors leads to formation of virus-like particles (VLPs) with a size and morphology seemingly identical to those of IBDV virions (8, 39). Our aim was to take advantage of the vaccinia virus (VV) expression system to gain information about the morphogenesis of IBDV particles as well as to generate new tools to analyze both the structure of the virus particle and the function of virus-encoded proteins.
In this report, we describe the molecular cloning of the region of the IBDV genome coding for VP1 and the generation and characterization of a recombinant vaccinia virus (rVV) inducibly expressing this protein. The use of this rVV along with another one expressing the IBDV polyprotein (8) has allowed us to demonstrate that VP1 is efficiently incorporated into newly formed VLPs in the absence of the IBDV genome. Immunofluorescence (IF), confocal laser scanning microscopy (CLSM), and immunoprecipitation (IP) analyses conclusively showed that VP1 and VP3 form stable complexes which are likely to play a key role during morphogenesis of IBDV particles.
MATERIALS AND METHODS
Cells and viruses.
The IBDV Soroa strain, a pathogenic serotype I virus isolated in Cuba, was propagated in chicken embryo fibroblasts (CEF). The rVVs VT7/POLY, VT7/VP3, and VT7/VP2 have been previously described (7, 8). rVVs were propagated and titrated in African green monkey kidney epithelial BSC-1 cells (American Type Culture Collection) as described previously (5). Experiments were carried out with either BSC-1 or human HeLa cells (American Type Culture Collection). Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% newborn calf serum (NCS).
Generation of the rVV VT7/VP1.
Genomic IBDV dsRNA segments were isolated from purified virus as previously described (35). A cDNA corresponding to the IBDV ORF B was generated by reverse transcription (RT)-PCR using IBDV genomic RNA as template and the primers 5′-GCGCATCGATGAGTGACGTTTTCAATAGTCC and 5′-GCGCGCGGCCGCCCATCATGGCTATTGGCGGCTC, following a previously described protocol (13). The resulting PCR product was subjected to digestion with ClaI and NotI. The DNA fragment was then ligated to the vector pBSK+, generating the derivative plasmid pBSK/VP1. The nucleotide sequence of the fragment was analyzed and shown to be correct. Thereafter, the VP1 fragment was isolated from pBSK/VP1 by restriction with ClaI followed by treatment with Klenow fragment and restriction with SacI. This fragment was cloned into the VV insertion/expression vector pVOTE.2 (40) that had been digested with NdeI treated with Klenow, followed by restriction with SacI. The resulting plasmid, pVOTE/VP1, was purified and used to obtain the recombinant virus VT7/VP1. For this, BSC-1 cells were infected with the rVV VT7LacOI (40) and transfected with pVOTE/VP1. Selection and amplification of VT7/VP1 was carried out as previously described (5). The plasmid vector pVOTE.2 and the rVV VT7LacOI were kindly provided by Bernard Moss (National Institutes of Health, Bethesda, Md.).
Analysis of protein expression.
BSC-1 or HeLa cells were infected with the corresponding rVV at a multiplicity of infection (MOI) of 5 PFU/cell and maintained either in the presence or absence of the inducer isopropyl-β-d-thiogalactopyranoside (IPTG) (4 mM final concentration). Double infections were carried out with 5 PFU of each rVV/cell. At 18 h postinfection, (p.i.), cells were washed twice with methionine-free DMEM and metabolically labelled for 30 min with 50 μCi of [35S]methionine/ml, washed twice with phosphate-buffered saline (PBS), and resuspended in 1× sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 0.25% bromophenol blue, 5% glycerol, and 5% β-mercaptoethanol). Protein samples (5 to 10 μg/sample) were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) followed by autoradiography. For densitometric analysis, the autoradiograms were scanned with a Powerlook 2000 apparatus (UMAX Data Systems Inc., Taipei, Taiwan), and the resulting images were analyzed with NIH Image software (30a).
Western blotting and preparation of rabbit anti-VP2/VPX and -VP3 antisera were carried out as previously described (8). Rat antisera against VP2 and VP3 were generated by using the same peptides described for the production of rabbit antisera against these proteins (8). Anti-VP1 antisera were obtained from rabbits and rats immunized with a His-tagged fusion protein that contains a region spanning Ala 45 to Leu 195 of VP1. The fusion protein was generated by cloning a 447-bp BamHI restriction fragment from the VP1 ORF into the unique BglII site of the expression vector pRSET-C (36). The resulting plasmid, pRSET-C-VP1BHIf, was used to transform BL21 Escherichia coli cells (36) that were subsequently used to express and purify the VP1 fusion protein, following standard protocols (19).
Purification of VLPs and IBDV.
Cultures of HeLa cells were infected with VT7LacOI/POLY (8) or coinfected with VT7LacOI/POLY and VT7LacOI/VP1 at an MOI of 5 PFU/cell and maintained in the presence of IPTG and rifampin (100 μg/ml) to prevent VV morphogenesis (10, 30). At 20 h p.i., cells were collected, resuspended in PES buffer (25 mM PIPES [pH 6.2], 150 mM NaCl, and 20 mM CaCl2), and subjected to three cycles of freeze-thawing. After removing the cell debris by low-speed centrifugation (10,000 × g for 15 min at 4°C), the supernatant (8 ml) was recovered, loaded on top of a 4-ml cushion of 25% (wt/wt) sucrose in PES buffer, and spun at 125,000 × g for 3 h at 4°C. After centrifugation, the pellet was resuspended in 1 ml of PES buffer, loaded on top of an 11-ml continuous 25 to 50% sucrose gradient and spun at 125,000 × g for 1 h at 4°C. Gradients were fractionated (0.5 ml per fraction), and the fractions were analyzed by Western blotting. Appropriate fractions were dialyzed overnight against 5,000 volumes of PES buffer and were then concentrated to 2 to 5 mg/ml by centrifugation at 125,000 × g for 3 h at 4°C and resuspension of the pellet in 100 μl of PES buffer.
For purification of IBDV particles, CEF were infected at an MOI of 0.1 PFU/cell. When cytopathic effect was complete, the cell medium was harvested. After removing cell debris by low-speed centrifugation, the supernatant was supplemented with polyethylene glycol 6000 and NaCl to reach a final concentration of 3.5% and 500 mM. After the mixture was incubated for 18 h at 4°C, the virus was sedimented by centrifugation (10,000 × g for 15 min at 4°C). The virus pellet was resuspended in PES and then purified following a protocol identical to that described for the purification of VLPs.
For protein analysis, samples were subjected to SDS-PAGE. After electrophoresis, gels were silver stained and scanned, and the resulting images were analyzed by densitometry as described above.
IF and CLSM analyses.
HeLa cells seeded onto glass coverslips were infected with the appropriate rVV at an MOI of 5 PFU/cell and maintained in the presence or the absence of IPTG. At 24 h p.i., cells were washed twice with PBS and fixed in methanol-acetone (1:1) at −20°C for 7 min. After fixation, coverslips were air dried, blocked in PBS containing 20% NCS for 1 h, and incubated with anti-VP1, -VP2/VPX, or -VP3 antisera diluted in PBS supplemented with 5% NCS for 45 min at room temperature. Immunoglobulin G (IgG) was visualized with goat anti-rabbit antibodies conjugated to Texas red (TXRD) (Jackson ImmunoResearch Laboratories Inc., West Grove, Pa.). Coverslips were dehydrated with ethanol, mounted, and viewed by epifluorescence with a Zeiss Axiophot microscope at a magnification of ×630. Images were captured with a MicroMAX digital camera (Princeton Instruments Inc., Trenton, N.J.) using the IPLab Spectrum software (Signal Analytics Corporation, Vienna, Va.). CEF grown in coverslips were infected with IBDV at an MOI of 5 PFU/cell. At 24 h p.i., coverslips were collected, cells were processed and viewed by IF as described above, and images were captured as described above.
For CLSM, after methanol fixation, coverslips were incubated with either rabbit anti-VP2/VP2 or -VP3 and with rat anti-VP1 antisera. Rabbit Ig was detected with an affinity-purified goat anti-rabbit Ig conjugated to fluorescein isothiocyanate (FITC) (Jackson ImmunoResearch Laboratories Inc.). Rat Ig was visualized with an affinity-purified goat anti-rat Ig conjugated to TXRD (Jackson ImmunoResearch Laboratories Inc.). The images were captured with a Leica TCS4D confocal microscope interfaced with argon and krypton lasers with a 63× oil immersion Planapochromatic objective (numerical aperture, 1.4). FITC and TXRD signals were captured separately and merged into a single image with TCS-Merge software (Leica Nussloch, Heidelberg, Germany).
IP analysis.
HeLa cells monolayers were infected with the corresponding rVV at an MOI of 5 PFU/cell and maintained either in the presence or the absence of IPTG. Double infections were carried out with 5 PFU/cell of each recombinant virus. At 4 h p.i., cells were washed twice with methionine-free DMEM and metabolically labelled for 20 h. For this, the medium was replaced with overnight labelling medium (DMEM containing 1.5 mg of l-methionine/ml) supplemented with 100 μCi of [35S]methionine/ml. At 24 h p.i., cells were harvested, washed twice with PBS, and resuspended in lysis buffer (50 mM Tris [pH 8], 150 mM NaCl, 1% Nonidet P-40) supplemented with a mixture of protease inhibitors (Complete; Boehringer Mannheim GmbH, Mannheim, Germany). Cells were harvested, washed twice with ice-cold PBS, and lysed with 25 strokes of a tight-fitting Dounce homogenizer. IPs were carried out with protein A-Sepharose CL-4B (Pharmacia Biotech AB, Uppsala, Sweden) after incubation of the extracts with specific rabbit anti-VP1 or -VP3 antisera in accordance with a previously described protocol (12). After IP, samples were resuspended in sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 0.25% bromophenol blue, 5% glycerol, and 5% β-mercaptoethanol) and subjected to SDS–10% PAGE. After electrophoresis, gels were electroblotted onto nitrocellulose filters that were subjected to autoradiography. To confirm the position of bands corresponding to VP1 and VP3, the filters were incubated with rat anti-VP1 or -VP3 antisera followed by incubation with peroxidase-labelled goat anti-rat IgG. The signal was revealed by enhanced chemiluminescence (ECL) using a commercial kit (Amersham International, Little Chalfont, United Kingdom).
Electron microscopy.
Samples (5 μl) were applied to glow-discharged carbon-coated grids for 1 to 2 min. Samples were negatively stained with 2% aqueous uranyl acetate. Micrographs were recorded with a JEOL 1200 EXII electron microscope operating at 100 kV at a nominal magnification of ×40,000. Immunodecoration of VLPs was carried out as previously described (8).
RESULTS
Inducible expression of VP1 in mammalian cells.
A cDNA corresponding to the ORF encoding VP1 generated by RT-PCR was cloned into the plasmid pBSK+. After assessing its nucleotide sequence, the VP1 gene was subcloned into the VV insertion/expression vector pVOTE.2 (40). The resulting plasmid was used to generate VT7/VP1, an rVV containing the VP1 gene under the control of a T7 bacteriophage RNA polymerase inducible promoter within the hemagglutinin locus of the rVV VT7LacOI (40).
To characterize the expression of VP1, BSC-1 cell monolayers were infected with VT7/VP1 and, after the adsorption, cultures were incubated either in normal medium or in medium supplemented with the inducer IPTG. At 18 h p.i., cells were metabolically labelled with [35S]methionine for 1 h, and protein samples were collected and analyzed by SDS-PAGE followed by autoradiography.
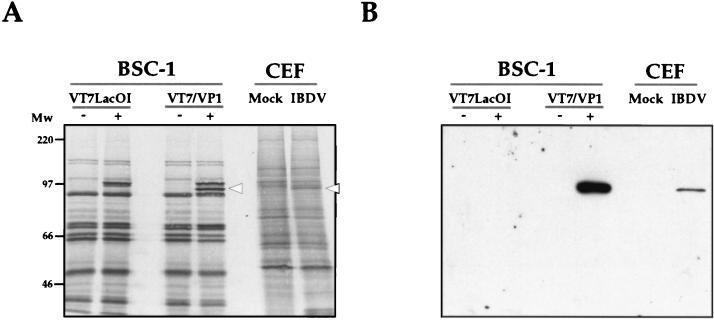
As shown in Fig. 1A, induction with IPTG resulted in the synthesis of two polypeptides of approximately 95 and 100 kDa. The 100-kDa product, detected in the lane from IPTG-induced cells infected with both the parental virus VT7LacOI and VT7/VP1, corresponds to the T7 polymerase (40). In contrast, the 95-kDa product was present only in IPTG-induced VT7/VP1-infected cells. This polypeptide, which had the Mr expected for VP1, was specifically recognized by the anti-VP1 antiserum and comigrated with protein specifically induced in CEF upon infection with IBDV, which was also detected by the anti-VP1 antiserum (Fig. 1A and B). Accumulation of VP1 in IBDV-infected CEF was much lower than that observed in cells infected with VT7/VP1.
FIG. 1.
Characterization of the expression of IBDV VP1 in cells infected with the rVV VT7/VP1. (A) BSC-1 cells infected with VT7/VP1 or the parental virus VT7LacOI, either treated (+) or untreated (−) with the inducer IPTG, were metabolically labelled with [35S]methionine. CEF mock-infected (Mock) or infected with IBDV (IBDV) were metabolically labelled with [35S]methionine at 48 h p.i. Protein samples from the different cultures were analyzed by SDS-PAGE. After electrophoresis, the gel was electroblotted onto nitrocellulose. The filter was air-dried and subjected to autoradiography. The positions of molecular weight markers are indicated (Mw). The positions of bands corresponding to the VP1 polypeptide are indicated by open arrowheads. (B) Western blot analysis of proteins expressed in cells infected with the rVV VT7/VP1 and CEF infected with IBDV. To determine the position of the VP1 polypeptide, after autoradiography, the nitrocellulose filter was rehydrated and incubated with a rabbit anti-VP1 antisera followed by the addition of horseradish peroxidase-conjugated goat anti-rabbit Ig. The signal was detected by ECL.
These results demonstrate that cells infected with VT7/VP1 inducibly express a protein with an Mr and an antigenic reactivity identical to those of the VP1 polypeptide synthesized in IBDV-infected cells.
VP1 is efficiently encapsidated into IBDV VLPs.
We have shown that expression of the 109-kDa IBDV polyprotein in mammalian cells leads to the formation of VLPs (8). Therefore, we were interested in determining whether the VP1 protein expressed by VT7/VP1 would encapsidate into IBDV VLPs.
BSC-1 monolayers were infected with either VT7/VP1 or VT7/POLY or were coinfected with both rVVs. After the adsorption, cells were induced with IPTG and maintained in the presence of rifampin to prevent VV maturation. Cells were metabolically labelled overnight with [35S]methionine. At 24 h p.i., cells were harvested and processed for purification of VLPs as described in Materials and Methods. Briefly, cells were homogenized, and after removing large debris by low-speed centrifugation, the extracts were passed through a 25% sucrose cushion. The pellets were resuspended and loaded on 25 to 50% sucrose gradients. After centrifugation, the gradients were fractionated, and the acid-precipitable radioactivity present in each fraction was determined.
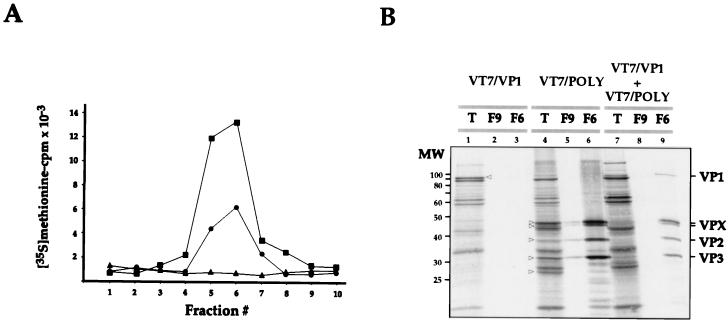
As expected, the distributions of acid-precipitable radioactivity in gradients from cells infected with VT7/POLY or coinfected with VT7/POLY and VT7/VP1 were similar (Fig. 2A). In both cases, a radioactive peak was detected within the central part of the gradient, where VLPs were expected to accumulate. A similar peak was not detected in the gradient from cells infected with VT7/VP1 alone. Most of the radioactivity of this sample was lost during the centrifugation through the sucrose cushion. This result ruled out the possibility that VP1 might self-assemble to form structures copurifying with VLPs.
FIG. 2.
Characterization of VLPs produced in cultures coinfected with the rVVs VT7/POLY and VT7/VP1. (A) Cultures infected with VV VT7/POLY (■) or VT7/VP1 (▴) or coinfected with VV VT7/POLY and VT7/VP1 (●) were maintained in medium supplemented with IPTG and rifampin. Cells were metabolically labelled with [35S]methionine. At 24 h p.i., cells were harvested and used to isolate VLPs by velocity sedimentation in sucrose gradients. Gradients were fractionated, and the total amount of acid-precipitable radioactivity was determined by scintillation counting. Fraction 1 corresponds to the bottom part of the gradient. (B) Samples corresponding to the total-cell extract (T) and gradient fractions 9 (F9) and 6 (F6) of samples from cells infected with VT7/VP1 or VT7/POLY or coinfected with VVs VT7/POLY and VT7/VP1 were analyzed by SDS-PAGE and autoradiography. Open arrowheads denote the positions of radioactive bands corresponding to VP1 in lane T from VT7/VP1-infected cells and to the VPX doublet, VP2, VP3, and VP4 in lane T from VT7/POLY-infected cells. The positions of molecular weight markers (MW) and bands corresponding to proteins VP1, VPX, VP2, and VP3 are indicated.
Samples from total-cell extracts and from fractions 6 and 9 of each gradient, corresponding to the radioactive peaks, were analyzed by SDS-PAGE followed by autoradiography. The extract from cells infected with the recombinant VT7/VP1 (Fig. 2B, lane 1) showed the presence of an intense radioactive band of 95 kDa corresponding to VP1. As expected from the results obtained after radioactivity counting of the gradient fractions, no radioactive bands were detected in fractions collected from the corresponding gradient (Fig. 2B, lanes 2 and 3).
The extract from VT7/POLY-infected cells (Fig. 2B, lane 4) contained five major IPTG-induced bands: (i) VPX (a doublet of 47 and 45 kDa), (ii) VP2 (37 kDa), (iii) VP3 (32 kDa), and (iv) VP4 (30 kDa). These polypeptides are generated by proteolytic processing of the 109-kDa polyprotein precursor. Fraction 6, corresponding to the radioactive peak of the gradient (Fig. 2A, lane 6), showed the presence of four major bands corresponding to the VPX doublet, VP2, and VP3. A densitometric analysis showed that these four bands accounted for over 90% of the total radioactivity detected in this fraction. These results indicated that the radioactive peak was due to the presence of VLPs. The top of the gradient (Fig. 2B, lane 5) contained a small amount of structural IBDV polypeptides, most likely representing denatured VLPs.
The extract from cells coinfected with VT7/VP1 and VT7/POLY contained VP1, VPX, VP2, VP3, and VP4 (Fig. 2B, lane 7). However, accumulation of the recombinant IBDV proteins was lower than that detected in samples from single infections. The reason for this reduced accumulation, which has been consistently observed, is, as yet, unknown. Analysis of fraction 6 from the corresponding sucrose gradient (Fig. 2B, lane 9) showed that, in addition of the structural IBDV proteins VPX, VP2, and VP3, this sample contained a significant amount of VP1.
These results strongly suggested that the VP1 expressed in cells coinfected with VT7/VP1 and VT7/POLY was specifically incorporated into the VLPs. To further confirm this result, purification of VLPs from cells infected with either VT7/POLY alone or coinfected with VT7/POLY and VT7/VP1 was carried out as described above. To minimize the presence of contaminant proteins after the sucrose gradient, purified VLPs were pelleted, resuspended in PES buffer, and subjected to a second sucrose gradient. Fractions of this gradient were analyzed by SDS-PAGE and electron microscopy (EM) after negative staining.
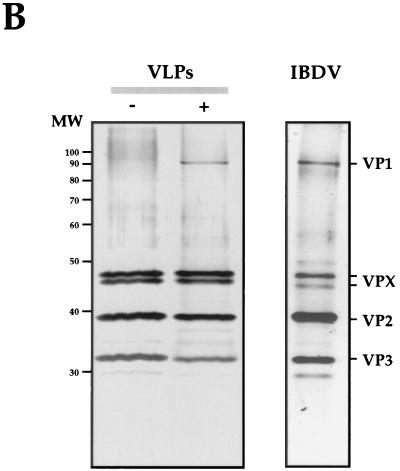
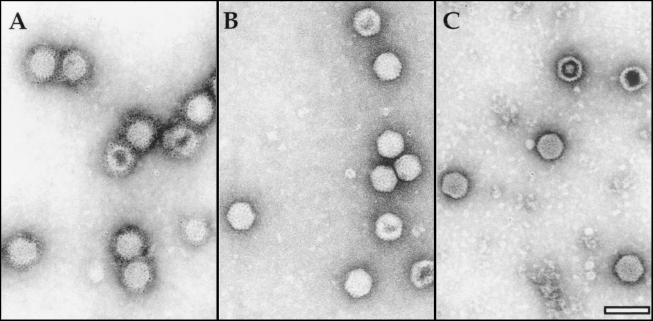
Examination of EM images of negatively stained VLPs showed the characteristic polyhedral contour and did not reveal size or morphological differences between the two types of VLPs and purified IBDV (Fig. 3A). Silver-staining of SDS-PAGE gels (Fig. 3B) showed that VLPs were highly purified (over 95% as determined by densitometry) and that, with the exception of the presence of VP1 detected in the sample from coinfected cells, both VLP preparations had almost identical protein compositions (Fig. 3B). The identity of these proteins was confirmed by Western blot analysis (data not shown).
FIG. 3.
Characterization of VLPs produced in the cells infected with VT7/POLY or coinfected with VT7/POLY and VT7/VP1 and purified IBDV particles. (A) EM images of VLPs purified from cultures infected with VT7/POLY (−VP1) or coinfected with VT7/POLY and VT7/VP1 (+VP1), and purified IBDV particles (IBDV). Bar represents 200 nm. (B) VLPs (5 μg/lane) purified from cultures infected with VT7/POLY (−) or coinfected with VT7/POLY and VT7/VP1 (+), and purified IBDV particles (IBDV) were subjected to SDS-PAGE. After electrophoresis, the gel was fixed and silver stained. The positions of molecular weight markers (MW) and bands corresponding to proteins VP1, VPX, VP2, and VP3 are indicated.
The polypeptide patterns of VP1-containing VLPs, and purified IBDV particles were also very similar (Fig. 3B). The results of a quantitative densitometric analysis of the polypeptides present in these two samples, shown in Table 1, demonstrated that the main difference between their protein profiles was related to the VPX/VP2 ratio. While in VP1-containing VLPs, the VPX/VP2 ratio was approximately 1.6:1, in purified virus it was 1:3. However, the overall VP1/VPX-VP2/VP3 ratios determined in preparations from VP1-containing VLPs (1:6.8:2.2) and purified IBDV were very similar (1:5.8:2).
TABLE 1.
Protein composition of VP1-containing VLPs and purified IBDV particlesa
| Sample | % (mean ± SE) of sample that consisted of:
|
|||
|---|---|---|---|---|
| VP1 | VPX | VP2 | VP3 | |
| VLPs plus VP1 | 10.2 ± 0.9 | 41.3 ± 3.1 | 26.4 ± 1.9 | 22.1 ± 1.3 |
| Purified virus | 11.3 ± 0.8 | 16.5 ± 1.3 | 49.4 ± 3.2 | 22.8 ± 1.9 |
Samples from VP1-containing VLPs and purified IBDV were subjected to SDS-PAGE. Gels were silver stained and analyzed by densitometry. The values shown are the percentages of each polypeptide with respect to the total amount of protein in each sample obtained after examination of samples from three independent VLP and virus particle preparations.
It is interesting to note the presence of a minor band of approximately 28 kDa that was not recognized by anti-VP1, -VP2/VPX, or -VP3 antisera in preparations of both purified IBDV and VLPs. This band might correspond to VP4, the putative protease, which is considered a nonstructural polypeptide.
The possibility existed that the VP1 protein detected in the VLP preparations might be associated to the external surface of the VLPs. To analyze it, reconstitution experiments were carried out. Metabolically labelled extracts from cells infected with VT7/VP1 or VT7/POLY were mixed and incubated for 1 h at 37°C. After incubation, VLPs were purified and analyzed by SDS-PAGE and autoradiography. VLPs obtained under these conditions did not contain detectable levels of VP1 (data not shown).
VP1-containing VLPs were analyzed by immunoelectron microscopy using anti-VP2/VPX, -VP1, and -VP3 antisera. Specific immunodecoration of VLPs was detected only after treatment with anti-VP2/VPX antiserum (Fig. 4). As shown in Fig. 4A, VLPs incubated with anti-VP2/VPX antiserum were surrounded by a dense material, corresponding to Igs bound to their external surface, decorated with 5-nm colloidal gold particles.
FIG. 4.
Immunoelectron microscopy analysis of VP1-containing VLPs. Purified VP1-containing VLPs were attached to coated EM grids and incubated with rabbit anti-VP2/VPX (A), -VP1 (B), and -VP3 (C), respectively. Bound antibody was detected with goat anti-rabbit Ig conjugated to 5-nm colloidal gold particles. Bar represents 100 nm.
These results demonstrated that under the described experimental conditions, VP2 is readily accessible to antibodies while VP3 and VP1 are either hidden or not recognized by their specific antisera.
The subcellular distribution of VP1 is modified by expression of the polyprotein.
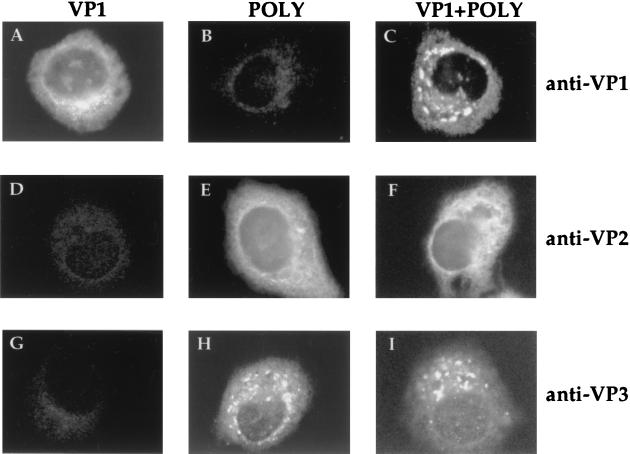
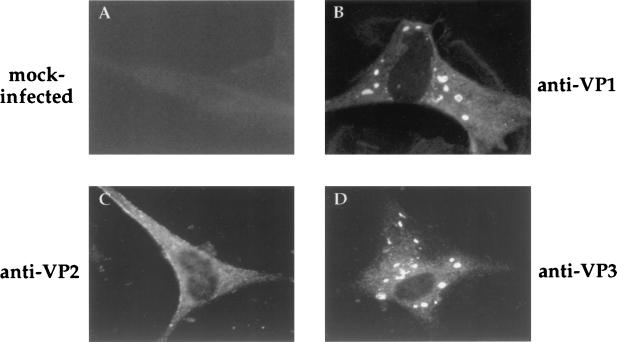
The results described above demonstrated that incorporation of VP1 into VLPs does not require the presence of IBDV RNAs. Hence, it seemed likely that VP1 incorporation might require interactions with VP2, VP3, or both. To study this, we first analyzed the distribution of VP1 in HeLa cells infected with either VT7/VP1 alone or coinfected with VT7/VP1 and VT7/POLY. The anti-VP1 IF signal observed in VT7/VP1-infected cells was exclusively cytosolic and formed by fine fluorescent grains concentrated within the perinuclear region (Fig. 5A). This pattern was dramatically modified in cells coinfected with VT7/VP1 and VT7/POLY. In these cells, the anti-VP1 IF signal was also cytoplasmic but formed by large, irregularly shaped fluorescent spots (Fig. 5C). This altered pattern was observed in more than 80% of the cells, indicating that expression of the polyprotein alters the subcellular distribution of VP1. Notably, the subcellular distribution of VP1, VP2, and VP3 in IBDV-infected CEF was very similar to that observed in HeLa cells coinfected with VT7/VP1 and VT/POLY (Fig. 6).
FIG. 5.
Subcellular distribution of IBDV proteins in cells infected with rVV. HeLa cell monolayers were infected with VT7/VP1 (VP1) or VT7/POLY (POLY) or were coinfected with both rVVs (VP1+POLY). At 24 h p.i., cells were fixed and incubated with rabbit anti-VP1 (anti-VP1), -VP2/VPX (anti-VP2/VPX), or -VP3 (anti-VP3) antisera. Thereafter, samples were incubated with FITC-conjugated goat anti-rabbit Ig and viewed by epifluorescence.
FIG. 6.
Subcellular distribution of IBDV proteins in IBDV-infected CEF. At 24 h p.i., cells were fixed and incubated with rabbit anti-VP1 (anti-VP1), -VP2/VPX (anti-VP2), or -VP3 (anti-VP3) antisera. Thereafter, samples were incubated with FITC-conjugated goat anti-rabbit Ig and viewed by epifluorescence.
A comparison of the distribution of the processing products of the polyprotein, VP2 and VP3, was also carried out. As shown in Fig. 5E and F, the anti-VP2/VPX IF signal was formed by fine grains within the cell cytoplasm. A specific anti-VP2/VPX IF signal was also detected within the nucleus. In more than 90% of the cells infected with either VT7/POLY or coinfected with VT7/VP1 and VT7/POLY, the anti-VP3-specific signal was formed by large cytoplasmic fluorescent spots (Fig. 5H and I). This IF pattern strongly resembled that observed with anti-VP1 in cells coinfected with VT7/VP1 and VT7/POLY. Interestingly, neither the VP2 (Fig. 5E and F) nor the VP3 (Fig. 5H and I) signals were significantly altered by coexpression of VP1.
VP1 and VP3 expressed in coinfected cells colocalize.
The similar IF patterns observed with anti-VP1 and -VP3 antibodies in coinfected cells suggested a possible interaction between VP3 and VP1. To further assess this possibility, distribution of VP1 and VP3 in coinfected cells was characterized by CLSM. As shown in Fig. 7, CLSM images indicated that expression of VP3 was, as expected, slightly higher than that of VP1. Despite this, the VP1-signal was consistently matched by VP3-specific fluorescence, thus demonstrating that, in cells coexpressing VP1 and the polyprotein, VP1 and VP3 colocalize. A similar analysis, carried out with anti-VP1 and -VP2/VPX antisera, showed that, under these experimental conditions, VP1 and VP2 had different subcellular distributions (data not shown).
FIG. 7.
Colocalization of VP1 and VP3 in cells coinfected with VT7/VP1 and VT7/POLY. Coinfected cells were fixed and incubated with rat anti-VP1 and rabbit anti-VP3 antisera. Thereafter, cells were incubated with goat anti-rat Ig antibodies conjugated to TXRD and with a goat anti-rabbit Ig conjugated to FITC as described in Materials and Methods. Green FITC (VP3) and red TXRD (VP1) fluorescence signals were recorded separately by using appropriate filters. Panel A shows, in yellow, the overlay of the VP3 and VP1 fluorescence signals. Panels B to I show a series of 500-nm sections of the analyzed cell.
VP1 and VP3 interact in coinfected cells.
We had previously observed (Fig. 2 and 3) the formation of IBDV VLPs containing VP1 in cells coinfected with VT7/VP1 and VT7/POLY. Although specific information is not yet available, the formation of IBDV capsids should require interactions between VP2/VPX and VP3. It seemed feasible that interaction between VP1 and VP3 might be dependent upon the presence of VP2/VPX. Therefore, it was interesting to analyze the subcellular localization of VP1 and VP3 in the absence of VP2. To this end, cells were coinfected with VT7/VP1 and VT7/VP3, an rVV expressing a 3′-terminal fragment of the polyprotein ORF corresponding to VP3 (7). Subcellular localization of VP1 and VP3 in these cells (Fig. 8A) was identical to that observed in cells coinfected with VT7/VP1 and VT7/POLY (Fig. 7). According to these data, colocalization of VP1 and VP3 is independent of the presence of other IBDV-encoded polypeptides.
FIG. 8.
(A) Colocalization of VP1 and VP3 in cells coinfected with VT7/VP1 and VT7/VP3. Coinfected cells were fixed and incubated with rat anti-VP1 and rabbit anti-VP3 antisera. Thereafter, cells were incubated with goat anti-rat Ig antibodies conjugated to TXRD and with a goat anti-rabbit Ig conjugated to FITC as described in Materials and Methods. Green FITC (VP3) and red TXRD (VP1) fluorescence signals were recorded separately by using appropriate filters. Panel i shows, in yellow, the overlay of the VP3 and VP1 fluorescent signals. Panels ii and iii show the VP3 and VP1 signals, respectively. (B) Colocalization of VP1 and VP2 in cells coinfected with VT7/VP1 and VT7/VP2. Coinfected cells were fixed and incubated with rat anti-VP1 and rabbit anti-VP2/VPX antisera. Thereafter, cells were incubated with goat anti-rat Ig antibodies conjugated to TXRD and with a goat anti-rabbit Ig conjugated to FITC as described in Materials and Methods. Green FITC (VP2/VPX) and red TXRD (VP1) fluorescence signals were recorded separately by using appropriate filters. Panel i shows, in yellow, the overlay of the VP1 and VP2 fluorescent signals. Panels ii and iii show the VP2 and VP1 signals, respectively.
A CLSM analysis was also carried out with cells coinfected with VT7/VP1 and VT7/VP2, an rVV expressing a 5′-terminal fragment of the polyprotein ORF corresponding to VP2 (7). Although both proteins were detected within the cell cytoplasm, no extensive colocalization was observed. Interestingly, the subcellular distribution of VP1 was similar to that observed in cells infected with only VT7/VP1 (Fig. 5A).
Detection of VP1-VP3 complexes.
Data from IF and CLSM analyses indicated the existence of interactions between VP1 and VP3. To confirm this, we investigated the presence of VP1-VP3 complexes in cells coinfected with VT7/VP1 and VT7/VP3. The approach used for these experiments was based on IP of protein extracts from cells infected with either VT7/VP1 or VT7/VP3, or coinfected with both rVVs. IP reactions were carried out with either anti-VP1 or -VP3 specific rabbit antisera. After IP, the samples were subjected to SDS-PAGE followed by electroblotting onto nitrocellulose filters and subjected either to autoradiography or Western blot analysis using anti-VP1 or -VP3 antisera.
IP of the extract from VT7/VP3-infected cells with anti-VP3 antiserum rendered a unique band of 32 kDa that corresponds to VP3 (Fig. 9A, lane 6). Notably, IP of the extract from coinfected cells with anti-VP3 antiserum rendered two polypeptides of 32 and 95 kDa, respectively (Fig. 9A, lane 4). The 32-kDa band corresponds to VP3, while the 95-kDa band comigrates with the VP1 band present in total extracts from cells infected with either VT7/VP1 (Fig. 9A, lane 2) or coinfected with both rVVs (Fig. 9A, lane 1). The anti-VP3 antiserum did not pull down specific polypeptides in the extract from cells infected with VT7/VP1 alone.
FIG. 9.
Detection of VP1-VP3 complexes. (A) HeLa cells were infected with VT7/VP1 (VP1) or VT7/VP3 (VP3) or were coinfected with both rVVs (VP1+VP3) and metabolically labelled with [35S]methionine. At 24 h p.i., cells were harvested, and the corresponding extracts (Total extract) were subjected to IP using anti-VP1 (IP Anti-VP1) or -VP3 (IP Anti-VP3) rabbit antisera. The resulting samples were subjected to SDS-PAGE and electroblotted onto nitrocellulose. The filter was subjected to autoradiography. The positions of molecular weight markers are indicated (Mw). The positions of the VP1 and VP3 polypeptides are indicated by open arrowheads. (B) Western blot analysis of the immunoprecipitated products. To determine the position of the VP1 polypeptide the nitrocellulose filter was incubated with anti-VP1 rat antisera followed by addition of horseradish peroxidase-conjugated goat anti-rat Ig. The signal was detected by ECL. (C) Western blot analysis of the immunoprecipitated products. To determine the position of the VP3 polypeptide, the filter was incubated with anti-VP3 rat antisera followed by the addition of horseradish peroxidase-conjugated goat anti-rat Ig. The signal was detected by ECL.
As expected, IP of the extract obtained from VT7/VP1-infected cells with the anti-VP1 produced a single band of 95 kDa, corresponding to VP1 (Fig. 9A, lane 8), that comigrated with the polypeptide observed after IP with anti-VP3 of the extract from cells coinfected with VT7/VP1 and VT7/VP3 (Fig. 9A, lane 4). A band with an identical electrophoretic mobility was also detected in the sample from cells coinfected with VT7/VP1 and VT7/VP3 (Fig. 9A, lane 7). The presence of a weak 32-kDa band after IP of the extract from VT7/VP3-infected cells (Fig. 9A, lane 9) with anti-VP1 indicates that this serum weakly cross-reacts with VP3. A minor band of approximately 50 kDa, detected in samples containing immunoprecipitated VP1 (Fig. 9A, lanes 4, 7, and 8), most likely represents a fragment produced by proteolytic degradation of VP1.
To confirm the identity of the immunoprecipitated 32- and 95-kDa polypeptides, after autoradiography, the filters were incubated with either rat anti-VP1 or -VP3 antisera followed by incubation with peroxidase-labelled goat anti-rat Ig and revealed by ECL. The immunoprecipitated 95-kDa protein was recognized by the anti-VP1 antibody (Fig. 9B, lanes 1, 2, 4, 7, and 8). Similarly, the 32-kDa immunoprecipitated protein was recognized by the anti-VP3 antiserum (Fig. 9C, lanes 1, 3, 4, and 6). These results unambiguously established the identity of the 32- and 95-kDa immunoprecipitated proteins as VP3 and VP1, respectively.
Similar results were obtained after IP analysis of extracts from cells coinfected with VT7/VP1 and VT7/POLY (data not shown). An IP analysis with extracts from cells coinfected with VT7/VP1 and VT7/VP2 was also carried out. As expected from the IF and CLSM data, VP1-VP2/VPX complexes were not detected (data not shown).
DISCUSSION
In all dsRNA viruses studied, transcription and replication of the genome takes place within the virus capsid. Seclusion of these processes in the interior of the capsid or inner-core structures exerts a considerable influence on the architecture of the virion (4, 41). With the exception of birnaviruses, dsRNA viruses of higher eukaryotes have a highly complex particle structure containing two or more shells (14, 32, 33, 37). This complexity has been highlighted recently by the determination by X-ray analysis of the structure of the bluetongue virus core (9). Birnaviruses are far simpler, with a capsid formed by a single protein layer (1). This feature makes members of the Birnaviridae excellent models for studying the molecular biology of dsRNA viruses and, at the same time, raises interesting questions about their life cycle.
The goals of our study were (i) to characterize the expression of the putative RdRp VP1 in mammalian cells, (ii) to analyze the requirements for the incorporation of this protein into the virus particle, and (iii) to define possible interactions between VP1 and other IBDV polypeptides that might be important for virus morphogenesis.
The results presented above show that the molecular mass and antigenic reactivity of the VP1 expressed in mammalian cells, using a VV expression inducible system, are similar to those of the product expressed in CEF upon infection with IBDV. Expression of the putative RdRp from infectious pancreatic necrosis virus (IPNV), the prototype member of the Birnaviridae in insect cells, also rendered a protein identical to that expressed in fish cells during the IPNV natural infection (21). Notably, previous attempts to express IBDV VP1 in E. coli and yeast cells resulted in the synthesis of polypeptides with abnormal electrophoretic mobilities (20). Experiments to demonstrate the RNA polymerase activity of VP1 by using extracts from E. coli and yeast cells expressing these altered proteins were unsuccessful. Although we have not studied the enzymatic activity of the VP1 polypeptide, it seems likely that the inducible VV expression system might provide a good starting point for future functional analyses.
Information about the morphogenesis of IBDV is very limited. Indeed, the finding that IBDV VLPs are readily formed in the absence of VP1 (8, 39) was the first demonstration that this polypeptide is not required for the assembly of the capsid. The data presented here show that VP1 is efficiently incorporated into newly formed VLPs when both VP1 and the polyprotein are coexpressed in a heterologous mammalian system. The possibility that the VP1 molecules detected in purified VLPs might be bound to the external surface of the particle was ruled out by the results obtained from reconstitution and immunoelectron microscopy analyses.
Our results are consistent with the structural model proposed by Bötcher et al. (1) in which the external surface of the particle is formed by trimers of VP2, while VP3 faces the inner cavity of the capsid. According to our data, VP3 would act as an anchor, interacting with both VP2 and VP1. The VP1-VP3 complexes would be located at the inner cavity of the capsid. This would explain the inaccessibility of both polypeptides to their specific antibodies. Notably, formation of VP3-derived shell-like structures in cells infected with VT7/VP3 has not been detected (2a).
We are currently characterizing, by cryoelectron microscopy, the three-dimensional structure of VLPs and VP1-containing VLPs. Hopefully, a comparison of their corresponding density maps will yield relevant information about the precise location of the structural IBDV elements.
The results presented here demonstrate that the relative amounts of VP1 in VLPs and purified virus are very similar. The finding that VLPs and purified virus possess different VPX/VP2 ratios is intriguing. VLPs produced in rVV-infected cells remain inside the cell cytoplasm (8). In contrast, most of the progeny produced in IBDV-infected cells is released to the extracellular medium. It seems feasible that the proteolytic processing of VPX might be associated with the maturation and/or the release process. It is exciting to speculate on the possibility that this process(es) requires the presence of the nonstructural polypeptide VP5 and/or the virus genome. The recent finding that mutational inactivation of VP5 causes a major reduction in virus virulence adds more interest to this hypothesis (28).
Although we had no information about the mechanism(s) responsible for incorporation of VP1 into the VLPs, IF analyses showed that in cells coexpressing the polyprotein, the subcellular distribution of VP1 was deeply modified. Under these conditions, the distribution of the structural IBDV proteins was akin to that observed in IBDV-infected CEF. When the distribution of VP1, VP2, and VP3 was analyzed by CLSM, an extensive colocalization of VP1 and VP3 was detected. In addition, colocalization of these two proteins was found to be independent of the presence of other polyprotein-derived products, namely VP2 and VP4. Taken together, these data indicated that VP1 and VP3 were interacting. This was confirmed by the results of the IP analyses that demonstrated that VP1 and VP3 form stable complexes that are efficiently immunoprecipitated with anti-VP3 antibodies. Somewhat surprisingly, although VP1 was readily immunoprecipitated with anti-VP1 antisera, the VP1-VP3 complexes were not. The reason for this is, as yet, unknown. The anti-VP1 antiserum was raised against a 151-amino-acid fragment corresponding to a highly hydrophilic stretch (Ala 45 to Leu 195) from the N-terminal region of VP1. It seems feasible that this region might be either directly involved or modified by the formation of the complex. Hence, under native conditions, the binding of anti-VP1 antibodies might be reduced. Additionally, under native conditions, the binding of anti-VP1 antibodies might strongly affect the stability of the VP1-VP3 complexes. Experiments to test these hypotheses are under way.
In purified virions, VP1 is found as a “free” polypeptide as well as bound to the dsRNA genome segments. Free VP1 is linked to short RNA sequences, probably corresponding to the terminal regions of the IBDV RNA molecules (17). Our data show that, in the absence of IBDV genomic RNAs, VP1 is incorporated into newly formed VLPs. Although the expression system used in this work is absent from IBDV RNA terminal sequences, the possibility that VP1 might bind small RNA sequences from either cellular or rVV origin cannot be ruled out at this time.
Formation of the VP3-VP1 complexes might be a critical step for the generation of infectious IBDV. Therefore, it seems feasible that blocking this interaction could strongly interfere with the virus replication process. The characterization of this interaction might indicate new ways to control the spread of IBDV.
ACKNOWLEDGMENTS
This work was supported by grants BIO-97-0576 from the Comisión Interministerial de Ciencia y Tecnología and 07B/0032/1998 from the Subdirección General de Investigación of the Comunidad Autónoma de Madrid. A.M. was the recipient of a fellowship from Ministerio de Educación.
We thank Diane Wilcock for critically reading the manuscript and Bernard Moss for the VOTE expression system. We also thank Inés Poveda and Angel Sanz for excellent photography work.
REFERENCES
- 1.Bötcher B, Kiselev N A, Maschuck V Y, Perevozchicova N A, Borisov A V, Crowther R A. Three-dimensional structure of bursal disease virus determined by electron cryomicroscopy. J Virol. 1997;71:330–335. doi: 10.1128/jvi.71.1.325-330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruenn J A. Relationships among positive strand and double-strand RNA viruses as viewed through their RNA-dependent RNA polymerases. Nucleic Acids Res. 1991;19:217–226. doi: 10.1093/nar/19.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Castón, J. R., and J. F. Rodriguez. Unpublished results.
- 3.Dobos P, Berthiaume L, Leong J A, Kibenge K S B, Muller H, Nicholson B L. Family Birnaviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy. Sixth report of the International Committee on Taxonomy of Viruses. New York, N.Y: Springer-Verlag; 1995. pp. 240–244. [Google Scholar]
- 4.Dryden K A, Wang G, Yeager M, Nibert M L, Coombs K M, Furlong D B, Fields B N, Baker T S. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Earl P L, Moss B. Generation of recombinant vaccinia viruses. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1993. pp. 16.17.1–16.18.10. [Google Scholar]
- 6.Fahey K J, O’Donnell I J, Azad A A. Characterization by Western blotting of the immunogens of infectious bursal disease virus. J Gen Virol. 1985;66:1479–1488. doi: 10.1099/0022-1317-66-7-1479. [DOI] [PubMed] [Google Scholar]
- 7.Fernández-Arias A, Martínez S, Rodriguez J F. The major antigenic protein of infectious bursal disease virus, VP2, is an apoptotic inducer. J Virol. 1997;71:8014–8018. doi: 10.1128/jvi.71.10.8014-8018.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernández-Arias A, Risco C, Martínez S, Albar J P, Rodriguez J F. Expression of ORF A1 of infectious bursal disease virus results in the formation of virus-like particles. J Gen Virol. 1998;79:1047–1054. doi: 10.1099/0022-1317-79-5-1047. [DOI] [PubMed] [Google Scholar]
- 9.Grimes J M, Burroughs J N, Gouet P, Diprose J M, Malby R, Zientara S, Mertens P P, Stuart D I. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- 10.Grimley P M, Rosenblum E N, Mims S J, Moss B. Interruption by rifampin of an early stage in vaccinia virus morphogenesis: accumulation of membranes which are precursors of virus envelopes. J Virol. 1970;6:519–533. doi: 10.1128/jvi.6.4.519-533.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harkness J W, Alexander D J, Pattison M, Scott A C. Infectious bursal disease agent: morphology by negative stain electron microscopy. Arch Virol. 1975;48:63–73. doi: 10.1007/BF01320566. [DOI] [PubMed] [Google Scholar]
- 12.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 13.Heine H G, Haritou M, Failla P, Fahey K, Azad A A. Sequence analysis and expression of the host-protective immunogen VP2 of a variant strain of infectious bursal disease virus which can circumvent vaccination with standard type I strains. J Gen Virol. 1991;22:1835–1843. doi: 10.1099/0022-1317-72-8-1835. [DOI] [PubMed] [Google Scholar]
- 14.Hewat E A, Booth T F, Roy P. Structure of bluetongue virus particles by cryoelectron microscopy. J Struct Biol. 1992;109:61–69. doi: 10.1016/1047-8477(92)90068-l. [DOI] [PubMed] [Google Scholar]
- 15.Hirai K, Shimakura S. Structure of infectious bursal disease virus. J Virol. 1974;14:957–964. doi: 10.1128/jvi.14.4.957-964.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hudson P J, McKern N M, Power B E, Azad A A. Genomic structure of the large RNA segment of infectious bursal disease virus. Nucleic Acids Res. 1986;14:5001–5012. doi: 10.1093/nar/14.12.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kibenge F S B, Dhama V. Evidence that virion-associated VP1 of avibirnaviruses contains viral RNA sequences. Arch Virol. 1997;142:1227–1236. doi: 10.1007/s007050050154. [DOI] [PubMed] [Google Scholar]
- 18.Kibenge F S, Qian B, Cleghorn J R, Martin C K. Infectious bursal disease virus polyprotein processing does not involve cellular proteases. Arch Virol. 1997;142:2401–2419. doi: 10.1007/s007050050251. [DOI] [PubMed] [Google Scholar]
- 19.Kroll D J, Abdel-Malek Abdel-Hafiz H, Marcell T, Simpson S, Chen C Y, Gutierrez-Hartmann A, Lustbader J W, Hoeffler J P. A multifunctional prokaryotic protein expression system: overproduction, affinity purification, and selective detection. DNA Cell Biol. 1993;12:441–453. doi: 10.1089/dna.1993.12.441. [DOI] [PubMed] [Google Scholar]
- 20.Macreadie I, Azad A A. Expression and RNA dependent RNA polymerase activity of birnavirus VP1 protein bacteria and yeast. Biochem Mol Biol Int. 1993;6:1169–1178. [PubMed] [Google Scholar]
- 21.Magyar G, Dobos P. Expression of infectious pancreatic necrosis virus polyprotein and VP1 in insect cells and detection of the polyprotein in purified virus. Virology. 1994;198:437–445. doi: 10.1006/viro.1994.1055. [DOI] [PubMed] [Google Scholar]
- 22.Magyar G, Chung H K, Dobos P. Conversion of VP1 to VPg in cells infected by infectious pancreatic necrosis virus. Virology. 1998;245:142–150. doi: 10.1006/viro.1998.9137. [DOI] [PubMed] [Google Scholar]
- 23.Müller H, Becht H. Biosynthesis of virus-specific proteins in cells infected with infectious bursal disease virus and their significance as structural elements for infectious virus and incomplete particles. J Virol. 1982;44:384–392. doi: 10.1128/jvi.44.1.384-392.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller H, Nitschke R. The two segments of the infectious bursal disease virus genome are circularized by a 90,000-Da protein. Virology. 1987;159:174–177. doi: 10.1016/0042-6822(87)90363-1. [DOI] [PubMed] [Google Scholar]
- 25.Müller H, Schnitzler D, Bernstein F, Becht H, Cornelissen D, Lutticken D H. Infectious bursal disease of poultry: antigenic structure of the virus and control. Vet Microbiol. 1992;33:175–183. doi: 10.1016/0378-1135(92)90045-u. [DOI] [PubMed] [Google Scholar]
- 26.Mundt E, Beyer J, Müller H. Identification of a novel viral protein in infectious bursal disease virus-infected cells. J Gen Virol. 1995;76:437–443. doi: 10.1099/0022-1317-76-2-437. [DOI] [PubMed] [Google Scholar]
- 27.Mundt E, Vakharia V N. Synthetic transcripts of double-stranded birnavirus genome are infectious. Proc Natl Acad Sci USA. 1996;93:11131–11136. doi: 10.1073/pnas.93.20.11131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mundt E, Köllner B, Kretzschmar D. VP5 of infectious bursal disease virus is not essential for virus replication in cell culture. J Virol. 1997;71:5647–5651. doi: 10.1128/jvi.71.7.5647-5651.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagarajan M M, Kibenge F S B. Infectious bursal disease virus: a review of molecular basis for variations in antigenicity and virulence. Can J Vet Res. 1995;61:81–88. [PMC free article] [PubMed] [Google Scholar]
- 30.Nagayma A, Pogo B G T, Dales S. Biogenesis of vaccinia: separation of early stages from maturation by means of rifampin. Virology. 1970;40:1039–1051. doi: 10.1016/0042-6822(70)90150-9. [DOI] [PubMed] [Google Scholar]
- 30a.National Institutes of Health. 14 April 1999, revision date. [Online.] NIH Image software. National Institutes of Health, Bethesda, Md. http://rsb.info.nih.gov/nih-image/. [20 December 1998, last date accessed.]
- 31.Özel M, Gelderblom H. Capsid symmetry of viruses of the proposed birnavirus group. Arch Virol. 1985;84:149–161. doi: 10.1007/BF01378968. [DOI] [PubMed] [Google Scholar]
- 32.Prasad B V, Wang G J, Clerx J P M, Chiu W. Three-dimensional structure of rotavirus. J Mol Biol. 1988;199:269–275. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- 33.Prasad B V, Rothnagel R, Zeng C Q-Y, Jakana J, Lawton J A, Chiu W, Estes M K. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature. 1996;382:471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- 34.Saif Y M. Infectious bursal disease and hemorrhagic enteritis. Poult Sci. 1998;77:1186–1189. doi: 10.1093/ps/77.8.1186. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Schoepfer R. The pRSET family of T7 promoter expression vectors for Escherichia coli. Gene. 1993;124:83–85. doi: 10.1016/0378-1119(93)90764-t. [DOI] [PubMed] [Google Scholar]
- 37.Shaw A L, Samal S K, Subramanian K, Prasad B V. The structure of aquareovirus shows how the different geometries of the two layers of the capsid are reconciled to provide symmetrical interactions and stabilization. Structure. 1996;4:957–967. doi: 10.1016/s0969-2126(96)00102-5. [DOI] [PubMed] [Google Scholar]
- 38.Stuart D I, Gouet P, Grimes J, Malby R, Diprose J, Zientara S, Burroughs J N, Mertens P P. Structural studies of orbivirus particles. Arch Virol Suppl. 1998;14:235–250. doi: 10.1007/978-3-7091-6823-3_21. [DOI] [PubMed] [Google Scholar]
- 39.Vakharia V N. Development of recombinant vaccines against infectious bursal disease. Biotech Annu Rev. 1997;3:151–168. [Google Scholar]
- 40.Ward G A, Stover C K, Moss B, Fuerst T R. Stringent chemical and thermal regulation of recombinant gene expression by vaccinia virus vectors in mammalian cells. Proc Natl Acad Sci USA. 1995;92:6773–6777. doi: 10.1073/pnas.92.15.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wickner R B. Double-stranded RNA virus replication and packaging. J Biol Chem. 1993;268:3797–3800. [PubMed] [Google Scholar]
- 42.Yao K, Goodwin M A, Vakharia V N. Generation of a mutant infectious bursal disease virus that does not cause bursal lesions. J Virol. 1998;72:2647–2654. doi: 10.1128/jvi.72.4.2647-2654.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]