Abstract
Human immunodeficiency virus type 1 (HIV-1) genomic RNA segments at nucleotide (nt) positions +240 to +274 are thought to form a stem-loop secondary structure, termed SL1, that serves as a dimerization initiation site for viral genomic RNA. We have generated two distinct deletion mutations within this region, termed BH10-LD3 and BH10-LD4, involving nt positions +238 to +253 and +261 to +274, respectively, and have shown that each of these resulted in significant diminutions in levels of viral infectiousness. However, long-term culture of each of these viruses in MT-2 cells resulted in a restoration of infectiousness, due to a series of compensatory point mutations within four distinct proteins that are normally cleaved from the Gag precursor. In the case of BH10-LD3, these four mutations were MA1, CA1, MP2, and MNC, and they involved changes of amino acid Val-35 to Ile within the matrix protein (MA), Ile-91 to Thr within the capsid (CA), Thr-12 to Ile within p2, and Thr-24 to Ile within the nucleocapsid (NC). The order in which these mutations were acquired by the mutated BH10-LD3 was MNC > CA1 > MP2 > MA1. The results of site-directed mutagenesis studies confirmed that each of these four substitutions contributed to the increased viability of the mutated BH10-LD3 viruses and that the MNC substitution, which was acquired first, played the most important role in this regard. Three point mutations, MP2, MNC, and MA2, were also shown to be sequentially acquired by viruses that had emerged in culture from the BH10-LD4 deletion. The first two of these were identical to those described above, while the last involved a change of Val-35 to Leu. All three of these substitutions were necessary to restore the infectiousness of mutated BH10-LD4 viruses to wild-type levels, although the MP2 mutation alone, but neither of the other two substitutions, was able to confer some viability on BH10-LD4 viruses. Studies of viral RNA packaging showed that the BH10-LD4 deletion only marginally impaired encapsidation while the BH10-LD3 deletion caused a severe deficit in this regard.
The RNA genome of human immunodeficiency virus type 1 (HIV-1) contains a 5′-end noncoding region that includes a long terminal repeat consisting of the R and U5 regions and the primer binding site, as well as exon 1 leader sequences downstream of U5 (13). Each of these elements plays a crucial role in viral replication. The R region includes cis-acting elements, such as TAR sequences that are required for transcriptional transactivation by Tat and the poly(A) signal needed for termination of transcription. The U5 region is involved in both regulation of reverse transcription and integration of viral cDNA into host cell chromosomal DNA. The primer binding site is an 18-nucleotide (18-nt) segment that binds to tRNA3Lys, which serves as the cognate primer of reverse transcription. The exon 1 leader sequences downstream of U5 include the splice donor site and are also involved in HIV-1 gene transcription as well as the dimerization and selective encapsidation of full-length viral genomic RNA (3, 6, 23, 27–29, 31).
The secondary structure of the exon 1 leader sequence downstream of U5 includes four distinct stem-loop RNA motifs, termed SL1, SL2, SL3, and SL4, that were initially predicted on the basis of computer modelling and biochemical analysis (4, 10, 20). Mutagenesis studies showed that the SL1, SL3, and SL4 elements were involved in the selective packaging of viral genomic RNA (33, 34). This is supported as well by cell-free studies that documented that the viral nucleocapsid (NC) protein, which acts in trans to promote viral RNA encapsidation, can also bind to the SL1, SL3, and SL4 structures with high affinity (7, 8, 10, 19, 39). SL1 contains a palindromic loop sequence (GCGCGC) that is thought to represent the dimerization initiation site for viral genomic RNA on the basis of cell-free experiments (3, 12, 25, 32, 36, 41). The importance of this region is also highlighted by the fact that mutations within SL1 severely decreased viral infectiousness (30, 35). However, the mechanisms involved are still poorly understood; for example, viruses containing a mutated SL1 element were only moderately affected in regard to incorporation of viral genomic RNA. Furthermore, since SL1 is located upstream of the splice donor site, it is still unclear how the former might exclude spliced viral RNAs from being incorporated into virions. Contradictory results have also been reported in regard to the role of SL1 in viral RNA dimerization in viral replication studies (5, 11, 26, 40).
To shed further light on the role of SL1 in viral replication, we selectively deleted a number of sequences in the SL1 region and cultured the recombinant viruses thus generated in MT-2 cells over long periods to determine whether and how compensation for viral replication and viral RNA encapsidation might occur. Two deletion mutations, BH10-LD3 and BH10-LD4, involving deletions of sequences at nt positions +238 to +253 and +261 to +274, respectively, within the SL1 region, were extensively analyzed (Fig. 1A).
FIG. 1.
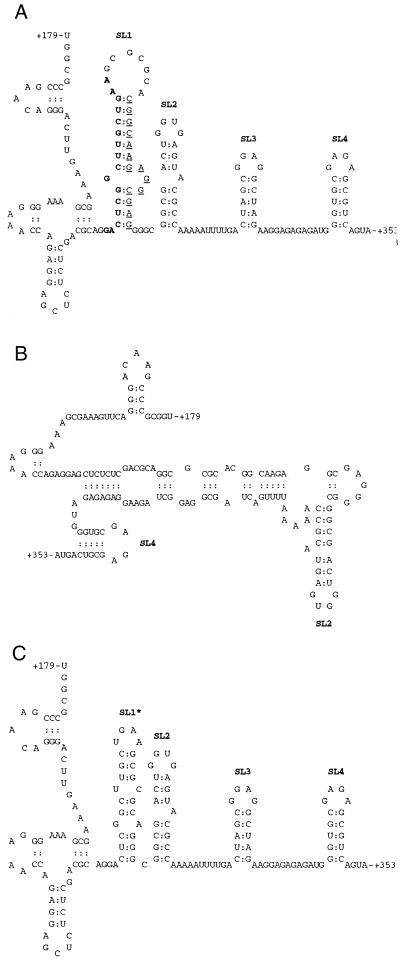
Schematic illustrations of the BH10-LD3 and BH10-LD4 deletions and their effects on the formation of RNA secondary structure. Only structures with the lowest levels of free energy are shown. (A) Secondary structures of wild-type HIV-1 RNA fragment (nt +179 to +353). Nucleotides removed in the BH10-LD3 and BH10-LD4 deletions are shown by bold and underlined letters, respectively. Secondary structures were predicted by the MFold program, using a calculated stability of −43.5 kcal/mol. The SL1, SL2, SL3, and SL4 RNA structures are consistent with previous reports (10). (B) Effects of the BH10-LD3 deletion on the formation of RNA secondary structures. The calculated stability of the secondary structures shown is −37.1 kcal/mol, and maintenance of SL2 and SL4 is indicated. (C) Effects of the BH10-LD4 deletion on the formation of RNA secondary structures at a stability of −38.0 kcal/mol. Formation of an SL1-like structure (SL1∗) is indicated.
Mutations termed MA1 (V35I), CA1 (I91T), MP2 (T12I), and MNC (T24I) are involved in compensation for the BH10-LD3 deletion.
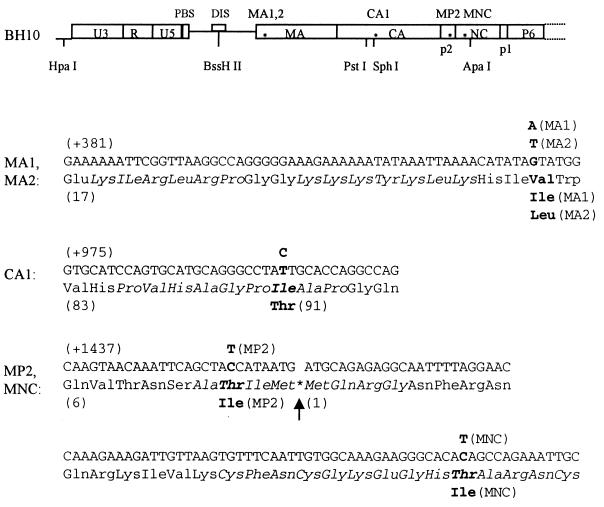
Previous work by members of our group has shown that mutated BH10-LD3 viruses, containing a deletion of the genomic segment from nt positions +238 to +253, were able to revert to near wild-type replication kinetics after 18 passages in MT-2 cells, due to two point mutations, MP2 (T12I) and MNC (T24I), located within the p2 peptide and NC protein, respectively (30). However, the reverted viruses, termed BH10-LD3-18, still showed a 2- to 3-day lag in growth. Therefore, we maintained the BH10-LD3-18 viruses in MT-2 cells for an additional 11 passages and showed that the virus thus generated, BH10-LD3-29, had reacquired full replication capacity (data not shown). To identify additional mutations responsible for this increased infectiousness, we amplified a 2-kb HIV-1 DNA fragment (nt positions −454 to +1548) that contained the BH10-LD3 deletion, using the primer pair Hpa-S–Apa-A and viral DNA extracted from MT-2 cells. The results of cloning and sequencing experiments showed that (i) the BH10-LD3 deletion and the previously described MP2 and MNC point mutations still existed within the genome of BH10-LD3-29 virus and that (ii) new point mutations MA1 [G(+430)A, i.e., V35I] and CA1 [T(+1000)C, i.e., I91T] were also present within the matrix protein (MA) and the capsid (CA), respectively (Fig. 2).
FIG. 2.
Compensatory point mutations MA1, MA2, CA1, MP2, and MNC within the HIV-1 genome. Locations are illustrated by asterisks. The mutations are as follows: MA1, Val-35 to Ile within MA; MA2, Val-35 to Leu within MA; CA1, Ile-91 to Thr within CA; MP2, Thr-12 to Ile within p2; and MNC, Thr-24 to Ile within NC. Letters in bold indicate original nucleotides and amino acids and their replacements. HpaI, BssHII, PstI, SphI, and ApaI are unique restriction sites within the HIV-1 genome that were employed in cloning experiments. The CA1, MA1, and MA2 substitutions were generated by means of PCR with primer pairs CA1–Apa-A (5′-CCAGTGCATGCAGGGCCTACTGCACCAGGCCAGATC-3′ [+981 to +1016] and 5′-CCTAGGGGCCCTGCAATTTCTG-3′ [+1559 to +1538]), MA1-S–MA1-A (5′-AATTAAAACATATAATATGGGCAAGC-3′ [+421 to +446] and 5′-GCTTGCCCATATTATATGTTTTAATT-3′ [+446 to +421]), and MA2-S–MA2-A (5′-AATTAAAACATATATTATGGGCAAGC-3′ [+421 to +446] and 5′-GCTTGCCCATAATATATGTTTTAATT-3′ [+446 to +421]), respectively (mutated nucleotides are underlined). PBS, primer binding site; DIS, dimerization initiation site.
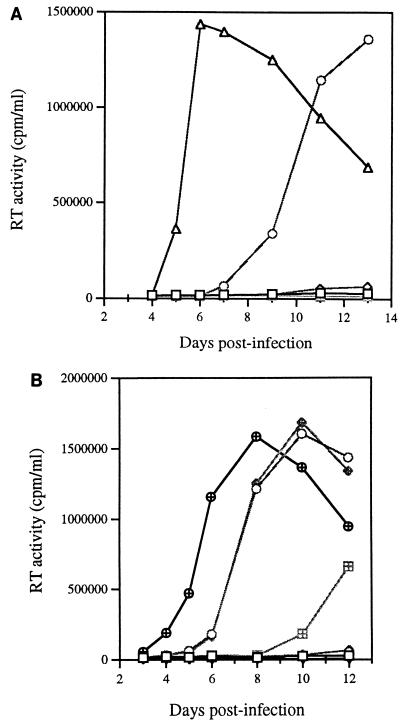
To evaluate the roles of the newly identified MA1 and CA1 point mutations in rescuing viruses containing the BH10-LD3 deletion, the above-mentioned substitutions were inserted into the BH10-LD3 construct individually or in the context of viruses also containing the MP2 and MNC substitutions. The results of infection assays (Fig. 3) showed that (i) MA1 and CA1 did not alone or together confer infectiousness on mutated BH10-LD3 virus (i.e., BH10-LD3-MA1, BH10-LD3-CA1, and BH10-LD3-MA1-CA1), (ii) MP2 did not help MA1 and CA1 to rescue viruses containing the BH10-LD3 deletion (i.e., BH10-LD3-MA1-MP2 and BH10-LD3-CA1-MP2), and (iii) association of MNC with either MA1 or CA1 gave rise to viruses able to generate high levels of reverse transcriptase (RT) activity in MT-2 cells (i.e., BH10-LD3-MA1-MNC and BH10-LD3-CA1-MNC). Since MNC could not by itself rescue BH10-LD3 virus in regard to replication competence (30), these data suggest that the ability of both the BH10-LD3-MA1-MNC and BH10-LD3-CA1-MNC viruses to infect and replicate in MT-2 cells was attributable to the presence of MA1 and CA1 mutations. These abilities are further demonstrated by the increase in infectiousness of the recombinant BH10-LD3 viruses containing various combinations of the MP2, MNC, MA1, and CA1 substitutions (Table 1).
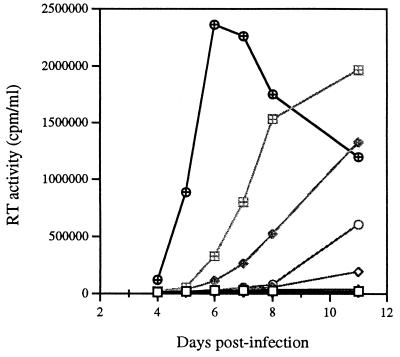
FIG. 3.
Roles of the MA1 and CA1 point mutations in compensation for the BH10-LD3 deletion mutation. (A) Growth curves of viruses formed from recombination of the BH10-LD3 deletion either with MA1, with both MA1 and MP2, or with both MA1 and MNC to yield BH10-LD3-MA1 (□), BH10-LD3-MA1-MP2 (◊), and BH10-LD3-MA1-MNC (○), respectively. The clones thus generated were transfected into COS-7 cells, and the progeny viruses were used to infect MT-2 cells. Levels of RT activity were then monitored in culture fluids. MT-2 cells were exposed to heat-inactivated wild-type virus as a negative control (⊞). ▵, BH10. (B) Growth curves of the recombinant viruses BH10-LD3-CA1 (□), BH10-LD3-CA1-MP2 (◊), BH10-LD3-CA1-MNC (○), BH10-LD3-MA1-CA1 (▵), BH10-LD3-MA1-CA1-MP2 (⊞), and BH10-LD3-MA1-CA1-MNC (◊+) in MT-2 cells. ▿, control; ⊕, BH10.
TABLE 1.
TCID50s of wild-type and various mutated viruses, determined by infection of MT-2 cells
| Virus | TCID50/mla |
|---|---|
| BH10-LD3-MP2-MNC | 1.6 × 102 ± 1.8 × 10 |
| BH10-LD3-MA1-MP2-MNC | 4.0 × 102 ± 2.1 × 10 |
| BH10-LD3-CA1-MP2-MNC | 4.5 × 102 ± 2.3 × 10 |
| BH10-LD3-MA1-CA1-MP2-MNC | 1.0 × 103 ± 3.2 × 10 |
| BH10-LD3-MA2-MP2-MNC | 6.4 × 102 ± 2.4 × 10 |
| BH10-LD4-MP2-MNC | 4.3 × 102 ± 2.0 × 10 |
| BH10-LD4-MA2-MP2-MNC | 2.6 × 103 ± 4.1 × 10 |
| BH10-LD4-MA1-MP2-MNC | 1.5 × 103 ± 3.7 × 10 |
| BH10 | 4.1 × 103 ± 6.2 × 10 |
| BH10-LD3b | <10 |
| BH10-LD4b | <10 |
Determination of TCID50s was as described previously (16). Results shown are the averages of three independent experiments.
Examined as negative controls and very poorly infectious in these assays.
The MNC mutation appeared to be unique in that its presence in concert with MA1, CA1, or MP2 was able to compensate for the LD3 deletion in regard to restoration of infectiousness. In addition, the recombinant virus BH10-LD3-MA1-CA1-MP2 replicated much less efficiently than did BH10-LD3-MA1-CA1-MNC (Fig. 3B), further highlighting the importance of MNC.
We also sought to determine the order in which the four point mutations were acquired by BH10-LD3 viruses with deletions during long-term culture in MT-2 cells. Analysis of nine passages revealed that the MNC substitution was generated first (i.e., in one of six clones after passage 6 and six of six clones after passage 10) (Table 2). This finding is consistent with the result described above showing that the MNC substitution is essential to compensation for the BH10-LD3 deletion. The next mutation to be detected was CA1 in two of six clones after 10 passages under conditions of high levels of RT activity and virus-induced cytopathology. The MP2 substitution was observed only after 15 passages in three of six clones, and MA1 appeared last (in five of six clones after 23 passages).
TABLE 2.
Sequential appearance of various point mutations within mutated BH10-LD3 virus after long-term culture in MT-2 cellsa
| Mutation | No. of clones with mutation during passage:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 5 | 6 | 7 | 10 | 12 | 15 | 18 | 23 | 29 | |
| MA1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 6 |
| CA1 | 0 | 0 | 0 | 2 | 4 | 4 | 3 | 4 | 5 |
| MP2 | 0 | 0 | 0 | 0 | 0 | 3 | 4 | 6 | 6 |
| MNC | 0 | 1 | 2 | 6 | 6 | 6 | 6 | 6 | 6 |
| LD3 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 |
Six positive clones were sequenced per passage. Bold indicates the first appearance of the given mutation.
Long-term culture of BH10-LD4 viruses in MT-2 cells yielded compensatory point mutations similar to those observed for BH10-LD3 viruses.
Both the BH10-LD3 and BH10-LD4 deletion mutations involve disruption of the SL1 RNA structure and diminish HIV replication capacity (30). We were therefore anxious to determine whether compensatory point mutations similar to those seen with BH10-LD3 viruses might arise after long-term culture of BH10-LD4 viruses in MT-2 cells. After 13 passages, a virus termed BH10-LD4-13 that possessed wild-type replication properties had emerged (data not shown). In order to detect the presence of any compensatory mutations, a 2-kb DNA fragment (nt −454 to +1548) containing the BH10-LD4 deletion was amplified from the DNA of infected MT-2 cells as described above. The DNA segment thus generated was used to replace the equivalent region in BH10 to generate a clone termed BH10-LD4-HA. The results of infectivity assays showed that BH10-LD4-HA was as replication competent as wild-type virus (data not shown). Therefore, we sequenced this 2-kb DNA fragment and observed that the BH10-LD4 deletion was still present alongside three point mutations termed MA2 (V35L), MP2 (T12I), and MNC (T24I) (Fig. 2). Both the MP2 and MNC point mutations were identical to those previously identified in the reverted BH10-LD3 virus, while the MA2 substitution involved a change of Val-35 to Leu instead of to Ile as was observed in the case of MA1. Therefore, near-identical profiles of these point mutations were observed in the genomes of viruses that had emerged from long-term growth of both the BH10-LD3 and BH10-LD4 viruses.
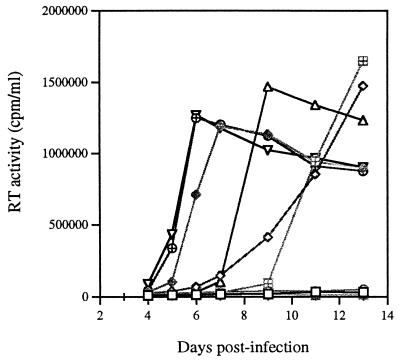
The ability of each of these three point mutations, MA2, MP2, and MNC, to compensate for the BH10-LD4 deletion mutation was next investigated by introducing them individually or collectively into the mutated BH10-LD4 DNA clone to generate the following constructs: BH10-LD4-MA2, BH10-LD4-MP2, BH10-LD4-MNC, BH10-LD4-MA2-MP2, BH10-LD4-MA2-MNC, BH10-LD4-MP2-MNC, and BH10-LD4-MA2-MP2-MNC. After transfection into COS-7 cells, the infectiousness of the viruses thereby produced was examined in MT-2 cells. The results of Fig. 4 show the following. (i) Neither the MA2 nor the MNC point mutation alone (i.e., neither BH10-LD4-MA2 nor BH10-LD4-MNC) was able to restore infectiousness to the BH10-LD4 mutant virus. However, the MP2 point mutation (i.e., BH10-LD4-MP2) did potentiate viral replication and it yielded high levels of RT activity in culture fluids after 13 days. (ii) Recombining the MA2 and MNC point mutations into the same construct, i.e., BH10-LD4-MA2-MNC, led to a renewal of viral replication, as seen with BH10-LD3 virus; similar results were seen with combinations of MP2 and MNC (i.e., BH10-LD4-MP2-MNC) or MA2 and MP2 (i.e., BH10-LD4-MA2-MP2) within the BH10-LD4 construct). (iii) Recombination of the three point mutations, i.e., MA2, MP2, and MNC, within BH10-LD4 (i.e., BH10-LD4-MA2-MP2-MNC) yielded even higher levels of infectiousness, as shown by both growth curves and 50% tissue culture infective doses (TCID50s) (Fig. 4; Table 2). Therefore, each of these three point mutations, MA2, MP2, and MNC, was able to contribute to the restoration of viral replication competence, and the MP2 point mutation played a key role in this regard.
FIG. 4.
Roles of the point mutations MA2, MP2, and MNC combined either separately or together with the BH10-LD4 deletion mutation to yield BH10-LD4-MA2 (□), BH10-LD4-MP2 (◊), BH10-LD4-MNC (○), BH10-LD4-MA2-MP2 (▵), BH10-LD4-MA2-MNC (⊞), BH10-LD4-MP2-MNC (◊+), and BH10-LD4-MA2-MP2-MNC (⊕). These constructs and wild-type BH10 (▿) were transfected into COS-7 cells, and the viruses thus generated were used to infect MT-2 cells. Viral replication was assessed by monitoring RT levels in culture fluids. ┌, control.
To further evaluate the importance of each of these point mutations in the rescue of BH10-LD4 virus, we determined the order in which they appeared during long-term culture of BH10-LD4 virus in MT-2 cells by sequencing cDNA from infected cells at passages 3, 4, 5, 7, 10, and 13. Table 3 shows that the MP2 point mutation appeared first and was present in two of six clones after three passages. Next to be detected was MNC in one of six clones at passage 4, followed by MA2 in one of six clones at passage 8. All three point mutations were present in all clones sequenced after 13 passages. These findings are consistent with the notion that MP2 was the most important mutation involved in the rescue of mutated BH10-LD4 virus.
TABLE 3.
Order of accumulation of the point mutations MA2, MP2, and MNC within mutated BH10-LD4 virus when cultured in MT-2 cellsa
| Mutation | No. of clones with mutation during passage:
|
|||||
|---|---|---|---|---|---|---|
| 3 | 4 | 5 | 7 | 10 | 13 | |
| MA2 | 0 | 0 | 0 | 1 | 3 | 6 |
| MP2 | 2 | 5 | 6 | 6 | 6 | 6 |
| MNC | 0 | 1 | 6 | 6 | 6 | 6 |
| LD4 | 6 | 6 | 6 | 6 | 6 | 6 |
Six clones were sequenced per passage. Bold indicates the first appearance the given mutation.
Comparison of the MA1 and MA2 point mutations in ability to compensate for the BH10-LD3 and BH10-LD4 deletions.
The above data indicate that two different amino acid substitutions were detected at position 35 in MA after long-term growth of the BH10-LD3 and BH10-LD4 viruses and that the two substitutions involved changes from Val to Ile and Leu, respectively, i.e., MA1 and MA2. Since Ile and Leu are similar in structure, it was conceivable that the MA1 and MA2 point mutations might function interchangeably in compensating for the BH10-LD3 and BH10-LD4 deletions.
MA2 and MA1 mutations were also introduced into viruses with BH10-LD3 and BH10-LD4 deletions to yield the following constructs, which also variably contained the MP2 and MNC point mutations: BH10-LD3-MA2, BH10-LD3-MA2-MP2, BH10-LD3-MA2-MNC, BH10-LD3-MA2-MP2-MNC, BH10-LD4-MA1, BH10-LD4-MA1-MP2, BH10-LD4-MA1-MNC, and BH10-LD4-MA1-MP2-MNC.
Infectivity assays were performed as described above and showed that neither the BH10-LD3-MA2 nor the BH10-LD3-MA2-MP2 virus was able to generate high levels of RT activity, while, in contrast, the BH10-LD3-MA2-MNC virus was able to replicate at a reasonably high level (Fig. 5). Introduction of the MA2 point mutation into BH10-LD3-MP2-MNC resulted in an increase in TCID50 (Table 1). Therefore, MA2 was able to function in place of MA1 in regard to compensating for the BH10-LD3 deletion. Similar experiments revealed that BH10-LD4-MA1-MNC generated high levels of RT activity after 11 days in culture, as did BH10-LD4-MA1-MP2 (Fig. 5). Finally, the presence of MA1 increased the TCID50 of BH10-LD4-MP2-MNC (Table 1). Therefore, the MA1 and MA2 point mutations play similar roles in compensating for either the BH10-LD3 or the BH10-LD4 deletion.
FIG. 5.
Roles of the MA1 and MA2 point mutations in compensation for both the BH10-LD3 and BH10-LD4 deletion mutations through generation of BH10-LD3-MA2 (□), BH10-LD3-MA2-MP2 (◊), BH10-LD3-MA2-MNC (○), BH10-LD4-MA1 (▵), BH10-LD4-MA1-MP2 (⊞), and BH10-LD4-MA1-MNC (◊+). These clones were transfected into COS-7 cells, and the progeny viruses obtained were used to infect MT-2 cells. ⊕, BH10; ▿, control.
The BH10-LD4 deletion only slightly affects encapsidation of viral genomic RNA.
Previous studies showed that the MNC point mutation was able to correct for the diminished levels of viral genomic RNA encapsidation that occurred in the case of the BH10-LD3 deletion (30). Since MA2 and MP2 were together able to restore high levels of infectiousness to BH10-LD4 mutant virus in the absence of MNC (see above), it was suggested that BH10-LD4 might not be severely impaired in regard to packaging of viral RNA. This was assessed by means of slot blot experiments that showed that both BH10-LD4 and BH10-LD4-MP2-MNC viruses packaged levels of viral genomic RNA similar to those packaged by the wild-type BH10 virus (Fig. 6). Therefore, those sequences that were deleted from BH10-LD3 viruses probably play a key role in packaging of viral genomic RNA while those that were deleted from BH10-LD4 viruses do not.
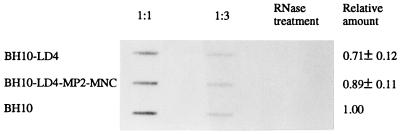
FIG. 6.
Encapsidation of viral genomic RNA into wild-type (BH10) and mutated (BH10-LD4 and BH10-LD4-MP2-MNC) viruses. Slot blot experiments were performed with viral RNA purified from viruses containing either 100 or 33.3 ng of p24. Viral RNA digested with RNase served as a negative control. Amounts of viral RNA packaged by either wild-type or mutated viruses were determined by arbitrarily setting a value of 1.0 for the former.
These studies have examined the abilities of two distinctly mutated viruses, BH10-LD3 and BH10-LD4, both involving deletions within SL1, to revert to wild-type replication kinetics after long-term culture in a permissive cell system. Surprisingly, all of the compensatory mutations were identified in the Gag protein and not within the 5′-end noncoding RNA leader region. We have observed that the revertants possessed similar mutations in the gag gene in each case and that the MP2 and MNC point mutations appeared first and were able to restore viral replication ability to fairly high levels. Although mutations at position 35 in MA were observed for both BH10-LD3 and BH10-LD4 at later stages, they involved replacements of amino acids by Ile and Leu, respectively. It is noteworthy that Ile and Leu are structurally similar and were functionally interchangeable in regard to compensation for the BH10-LD3 and BH10-LD4 deletions. Thus, deletions of the sequences from +238 to +253 and from +261 to +274 within SL1 can be compensated for by similar point mutations within gag, and therefore, the two sequences probably possess similar roles in viral replication. This work provides important in vivo confirmation for the existence of an SL1 functional structure within the HIV-1 RNA sequence from +238 to +274. However, our results also indicate that the two deleted segments are not functionally equivalent. First, an additional point mutation, CA1, was detected in mutated BH10-LD3 but not BH10-LD4 viruses during long-term culture. Second, the MNC and MP2 point mutations were observed to have different roles in compensation of the deletions in BH10-LD3 and BH10-LD4 viruses. For example, MNC was a major factor in regard to the replication kinetics of BH10-LD3 and was able to confer infectiousness on this construct when combined with each of the MA1, CA1, and MP2 point mutations. Furthermore, MNC was the first substitution to be acquired by BH10-LD3 virus during long-term culture.
In contrast, MP2 played a more important role than MNC in regard to the BH10-LD4 deletion. For example, MP2 alone was able to confer infectiousness on BH10-LD4 and was the first point mutation to be identified in BH10-LD4 virus during long-term culture in MT-2 cells. Biochemical analysis showed that BH10-LD3 but not BH10-LD4 virus was severely compromised in the ability to encapsidate viral genomic RNA, helping to explain why the MNC mutation, important for restoration of packaging activity, was crucial to compensate for BH10-LD3 but not for BH10-LD4 (30). Hence, the sequences from +238 to +253 and from +261 to +274 together constitute a functional element, yet at the same time, they play distinct roles in viral replication. Ongoing studies are being performed in regard to the effects of the BH10-LD3 and BH10-LD4 deletions as well as of the newly identified compensatory point mutations on viral RNA dimerization.
The different effects of the BH10-LD3 and BH10-LD4 deletions on viral RNA encapsidation might be due to their ability to modulate stem-loop secondary structures in different ways. In the case of wild-type viral RNA, cis-acting RNA packaging signals result in four distinct stem-loop structures (i.e., SL1 to SL4) (Fig. 1A). Deletion of the fragment from nt +238 to +253 (i.e., BH10-LD3) eliminates SL1 and SL3, both of which can bind to Gag protein and are involved in RNA packaging (Fig. 1B) (10). In contrast, the BH10-LD4 deletion (nt +261 to +274) does not disrupt the formation of SL2, SL3, or SL4 and, moreover, leads to the formation of an SL1-like RNA structure (Fig. 1C). This explains how the BH10-LD3 but not the BH10-LD4 deletion severely compromises viral RNA packaging.
The mechanisms involved in compensation for the BH10-LD3 and BH10-LD4 deletions can be inferred from the amino acids involved in the various point mutations that have been identified. For example, MNC involves a change of Thr-24 to Ile within the first Zn finger motif of NC, i.e., CysPheAsnCysGlyLysGluGlyHisThrAlaArgAsnCys (Fig. 2). NC is known to play a dominant role in regulation of packaging of viral genomic RNA, and mutagenesis studies have demonstrated that both the Zn fingers themselves as well as the basic amino acids that flank them are involved in this process. Furthermore, the first Zn finger domain is functionally more important than the second (2, 7–9, 14, 15, 18, 38). Therefore, it is consistent with these observations that the MNC point mutation was able to restore levels of packaging of viral RNA in the case of the BH10-LD3-MNC recombinant virus.
The MP2 point mutation involves a substitution of Ile for Thr at position 12 in p2, a peptide that is only 14 amino acids long. It is interesting that Thr-12 is located at the P3 position at the initial site that is cleaved by protease (PR), i.e., AlaThrIleMet*MetGlnArgGly, located between the C terminus of p2 and the N terminus of NC (Fig. 2). Therefore, replacement of Thr by Ile may also have changed the nature of this cleavage site and/or the ability of Gag and/or Gag-Pol to serve as the substrate for PR. Previous results have shown that p2 plays an important role in the processing of the Gag precursor protein and in viral maturation (1, 24, 37, 42). Recent work also implicated the p2 domain in the specific packaging of HIV-1 genomic RNA (22). The fact that the MP2 mutation can compensate for deletions in SL1 suggests that the latter RNA element may also participate in the processing of Gag protein.
It is also interesting that mutations within MA (V35I and V35L) were able to compensate for deletion mutations within the SL1 structure. The amino acid stretch within MA from positions 30 to 43, within which lies Val35, forms an α-helix (i.e., α-H2) that constitutes an N-terminal globular structure (21). Others have shown that the SL1 RNA hairpin region can bind to Gag protein with high affinity in cell-free experiments (10). Thus, such interactions may conceivably play important roles in Gag protein assembly and in the processing of Gag by PR in vivo. Our deletions within SL1 probably affected interactions between Gag protein and viral genomic RNA and may have thereby compromised the assembly of Gag proteins. The MA1 and MA2 point mutations may help to compensate for this defect by modulating the assembly of Gag proteins in the case of the viral deletion mutants.
Another point mutation at position 91 (I91T) in CA termed CA1 was also identified as involved in compensation for the BH10-LD3 deletion. Interestingly, the Ile at position 91 is one of only nine amino acids (i.e., 85-ProValHisAlaGlyProIleAlaPro-93) within the CA protein that comprises the binding site for cyclophilin A (CypA). Crystal structure analysis of the CA-CypA complex shows that the side chain of Ile-91 projects directly out from the CypA active site and has no intermolecular contact (17). Mutagenesis studies also showed that replacement of Ile-91 by either Ala or Val had no measurable effect on the efficiency of CypA binding (43). Therefore, the CA1 point mutation may not affect the binding of CypA to CA but may instead function through some other mechanism to help rescue the BH10-LD3 deletion. In addition, site-directed mutagenesis studies in our laboratory showed that the CA1 point mutation does not diminish the infectiousness of wild-type virus (data not shown). Studies of the effects of the MA1 and MA2 point mutations on viral infectivity are ongoing.
In summary, two distinct deletions within the noncoding viral RNA sequence, SL1, can be compensated for by substitutional mutations within four different Gag proteins. Analysis of these mutations suggests that the SL1 region is involved in Gag protein assembly and processing in addition to its well-documented roles in viral RNA packaging and dimerization.
Acknowledgments
This work was supported by grant R01 AI43878-01 from the National Institute of Allergy and Infectious Diseases and by the Medical Research Council of Canada.
We thank Maureen Oliveira for technical assistance.
REFERENCES
- 1.Accola M A, Höglund S, Göttlinger H G. A putative α-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72:2072–2078. doi: 10.1128/jvi.72.3.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Awang G, Sen D. Mode of dimerization of HIV-1 genomic RNA. Biochemistry. 1993;32:11453–11457. doi: 10.1021/bi00093a024. [DOI] [PubMed] [Google Scholar]
- 4.Baudin G, Marquet R, Isel C, Darlix J L, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout B, van Wamel J L B. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 7.Berkowitz R D, Goff S P. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology. 1994;202:233–246. doi: 10.1006/viro.1994.1339. [DOI] [PubMed] [Google Scholar]
- 8.Berkowitz R D, Luban J, Goff S P. Specific binding of human immunodeficiency virus type 1 gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J Virol. 1993;67:7190–7200. doi: 10.1128/jvi.67.12.7190-7200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkowitz R D, Ohagen Å, Höglund S, Goff S P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clever J, Sassetti C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clever J L, Wong M L, Parslow T G. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J Virol. 1996;70:5902–5908. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffin J M, Hughes S H, Varmus H E. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 14.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1500. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darlix J L, Lapadat-Tapolsky M, de Rocquigny H, Roques B. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 16.Dulbecco R. The nature of viruses. In: Dulbecco R, Ginsberg H S, editors. Virology. 2nd ed. Philadelphia, Pa: J. B. Lippincott; 1988. pp. 22–25. [Google Scholar]
- 17.Gamble T R, Vajdos F F, Yoo S, Worthylake D K, Houseweart M, Sundquist W I, Hill C P. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 18.Gorelick R J, Chabot D J, Rein A, Henderson L E, Arthur L O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzman R N, Wu Z R, Stalling C C, Pappalardo L, Borer P N, Summers M F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 ψ-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 20.Harrison G P, Lever A M L. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaye J F, Lever A M L. Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the p2 domain of Gag in RNA encapsidation. J Virol. 1998;72:5877–5885. doi: 10.1128/jvi.72.7.5877-5885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim H J, Lee K, O’Rear J J. A short sequence upstream of the 5′ major splice site is important for encapsidation of HIV-1 genomic RNA. Virology. 1994;198:336–340. doi: 10.1006/viro.1994.1037. [DOI] [PubMed] [Google Scholar]
- 24.Kräusslich H-G, Fäcke M, Heuser A-M, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 26.Laughrea M, Jetté L, Mak J, Kleiman L, Liang C, Wainberg M A. Mutations in the kissing-loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA packaging and dimerization. J Virol. 1997;71:3397–3406. doi: 10.1128/jvi.71.5.3397-3406.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenz C, Scheid A, Schaal H. Exon 1 leader sequences downstream of U5 are important for efficient human immunodeficiency virus type 1 gene expression. J Virol. 1997;71:2757–2764. doi: 10.1128/jvi.71.4.2757-2764.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang C, Li X G, Quan Y D, Laughrea M, Kleiman L, Hiscott J, Wainberg M A. Sequence elements downstream of the human immunodeficiency virus type 1 long terminal repeat are required for efficient viral gene transcription. J Mol Biol. 1997;272:167–177. doi: 10.1006/jmbi.1997.1239. [DOI] [PubMed] [Google Scholar]
- 30.Liang C, Rong L, Laughrea M, Kleiman L, Wainberg M A. Compensatory point mutations in the human immunodeficiency virus type 1 Gag region that are distal from deletion mutations in the dimerization initiation site can restore viral replication. J Virol. 1998;72:6629–6636. doi: 10.1128/jvi.72.8.6629-6636.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luban J, Goff S P. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J Virol. 1994;68:3784–3793. doi: 10.1128/jvi.68.6.3784-3793.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marquet R, Paillart J C, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus type 1 RNA involves sequences located upstream of the splice donor site. Nucleic Acids Res. 1994;22:145–151. doi: 10.1093/nar/22.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McBride M S, Panganiban A T. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol. 1997;71:2050–2058. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paillart J-C, Berthoux L, Ottmann M, Darlix J-L, Marquet R, Ehresmann B, Ehresmann C. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J Virol. 1996;70:8348–8354. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paillart J C, Marquet R, Skripkin E, Ehresmann B, Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J Biol Chem. 1994;269:27486–27493. [PubMed] [Google Scholar]
- 37.Pettit S C, Moody M D, Wehbie R S, Kaplan A H, Nantermet P V, Klein C A, Swanstrom R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poon D T K, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakaguchi K, Zambrano N, Baldwin E T, Shapiro B A, Erickson J W, Omichinski J G, Clore G M, Gronenborn A M, Appella E. Identification of a binding site for the human immunodeficiency virus type 1 nucleocapsid protein. Proc Natl Acad Sci USA. 1993;90:5219–5223. doi: 10.1073/pnas.90.11.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakuragi J-I, Panganiban A T. Human immunodeficiency virus type 1 RNA outside the primary encapsidation and dimer linkage region affects RNA dimer stability in vivo. J Virol. 1997;71:3250–3254. doi: 10.1128/jvi.71.4.3250-3254.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skripkin E, Paillart J C, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Kräusslich H-G. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J Virol. 1998;72:2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoo S, Myszka D G, Yeh C-Y, McMurray M, Hill C P, Sundquist W I. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J Mol Biol. 1997;269:780–795. doi: 10.1006/jmbi.1997.1051. [DOI] [PubMed] [Google Scholar]