Abstract
A complete endogenous type C viral genome has been isolated from a baboon genomic library. The provirus, Papio cynocephalus endogenous retrovirus (PcEV), is 8,572 nucleotides long, and 38 to 59 proviral copies per baboon genome are found. The PcEV provirus possesses the typical simple retroviral gene organization, including two long terminal repeats and genes encoding gag, pol, and env proteins. The open reading frames for gag-pol and env are complete but have premature stop codons or frameshift mutations. The primer binding site of PcEV is complementary to tRNAGly. The gag and pol genes of PcEV are closely related to those of the baboon endogenous virus (BaEV). The env coding region of PcEV is related to the env genes of type C retroviruses. This suggests that PcEV is one of the ancestors of BaEV contributing the type C gag-pol genome fragment to the type C/D recombinant virus BaEV. Earlier it was shown that another endogenous type D virus (simian endogenous retrovirus) provided the env gene for BaEV (A. C. van der Kuyl et al., J. Virol. 71:3666–3676, 1997).
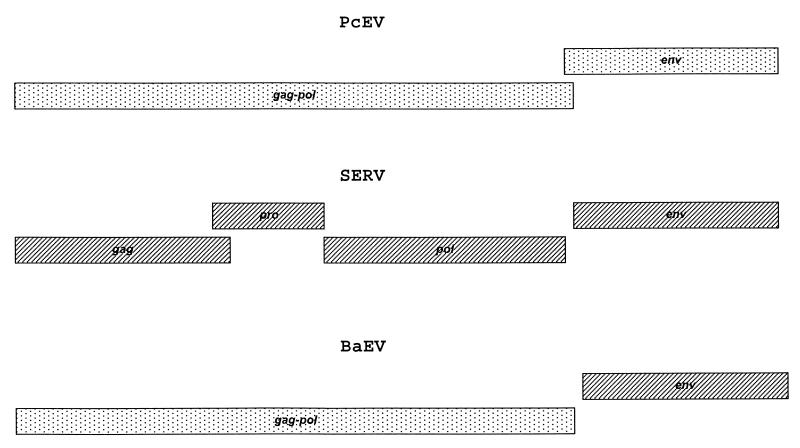
Two complete endogenous retroviral sequences, those of baboon endogenous virus (BaEV) (15) and simian endogenous retrovirus (SERV) (39), have been isolated from the genome of the baboon (Papio cynocephalus). Originally, BaEV was isolated from baboon tissue by cocultivation with permissive cell lines (3, 34). The proviruses of BaEV are present only in the genomes of the Papionini tribe and in African green monkeys (Cercopithecus aethiops). Analysis of viral sequences suggested that BaEV was repeatedly introduced in the germ line of these species between 24,000 and 400,000 years ago and was not inherited from an early common ancestor of these African monkeys (36). BaEV proviruses appear to be chimeric, containing type C gag and pol and type D env genes (see Fig. 1) (15), which suggested that BaEV is the result of a recombination event between two retroviruses in the past. It is well-known that recombination in retroviruses is common during retroviral evolution. For human immunodeficiency virus (HIV), this process may account for novel HIV genotypes arising within human populations. It has been shown that up to 10% of the HIV type 1 genomes studied have mosaic structures (24, 25). Previously, members of our team were involved in sequencing the first putative ancestor of BaEV, SERV, the first complete endogenous type D virus sequenced from primates (39). The gag and pol proteins of SERV are closely related to SRV1, SRV2, and SRV3, which are exogenous type D retroviruses causing simian AIDS in captive macaques (19). The env gene, coding the gp70 and p20 proteins of SERV, is closely related to that of BaEV (see Fig. 1). PCR analysis of primate DNA showed that SERV-related proviral sequences are present in all Old World monkeys of the subfamily Cercopithecinae but not in those of Colobinae and Hominoidea (39). This suggested that SERV entered the germ line of a common ancestor at least 9 million years ago, which is the estimated time of the split between the Cercopithecinae and Colobinae (18).
FIG. 1.
Genetic organization of PcEV, SERV, and BaEV. Rectangles indicate ORFs for the genes marked. C-type genes are indicated by dots. D-type genes are indicated by stripes. PcEV contains two type C ORFs for the gag-pol and env genes, which overlap. SERV contains four type D ORFs, for the gag, pro, pol, and env genes. BaEV is a type C/D chimeric virus which contains a type C ORF for gag-pol and a type D env gene.
During our initial studies of BaEV integrations in the baboon genome with BaEV-specific probes (37), a lambda clone (named 30.1) which contained the gag gene of a novel type C virus with 80% homology to BaEV gag was obtained. Unfortunately, sequencing revealed that clone 30.1 contained a truncated proviral genome starting in the gag gene, due to the method of library construction. The present study was designed to obtain the complete genome of this novel type C virus.
A baboon genomic library in the lambda DASH II vector, constructed from kidney tissue of a healthy 18-year-old male baboon, was obtained from Stratagene (La Jolla, Calif.), and approximately 64,000 plaques were screened with a probe homologous to the clone 30.1 gag gene. A total of 32 positive plaques were obtained and purified. After a second screening with BaEV reverse transcriptase (RT) (type C) and BaEV env (type D) probes, a clone named E7, which hybridized with both the gag and RT probes, but not with the BaEV env probe, was obtained. Lambda DNA of E7 was isolated by using the Wizard Lambda Preps DNA purification system from Promega (Madison, Wis.) and was digested by BamHI, XbaI, and HindIII. Southern blots were probed with 32P-labelled fragments homologous to the 30.1 gag and BaEV pol genes, respectively. Subcloning was done with pBS-SK vector (Stratagene). Plasmid DNA was prepared by using the QIAprep Spin Miniprep kit (Qiagen, Hilden, Germany) and sequenced from both directions with an Applied Biosystems 373A or 377 automated sequencer with M13 reverse and T7 dye primers, following the manufacturer’s protocols. To fill in some gaps between the cloned fragments, phage DNA was sequenced directly with purified specific primers and the ABI Prism Big-Dye terminator cycle sequencing kit (Perkin-Elmer Applied Biosystems, Foster City, Calif.). Analysis of the obtained sequences showed that clone E7 contained the complete genome of a novel endogenous type C virus, which we named Papio cynocephalus endogenous retrovirus (PcEV).
Alignment of the sequences was done with the PCGENE software package. The phylogenetic analyses were done by the neighbor-joining method, as implemented in the TREECON package (35). Evolutionary distances were estimated by Kimura’s methods (16) for both nucleotide and amino acid sequences. One hundred bootstrap replicates were analyzed. Other methods for distance determination did not influence the trees. Gaps introduced for optimal alignment were not considered informative and were not included in the analysis. The GenBank accession number of PcEV is provided below, and the accession numbers of the sequences used for comparison were D10032 (BaEV), M18247 (feline leukemia virus [FeLV]), U60065 (gibbon ape leukemia virus [GaLV]), Y17013 (porcine endogenous virus [PERV]), J01998 (murine leukemia virus [MuLV]), M77194 (rat leukemia virus), AF038599 (Sus scrofa porcine endogenous retrovirus), and AF053745 (Mus dunni endogenous virus).
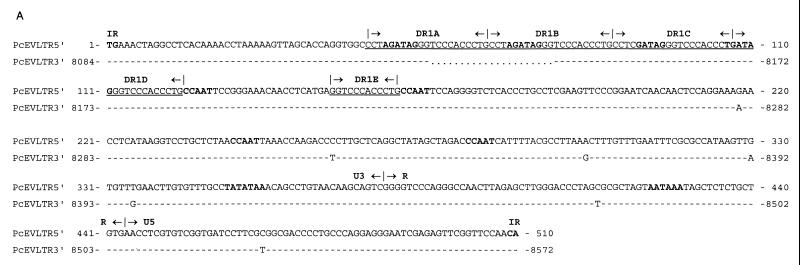
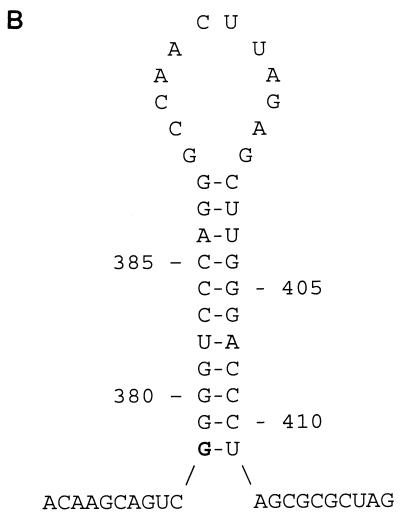
The baboon genomic clone E7 contained a complete retroviral sequence with a length of 8,572 nucleotides (nt) and a genomic organization identical to that of known type C retroviruses (Fig. 1). Two main open reading frames (ORFs) are present, with coding regions for gag-pol and env genes. The env gene overlaps the pol gene in the manner reported for other type C viruses, such as FeLV (11) and MuLV (31). The coding region is flanked by two long terminal repeats (LTRs) of 510 and 489 nt, respectively, including 377 and 356 nt in the U3 regions of the 5′ and 3′ LTRs, respectively, 66 nt in the R region, and 67 nt in the U5 region (Fig. 2A). Through comparison of the 5′ and 3′ LTR sequences, the 5′ boundary of the U3 region was found. The 3′ boundary of the 5′ LTR U5 region is located at nt 510. This corresponds with the common pattern of LTRs being bound by TG/CA inverted repeats (33). The boundary between the U3 and the R regions was determined by the start of a hairpin structure (Fig. 2B), like the TAR hairpin in HIV RNA. A hairpin with a stable structure (ΔG = −20.6 kcal) was predicted between nt 378 and 411, so it was assumed that the first G (nt 378) of the stem of the hairpin structure was the beginning of PcEV viral RNA. The boundary between the R and the U5 regions was found by comparison of the LTR sequences from PcEV, BaEV, and RD114. RD114 is an endogenous virus of domestic cats which has a high level of homology to BaEV and supposedly arose from a cross-species transmission (4, 29).
FIG. 2.
(A) Nucleotide sequence alignment of the 5′ and 3′ LTRs of PcEV. Identical nucleotides are indicated by dashes; gaps introduced for optimal alignment are indicated by dots. The LTRs of PcEV differ in length due to the deletion of a direct repeat sequence in the 3′ LTR. The boundaries between U3, R, and U5 are indicated by vertical lines and arrows. The four GATA-1 binding sites ([A/T]GATA[A/G]), the CAAT box (CCAAT), the TATA box (TATATAA), the polyadenylation site (AATAAA), and the flanking indirect repeats (IR) are indicated by bold letters. The direct repeat sequences DR1A to DR1E are indicated by underlining and demarcated by vertical lines. (B) Predicted RNA secondary structure at the 5′ end of the R region in the LTRs of PcEV. The length of the stem is 11 nt, the size of the loop is 12 nt, and the free energy value (at 25°C) is −20.6 kcal. The predicted first nucleotide of viral RNA is indicated by bold type.
Several regulatory sequence motifs are present in the U3 and the R regions of the PcEV LTR, including one consensus TATA box (TATATAA) at nt 351, one consensus polyadenylation site (ATAAA) at nt 423, and four consensus CAAT boxes (CCAAT) located at nt 124, 161, 242, and 282 (Fig. 2A). A set of direct repeats, designated DR1, was found in the U3 region of the PcEV LTR. The length of the DR1 is 21 nt, and it has the motif 5′-CCTAGATAGGGTCCCACCCTG-3′, which contains the GATA-1 binding site ([A/T]GATA[A/G]). In the 5′ LTR, the DR1 is perfectly repeated three times (DR1A, DR1B, and DR1C); a fourth imperfect repeat (DR1D) contains 16 of the 21 nt and is adjacent to DR1C; 25 nt downstream of DR1D, another partial DR1 (DR1E), which is only 12 nt (5′-GGTCCCACCCTG-3′) long, was found. The 5′ and 3′ LTR sequences of PcEV are almost identical (98% at the nucleotide level), except that DR1B is missing from the 3′ LTR so that the 3′ LTR is 21 nucleotides shorter than the 5′ LTR (Fig. 2A). It has been shown that different DR sequences are present at the 5′ end of the U3 regions of several retroviruses (17, 29, 40). Since the U3 region of the LTR contains promoter and enhancer sequences, we examined the DR1 of PcEV LTR for the presence of consensus enhancer elements. It was found that the sequence between DR1A and DR1D contains four consensus sequences of the GATA-1 binding site ([A/T]GATA[A/G]) (Fig. 2A) (21). GATA-1 is the major erythroid transcription factor. It activates transcription in a synergistic fashion with two Krüppel family factors, Sp1 (recognizes GC) and EKLF (recognizes G[T/C]ACC), and their binding sites are often found in close association in the promoters and enhancers of numerous erythroid cell-expressed genes and appear to cooperate in directing their expression (22). A recent report showed that three GATA family members (GATA-1, -2, and -3) could bind to the GATA box within the U3 regions of Cas-Br-E and Graffi retroviruses in vitro and activate the respective viruses. These murine retroviruses can induce myeloid leukemia in mice (17). Besides the four GATA-1 binding sites in the LTR, there are also an EKLF binding consensus sequence (GCACC, at nt 33 of PcEV U3) and many GC stretches that could be binding sites for Sp1, which suggests that PcEV possibly infected hematopoietic cells.
The 5′ untranslated region of the PcEV genome extends 452 nt from the 3′ end of the U5 region of the LTR to the initiation codon for the gag protein. A stretch of 18 nt (5′-TGGTGCATTGGCCGGGAA-3′) located immediately downstream of the 5′ LTR constitutes the primer binding site (PBS) of PcEV. This PBS is perfectly complementary to the 3′ end of a tRNA isotype of human origin, tRNAGly (30). Interestingly, RD114 also uses tRNAGly to initiate viral gene amplification (29), unlike BaEV and other type C viruses, which commonly use tRNAPro. Downstream of the PBS, between nt 619 and 622, a splice donor site was found (AGGT) by comparison of the PcEV and BaEV genomes (15).
The gag coding region of PcEV was identified based on sequence homology to the gag gene of BaEV. These genes share 81% of their nucleotides. Sequence comparison of the PcEV and BaEV gag genes showed that there is a single nucleotide deletion located between nt 1019 and 1020, causing a frameshift in the gag ORF of PcEV. The deletion was confirmed by both dye-primer and dye-terminator sequencing methods to eliminate the possibility that it was introduced during PCR. Another lambda clone isolated at the same time did not contain this single nucleotide deletion. Except for the deletion, the reading frame for the gag gene of PcEV is completely open. The predicted precursor protein is 543 amino acids (aa) long, and the homology between PcEV and BaEV gag proteins is 84% at the amino acid level. A Cys-His box (C-X-X-C-X-X-X-X-H-X-X-X-X-C), required for efficient RNA packaging (13, 20), is present in the predicted p10 polypeptide of the PcEV gag protein. Three potential N-linked glycosylation sites (N-X-[T/S]) (1) were found in the PcEV gag protein. One was found at the N terminus of p12; the other two are located at the N terminus of p30.
The gag-pro junction in PcEV was identified based on sequence homology to BaEV. A putative protease gene is present between nt 2570 and nt 2955, starting 21 nt upstream of the gag stop codon. The organization of the PcEV gag, pro, and pol OFRs is the same as those described for BaEV and other type C viruses, like Moloney MuLV (41). A suppressor tRNA is responsible for the readthrough of the gag stop codon (14). The homology between PcEV and BaEV protease genes is 87% at the nucleotide level. A 10-nt poly(C) stretch was found at the 5′ end of the protease gene, which caused the PcEV protease gene to be out of frame. In BaEV, this stretch is only 8 nt long. After correcting for the two cytosine insertions, an ORF of 128 codons which has 92% homology to the BaEV protease at the amino acid level was identified. A 3-amino-acid motif (DTG, encoded by nt 2674 to 2682) is located at the N terminus of the protease in PcEV. This motif is part of the activation domain of the retroviral protease and is highly conserved among the proteases of different members of the retrovirus family (32).
The putative pol gene of PcEV is 3,210 nt long (from nt 2956 to 6165), encoding 1,069 amino acid residues of both the putative reverse transcriptase and the endonuclease. The pol ORF is completely open in clone E7 of PcEV and is closely related to the BaEV pol gene, with 92% homology at the nucleotide level and 96% homology at the amino acid level. Two highly conserved sequences found in reverse transcriptases, LPQGFK and QY(V/M)DD (12), were also present in the PcEV pol protein. Highly conserved sequences were also found in the endonuclease domain. The first one is composed of a pair of histidine residues (encoded by nt 5128 to 5130) and a pair of cysteine residues (encoded by nt 5233 to 5235) that are separated by 30 aa residues, H-X3-H-X30-C-X2-C; the general form of the second motif is D-X39–58-D-X35-E, which is the catalytic core of the enzyme (32). In the endonuclease domain of PcEV, the first aspartic acid is encoded by nt 5383 to 5385 and the second aspartic acid is encoded by nt 5503 to 5505, and they are separated by 39 aa residues, but the last glutamic acid is changed to lysine due to a G→A mutation at nt 5611.
The ORF encoding the PcEV env is 1,947 nt long (nt 6111 to 8057 of the complete genome) and encodes 648 aa. Like other mammalian type C retroviruses, such as Moloney MuLV, FeLV, and GaLV, the 5′ end of PcEV env gene overlaps the 3′ end of the pol gene by 55 nt. The ORF of PcEV env is interrupted by two premature termination codons located at codons 203 and 582. The putative cleavage site separating the gp70 and p15E proteins is the amino acid sequence A-L-V-H (aa 479 to 482). The region upstream of this cleavage site is part of the surface peptide (SU) gp70 and contains five potential glycosylation sites (N-X-[T/S]) (1), located at amino acid residues 60, 310, 342, 345, and 381. In the transmembrane protein (TM) p15E of PcEV, a putative immunosuppressive peptide of 26 aa residues (9) in which 23 out of 26 aa (aa 521 to 546) are identical with the immunosuppressive domain of other mammalian retroviruses is identified.
Downstream of the env stop codon (located at nt 8057), a purine-rich stretch (5′-AAAAAGAGGAGGG-3′) was found between nt 8069 and 8081. It is separated from the 5′ end of 3′ LTR (nt 8084) by two adenines. Because of its genomic location and sequence, it has been speculated that this element is an initiation site for positive-strand DNA synthesis (8).
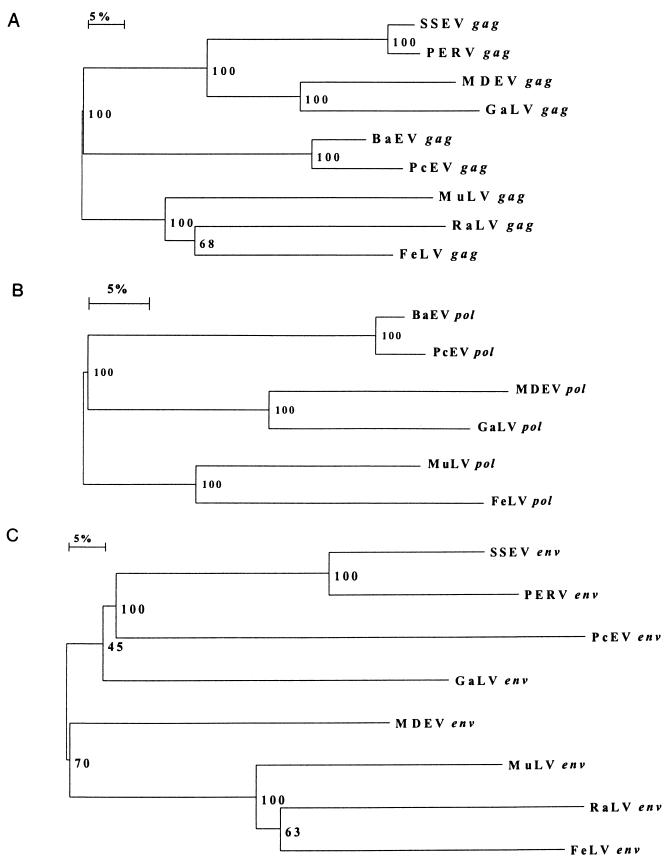
To estimate the level of gene divergence between PcEV and other type C viruses, including BaEV, gag, pol, and env amino acid sequences from different mammalian type C retrovirus were aligned and phylogenetic analyses were performed (Fig. 3A, B and C, respectively). The results of these analyses showed that PcEV gag and pol are closely related to BaEV gag and pol. Furthermore, the cluster of gag proteins from PcEV and BaEV is more closely related to a cluster of proteins from M. dunni endogenous virus (40) and GaLV (10) than to a cluster consisting of gag proteins from MuLV (28) and FeLV (11) (Fig. 3A). The same was found to be true for the pol gene (Fig. 3B). Since BaEV is a chimeric virus which contains type C gag and pol genes but a type D env gene, the BaEV and PcEV env proteins are only distantly related. Phylogenetic analysis of aligned type C env proteins showed that the PcEV env is different from all env proteins included in the analysis but is most closely related to PERV. S. scrofa porcine endogenous retrovirus, and GaLV (Fig. 3C).
FIG. 3.
Neighbor-joining trees based upon derived protein sequences for the gag (A), pol (B), and env (C) genes of type C retroviruses. Bootstrap values for 100 replicated trees are indicated. MDEV, M. dunni endogenous virus; RaLV, rat leukemia virus; SSEV, S. scrofa endogenous retrovirus.
To estimate the copy number of PcEV in the baboon genome, a genomic DNA sample of Papio hamadryas was obtained from the Zoologischer Garten Leipzig (Leipzig, Germany) through the European Gene Bank of Primates (Munich, Germany). A limiting dilution of genomic DNA and nested PCR were performed. The outer primers for the PCR were 5′-CGCACTCAAGGACTAGAGCC-3′ (upstream) and 5′-CTTGATGCGGACCAGGTTGC-3′ (downstream); the nested primers were 5′-ACGCTCCGCGAACCCGCTCAAG-3′ (upstream) and 5′-AAGGACATGGTTATGTACCA-3′ (downstream). The nested PCR was optimized to amplify a single copy of target DNA. Baboon genomic DNA of P. hamadryas (10 ng) was diluted in 10-fold increments for the nested PCR, and the last two positive samples were used for additional twofold dilutions. For each twofold dilution, 10 nested PCRs were performed twice. The copy number of PcEV proviral genomes input in the PCR was calculated by a computer program called QUALITY (for “quantitation using a limiting dilution assay”) (26). In the calculations it was assumed that a baboon cell contains the same amount of genomic DNA as a human cell (6 pg of DNA/cell). The copy number of PcEV proviruses in the P. hamadryas diploid genome was thus estimated to be in the range of 38 to 59 copies.
In conclusion, we have isolated a full-length proviral genome of an endogenous type C retrovirus, PcEV, from a baboon genomic library. The proviral genome of PcEV contains two LTRs and two ORFs encoding gag-pol and env. However, the ORFs are all interrupted by either frameshift mutations or premature stop codons. The gag, pro and pol proteins of PcEV are closely related to BaEV gag, pro and pol. The env protein of PcEV is related to the env proteins of type C retroviruses, including the pig virus PERV, and the primate virus GaLV.
Baboon endogenous virus is one of the first-isolated and best-characterized complete endogenous retroviruses of primates. By using DNA hybridization techniques, preliminary studies indicated that BaEV genomes were present in all Old World monkey species in approximately 50 to 100 copies per cell (2, 5, 7, 27). However, based on PCR results, a previous study from our laboratory showed that BaEV was present in the genome of only a limited set of African monkeys, including baboons, geladas, mangabeys, mandrills, and African green monkeys. The germ line integrations were estimated to have occurred only 24,000 to 400,000 years ago, which is quite recent in evolutionary terms. The copy number of BaEV proviruses was found to be significantly lower (10 to 30 per cell) than previously reported (6, 36). BaEV is a chimeric type C/type D virus (15), suggesting that BaEV is the product of a recombination event following coinfection by a type C and a type D virus. An endogenous type D retrovirus, SERV, was isolated from a baboon genomic library with an env gene closely related to the BaEV env. As SERV proviruses can be found in all species of the Papionini and Cercopithecini tribes, SERV is older than BaEV and thus an ancestor of the type D env gene of BaEV (39). Phylogenetic analysis showed that the PcEV gag-pol region is closely related to that of BaEV. As a general rule, endogenous proviruses increase in copy number with time. The proviral copy number of PcEV in baboon genome (∼38 to 59 copies/genome) is significantly higher than the copy number of BaEV. So, it is most likely that PcEV is older than BaEV and that it provided the type C gag-pol to BaEV.
PERV, a type C virus of pigs, was shown to be able to infect and replicate in human cells (23). Phylogenetic analysis of env proteins showed that among type C retroviral env proteins known at present, PcEV env is most closely related to PERV env. In the E7 isolate of PcEV, not all ORFs are open, due to the presence of either frameshift mutations or premature stop codons. However, different PcEV genomic clones isolated at the same time were shown to contain open gag and pol genes. As the PcEV genome is surprisingly intact and the integration number of PcEV in the baboon genome is sufficiently large (∼38 to 59 copies/diploid cell), an infectious type C virus could arise by recombination. The presence of PcEV, an almost intact type C retrovirus in the baboon genome, could constitute another problem when baboon organs or tissues are used in xenotransplantation (38).
Nucleotide sequence accession number.
The GenBank accession number of PcEV is AF142988.
Acknowledgments
We thank John Dekker and Jolanda Maas for helpful technical support and Vladimir Lukashov for stimulating discussions.
This study was partly supported by Amsterdam Support Diagnostics, Inc.
REFERENCES
- 1.Bause E. Structural requirements of N-glycosylation of proteins. Studies with proline peptides as conformational probes. Biochem J. 1983;209:331–336. doi: 10.1042/bj2090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benveniste R E, Heinemann R, Wilson G L, Callahan R, Todaro G J. Detection of baboon type C viral sequences in various primate tissues by molecular hybridization. J Virol. 1974;14:56–67. doi: 10.1128/jvi.14.1.56-67.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benveniste R E, Lieber M M, Livingston D M, Sherr C J, Todaro G J, Kalter S S. Infectious C-type virus isolated from a baboon placenta. Nature. 1974;248:17–20. doi: 10.1038/248017a0. [DOI] [PubMed] [Google Scholar]
- 4.Benveniste R E, Todaro G J. Evolution of C-type viral genes: inheritance of exogenously acquired viral genes. Nature. 1974;252:456–459. doi: 10.1038/252456a0. [DOI] [PubMed] [Google Scholar]
- 5.Benveniste R E, Todaro G J. Evolution of type C viral genes. I. Nucleic acid from baboon type C virus as a measure of divergence among primate species. Proc Natl Acad Sci USA. 1974;71:4513–4518. doi: 10.1073/pnas.71.11.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benveniste R E, Todaro G J. Multiple divergent copies of endogenous C-type virogenes in mammalian cells. Nature. 1974;252:171–173. doi: 10.1038/252170a0. [DOI] [PubMed] [Google Scholar]
- 7.Bonner T I, Todaro G J. The evolution of baboon endogenous type C virus: related sequences in the DNA of distant species. Virology. 1980;103:217–227. doi: 10.1016/0042-6822(80)90139-7. [DOI] [PubMed] [Google Scholar]
- 8.Chen H R, Barker W C. Nucleotide sequences of the retroviral long terminal repeats and their adjacent regions. Nucleic Acids Res. 1984;12:1767–1778. doi: 10.1093/nar/12.4.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cianciolo G J, Kipnis R J, Snyderman R. Similarity between p15E of murine and feline leukaemia viruses and p21 of HTLV. Nature. 1984;311:515. doi: 10.1038/311515a0. [DOI] [PubMed] [Google Scholar]
- 10.Delassus S, Sonigo P, Wain-Hobson S. Genetic organization of gibbon ape leukemia virus. Virology. 1989;137:205–213. doi: 10.1016/0042-6822(89)90236-5. [DOI] [PubMed] [Google Scholar]
- 11.Donahue P R, Hoover E A, Beltz G A, Riedel N, Hirsch V M, Overbaugh J, Mullins J I. Strong sequence conservation among horizontally transmissible, minimally pathogenic feline leukemia viruses. J Virol. 1988;62:722–731. doi: 10.1128/jvi.62.3.722-731.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donehower L A, Bohannon R C, Ford R J, Gibbs R A. The use of primers from highly conserved pol regions to identify uncharacterized retroviruses by the polymerase chain reaction. J Virol Methods. 1990;28:33–46. doi: 10.1016/0166-0934(90)90085-t. [DOI] [PubMed] [Google Scholar]
- 13.Green L M, Berg J M. A retroviral Cys-Xaa2-Cys-Xaa4-Cys peptide binds metal ions: spectroscopic studies and a proposed three-dimensional structure. Proc Natl Acad Sci USA. 1989;86:4047–4051. doi: 10.1073/pnas.86.11.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harada F, Peters G G, Dahlberg J E. The primer tRNA for Moloney leukemia virus DNA synthesis. J Biol Chem. 1979;254:10979–10985. [PubMed] [Google Scholar]
- 15.Kato S, Matsuo K, Nishimura N, Takahashi N, Takano T. The entire nucleotide sequence of baboon endogenous virus DNA: a chimeric genome structure of murine type C and simian type D retroviruses. Jpn J Genet. 1987;62:127–137. [Google Scholar]
- 16.Kimura M A. The neutral theory of molecular evolution. Cambridge, England: Cambridge University Press; 1983. [Google Scholar]
- 17.Lee S-Y, Howard T M, Rasheed S. Genetic analysis of the rat leukemia virus: influence of viral sequences in transduction of the c-ras proto-oncogene and expression of its transforming activity. J Virol. 1998;72:9906–9917. doi: 10.1128/jvi.72.12.9906-9917.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin R D. Primate origins: plugging the gaps. Nature. 1993;363:223–234. doi: 10.1038/363223a0. [DOI] [PubMed] [Google Scholar]
- 19.Marx P A, Maul D H, Osborn K G, Lerche N W, Moody P, Lowenstine L J, Henrickson R V, Arthur L O, Gilden R V, Gravell M, London W T, Sever J L, Levy A, Munn R J, Gardner M J. Simian AIDS: isolation of a type D retrovirus and transmission of the disease. Science. 1984;223:1083–1086. doi: 10.1126/science.6695196. [DOI] [PubMed] [Google Scholar]
- 20.Méric C, Goff S P. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J Virol. 1989;63:1558–1568. doi: 10.1128/jvi.63.4.1558-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merika M, Orkin S H. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merika M, Orkin S H. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Krüppel family proteins Sp1 and EKLF. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patience C, Takeuchi Y, Weiss R A. Infection of human cells by an endogenous retrovirus of pigs. Nat Med. 1997;3:282–286. doi: 10.1038/nm0397-282. [DOI] [PubMed] [Google Scholar]
- 24.Robertson D L, Hahn B H, Sharp P M. Recombination in AIDS viruses. J Mol Evol. 1995;40:249–259. doi: 10.1007/BF00163230. [DOI] [PubMed] [Google Scholar]
- 25.Robertson D L, Sharp P M, McCutchan F E, Hahn B H. Recombination in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 26.Rodrigo A G, Goracke P C, Rowhanian K, Mullins J I. Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res Hum Retroviruses. 1997;13:737–742. doi: 10.1089/aid.1997.13.737. [DOI] [PubMed] [Google Scholar]
- 27.Shih A, Coutavas E E, Rush M G. Evolutionary implications of primate endogenous retroviruses. Virology. 1991;182:495–502. doi: 10.1016/0042-6822(91)90590-8. [DOI] [PubMed] [Google Scholar]
- 28.Sijts E J, Leupers C J, Mengede E A, Loenen W A, van den Elsen P J, Melief C J. Cloning of the MCF1233 murine leukemia virus and identification sequences involved in viral tropism, oncogenicity and T cell epitope formation. Virus Res. 1994;34:339–349. doi: 10.1016/0168-1702(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 29.Spodick D A, Ghosh A K, Parimoo S, Roy-Burman P. The long terminal repeat of feline endogenous RD-114 retroviral DNAs: analysis of transcription regulatory activity and nucleotide sequence. Virus Res. 1988;9:263–283. doi: 10.1016/0168-1702(88)90035-4. [DOI] [PubMed] [Google Scholar]
- 30.Sprinzl M, Hartmann T, Weber J, Blank J, Zeidler R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989;17(Suppl.):1–172. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoye J P, Coffin J M. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J Virol. 1987;61:2659–2669. doi: 10.1128/jvi.61.9.2659-2669.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swanstrom R, Wills J W. Synthesis, assembly, and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor Laboratory, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 33.Temin H M. Structure, variation and synthesis of retrovirus long terminal repeat. Cell. 1981;27:1–3. doi: 10.1016/0092-8674(81)90353-6. [DOI] [PubMed] [Google Scholar]
- 34.Todaro G J, Sherr C J, Benveniste R E, Lieber M M. Type C viruses of baboons: isolation from normal cell cultures. Cell. 1974;2:55–61. doi: 10.1016/0092-8674(74)90008-7. [DOI] [PubMed] [Google Scholar]
- 35.van de Peer Y. TREECON for Windows user manual (1.3b). Antwerp, Belgium: University of Antwerp; 1998. [Google Scholar]
- 36.van der Kuyl A C, Dekker J T, Goudsmit J. Distribution of baboon endogenous virus among species of African monkeys suggests multiple ancient cross-species transmissions in shared habitats. J Virol. 1995;69:7877–7887. doi: 10.1128/jvi.69.12.7877-7887.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Kuyl A C, Dekker J T, Goudsmit J. Full-length proviruses of baboon endogenous virus (BaEV) and dispersed BaEV reverse transcriptase retroelements in the genome of baboon species. J Virol. 1995;69:5917–5924. doi: 10.1128/jvi.69.9.5917-5924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Kuyl A C, Goudsmit J. Xenotransplantation: about baboon hearts and pig livers. Trends Microbiol. 1998;6:431–432. doi: 10.1016/s0966-842x(98)01387-0. [DOI] [PubMed] [Google Scholar]
- 39.van der Kuyl A C, Mang R, Dekker J T, Goudsmit J. Complete nucleotide sequence of simian endogenous type D retrovirus with intact genome organization: evidence for ancestry to simian retrovirus and baboon endogenous virus. J Virol. 1997;71:3666–3676. doi: 10.1128/jvi.71.5.3666-3676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolgamot G, Bonham L, Miller A D. Sequence analysis of Mus dunni endogenous virus reveals a hybrid VL30/gibbon ape leukemia virus-like structure and a distinct envelope. J Virol. 1998;72:7459–7466. doi: 10.1128/jvi.72.9.7459-7466.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshinaka Y, Katoh I, Copeland T D, Oroszlan S. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc Natl Acad Sci USA. 1985;82:1618–1622. doi: 10.1073/pnas.82.6.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]