Abstract
The impact of cytomegalovirus (CMV) on human immunodeficiency virus type 1 (HIV-1) disease progression has been controversial. In this study, we sought to determine if CMV viral load is independent of HIV-1 viral load in predicting CMV disease and survival. Our findings indicate that in patients with advanced AIDS, CMV DNA load is an independent marker of CMV disease and survival and is more predictive than HIV-1 RNA load. Moreover, patients who respond to preemptive therapy with oral ganciclovir, with resulting undetectable levels of CMV DNA, in their plasma, have a significantly lower risk of developing CMV disease and higher rates of survival, despite stable or increasing HIV-1 RNA loads. These data provide support for CMV as an independent risk factor for mortality in persons with advanced AIDS and further suggest that effective preemptive therapy for CMV can improve patient survival rates.
Human immunodeficiency virus type 1 (HIV-1) RNA load within plasma has been identified as an important determinant of disease progression and survival in infected persons and is used to monitor the efficacy of antiretroviral therapy (4, 15, 22, 23). The impact of opportunistic pathogens, particularly cytomegalovirus (CMV), on the progression of HIV-1-related disease has been controversial. Although retinitis is the most common clinical disease associated with CMV in persons with advanced AIDS, CMV may cause a broad range of clinical diseases, including colitis, esophagitis, encephalitis, polyradiculitis, pneumonia, adrenalitis, and generalized wasting (7, 10, 13, 16–18, 24, 28, 31). Considerable research has examined the interaction between HIV-1 and CMV in vitro, identifying both positive and negative effects on HIV-1 replication (6, 12, 14, 20, 21, 30) and coinfection of the same cells (26). In almost all studies, the development of CMV disease has been correlated with shortened survival times (8, 11, 14, 20, 21, 34). However, the independent role of CMV in HIV-1-related disease progression has remained under dispute, because CMV disease occurs in persons with low CD4+-lymphocyte counts and high plasma HIV-1 RNA loads. Thus, it has been difficult to establish CMV infection as an independent risk factor for mortality.
Recently, we demonstrated that the presence and quantity of CMV DNA within the plasma of patients with advanced AIDS are predictive of CMV disease and survival (33). In the present study, we hypothesized that if CMV DNA load within plasma was, in fact, an independent predictor of CMV disease and survival, then (i) CMV DNA load would exhibit variations that were independent of HIV-1 RNA load and (ii) patients responding to oral ganciclovir preemptive therapy against CMV would be at reduced risk of developing CMV disease and would have improved rates of survival, even with stable HIV-1 RNA levels.
Plasma samples for viral quantification were collected at 19 participating sites, frozen, and shipped on dry ice to the central repository. CMV DNA was quantified by procedures previously described (1, 29, 33). Briefly, 100 μl of each plasma sample was treated with 900 μl of lysis buffer (4.23 M guanidinium thiocyanate, 20 mM disodium EDTA, and 1.0% Triton X-100 in 0.1 M Tris-HCl [pH 6.4]) together with 40 μl of silica suspension for 10 min at room temperature in a 2-ml microtube. The nucleic acid-silica complex was spun down for 15 s at 12,000 × g in a microcentrifuge. the pellet was washed twice with 70% ethanol and once with acetone. The pellet was dried at 56°C for 10 min in a heat block, resuspended with 200 μl of sterile water (Millipore), and incubated for 10 min at 56°C. The nucleic acid supernatant was recovered after centrifugation for 2 min at 12,000 × g in a microcentrifuge. The supernatant was then clarified and stored at −20°C until used in PCR. The assay could quantitate the presence of ≥25 copies of CMV DNA per 10 ml of plasma (≥2,500 copies/ml). Plasma specimens positive for CMV DNA were divided into two categories, those with values below 2,500 copies/ml and those with values above 2,500 copies/ml; the latter were quantified further.
HIV-1 RNA was quantified with the standard Roche Amplicor assay (25). Values below 400 copies/ml were considered undetectable.
Event rates were estimated by the Kaplan-Meier product limit method (19). The relative risk of CMV disease (or death) and 95% confidence intervals for relative risk were computed from a Cox model, with conversion from a PCR-positive to PCR-negative status at baseline as the single covariate (5). The Pearson correlation coefficient was used to measure the association between log-transformed plasma CMV DNA and HIV-1 RNA loads. Multivariate logistic regression was used to model the rates of CMV disease and survival as functions of both baseline log-transformed plasma CMV DNA and HIV-1 RNA loads. All analyses were two tailed.
The plasma specimens used in this study were obtained from participants in a trial that evaluated the efficacy of oral ganciclovir in preventing CMV disease (32). The original study included 725 participants who were randomized in a 2:1 ratio to receive either oral ganciclovir (1,000 mg) three times daily or a placebo. For this study, baseline plasma specimens were available from 619 study participants whose demographic data did not differ significantly from those for the original cohort of 725 participants (demographic data for both cohorts have been described previously [32, 33]). Of note, the median baseline CD4+-lymphocyte counts for the ganciclovir and placebo groups were 21 and 23 cells/μl, respectively. The median age of the participants was 38 years, and 99% of the participants were male. This study was conducted prior to the availability of antiretrovirals with activity against the HIV-1 protease.
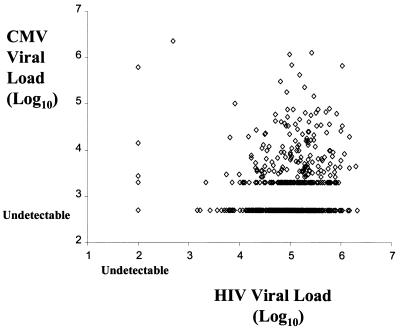
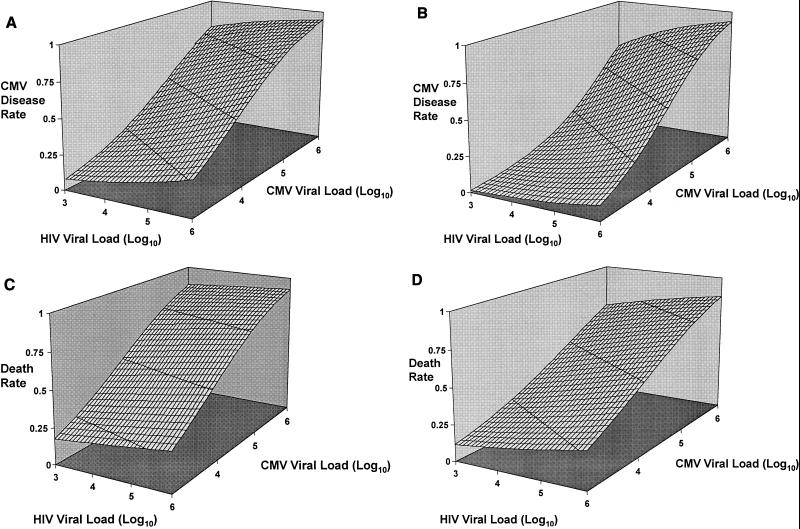
As seen in Fig. 1, there was only a weak correlation between levels of CMV DNA and HIV-1 RNA in plasma (r = 0.12, P = 0.0037). By using a multivariate logistic regression model, the rate of CMV disease was examined as a function of both baseline CMV and HIV-1 plasma loads (Fig. 2A and B). Predicted CMV disease rates were highest among patients with high circulating levels of both CMV and HIV-1 and lowest among patients with low levels of both viruses in plasma. Although both viruses influenced CMV disease rates CMV viral load was a better predictor, as indicated by the relative steepness of the logistic regression curves in the direction of the CMV axis. The impact of the CMV DNA load was observed for both ganciclovir- and placebo-treated patients.
FIG. 1.
Scatter plot comparing CMV DNA and HIV-1 RNA in plasma specimens obtained at baseline. CMV DNA was considered undetectable at copy numbers below 500/ml, while HIV-1 RNA was considered undetectable at copy numbers below 400/ml. r = 0.12, P = 0.0037.
FIG. 2.
Multivariate logistic regression modeling of the rates of CMV disease and survival as functions of both baseline plasma CMV DNA and HIV-1 RNA loads. (A) CMV disease in placebo recipients. (B) CMV disease in ganciclovir recipients. Predicted CMV disease rates are highest among patients with high circulating levels of both CMV and HIV-1 and lowest among patients with low levels of both viruses. Although both viruses influence the CMV disease rate, plasma CMV load is a better predictor for development of disease, as indicated by the relative steepness of the logistic curve in the direction of the CMV axis. (C) Survival in placebo recipients. (D) Survival in ganciclovir recipients. Predicted death rates are highest among patients with high levels of CMV DNA and HIV-1 RNA in plasma. However, the predictive value of HIV load in this population of patients with advanced AIDS is weak compared to that of CMV load, as demonstrated by the relative steepness of the logistic curve in the direction of the CMV axis.
Figures 2C and D show the results of examining, by the same multivariate logistic regression model, survival for the patient cohort. Predicted death rates were highest among patients with high circulating levels of both viruses in plasma. Again, the steepness of the curves indicates that CMV DNA levels in plasma are much stronger predictors of survival than HIV-1 RNA levels for both placebo- and ganciclovir-treated patients.
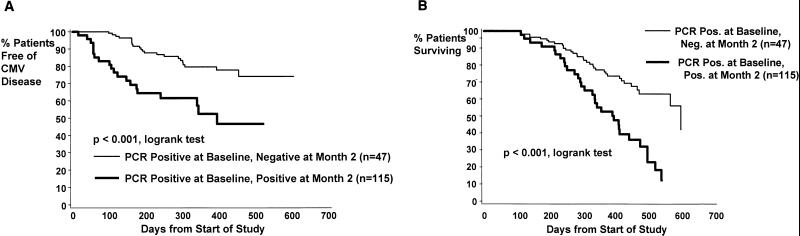
Figure 3A shows the Kaplan-Meier estimates for the CMV disease rates in patients receiving ganciclovir who had detectable levels of CMV DNA in their plasma at baseline. This group was further stratified according to CMV DNA PCR status at 2 months. Patients who went from CMV DNA-positive- to -negative status were considered responders, while those who continued to have detectable levels of CMV DNA in their plasma at 2 months were considered nonresponders. The median follow-up times were 13 and 14 months, respectively, for nonresponders and responders. Responders had a significantly lower rate of CMV disease than nonresponders. By Kaplan-Meier estimates, 20% of responders had developed CMV disease at 12 months, compared to 48% of nonresponders. The relative risk of CMV disease was 0.326 (95% confidence interval, 0.178 to 0.598; P < 0.001) among responders relative to nonresponders.
FIG. 3.
Kaplan-Meier survival curves for study participants receiving ganciclovir who were initially plasma CMV DNA PCR positive (≥500 copies/ml) at baseline, stratified by whether they became PCR negative at 2 months. (A) Conversion from PCR-positive to PCR-negative status at 2 months, with time to the development of CMV disease shown. (B) Conversion from PCR-positive to PCR-negative status at 2 months, with time to death shown.
Responders and nonresponders were monitored for rates of survival during the course of the study. As shown in Fig. 3B, responders had a significantly lower mortality rate than nonresponders (P < 0.001; relative risk, 0.363; 95% confidence interval, 0.222 to 0.596). At 12 months, the Kaplan-Meier estimate of mortality in responders was 24%, compared to 47% in the nonresponder group. Thus, a decrease in CMV DNA load to undetectable levels in plasma at 2 months resulted in improved rates of survival as well as decreased rates of CMV disease.
Although ganciclovir has no known activity against HIV-1, we investigated whether the decline of CMV DNA to undetectable levels was associated with a decline in plasma HIV-1 RNA levels. For this analysis, baseline and 2-month-study plasma samples were available from 92 patients who had detectable levels of plasma CMV DNA at baseline that, during ganciclovir treatment, had declined to undetectable levels at 2 months. There was a poor correlation between a decline in plasma CMV DNA and a change in plasma HIV-1 RNA (r = 0.117, P = 0.27). Of the 92 study participants, only 28 (30%) experienced any decline in plasma HIV-1 RNA levels, while the remainder had increased HIV-1 RNA loads at 2 months. Because a log10 change (10-fold) is often considered biologically significant, we examined those patients who experienced a ≥10-fold decline in detectable levels of CMV DNA in plasma. Of the 20 patients with a ≥10-fold decline in CMV DNA, only 5 (25%) had a ≥10-fold decline in HIV-1 RNA. The mean plasma HIV-1 RNA change ± standard deviation from baseline to 2 months increased by 1.46 × 105 ± 4.84 × 105 copies/ml. Thus, we could identify no correlation between a decrease in CMV DNA load, associated with preemptive ganciclovir treatment, and a decline in HIV-1 RNA load in plasma.
Our data indicate that the presence and quantity of CMV DNA in plasma are independent predictors of CMV disease and survival in persons with advanced AIDS. In these patients, CMV load is a better predictor of survival than circulating HIV-1 RNA load. It is possible that other opportunistic pathogens may also influence the mortality of AIDS patients. These could include other herpesviruses, including human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus), Epstein-Barr virus, and herpes simplex virus, all of which can reactivate during immunosuppression and are sensitive to ganciclovir. However, these data combined with previously published reports (2, 29, 33) support a strong and independent role for CMV in disease progression in patients with AIDS. These findings include the following. (i) The mortality of patients with advanced AIDS is 2.5-fold greater if CMV DNA is detected in plasma. (ii) For each log10 increase in CMV DNA, there is a 2.2-fold increase in risk of death. (iii) Treatment with ganciclovir, resulting in a decline of plasma CMV DNA to undetectable levels, is associated with improved survival. (iv) In advanced AIDS, CMV DNA load is a stronger predictor of survival than CD4+-lymphocyte counts. (v) CMV DNA load is independent of plasma HIV-1 RNA levels before and after ganciclovir treatment. The finding that persons who respond to preemptive treatment (defined as those having undetectable levels of plasma CMV DNA after 2 months of receiving ganciclovir) have higher survival rates further supports an important and independent role for CMV in patient outcome.
A possible concern regarding the data presented in this report is that there may be something unique about the qualitative and quantitative PCR assays used to detect CMV DNA in plasma. We believe that this is unlikely to be the case. First, several other studies using different assays have shown that the presence and quantity of CMV in blood specimens are correlated with the development of CMV disease (2, 8, 13, 26). Additionally, previous reports suggested a correlation between the presence of CMV and patient survival. Moreover, we have performed two evaluations of the assay used for the data presented in this study on blinded specimens within the AIDS Clinical Trials Group CMV Quantitation Working Group. In the first study, our quality control PCR assay had correlations of 0.997 with the plasma CMV DNA PCR assay developed by Roche Molecular Systems (Alameda, Calif.), 0.75 with a branch chain DNA assay being developed by Chiron Corporation (Emeryville, Calif.), and 0.75 with a CMV antigenemia assay (INCSTAR Corporation, Stillwater, Minn.). In a second study comparing our quantitative competitive PCR assay with the Roche assay, there was again a high correlation between the two assays; however, the PCR assay used for our studies gave CMV DNA values a mean of 0.8 log unit above those of the Roche assay (10a). It is likely that the difference in the absolute values given by the two assays is related to the standards used for quantification in the assays. Thus, there is considerable evidence that although absolute values for the CMV load may vary among different assays, the important concepts of this research (i.e., that CMV DNA load is independent of HIV RNA load and is an independent predictor of survival in patients with advanced AIDS) would be observed with any sensitive and well-standardized quantitative assay for CMV DNA in blood specimens.
Although this study was conducted prior to the availability of drugs that inhibit the HIV-1 protease, the findings are of importance for understanding the pathogenesis of HIV-1-related disease progression as well as for clinical management. Persons with AIDS who are at risk for CMV disease generally have CD4+-lymphocyte counts below 50/μl. The risk of reactivation of CMV with the subsequent development of CMV disease may exist for patients who fail combination therapies, including therapies with antiprotease compounds, once their CD4+-lymphocyte counts fall below 50/μl and they do not continue to improve with additional antiretroviral therapy. In previous studies, we and others demonstrated that CMV reactivates months prior to the development of clinical disease (3, 9, 27, 29, 32). These data provide strong support for preemptive treatment strategies for AIDS patients who have failed combination antiretroviral therapy, have sustained low CD4+-lymphocyte counts, and have detectable levels of CMV in their blood. Specific data on the utility of combination therapy are needed before specific recommendations can be made. It is likely, however, that preventing acute CMV infection not only will decrease the incidence of CMV disease but also will improve survival in patients with advanced AIDS.
Acknowledgments
This research was supported by grants from Roche Global Development and National Institutes of Health grants AI-27670 and AI-36214 (to the UCSD Center for AIDS Research).
REFERENCES
- 1.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowen E F, Wilson P, Cope A, Sabin C, Griffiths P, Davey C, Johnson M, Emery V. Cytomegalovirus retinitis in AIDS patients: influence of cytomegaloviral load on response to ganciclovir, time to recurrence and survival. AIDS. 1996;10:1515–1520. doi: 10.1097/00002030-199611000-00009. [DOI] [PubMed] [Google Scholar]
- 3.Bowen E F, Sabin C A, Wilson P, Griffiths P D, Davey C C, Johnson M A, Emery V C. Cytomegalovirus (CMV) viraemia detected by polymerase chain reaction identifies a group of HIV-positive patients at high risk of CMV disease. AIDS. 1997;11:889–893. doi: 10.1097/00002030-199707000-00008. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Guidelines for the use of antiretroviral agents in HIV-infected adults and adolescents. Morbid Mortal Weekly Rep. 1998;47(IRR-5):39–82. [Google Scholar]
- 5.Cox D R, Oakes D. Analysis of survival data. London, England: Chapman and Hall; 1984. [Google Scholar]
- 6.Dal Monte P, Landini M-P, Sinclair J, Virelizier J-L, Michelson S. TAR and Sp1-independent transactivation of HIV long terminal repeat by the Tat protein in the presence of human cytomegalovirus IE1/IE2. AIDS. 1997;11:297–303. doi: 10.1097/00002030-199703110-00006. [DOI] [PubMed] [Google Scholar]
- 7.d’Arminio Monforte A, Mainini F, Testa L, Vago L, Balotta L, Nebuloni M, Antinori S, Bini T, Moroni M. Predictors of cytomegalovirus disease, natural history and autopsy findings in a cohort of patients with AIDS. AIDS. 1997;11:517–524. doi: 10.1097/00002030-199704000-00016. [DOI] [PubMed] [Google Scholar]
- 8.Detels R, Leach C T, Hennessey K, Liu Z, Visscher B R, Cherry J D, Giorgi J V. Persistent cytomegalovirus infection of semen increases risk of AIDS. J Infect Dis. 1994;169:766–768. doi: 10.1093/infdis/169.4.766. [DOI] [PubMed] [Google Scholar]
- 9.Dodt K K, Jacobsen P H, Hofmann B, Meyer C, Kolmos H J, Skinhoj P, Norrild B, Mathiesen L. Development of cytomegalovirus (CMV) disease may be predicted in HIV-infected patients by CMV polymerase chain reaction and the antigenemia test. AIDS. 1997;11:F21–F28. doi: 10.1097/00002030-199703110-00001. [DOI] [PubMed] [Google Scholar]
- 10.Drew W L. Cytomegalovirus infection in patients with AIDS. Clin Infect Dis. 1992;14:608–615. doi: 10.1093/clinids/14.2.608-a. [DOI] [PubMed] [Google Scholar]
- 10a.Erice, A., et al. Unpublished data.
- 11.Frenkel L D, Gaur S, Tsolia M, Scudder R, Howell R, Kesarwala H. Cytomegalovirus infection in children with AIDS. Rev Infect Dis. 1990;12:S820–S826. doi: 10.1093/clinids/12.supplement_7.s820. [DOI] [PubMed] [Google Scholar]
- 12.Ghazal P, Nelson J A. Interactions between cytomegalovirus immediate-early proteins and the long terminal repeat of human immunodeficiency virus. Rev Med Virol. 1993;3:47–55. [Google Scholar]
- 13.Goodgame R W. Gastrointestinal cytomegalovirus disease. Ann Intern Med. 1993;119:924–935. doi: 10.7326/0003-4819-119-9-199311010-00010. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths P D. Studies of viral co-factors for human immunodeficiency virus in vitro and in vivo. J Gen Virol. 1998;79:213–220. doi: 10.1099/0022-1317-79-2-213. [DOI] [PubMed] [Google Scholar]
- 15.Hughes M D, Johnson V A, Hirsch M S, Bremer J W, Elbeik T, Erice A, Kuritzkes D R, Scott W A, Spector S A, Basgoz N, Fischl M A, D’Aquila R T. Monitoring plasma HIV-1 RNA levels in addition to CD4+ lymphocyte count improves assessment of antiretroviral therapeutic response. Ann Intern Med. 1997;126:929–938. doi: 10.7326/0003-4819-126-12-199706150-00001. [DOI] [PubMed] [Google Scholar]
- 16.Jabs D A, Enger C, Bartlett J G. Cytomegalovirus retinitis and acquired immunodeficiency syndrome. Arch Ophthalmol. 1989;197:75–80. doi: 10.1001/archopht.1989.01070010077031. [DOI] [PubMed] [Google Scholar]
- 17.Jacobson M A, Mills J. Serious cytomegalovirus disease in the acquired immunodeficiency syndrome (AIDS) Ann Intern Med. 1988;108:585–594. doi: 10.7326/0003-4819-108-4-585. [DOI] [PubMed] [Google Scholar]
- 18.Kalayjian R C, Choen M L, Bonomo R A, Flanigan T P. Cytomegalovirus ventriculoencephalitis in AIDS: a syndrome with distinct clinical and pathologic features. Medicine. 1993;72:67–77. [PubMed] [Google Scholar]
- 19.Kaplan E L, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 20.Koval V, Clark C, Vaishnav M, Spector S A, Spector D H. Human cytomegalovirus inhibits human immunodeficiency virus replication in cells productively infected by both viruses. J Virol. 1991;65:6969–6978. doi: 10.1128/jvi.65.12.6969-6978.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lathey J L, Spector D H, Spector S A. Human cytomegalovirus-mediated enhancement of human immunodeficiency virus type-1 production in monocyte-derived macrophages. Virology. 1994;199:98–104. doi: 10.1006/viro.1994.1101. [DOI] [PubMed] [Google Scholar]
- 22.Mellors J W, Muanoz A, Giorgi J V, Margolick J B, Tassoni C J, Gupta P, Kingsley L A, Todd J A, Saah A J, Detels R, Phair J P, Rinaldo C R., Jr Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 23.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126:946–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 24.Morgello S, Cho E S, Nielsen S, Devinsky O, Petito C K. Cytomegalovirus encephalitis in patients with acquired immunodeficiency syndrome: an autopsy study of 30 cases and a review of the literature. Hum Pathol. 1987;18:289–297. doi: 10.1016/s0046-8177(87)80012-6. [DOI] [PubMed] [Google Scholar]
- 25.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson J A, Reynolds-Kohler C, Oldstone M B A, Wiley C A. HIV and HCMV coinfect brain cells in patients with AIDS. Virology. 1990;165:286–290. doi: 10.1016/0042-6822(88)90685-x. [DOI] [PubMed] [Google Scholar]
- 27.Rasmussen L, Morris S, Zipeto D, Fessel J, Wolitz R, Dowling A, Merigan T C. Quantitation of human cytomegalovirus DNA from peripheral blood cells of human immunodeficiency virus-infected patients could predict cytomegalovirus retinitis. J Infect Dis. 1995;171:177–182. doi: 10.1093/infdis/171.1.177. [DOI] [PubMed] [Google Scholar]
- 28.Rodriquez-Barradas M C, Stool E, Musher D M, Gathe J, Jr, Goldstein J, Genta R M, Yoffe B. Diagnosing and treating cytomegalovirus pneumonia in patients with AIDS. Clin Infect Dis. 1996;23:76–81. doi: 10.1093/clinids/23.1.76. [DOI] [PubMed] [Google Scholar]
- 29.Shinkai M, Bozzette S A, Powderly W, Frame P, Spector S A. Utility of urine and leukocyte cultures and plasma DNA PCR for identification of AIDS patients at risk for developing human cytomegalovirus disease. J Infect Dis. 1997;175:302–308. doi: 10.1093/infdis/175.2.302. [DOI] [PubMed] [Google Scholar]
- 30.Spector D H, Koval V, Jault F M, Lathey J, Spector S A. Positive and negative effects of human cytomegalovirus on HIV replication. In: Kung H-J, editor. Interactions between retroviruses and herpesviruses. River Edge, N.J: World Scientific Publishing Co., Inc.; 1994. pp. 65–89. [Google Scholar]
- 31.Spector S A. Spectrum and treatment of cytomegalovirus disease in persons with AIDS. J Int Assoc Physicians AIDS Care. 1996;2:9–22. [PubMed] [Google Scholar]
- 32.Spector S A, McKinley G F, Lalezari J P, Samo T, Andruczk R, Follansbee S, Sparti P D, Havlir D V, Simpson G, Buhles W, Wong R, Stempien M J for the Roche Cooperative Oral Ganciclovir Study Group. Oral ganciclovir for the prevention of cytomegalovirus disease in persons with AIDS. N Engl J Med. 1996;334:1491–1497. doi: 10.1056/NEJM199606063342302. [DOI] [PubMed] [Google Scholar]
- 33.Spector S A, Wong R, Hsia K, Pilcher M, Stempien M J. Plasma cytomegalovirus (CMV) DNA load predicts CMV disease and survival in AIDS patients. J Clin Investig. 1998;101:497–502. doi: 10.1172/JCI1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webster A, Phillips A N, Lee C A, Janossy G, Kernoff P B, Griffiths P D. Cytomegalovirus (CMV) infection, CD4+ lymphocyte counts, and the development of AIDS in HIV-1-infected hemophiliac patients. Clin Exp Immunol. 1992;88:6–9. doi: 10.1111/j.1365-2249.1992.tb03030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]