Abstract
Ovarian cancer (OC) is a common and lethal gynaecological malignancy. RNA-binding proteins (RBPs) play a crucial role in governing RNA metabolism and have been implicated in the development and progression of diverse cancer types. Slight alterations in RBPs' expression or activity can induce substantial modifications in the regulatory network. THUMPD2, as member of the RBP family, was found to have differential expression in ovarian cancer, with the mechanism has not been studied yet. In this study, THUMPD2 protein was found to be weakly expressed in the early (I + II) stages of OC (P = 0.013), with a low expression rate of 78.6 %, and highly expressed in late (III + IV) stages (P = 0.009), with a high expression rate of 84.8 %. The shRNA-mediated knockdown of THUMPD2 in OVCAR3 and SKOV3 cells resulted in increased cell proliferation but inhibited metastasis, whereas THUMPD2 overexpression had the opposite effect. THUMPD2 overexpression suppressed tumour growth in vivo. Conversely, low THUMPD2 expression promoted tumour growth. Furthermore, we identified the potential target genes and pathways of THUMPD2 using GO and KEGG analyses, which were related to the centrosome, microtubules, cell cycle, and extracellular matrix. We demonstrated that low expression of THUMPD2 in the early stage promoted tumour growth and high expression in the late stage promoted tumour metastasis. Our findings reveal the dual function of THUMPD2 in OC and suggest that THUMPD2 may serve as a therapeutic target for the treatment of OC.
Keywords: Ovarian cancer, THUMPD2, Proliferation & metastasis, Centrosome, Extracellular matrix (ECM)
1. Introduction
Ovarian cancer(OC) ranks second globally in terms of mortality among gynaecological malignancies, following cervical cancer. In developed countries, it is the leading cause of death, with approximately 200,000 deaths worldwide 2020 [1]. OC refers to a highly heterogeneous group of diseases with almost no specific symptoms in the early stages, and a considerable proportion of patients diagnosed at an advanced stage. For many years, the mainstay of ovarian cancer treatment has been cytoreductive surgery combined with carboplatin and paclitaxel as monotherapy for surgery has been proven to be inefficient. However, 70 % of tumors recur and eventually become platinum resistant [2]. Moreover, approximately 20 % of the patients do not respond to frontline platinum-based treatment [3].Bevacizumab treatment has side effects, including hypertension, proteinuria, haemorrhage, thrombosis, and bowel perforation, which restrict its use in some patient, Furthermore, it has no Level I evidence for additional improvement in efficacy [4]. The application of poly ADP-ribose polymerase (PARP) inhibitors may rely on Breast Cancer Susceptibility Gene (BRCA) and Homologous Recombination Deficiency (HRD) status [5]. In addition, a potential detrimental effect on OS has been found in patients with recurrent ovarian cancer who received PARP inhibitor monotherapy versus chemotherapy based on SOLO336 and ARIEL437 studies [6,7]. CAR-T cell therapy represents a significant advancement in cancer immunotherapy and holds promise for the treatment of various malignancies. However, it involves many challenges, including off-target effects, tumour antigen escape, the heterogeneity of ovarian tumors and the tumour microenvironment (TME) of immunosuppressive tumour cells [8]. Overall, most patients diagnosed with OC present with a late-stage malignancy, leading to a clinical conundrum because there are no established methods for early detection. Despite the fact that OC in most cases is responsive to platinum-based chemotherapy, the majority of patients experience relapse following initial surgery and chemotherapy, ultimately leading to death. Combination therapy and immunotherapy, with different toxicities, fail to completely meet the needs of ovarian treatments, highlighting the urgent need to develop new therapeutic strategies [9].
Efforts have been made to explore new strategies for ovarian cancer treatment. RNA-binding proteins (RBPs) function as key regulators of RNA processing, ultimately affecting gene expression and cellular behavior. RBPs play a crucial role in maintaining homeostasis of gene expression [10]. Several studies have demonstrated that RBPs are involved in cellular development, differentiation, metabolism, transport, localization, and post-transcriptional regulation [[11], [12], [13]]. Aberrant RBP expression is a common feature of OC development. Recent studies have reported that RBPs act a role as oncogenes or tumor inhibitors to regulate OC. YTHDF1 is highly expressed in HGSOC. It acts as an m6A reader to augment EIF3C translation and promote OC progression, which is related to adverse prognosis in patients [14]. HuR modulates the expression of TIMM44 by stabilizing its mRNA levels, and its expression is correlated with OC cell progression [15]. CircE2F2 can bind to HuR to stabilize E2F2 mRNA and consequently promote OC cell proliferation, glucose metabolism, and metastasis [16]. Circ-NOLC1 promotes EOC progression by binding to ESRP1 and modulating Cyclin-dependent kinase 1 (CDK1) and Ras homolog family member A (RhoA) expression [17]. The coiled-coil domain containing protein-124 (CCDC124) is upregulated in OC and is associated with a prolonged prognosis in OC patients [18]. These findings demonstrate that RBPs play an important role in OC development.
Among members of the RBP family, THUMPD2 has received little attention. It is a THUMP domain-containing protein, and its expression is related to the cell cycle [19,20]. The THUMP domain is essential for the formation of tRNA N4-acetylcysteine [21,22]. The modification site is located at the leucine and serine 12 positions of tRNA, which is important for tRNA stability. THUMP consists of ThiI, Tan1, and fTrm G10 proteins, which are implicated in site-specific modifications [23,24]. It is predicted to result in the delivery of serial RNA-modifying enzymes to their target molecules. THUMPD2 ensures RNA-binding, protein-binding, and methyltransferase activities. The protein shows broad tissue distribution, and is essential for function of vital organs such as the brain, heart, and kidneys, in addition to the digestive and immune systems [19]. Currently, there is very little research on the role of THUMPD2 in malignant tumors. It has reported that THUMPD2 induces resistance of human esophageal squamous carcinoma cells to cisplatin (CDDP) and 5-fluorouracil (5-FU) in vitro [20]. Several studies have been published on THUMPD1 and THUMPD3. For example, THUMPD1 acts as a conductor that, orchestrates metastasis and invasion of breast cancer cells [25]. Overexpression of THUMPD3-AS1 has emerged as a potential therapeutic strategy for gastric cancer, because it hinders the hallmarks of aggressive tumor behavior, including proliferation, migration, invasion, and ROS accumulation [26]. To investigate the role of RBPs in OC, we selected several genes in the RBP family, including THUMPD1, THUMPD2, THUMPD3, YTHDF1, YTHDF2, and YTHDF3 and we performed a Polymerase Chain Reaction (PCR) in OC tissues. We found that THUMPD2 showed significant differential expression in OC. We then constructed a plasmid to knock down these genes using the pGreenPuro vector and, transfected these plasmids into OC cell lines (OVCAR3 or SKOV3). First, we assayed the growth potential using a colony proliferation assay. We found that knockdown of THUMPD2 and YTHDF3 significantly increased cell growth and proliferation compared to those in the scramble group. These results prompted us to study the function of THUMPD2 in OC.
To meet the needs of OC management, we investigated the role of THUMPD2 in OC to identify new biological markers for OC prognosis and explore new ideas for treatment.
2. Materials and methods
2.1. Cell culture and reagents
OVCRA3 (ATCC, Procell Life Science & Technology Co. China, catalogue No: CL-0178, RRID: CVCL_0465) and SKOV3(ATCC, Procell Life Science & Technology Co. China, catalogue No: CL-0215, RRID: CVCL_0532) cell lines were maintained in Roswell Park Memorial Institute 1640 medium(01-100-1B, Biological Industries, USA) supplemented with 10 % fetal bovine serum (FBS, #1928703, Biological Industries, USA), 100 U/mL penicillin, and 50 μg/mL streptomycin (Beyotime Biotechnology, China). The cells were incubated at 37 °C in 5 % CO2 atmosphere.
2.2. Plasmid construction and cell transfection
THUMPD2 shRNA and Scramble validations were designed using the Sigma online database (https://www.sigmaaldrich.com/china-mainland/zh/life-science/functional-genomics-and-rnai/shrna/individual-genes.html). The shRNA fragments were inserted into the pGreenPuro shRNA Expressing Lentivector (#s SI505A-1, System Biosciences, USA) at the BamHI and EcoRI restriction sites through subcloning. The fragment of THUMPD2 CDS (810 bp) was sub-cloned between the EcoRI and BamHI sites of the pEGFP-N1 (enhanced green fluorescent protein plasmid-N1) mammalian expression plasmid (#60360, Addgene, USA). OVCAR3 and SKOV3 cells were transfected with the aforementioned plasmids using the jetPRIME Transfection Reagent (101000046, Polyplus, France) in accordance with the instructions provided by the manufacturer. Following transfection, the cells were treated with 1 μg/ml puromycin (sc-108071, Santa Cruz, USA) for the pGreenPuro shRNA plasmid and 100 μg/mL neomycin for the pEGFP-N1 expression plasmid 48 h to facilitate the selection of stably transfected cells. The cell culture media containing puromycin or neomycin was replaced every 2 days for up to 3 weeks. The shRNA and overexpression primer sequences are shown in Table 1 (THUMPD2 shRNA2 was used in this study after Validity verification).
Table 1.
Primer used in this study.
| gene | Sequences (5′-3′) |
|---|---|
| THUMPD2 shRNA1 |
Forward:gatccCAGAGAGATGATAACCAACTATTCAAGAGATAGTTGGTTATCATCTCTCTGtttttg Reverse:aattcaaaaaCAGAGAGATGATAACCAACTATCTCTTGAATAGTTGGTTATCATCTCTCTGg |
| THUMPD2 shRNA2 |
Forward:gatccGCCAGTAACCACAAATTCTTATTCAAGAGATAAGAATTTGTGGTTACTGGCtttttg Reverse:aattcaaaaaGCCAGTAACCACAAATTCTTATCTCTTGAATAAGAATTTGTGGTTACTGGCg |
| Scramble shRNA THUMPD2-up |
Forward:gatccTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAAtttttg Reverse:aattcaaaaaTTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAAg Forward: ggaattccATGAAACACTTTGGATGGA Reverse: cgggatcccgCTACAGTCCAGAAGAGTGCG |
| THUMPD2 |
Forward: CAGCTGGACTGCGATCTACA Revese: TCGCTGACATCAGCACCTAC |
| GAPDH |
Forward: ACAACTTTGGTATCGTGGAAGG Revese: GCCATCACGCCACAGTTTC |
2.3. qRT-PCR
Total RNA was extracted using Trizol reagent (B511311, Sangon Biotech) and reverse transcribed into cDNA with HiScript III kit (+gDNA wiper, R312, Vazyme). qPCR was performed on a StepOne/StepOnePlus Real-Time PCR Systems (Applied Biosystems, ThermoFisher, USA) using SYBR-Green (Q711, Vazyme, Nanjing, China). Gene expression was analyzed by qPCR on a StepOne/StepOnePlus system (Applied Biosystems) using SYBR-Green (Q711, Vazyme) and normalized to GAPDH (primer sequences in Table 1).
2.4. Immunohistochemistry (IHC)
All paraffin sections were collected from the Affiliated Hospital of Nantong University from January 2010 to December 2020, including 29 cases of serous OC, 10 cases of mucinous cystadenocarcinoma, and 8 cases of endometrioid adenocarcinoma. Primary cytoreductive surgery was performed on all patients with ovarian cancer in this study. No chemotherapy, biological therapy, or immunotherapy was administered before surgery. Cases with synchronous malignant tumors were excluded. The study was conducted with informed consent from all participants and got the ethical approval from the Ethics Committee of the Affiliated Hospital of Nantong University (No. 2021-L084).
After routine dewaxing, gradient ethanol hydration, and antigen retrieval, an antibody was added to the 5 μm slices of the paraffin blocks (dilution ratio 1:100) and then the slices were incubated overnight at 4 °C. Next, a secondary antibody drop was incubated for 30 min. After double antibody (DAB) staining, hematoxylin contrast staining, and neutral gum patching, the sections were observed according to the Beesley grading method. The proportion of brown granules in the nucleus was used as the standard for judging the THUMPD2 protein-positive staining. Cells that did not stain were considered negative, and a score of 0 point was recorded; however, cell membranes that presented as brownish-yellow particles were considered positive. Light yellow marks were scored 1 point, brownish-yellow marks were scored 2 points, and brownish brown marks were scored 3 points. The scoring as determined by the percentage of positive cells was carried out as follows: 0 point if less than 5 %, 1 point if 5–25 %, 2 points if 26–50 %, and 3 points if ≥ 51 %. The sum of the two preceding scores was calculated to determine the degree of staining: 0–2 points were considered to indicate low expression; 3 points were considered to indicate moderate expression, and scores ≥4 points were considered to indicate high expression. The expression of THUMPD2 was independently evaluated by two senior medical doctors.
2.5. Western blot
Cells were lysed in RIPA buffer containing protease and phosphatase inhibitors. Lysates (40 μg protein) were subjected to SDS-PAGE and transferred to PVDF membranes (IPVH00010, Merck, USA). After blocking with 5 % skim milk, the membranes were incubated with anti-THUMPD2 antibody (1:500, sc-393018, Santacruz, Texas, USA), CDK6 (1:1000, #13331, CST, USA), MIB1(1:500, sc-393811, Santa Cruz, USA), APC (1:1000, ab40778, Abcam, UK), RB1(1:500, 10048-2-Ig, Proteintech, USA), PTPN11 (1:500, 24570-1-AP, Proteintech, USA), or anti-β-actin (1:1000, 81115-1-RR, Proteintech, USA) antibodies were incubated overnight at 4 °C. The membranes were washed with TBST and incubated with HRP-conjugated secondary antibodies for 1 h at room temperature, respectively. The blots were visualized using an enhanced chemiluminescence kit (WBULS0500, Millipore, USA). All experiments were performed in triplicates. The densitometric analysis of Western blots was performed utilizing Image-Pro Plus software.
2.6. Cell proliferation assay
Cell viability was assessed at 0, 24, 48, and 72 h using the Cell Counting Kit-8 (CCK-8) method (CK04, Dojindo, Japan). The microplate reader was used to measure the absorbance at 450 nm. Colony formation assays were conducted in six-well plates seeded with 150 cells each of the stably transfected lines. These were cultured for a 14-day period. After the incubation period, the formed colonies were fixed with 4 % paraformaldehyde and subsequently stained with 0.5 % crystal violet. Images of the plates were captured for documentation purposes.
2.7. Wound-healing assay
In the wound healing assay for cell migration, OVCAR3 or SKOV3 cells transfected with stable vectors were cultured in a medium containing low fetal bovine serum (FBS) concentration (<2 %) and incubated for 48 h. Once the cells reached confluence, a straight-line scratch was made on the cell layer using a 100 μL pipette tip. Subsequently, the scratched cell monolayer was gently washed with PBS to eliminate any detached cells and then replenished with fresh medium containing a low FBS concentration (<2 %). Images were captured at 0, 24, and 48 h using a phase contrast microscope (Nikon, Japan).
2.8. Transwell invasion and migration assays
Extracellular matrix (ECM) gel was added to the upper chambers of the insert. To promote Matrigel polymerization, the plate containing the insert and ECM gel was immediately incubated at 37 °C for 2 h. The lower chamber was filled with RPMI-1640 medium containing 20 % FBS. Stably transfected cells in serum-free RPMI-1640 (no more than 0.2 mL) were seeded gently in the upper chambers. The plate was then placed in an incubator set at 37 °C for a period of 24 h. The cells that had migrated to the lower side of the membrane were immobilized using 4 % paraformaldehyde, followed by staining with a solution of 1 % crystal violet in 2 % ethanol for an additional duration of 20 minutes. A wet cotton swab was used to wipe the upper side of the membrane and remove the cells and gel in the upper chamber. Under a microscope, the cells on the lower side of the filter were counted. Unlike the invasion assay, the insert in the upper chamber of the migration assay did not require incubation with Matrigel. Images of the cells were taken from random microscopic fields.
2.9. Xenograft model assay
Twelve female BALB/c nude mice (6 weeks old, 18–22 g) were randomly assigned to either the control or experimental group after being housed under standard laboratory conditions with a 12-h light/dark cycle. To establish the tumors, the mice were subcutaneously injected in the axilla with THUMPD2 knockdown (n = 4) and overexpressed stable OVCAR3 cells (2 × 106 cells in 200 μL PBS) (n = 4), or stable OVCAR3 cells transfected with the scramble shRNA (n = 4). The scramble group served as the control for both the THUMPD2 overexpression and the THUMPD2 knockdown groups. The in vivo xenograft assay was conducted for 28 days. The mice were subsequently sacrificed by carbon dioxide asphyxiation and the xenografts were completely dissected for visual examination and weight detection.
All animal experiments were conducted with ethical approval from the Nantong University Institutional Animal Care and Use Committee (IACUC) (approval No. IACUC20200921-1001). The ethical review of laboratory animal welfare was performed following the guidelines outlined in the People's Republic of China National Standard GB/T35892−2018. According to the ethical guidelines, the maximum allowable tumor burden for the mice in this study was set at 2000 mm3. Any mice found to have tumors exceeding this limit were euthanized as per the approved IACUC protocol.
2.10. Data collection and preprocessing
Analysis of GEPIA2 (http://gepia2.cancer-pku.cn/#analysis) was used to evaluate THUMPD2 expression in TCGA (The Cancer Genome Atlas)/Genotype-Tissue Expression (GTEx) pan-cancer database.
2.11. Survival curves analysis
We analyzed data from The Cancer Genome Atlas (TCGA) to obtain Overall Survival (OS), Post-Progression Survival (PPS), and Progression-Free Survival (PFS) for THUMPD2 expression in various cancers. Kaplan-Meier plots (https://kmplot.com/analysis/) were used for this analysis.
2.12. Construction and evaluation of the nomogram
A nomogram is a visual risk assessment tool that can be used to estimate the probability of an event. It is a popular tool for predicting clinical efficacy of various diseases [27]. A nomogram was developed for individualized and multivariate analysis of predicted survival probabilities at 1, 3, and 5 years. The RMS (Recession Modeling Strategy) package in R was used to generate nomograms that incorporate clinical variables, a comprehensive approach was employed, ensuring a low repetition rate of 15 % or less. significantly associated with THUMPD2 and calibration maps. The calibration curve was evaluated by mapping predicted outcomes against observed outcomes, with the 45-degree line representing the ideal predicted value. The discrimination of the nomogram was evaluated using the concordance index (C-index) with a 1000-resample bootstrap method for validation. The C-index and receiver operating characteristic (ROC) curves were used to evaluate and compare the predictive accuracy of both the nomograms and individual prognostic factors. All statistical tests were conducted as two-tailed tests, with a significance threshold set at 0.05.
2.13. Clinical proteomic tumor analysis consortium (CPTAC) and UALCAN
We utilized UALCAN (http://ualcan.path.uab.edu/index.html), a cancer data analysis platform, to analyze protein expression in CPTAC datasets. The expression of THUMPD2 in different tumor stages can also be observed.
2.14. RNA sequencing
Total RNA was isolated from sh-THUMPD2 and the scramble cell lines. RNA sequencing was performed on an Illumina HiSeq 2500 platform at Novogene (Beijing, China), which also included quality control and analysis of the sequencing data.
2.15. Statistical analysis
Statistical representations are presented as the mean ± standard deviation (SD) and their significance was assessed using either the Mann-Whitney U test or Kruskal-Wallis test, followed by Tukey's post-hoc analysis. Statistical significance was determined at P values less than 0.05. All analyses were conducted utilizing GraphPad Prism software (San Diego, CA, USA).
3. Results
3.1. Abnormal expression of THUMNPD2 in OC
First, a pan-cancer analysis of THUMPD2 expression in tumor samples compared with normal samples from the Genotype-Tissue Expression (GTEx) and TCGA database was conducted using the GEPIA2 database. This analysis revealed significantly high THUMPD2 expression in cholangiocarcinoma (CHOL), diffuse large B-cell lymphoma (DLBCL), and Thymoma (THYM). Conversely, ovarian cancer (OC), kidney chromophobe (KICH), skin cutaneous melanoma (SKCM), and thyroid cancer (THCA) showed lower THUMPD2 expression. These findings provide valuable insights into the potential roles of THUMPD2 in different cancer types (Fig. 1A).
Fig. 1.
Analysis of THUMPD2 expression in ovarian cancer tissue. (A) The expression of THUMPD2 in tumor and normal tissues in pan-cancer data of TCGA/GTEx. (B) THUMPD2 mRNA expression in 26 tissue samples, including 18 samples of ovarian cancer and eight matched normal samples. (C) Low expression of THUMPD2 in ovarian cancer. (D) Moderate expression of THUMPD2 in benign ovarian tissue. (E) High expression of THUMPD2 in ovarian cancer. Representative microscopic images (200× magnification, scale bar 50 μm) of patients with IHC staining for THUMPD2. *P < 0.05; **P < 0.01; ***P < 0.001. TPM, Transcripts Per Million; Ctrl, Control; TCGA, The Cancer Genome Atlas.
To validate the abnormal mRNA expression of THUMPD2 in OC tissues, total RNA was extracted from 26 samples. The tumor tissues comprised 10 serous OC, 7 mucinous OC, and 1 endometrioid OC samples. The patients underwent cytoreduction between January 2019 and September 2020 (age:45–71 years; mean age: 54 years; standard deviation: 6 years). Eight matched normal samples contained five specimens from patients with ovarian serous cystadenoma who underwent unilateral accessory resection, and three specimens from patients with adenomyosis who underwent hysterectomy and bilateral adnexectomy. QRT-PCR was performed on the 26 tissue specimens. We found that THUMPD2 mRNA was downregulated in 10 patients and upregulated in 4 patients (Fig. 1B). Next, immunohistochemistry was performed to observe THUMPD2 protein expression in the paraffin cuts of 47 OC cases and 30 benign cases. We observed a moderate THUMPD2 expression in benign ovarian lesions (Fig. 1C) and low THUMPD2 expression in stage I–II OC (Fig. 1D). High THUMPD2 expression was observed in stage III–IV OC tissues (Fig. 1E).
To investigate the relationship between THUMPD2 expression and clinical characteristics of patients, statistical analysis was performed on sample information. The result showed no significant difference in THUMPD2 expression among age, histological type, grade and ascites formation. Expression of THUMPD2 was correlated with FIGO stage and tumor metastasis. Low THUMPD2 staining was observed in I & II stages (P = 0.013), and a high THUMPD2 expression in the advanced (III & IV) stages (P = 0.009) were observed with expression rates of 78.6 % and 84.8 %, respectively (Table 2).
Table 2.
Clinical-pathological analysis of THUMPD2 expression.
| Variables | Cases | THUMPD2 |
χ2 | P | |||
|---|---|---|---|---|---|---|---|
| low | moderate | high | |||||
| Age | <50 | 22 | 8(36.4) | 2(9.1) | 12(54.5) | 3.088 | 0.214 |
| ≥50 | 25 | 7(28.0) | 0(0) | 18(72.0) | |||
| Histologic classification | serous | 29 | 8(27.6) | 2(6.9) | 19(65.5) | 1.733 | 0.785 |
| mucinous | 10 | 4(40.0) | 0(0) | 6(60.0) | |||
| endometrioid | 8 | 3(37.5) | 0(0) | 5(62.5) | |||
| Differentiation degree | I | 10 | 3(30.0) | 1(10.0) | 6(60.0) | 2.037 | 0.729 |
| II | 17 | 6(35.3) | 1(5.9) | 10(58.8) | |||
| III | 20 | 6(30.0) | 0(0) | 14(70.0) | |||
| FIGO stage | I + II | 14 | 11(78.6) | 1(7.1) | 2(14.3) | 21.695 | 0.000 |
| III + IV | 33 | 4(12.1) | 1(3.0) | 28(84.8) | |||
| ascites | No ascites | 13 | 4(30.8) | 1(76.9) | 8(61.5) | 0.521 | 0.771 |
| ascites | 34 | 11(32.4) | 1(2.9) | 22(64.7) | |||
3.2. Survival rate analysis for THUMPD2 in OC
Having seen the different expression of THUMPD2 in ovarian cancer of different stages, we next set out to investigate whether THUMPD2 is related to survival outcomes. Then OS (Overall-Survival), PFS (Progression-Free-Survival), and PPS (Post-Progression-Survival) were analyzed using bioinformatics methods by utilizing public datasets (https://kmplot.com/analysis/index.php?p=service&cancer=ovar). The result showed that no significant difference was observed in the 5-year OS rate (Fig. 2A, P = 0.200) and PFS rate (Fig. 2B, P = 0.250); however, patients with lower THUMPD2 expression showed higher PPS rate compared to those with high THUMPD2 expression (Fig. 2C, P = 0.0249).
Fig. 2.
The association between THUMPD2 expression and the OS, PFS, and PPS of ovarian cancer patients. (A,B) Kaplan-Meier analysis of the 5-year OS and PFS in ovarian cancer; meaningless results were not shown. (C) Kaplan-Meier analysis of PPS for ovarian cancer (P = 0.029). (D–F) Kaplan-Meier analysis of OS, PFS, and PPS based on stage III clinical survival analysis. (G–I) Kaplan-Meier analysis of OS, PFS, and PPS based on stage IV clinical survival analysis. OS, overall survival; HR, hazard ratio; PPS, post-progression survival; PFS, progression-free survival.
Next, we analyzed the OS, PFS, and PPS in the different subgroups. Based on these findings, it was observed that high THUMPD2 expression demonstrated an adverse outcome in stage III (Fig. 2D–F, OS, P = 0.140; PFS, P = 0.026; PPS, P = 0.023). Similarly, patients with low THUMPD2 expression in stage IV had a higher 5-year PFS rate than those in the high expression group (Fig. 2G–I, OS, P = 0.093; PFS, P = 0.004; PPS, P = 0.200). These findings underscore the complex role of THUMPD2 in OC progression and warrant further investigation.
3.3. Overexpression of THUMPD2 inhibits cell proliferation and promotes migration and invasion in vitro
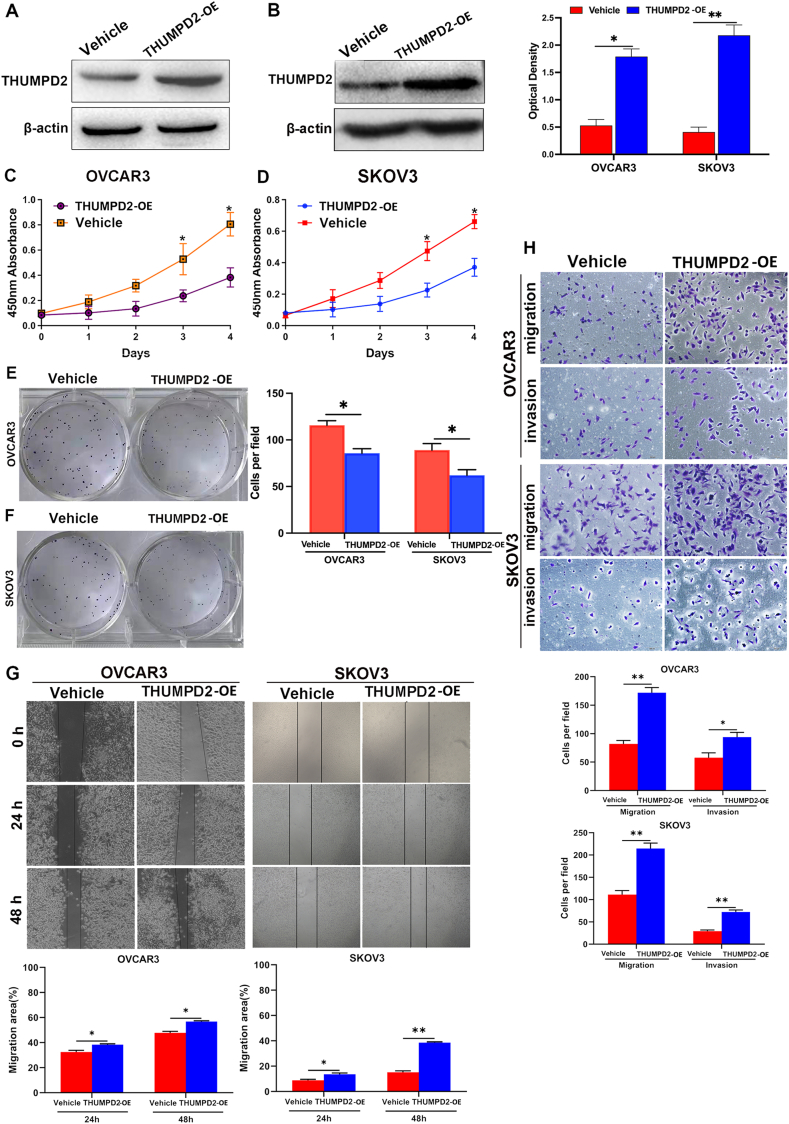
After we proved that the expression of THUMPD2 was differential in tissues of ovarian cancer patients, we conducted experiments on cell lines(SKOV3/OVCAR3) to understand the role of THUMPD2 in the biological behavior of ovarian cancer cells. We stably transfected OVCAR3 and SKOV3 cells with a THUMPD2 overexpression (OE) plasmid. Notably, the expression of THUMPD2 was upregulated in the THUMPD2 overexpression group (THUMPD2 OE) (Fig. 3A–B). CCK-8 (Fig. 3C–D) and colony formation (Fig. 3E–F) assays were performed. We observed that the proliferative ability was reduced in the THUMPD2 OE group compared with the empty vector (vehicle group) (Fig. 3C–F). In addition,the scratch assay revealed a higher migration rate in the THUMPD2 OE group (Fig. 3G). The transwell assay also demonstrated the significantly increased migration and invasion rates in the THUMPD2 OE group compared to those in the vehicle group (Fig. 3H). These results suggest that THUMPD2 promotes the migration and invasion of OVCAR3 and SKOV3 cells.
Fig. 3.
THUMPD2 decreased tumor proliferation and promoted metastasis. (A, B) Western blotting displayed the expression of THUMPD2 in the OVCAR3 and SKOV3 cell lines transfected with the vehicle and THUMPD2 overexpression plasmid, respectively. Optical densitometry of Western blot bands was performed using Image-Pro Plus. (C–F) CCK-8 and colony formation analysis showed that THUMPD2 overexpression decreased the proliferative capacity of OVCAR3 and SKOV3 cells. Cells in colony formation analysis were stained with 1 % crystal violet in 2 % ethanol. (G) The wound-healing assay detected increased migration of THUMPD2-expression cells. (H) The migration and invasion ability of the THUMPD2-overexpressing cells was higher than that of the Vehicle cells. Cells were stained with 1 % crystal violet in 2 % ethanol (scale bars: 100 μm). All experiments were repeated at least three times for reproducibility. *P < 0.05; **P < 0.01. CCK-8, Cell Counting Kit 8.
3.4. Knockdown of THUMPD2 promotes proliferation and inhibits cell migration and invasion in vitro
To further investigate the effect of low THUMPD2 expression on regulating the characteristics of tumor biology in OC, we transfected shRNA against THUMPD2 in OVCAR3 and SKOV3 cells and, identified that THUMPD2 was significantly downregulated compared to that in cells transfected with scramble shRNA (Fig. 4A–B). Based on the CCK-8 assay results, we observed that the downregulation of THUMPD2 significantly promoted cell growth (Fig. 4C–D). The cell proliferation assay showed that the colony formation ability was promoted following THUMPD2 downregulation (Fig. 4E–F). These results suggested that THUMPD2 downregulation increased the proliferation rate of OVCAR3 and SKOV3 cells.
Fig. 4.
Down-regulation of THUMPD2 expression promotes tumor proliferation and inhibits tumor metastasis. (A, B) The expression of THUMPD2 in OVCAR3 and SKOV3 cells was downregulated, as detected by Western blot analysis. (C, D). CCK-8 analysis demonstrated that knocking down THUMPD2 by shRNA significantly increased the multiplication capacity rate of OVCAR3 and SKOV3 cells on days 3 and 4. (E, F) Colony formation analysis indicated that the colony-forming ability was enhanced in the THUMPD2-shRNA group. Cells were stained with 1 % crystal violet in 2 % ethanol. (G) The wound-healing assay detected decreased migration of sh-THUMPD2 cells. (H) The role of THUMPD2 in the migration and invasion of OVCAR3 and SKOV3 cells was detected by transwell assay. Cells were stained with 1 % crystal violet in 2 % ethanol (scale bars: 100 μm). Each experiment was repeated three times. *P < 0.05; **P < 0.01. CCK-8, Cell Counting Kit 8.
To determine whether THUMPD2 knockdown affects the migration ability of cells, wound healing and Transwell assays were performed. We noticed that a high migration rate was only observed in the scramble group cells after 24 h and 48 h using low serum concentrations of the cell medium (Fig. 4G). To further verify the validity of this phenotype, we used transwell plates coated with or without Matrigel cell culture chambers for the migration (without Matrigel) and invasion (with Matrigel) assays. The results demonstrated that the migration and invasion rates were exhibited in the scramble group compared to the THUMPD2-shRNA group (Fig. 4H). Additionally, there was a marked reduction in the number of cells adhering to the bottom membrane in the sh-THUMPD2 group. These findings indicated that THUMPD2 affects the proliferation, migration, and invasion of OVCAR3 and SKOV3 cells, and regulates tumor cell proliferation and migration via a completely different mechanism.
3.5. THUMPD2 regulates tumor growth in a xenograft model of ovarian cancer
As THUMPD2 has showed an ability to inhibit proliferation and promote migration and invasion in vitro, xenograft models were established by subcutaneously injecting OVCAR3 scramble and stable transfected cells into the axilla (The scramble group served as the control for both the THUMPD2 up and sh-THUMPD2 groups) to study the impact of THUMPD2 on the biological behavior of ovarian cancer in vivo. The xenograft assay was conducted over a 28-day period. During this time, the mice were monitored for tumor growth and other relevant parameters. The xenografts were dissected at the end of the 28-day window for visual examination and weight determination. The tumor volume and net weight of the THUMPD2 overexpression group were significantly lower compared with the control group while those in the sh-THUMPD2 group were much higher. (Fig. 5A–D). These findings suggested that THUMPD2 inhibits tumor growth in vivo. This result indicated that THUMPD2 may be a potential therapeutic target in OC.
Fig. 5.
THUMPD2 affects the tumorigenicity of ovarian cancer cells in xenograft models. (A) A photograph of the tumor-bearing nude mouse from the control ((left), THUMPD2-overexpression (middle) group and sh-THUMPD2 group (right). (B) Photograph of stripped tumors from OVCAR3 control (left), THUMPD2 up (middle) and sh-THUMPD2 (right) groups 28 days after transplantation. (C) The tumor weights of the control and THUMPD2 overexpression groups. (D) The tumor weights of the control and sh-THUMPD2 overexpression groups. Scramble group was used as the control for both the THUMPD2 up and sh-THUMPD2 groups.
3.6. Enrichment analysis in gene ontology (GO) and Kyoto Encyclopaedia of genes and genomes (KEGG)
To further explore the involvement of downstream genes related to THUMPD2 in ovarian cancer, RNA sequencing was performed to investigate the genes that were co-expressed in OC cells (sh-THUMPD2 and scramble) (Supplementary Table S1). A total of 16,699 transcripts were obtained from the sequencing data in the study, while 3674 upregulated and 1946 downregulated differentially expressed genes (DEGs) were detected based on selection criteria (adjusted P < 0.05 and |log2foldchange| >1) were found between THUMPD2-shRNA and scramble in the OVCAR3 cell line (Supplementary Fig. S1A). GO hierarchy analysis showed that the function of THUMPD2 is associated with microtubule cytoskeleton organization (GO: 0070507), mitotic nuclear division (GO: 0140014), and microtubule organizing center organization (GO: 0031023) (Supplementary Fig. S1B).
GO and KEGG enrichment analyses revealed that THUMPD2 was related to the cell cycle checkpoint (109 DEGs, P = 1.69E-06), the DNA damage checkpoint (26 DEGs, P = 1.12E-05), and the regulation of microtubule cytoskeleton organization (83 DEGs, P = 3.08E-06) in the biological process (BP) category; the centrosome (223 DEGs, P = 2.82E-09) in the cellular component (CC) category; and DNA-dependent ATPase activity (48 DEGs, P = 3.16E-06) in the molecular function (MF) category (Fig. 6A). To further explore the differential genes at the MF level, we performed a separate enrichment analysis of all down-regulated and up-regulated DEGs. The results showed that up-regulated DEGs were predominantly enriched in BPs such as microtubule cytoskeleton organization (178 DEGs, P = 1.36E-11) and chromosome segregation (128 DEGs, P = 4.90E-11), CCs such as the centrosome (182 genes, P = 3.87E-15), and MFs such as DNA-dependent ATPase activity (45 genes, P = 1.57E-10) (Fig. 6B). Down-regulated DEGs were mainly involved in BPs such as ECM organization (63 DEGs, P = 2.83E-08) and extracellular structure organization (68 DEGs, P = 5.45E-08). CCs such as ECM (70 DEGs, P = 4.78E-07), and MFs including serine hydrolase activity (35 DEGs, P = 4.83E-06) (Fig. 6C). In the KEGG analysis, these genes were significantly enriched in apoptosis, sphingolipid signaling pathway, and viral infection (Fig. 6D). A total of 56 genes identified by the prediction model that were significantly associated with discrimination (|log2fold change| > 2.2) were subjected to pathway analysis to understand the molecular basis of the model's ability. Interestingly, these genes were enriched in biological functions related to DNA-dependent ATPase activity, centrosome, and regulation of microtubule cytoskeleton organization. (Fig. 6E). Our RNA-Seq results showed that the expression of AKAP9 (10.36-fold), ASPM (6.93-fold), BRBP1 (6.72-fold), APC (5.9-fold), ATR (5.68-fold), PTPN11 (5.46-fold), CDK6 (5.32-fold), ROCK2 (5.31- fold), MIB1 (5.25-fold), BRCA2 (5.09-fold), PPP1R12A (4.89-fold), ATM (4.75-fold), RB1 (4.61-fold) were increased, and the expression of ID1 (5.89-fold) was decreased after the knockdown of THUMPD2. Western blotting showed that the protein levels of MIB1, RB1, PTPN11, APC and CDK6 were significantly upregulated after the knockdown of THUMPD2 (Fig. 6F). These results provide a theoretical foundation for further exploring biological functions of THUMPD2. Taken together, these findings indicated that THUMPD2 was involved in cell proliferation and metastasis.
Fig. 6.
Functional enrichment and analysis of DEGs in ovarian cancer. (A) Significantly enriched GO analysis of DEGs in ovarian cancer. (B,C) Significantly enriched up-regulated and down-regulated DEGs in GO, respectively. (D) Significantly KEGG analysis in ovarian cancer. (E) A chord diagram was employed to visualize the complex relationships between enriched pathways identified through the enrichment analysis of 56 mRNAs used in the prediction model. (F) Western blotting displayed the expression of MIB1, RB1, PTPN11, APC and CDK6 in stable transfection OVCAR3 cell lines with knocked-down THUMPD2, respectively. (G) Optical densitometry of Western blot bands was performed using Image-Pro Plus. DEG, differentially expressed gene; CC, cellular component; BP, biological process; MF, molecular function.
4. Discussion
Ovarian cancer is the second most common cause of gynecologic cancer death in women around the world. It is diagnosed at a progressed stage in most occasions, encompasses multiple variants with unique physiological and genetic characteristics, and exhibits variability in the accessibility and availability of therapeutic interventions. To improve survival in this aggressive disease, we need to seek more treatment options, which calls for a deeper understanding of the pathogenesis and development mechanisms of ovarian cancer. Many studies have demonstrated that RBPs play an important role in ovarian cancer. In the present study, we identified THUMPD2 as an important pluripotency related RBP in OC. Among the 18 OC patient samples, 10 showed significant THUMPD2 downregulation, and 4 exhibited THUMPD2 upregulation. The abnormal expression of THUMPD2 in OC tissues sheds light on its critical role in OC pathogenesis and progression, highlighting it as a potential target for therapeutic interventions. To further confirm the differential expression of THUMPD2 in OC, IHC staining suggested that the expression of THUMPD2 decreased significantly in the early stage (I–II) but was higher in the advanced stage (III–IV). It seems to us that THUMPD2 had unique dual role in both tumorigenic and anti-tumorigenic processes. According to the results we speculated that due to the lack of effective screening methods for ovarian cancer, most patients with stage I-II ovarian cancer were unexpectedly discovered during surgery for enlarged ovarian tumors. We speculate that ovarian cancer cell is mainly characterized to be proliferative in this stage. As we demonstrated in the results that THUMPD2 inhibits proliferation, THUMPD2 weakly expressed in the early stages may indicate tumor proliferation.Increased expression of THUMPD2 in tissues of OC patients diagnosed in the late stage may indicate that the kind of tumor in these patients is characterized by migration and infiltration resulting metastasis rather than a significant increase in tumor volume.
Kaplan-Meier analysis indicated that elevated THUMPD2 expression was associated with an unfavorable survival rate in stage III OC, while low THUMPD2 expression improved the survival rate in IV stage OC patients. The reduced survival rate in stage III may be due to enhanced tumor cell metastasis and the improved survival rate in stage IV may benefit from reduced metastasis. Although the survival curve is somewhat inadequate for some potential limiting factors including the patient's treatment regimen, complete and incomplete cytoreduction, chemotherapy regimen, etc., we could still speculate that elevated levels of THUMPD2 expression were found to be positively associated with stronger metastatic capacity resulting poor clinical outcomes in OC patients. The results suggest that the prognosis of OC patients may depend more on migration and invasion rather than proliferation, which are regulated by THUMPD2.
Therefore, we designed and synthesized specific shRNAs against THUMPD2 and overexpressed THUMPD2 to study the effects of THUMPD2 on OVCAR3 and SKOV3 cells. These findings demonstrated a significant enhancement in the proliferative capacities of OVCAR3 and SKOV3 cells, whereas their invasion and migration abilities were significantly decreased by THUMPD2 knockdown. Similarly, the proliferative ability of the cells was inhibited, whereas overexpression of THUMPD2 in the cell line resulted in increased invasion and migration. We also verified the biological significance of THUMPD2 in vivo using xenograft experiments and observed that the subcutaneous tumor formation was greater in the low THUMPD2 expression group than in the control group. Inevitably, the tumor volume and net weight in the xenograft model with THUMPD2 overexpression were smaller than those of the control group. According to previous studies, THUMPD2 is expressed at low levels in most OC tissues but is highly expressed in a small proportion of OC tissues. These results led us to speculate that THUMPD2 may play different regulatory roles at various tumor stages. Low expression of THUMPD2 in the early stages of OC implies that it mediates high cancer cell proliferation as major a biological behavior. However, when THUMPD2 is highly expressed in the late stages of OC, tumors tend to metastasize and invade. This explains the results obtained by the bioinformatics method assay, which showed that patients with high THUMPD2 expression in stage I-II demonstrated a poor prognosis and those with low THUMPD2 expression had a higher survival rate in stage IV.
Currently, the interaction between THUMPD2 and its downstream target genes remains unclear. A previous report illustrated the coexistence of the regulator of G protein signaling 18 downstream of the intergenic region of anaplastic lymphoma kinase-THUMPD2 fusion in patients with stage IV LUAD accompanied by pleural metastasis [28]. This finding indicated a potential correlation between THUMPD2 and tumor metastasis. Another report demonstrated that THUMPD2 was downregulated in both cisplatin (CDDP)/5-FU human esophageal squamous cell carcinoma [20], and THUMPD2 was identified as a CDDP/5-FU-resistant gene in human esophageal squamous cell carcinoma in vitro. It is speculated that THUMPD2 is likely to influence downstream gene expression by affecting drug influx or efflux. From our RNA sequence results and GO analysis, we determined that THUMPD2 may be a pluripotency role gene mediating multiple biological processes; THUMPD2 knockdown in OC cells results in the up-regulation of genes involved in microtubule cytoskeleton organization, the centrosome, and DNA-dependent ATPase activity. However, the down-regulation of genes is associated with ECM organization, extracellular structure organization, and serine hydrolase activity.
The GO signaling network analysis revealed that alteration of the THUMPD2 target genes was mostly associated with the “centrosome”, “microtubule cytoskeleton organization”, “cell cycle”, “extracellular structure organization”, “ECM organization” and “collagen metabolic process” pathways. This finding was consistent with previous studies, that have provided evidence for the regulation of these signaling pathways in the progression of OC. Cancer cell metastasis is a complex process that involves dramatic reorganizations and remodeling of the microtubule cytoskeleton. Investigating the actin cytoskeleton as a target for developing novel therapies could offer an intriguing approach for treating metastatic cancer [29]. Microtubules are dynamic structures that play crucial roles in cell growth, vesicle trafficking, and mitosis. In interphase cells, microtubules originate from the centrosome to establish a hub-and-spoke network, that regulates vesicular transport [30]. The centrosome, which acts as the cell's microtubule-organizing center, undergoes a single replication during the cell cycle to orchestrate the formation of the bipolar mitotic spindle, essential for proper chromosome segregation. Centrosomes, which are crucial for cell division fidelity, regulate cellular organization, polarity, and interphase movement. Initiated in the G1/S phase, centrosome replication is tightly coupled with DNA replication. Notably, centrosome abnormalities are prevalent in ovarian tumors, exceeding 60 % of stage I cases, suggesting their early involvement in ovarian carcinogenesis. Furthermore, its increasing frequency with tumor stage implies a potential mechanistic role in OC progression [[31], [32], [33]]. Multiple lines of evidence have demonstrated that abnormalities in ECM remodeling play a profound role in tumor aggressiveness and metastasis [[34], [35], [36], [37]]. Elucidating the factors driving ECM organization and remodeling could yield promising therapeutic strategies for the prevention and eradication of tumor metastasis [38]. We summarized the possible roles of THUMPD2 in the tumorigenesis and treatment of OC (Fig. 7).
Fig. 7.
Schematic outline of THUMPD2-regulated tumorigenesis of ovarian cancer: reduced THUMPD2 may promote proliferation by negatively regulating genes related to ‘microtubule cytoskeleton organization’ and ‘cell cycle’; elevated THUMPD2 may promote metastasis by positively regulating genes related to ‘ECM organization’ and ‘extracellular organization’. ECM, extracellular matrix.
Our RNA-Seq analysis revealed that the downregulation of THUMPD2 resulted in significant alterations in the expression of several critical genes implicated in ovarian cancer development, such as BRBP1 [39], APC [40], ATR [41], PTPN11 (SHP2) [42], CDK6 [43], ATM [44], RB1 [45], AKAP9 [46], ROCK2 [47], ASPM [48], MIB1 [49], PPP1R12A [50] BRCA2 [51], ID1 [52]. These results were consistent with prior studies [[39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52]]. Although the RNA-sequence results have provided some inspiration for THUMPD2 downstream genes, The precise mechanisms governing the regulation of downstream genes by THUMPD2 have not been conclusively identified in this study. So precise regulatory pathways will require further investigation. Future research will focus on RNA immunoprecipitation sequencing (Rip-seq) test to assess the exact target genes of THUMPD2 and to identify the specific signaling pathways through which THUMPD2 regulates the proliferation, metastasis, and invasion of OC cells.
5. Conclusions
THUMPD2 is downregulated in early-stage OC; however, it is upregulated in advanced-stage epithelial OC. THUMPD2 suppressed cell proliferation. Conversely, it also promotes migration and invasion. High THUMPD2 expression is correlated with poor survival in epithelial OC patients. THUMPD2 may facilitate tumor metastasis by modulating the centrosome and ECM pathways (Fig. 7). The precise mechanisms governing the regulation of THUMPD2 have not been conclusively identified. So precise regulatory pathways will require further investigation.
Ethics and consent
Animal experiments were approved by the Animal Care and Use Committee of Nantong University (approval Number. S20210227-009).
Patient consent for publication
The study received approval from the Ethics Committee of Affiliated Hospital of Nantong University (Approval Number. 2021-L084). Written Informed Consent was obtained from all patients/participants to publish their data.
Funding
Minhui Hua was supported by Nantong social science and Technology Plan for People's livelihood Project,Science and Technology Bureau of Nantong, China (grant No.JCZ2023003) and the Research Physician Development Fund, Affiliated Hospital of Nantong University, China (YJXYY202204-YSC03). Yuquan Zhang was supported by the Research Physician Development Fund, Affiliated Hospital of Nantong University, China (YJXYY202204-YSB05).
Data availability statement
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
CRediT authorship contribution statement
Minhui Hua: Writing – review & editing, Conceptualization. Yujie Chen: Methodology, Investigation. Meiqun Jia: Visualization, Software. Wenxuan Lv: Resources, Investigation, Data curation. Yunzhao Xu: Writing – review & editing, Validation. Yuquan Zhang: Writing – original draft, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e33201.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Figs1.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Yang L., Xie H.J., Li Y.Y., Wang X., Liu X.X., Mai J. Molecular mechanisms of platinum-based chemotherapy resistance in ovarian cancer. Oncol. Rep. 2022;47 doi: 10.3892/or.2022.8293. (Review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colombo N., Sessa C., du Bois A., Ledermann J., McCluggage W.G., McNeish I., et al. ESMO-ESGO consensus conference recommendations on ovarian cancer: pathology and molecular biology, early and advanced stages, borderline tumours and recurrent diseasedagger. Ann. Oncol. 2019;30:672–705. doi: 10.1093/annonc/mdz062. [DOI] [PubMed] [Google Scholar]
- 4.Burger R.A., Brady M.F., Bookman M.A., Fleming G.F., Monk B.J., Huang H., et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N. Engl. J. Med. 2011;365:2473–2483. doi: 10.1056/NEJMoa1104390. [DOI] [PubMed] [Google Scholar]
- 5.Mirza M.R., Coleman R.L., Gonzalez-Martin A., Moore K.N., Colombo N., Ray-Coquard I., Pignata S. The forefront of ovarian cancer therapy: update on PARP inhibitors, Ann. Oncol. 2020;31:1148–1159. doi: 10.1016/j.annonc.2020.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Penson R., Valencia R.V., Colombo N., Leath C., Bidzinski M., Kim J.W., et al. Final overall survival results from SOLO3: phase III trial assessing olaparib monotherapy versus non-platinum chemotherapy in heavily pretreated patients with germline BRCA1-and/or BRCA2-mutated platinum-sensitive relapsed ovarian cancer. Gynecol. Oncol. 2022;166:S19–S20. [Google Scholar]
- 7.Oza A.M., Lisyanskaya A.S., Fedenko A.A., de Melo A.C., Shparik Y., Bondarenko I., et al. Overall survival results from ARIEL4: a phase III study assessing rucaparib vs chemotherapy in patients with advanced, relapsed ovarian carcinoma and a deleterious BRCA1/2 mutation, Ann. Oncol. 2022;33 doi: 10.1016/j.annonc.2022.07.646. S780–S780. [DOI] [Google Scholar]
- 8.Zhang X.W., Wu Y.S., Xu T.M., Cui M.H. CAR-T cells in the treatment of ovarian cancer: a promising cell therapy. Biomolecules. 2023;13 doi: 10.3390/biom13030465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konstantinopoulos P.A., Matulonis U.A. Clinical and translational advances in ovarian cancer therapy. Nat. Can. (Ott.) 2023;4:1239–1257. doi: 10.1038/s43018-023-00617-9. [DOI] [PubMed] [Google Scholar]
- 10.Matia-Gonzalez A.M., Laing E.E., Gerber A.P. Conserved mRNA-binding proteomes in eukaryotic organisms. Nat. Struct. Mol. Biol. 2015;22:1027–1033. doi: 10.1038/nsmb.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou H., Luo J., Guo Y., Liu Y., Wang Y., Deng L., Li P. RNA-binding protein complex LIN28/MSI2 enhances cancer stem cell-like properties by modulating Hippo-YAP1 signaling and independently of Let-7. Oncogene. 2022 doi: 10.1038/s41388-022-02198-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arandel L., Matloka M., Klein A.F., Rau F., Sureau A., Ney M., et al. Reversal of RNA toxicity in myotonic dystrophy via a decoy RNA-binding protein with high affinity for expanded CUG repeats. Nat. Biomed. Eng. 2022;6:207–220. doi: 10.1038/s41551-021-00838-2. [DOI] [PubMed] [Google Scholar]
- 13.Lin X., Zhou L., Zhong J., Zhong L., Zhang R., Kang T., Wu Y. RNA-binding protein RBM28 can translocate from the nucleolus to the nucleoplasm to inhibit the transcriptional activity of p53, J Biol Chem. 298. 2022;101524 doi: 10.1016/j.jbc.2021.101524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu T., Wei Q., Jin J., Luo Q., Liu Y., Yang Y., et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation, Nucleic Acids Res. 2020;48:3816–3831. doi: 10.1093/nar/gkaa048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X., Li Y., Ding Y., Zhang H., Ding N., Lu M. HuR promotes ovarian cancer cell proliferation by regulating TIMM44 mRNA stability. Cell Biochem. Biophys. 2020;78:447–453. doi: 10.1007/s12013-020-00939-w. [DOI] [PubMed] [Google Scholar]
- 16.Zhang M., Xu Y., Zhang Y., Li B., Lou G. Circular RNA circE2F2 promotes malignant progression of ovarian cancer cells by upregulating the expression of E2F2 protein via binding to HuR protein. Cell. Signal. 2021;84:110014. doi: 10.1016/j.cellsig.2021.110014. [DOI] [PubMed] [Google Scholar]
- 17.Chen S., Wu W., Li Q.H., Xie B.M., Shen F., Du Y.P., et al. Circ-NOLC1 promotes epithelial ovarian cancer tumorigenesis and progression by binding ESRP1 and modulating CDK1 and RhoA expression. Cell Death Dis. 2021;7:22. doi: 10.1038/s41420-020-00381-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arslan O., Soylu N.K., Akillilar P.T., Tazebay U.H. Coiled-coil domain-containing protein-124 (Ccdc124) is a novel RNA binding factor up-regulated in endometrial, ovarian, and urinary bladder cancers. Cancer Biomarkers. 2021;31:149–164. doi: 10.3233/CBM-200802. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y., Gorry M.C., Hart P.S., Pettenati M.J., Wang L., Marks J.J., Lu X., Hart T.C. Localization, genomic organization, and alternative transcription of a novel human SAM-dependent methyltransferase gene on chromosome 2p22-->p21. Cytogenet. Cell Genet. 2001;95:146–152. doi: 10.1159/000059337. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi M., Kawakubo H., Fukuda K., Mayanagi S., Nakamura R., Suda K., Hayashida T., Wada N., Kitagawa Y. THUMP domain containing 2 protein possibly induces resistance to cisplatin and 5-fluorouracil in in vitro human esophageal squamous cell carcinoma cells as revealed by transposon activation mutagenesis. J. Gene Med. 2019;21 doi: 10.1002/jgm.3135. [DOI] [PubMed] [Google Scholar]
- 21.Gabant G., Auxilien S., Tuszynska I., Locard M., Gajda M.J., Chaussinand G., et al. THUMP from archaeal tRNA:m22G10 methyltransferase, a genuine autonomously folding domain. Nucleic Acids Res. 2006;34:2483–2494. doi: 10.1093/nar/gkl145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansson M.J., Bystrom A.S. The Saccharomyces cerevisiae TAN1 gene is required for N4-acetylcytidine formation in tRNA, RNA. 2004;10:712–719. doi: 10.1261/rna.5198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waterman D.G., Ortiz-Lombardia M., Fogg M.J., Koonin E.V., Antson A.A. Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain. J. Mol. Biol. 2006;356:97–110. doi: 10.1016/j.jmb.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 24.Armengaud J., Urbonavicius J., Fernandez B., Chaussinand G., Bujnicki J.M., Grosjean H. N2-methylation of guanosine at position 10 in tRNA is catalyzed by a THUMP domain-containing, S-adenosylmethionine-dependent methyltransferase, conserved in Archaea and Eukaryota. J. Biol. Chem. 2004;279:37142–37152. doi: 10.1074/jbc.M403845200. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X., Jiang G., Sun M., Zhou H., Miao Y., Liang M., Wang E., Zhang Y. Cytosolic THUMPD1 promotes breast cancer cells invasion and metastasis via the AKT-GSK3-Snail pathway. Oncotarget. 2017;8:13357–13366. doi: 10.18632/oncotarget.14528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan Y., Liu L., Zhang X., Xue Y., Gao J., Zhao J., Chi N., Zhu Y. THUMPD3-AS1 is correlated with gastric cancer and regulates cell function through miR-1252-3p and CXCL17. Crit. Rev. Eukaryot. Gene Expr. 2022;32:69–80. doi: 10.1615/CritRevEukaryotGeneExpr.2022042848. [DOI] [PubMed] [Google Scholar]
- 27.Li W., Dong S., Wang H., Wu R., Wu H., Tang Z.R., Zhang J., Hu Z., Yin C. Risk analysis of pulmonary metastasis of chondrosarcoma by establishing and validating a new clinical prediction model: a clinical study based on SEER database. BMC Muscoskel. Disord. 2021;22:529. doi: 10.1186/s12891-021-04414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y.L., Wu Z.Z., Zhang H.R., Chen D.S., Zhao X. Coexistence of a novel RGS18 downstream intergenic region ALK fusion and a THUMPD2-ALK fusion in a lung adenocarcinoma patient and response to crizotinib. Lung Cancer. 2021;154:216–218. doi: 10.1016/j.lungcan.2021.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Fife C.M., McCarroll J.A., Kavallaris M. Movers and shakers: cell cytoskeleton in cancer metastasis. Br. J. Pharmacol. 2014;171:5507–5523. doi: 10.1111/bph.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jordan M.A., Wilson L. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer. 2004;4:253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 31.Hsu L.C., Kapali M., DeLoia J.A., Gallion H.H. Centrosome abnormalities in ovarian cancer. Int. J. Cancer. 2005;113:746–751. doi: 10.1002/ijc.20633. [DOI] [PubMed] [Google Scholar]
- 32.Piemonte K.M., Anstine L.J., Keri R.A. Centrosome aberrations as drivers of chromosomal instability in breast cancer. Endocrinology. 2021;162 doi: 10.1210/endocr/bqab208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kramer A., Ho A.D. Centrosome aberrations and cancer. Onkologie. 2001;24:538–544. doi: 10.1159/000055141. [DOI] [PubMed] [Google Scholar]
- 34.Lu P., Weaver V.M., Werb Z. The extracellular matrix: a dynamic niche in cancer progression, J. Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kai F., Drain A.P., Weaver V.M. The extracellular matrix modulates the metastatic journey, Dev. Cell. 2019;49:332–346. doi: 10.1016/j.devcel.2019.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilkes D.M., Semenza G.L., Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat. Rev. Cancer. 2014;14:430–439. doi: 10.1038/nrc3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Natarajan S., Foreman K.M., Soriano M.I., Rossen N.S., Shehade H., Fregoso D.R., et al. Collagen remodeling in the hypoxic tumor-mesothelial niche promotes ovarian cancer metastasis. Cancer Res. 2019;79:2271–2284. doi: 10.1158/0008-5472.CAN-18-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan Z., Li Y., Zhang S., Wang X., Dou H., Yu X., Zhang Z., Yang S., Xiao M. Extracellular matrix remodeling in tumor progression and immune escape: from mechanisms to treatments. Mol. Cancer. 2023;22:48. doi: 10.1186/s12943-023-01744-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramus S.J., Song H., Dicks E., Tyrer J.P., Rosenthal A.N., Intermaggio M.P., et al. Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J. Natl. Cancer Inst. 2015;107 doi: 10.1093/jnci/djv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhai Y., Kuick R., Tipton C., Wu R., Sessine M., Wang Z., Baker S.J., Fearon E.R., Cho K.R. Arid1a inactivation in an Apc- and Pten-defective mouse ovarian cancer model enhances epithelial differentiation and prolongs survival. J. Pathol. 2016;238:21–30. doi: 10.1002/path.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biegala L., Gajek A., Marczak A., Rogalska A. Olaparib-resistant BRCA2(MUT) ovarian cancer cells with restored BRCA2 abrogate olaparib-induced DNA damage and G2/M arrest controlled by the ATR/CHK1 pathway for survival. Cells. 2023;12 doi: 10.3390/cells12071038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fedele C., Ran H., Diskin B., Wei W., Jen J., Geer M.J., et al. SHP2 inhibition prevents adaptive resistance to MEK inhibitors in multiple cancer models. Cancer Discov. 2018;8:1237–1249. doi: 10.1158/2159-8290.CD-18-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dall'Acqua A., Sonego M., Pellizzari I., Pellarin I., Canzonieri V., D'Andrea S., et al. CDK6 protects epithelial ovarian cancer from platinum-induced death via FOXO3 regulation. EMBO Mol. Med. 2017;9:1415–1433. doi: 10.15252/emmm.201607012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schram A.M., Colombo N., Arrowsmith E., Narayan V., Yonemori K., Scambia G., et al. Avelumab Plus talazoparib in patients with BRCA1/2- or ATM-altered advanced solid tumors: results from JAVELIN BRCA/ATM, an open-label, multicenter, phase 2b, tumor-agnostic trial. JAMA Oncol. 2023;9:29–39. doi: 10.1001/jamaoncol.2022.5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L., Zhai Y., Wang Y., Fearon E.R., Nunez G., Inohara N., Cho K.R. Altering the microbiome inhibits tumorigenesis in a mouse model of oviductal high-grade serous carcinoma. Cancer Res. 2021;81:3309–3318. doi: 10.1158/0008-5472.CAN-21-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frank B., Wiestler M., Kropp S., Hemminki K., Spurdle A.B., Sutter C., et al. Association of a common AKAP9 variant with breast cancer risk: a collaborative analysis. J. Natl. Cancer Inst. 2008;100:437–442. doi: 10.1093/jnci/djn037. [DOI] [PubMed] [Google Scholar]
- 47.Wang H.F., Takenaka K., Nakanishi A., Miki Y. BRCA2 and nucleophosmin coregulate centrosome amplification and form a complex with the Rho effector kinase ROCK2. Cancer Res. 2011;71:68–77. doi: 10.1158/0008-5472.CAN-10-0030. [DOI] [PubMed] [Google Scholar]
- 48.Bruning-Richardson A., Bond J., Alsiary R., Richardson J., Cairns D.A., McCormack L., et al. ASPM and microcephalin expression in epithelial ovarian cancer correlates with tumour grade and survival. Br. J. Cancer. 2011;104:1602–1610. doi: 10.1038/bjc.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Neill C.J., Deavers M.T., Malpica A., Foster H., McCluggage W.G. An immunohistochemical comparison between low-grade and high-grade ovarian serous carcinomas: significantly higher expression of p53, MIB1, BCL2, HER-2/neu, and C-KIT in high-grade neoplasms. Am. J. Surg. Pathol. 2005;29:1034–1041. [PubMed] [Google Scholar]
- 50.Benvenuto G., Todeschini P., Paracchini L., Calura E., Fruscio R., Romani C., et al. Expression profiles of PRKG1, SDF2L1 and PPP1R12A are predictive and prognostic factors for therapy response and survival in high-grade serous ovarian cancer. Int. J. Cancer. 2020;147:565–574. doi: 10.1002/ijc.32935. [DOI] [PubMed] [Google Scholar]
- 51.Samuel D., Diaz-Barbe A., Pinto A., Schlumbrecht M., George S. Hereditary ovarian carcinoma: cancer pathogenesis looking beyond BRCA1 and BRCA2. Cells. 2022;11 doi: 10.3390/cells11030539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su Y., Gao L., Teng L., Wang Y., Cui J., Peng S., Fu S. Id1 enhances human ovarian cancer endothelial progenitor cell angiogenesis via PI3K/Akt and NF-kappaB/MMP-2 signaling pathways. J. Transl. Med. 2013;11:132. doi: 10.1186/1479-5876-11-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available from the corresponding author upon reasonable request.