Summary
Background
The diagnosis of hepatocellular carcinoma (HCC) often experiences latency, ultimately leading to unfavorable patient outcomes due to delayed therapeutic interventions. Our study is designed to develop and validate a model that employs triple-phase computerized tomography (CT)-based deep learning radiomics and clinical variables for early warning of HCC in patients with cirrhosis.
Methods
We studied 1858 patients with cirrhosis primarily from the PreCar cohort (NCT03588442) between June 2018 and January 2020 at 11 centres, and collected triple-phase CT images and laboratory results 3–12 months prior to HCC diagnosis or non-HCC final follow-up. Using radiomics and deep learning techniques, early warning model was developed in the discovery cohort (n = 924), and then validated in an internal validation cohort (n = 231), and an external validation cohort from 10 external centres (n = 703).
Findings
We developed a hybrid model, named ALARM model, which integrates deep learning radiomics with clinical variables, enabling early warning of the majority of HCC cases. The ALARM model effectively predicted short-term HCC development in cirrhotic patients with area under the curve (AUC) of 0.929 (95% confidence interval 0.918–0.941) in the discovery cohort, 0.902 (0.818–0.987) in the internal validation cohort, and 0.918 (0.898–0.961) in the external validation cohort. By applying optimal thresholds of 0.21 and 0.65, the high-risk (n = 221, 11.9%) and medium-risk (n = 433, 23.3%) groups, which covered 94.4% (84/89) of the patients who developed HCC, had significantly higher rates of HCC occurrence compared to the low-risk group (n = 1204, 64.8%) (24.3% vs 6.4% vs 0.42%, P < 0.001). Furthermore, ALARM also demonstrated consistent performance in subgroup analysis.
Interpretation
The novel ALARM model, based on deep learning radiomics with clinical variables, provides reliable estimates of short-term HCC development for cirrhotic patients, and may have the potential to improve the precision in clinical decision-making and early initiation of HCC treatments.
Funding
This work was supported by National Key Research and Development Program of China (2022YFC2303600, 2022YFC2304800), and the National Natural Science Foundation of China (82170610), Guangdong Basic and Applied Basic Research Foundation (2023A1515011211).
Keywords: Cirrhotic, Hepatocellular carcinoma, Computed tomography, Radiomics, Deep learning
Research in context.
Evidence before this study
We searched PubMed from database inception to April 2024, for publications on triple-phase CT-based deep learning radiomics and clinical variables for early warning of HCC in patients with cirrhosis. We used the search terms “artificial intelligence” or “machine learning” or “deep learning”, “radiomics”, “latency period” or “prediction”, “aMAP”, without language restrictions. We also reviewed reference lists of eligible articles. Our search did not identify any previous studies on the use of radiomics, deep learning techniques, and aMAP HCC risk score for predicting HCC occurrence of cirrhotic patient 3–12 months before HCC diagnosis.
Added value of this study
We developed and externally validated the first clinical decision tool, called ALARM model, capable of accurately early warning HCC development for patients with cirrhosis 3–12 months before the HCC clinical diagnosis in a large multicenter cohort of 1858 patients. ALARM, serving as a comprehensive model that integrates radiomics and deep learning scores along with aMAP HCC risk score, could identify the majority of individuals with HCC occurrence in advance.
Implications of all the available evidence
ALARM holds the potential for practical implementation in clinical settings, allowing for the early warning of tumorigenesis in cirrhotic patients. This novel addition is poised to significantly refine the precision of clinical decision-making, fostering proactive and personalized anti-tumor interventions. Moving forward, forthcoming efforts should concentrate on prospectively validating the model's clinical utility.
Introduction
Early diagnosis and effective treatment of hepatocellular carcinoma (HCC) represents a momentous challenge for global health organizations, placing an enormous burden on healthcare systems worldwide.1,2 In China, this is particularly significant for hepatitis B virus (HBV)-related cirrhotic patients, as HBV infection is the main etiology for HCC development. The diagnosis of HCC often has an latency period, and the lack of timely treatment measures ultimately leads to poor patient outcomes, thus early warning contributes to a favorable prognosis and opens up the possibility for curative treatment.3
Substantial efforts have been made towards developing a robust, sensitive, and non-invasive test for the detection of HCC.4 Radiomics as a promising technology for cancer detection that has gained significant attention in recent years.5, 6, 7, 8 It combines expertise from medical imaging, computer science, and statistics to analyze medical image findings using computer-assisted methods. Deep learning algorithms have improved the accuracy and efficiency of Radiomics, enabling the identification of subtle image signatures that were previously undetectable, resulting in better sensitivity and specificity in diagnosing HCC.9, 10, 11
Recently, a novel aMAP HCC risk score, which was calculated by five common clinical variables, was developed to predict the likelihood of HCC development in patients with chronic hepatitis. It has demonstrated remarkable accuracy in identifying patients with chronic hepatitis at high risk of developing HCC, thereby providing opportunities for early detection by intensive surveillance.12,13
Considering the recent advances of above methods, we conducted this nationwide multicenter study, aiming to develop and validate a novel model (called ALARM), by integrating deep learning-based radiomics and aMAP risk score, to identify early changes indicative of HCC in patients with cirrhosis, thereby providing early warning information to patients and physicians.
Methods
Study design and patients
This is a retrospective, multicentre, cohort study. The patients enrolled in this study were mainly from PreCar cohort. PreCar cohort is a prospective multicentre observational cirrhotic cohort conducted in mainland China (NCT03588442), in which 4692 adult liver cirrhotic patients were enrolled from June 2018 to January 2020 from 16 centres in 11 provinces across China.13 At screening period, they all had enhanced computerized tomography (CT) scans done to provide a thorough assessment of their baseline physical condition and detect any underlying abnormalities. The biannual protocol follow-up for all patients included ultrasound examinations as well as routine clinical assessments to monitor the occurrence of HCC. In the current study, inclusion criteria were as follows: (i) patients diagnosed with cirrhosis; (ii) for patients diagnosed with HCC during follow-up, having enhanced CT image taken within a 3–12 month window prior to HCC diagnosis; (iii) patients without HCC diagnosis, having enhanced CT images taken within a 3–12 month window prior to their last follow-up. In brief, all the CT images included in the analysis were those without clinically diagnosing HCC. Exclusion criteria included: (i) patients under the age of 18; (ii) patients who had undergone previous liver-related surgeries, such as hepatectomy or liver transplantation; (iii) patients who had undergone previous chemotherapy or immunotherapy; (iv) patients with poor-quality CT image. (v) partial or complete data is missing. Besides, due to the scarcity of HCC patients who met the above inclusion criteria, we additionally included HCC patients from the Search-B cohort (NCT02167503)13,14 and outpatient from Nanfang hospital in the current study. The diagnosis of cirrhosis and HCC were based on standard histological and/or compatible radiological findings. For detailed diagnostic information, please refer to Supplementary A1.
Ethics
This study was approved by the Ethics Committee of Nanfang Hospital, with reference number NFEC-201808-101, and was conducted in accordance with the guidelines of the Declaration of Helsinki and the principles of good clinical practice. All patients provided written informed consent to have their data used (anonymously) for research purposes.
CT imaging
All patients in the study underwent CT scans in the arterial phase, venous phase and delayed phase. Supplementary A2 provides further details regarding the specific parameters utilized for CT image acquisition. Before the analysis, the images underwent preprocessing, including resampling voxel size and discretization of Hounsfield Units. The voxel dimensions were resampled to 1 × 1 × 1 mm (x-, y-, and z-axes) to correct for acquisition-related voxel resolution variations. To emphasize the liver in abdominal CT scans and reduce interference from surrounding organs, we set the window width to 200 and the window level to 40 (Supplementary A3).
Regions of interest segmentation
For patients with cirrhosis, since the occurrence and development of HCC involve extensive areas of the liver and various complex factors, comprehensively understanding the condition of the liver helps to assess the risk more accurately. Therefore, accurately identifying the region of interest (ROI) encompassing the entire liver is crucial. In addition, to expedite the time-consuming and labor-intensive process of defining ROI, we employed the nnU-Net-based automatic delineation method (Supplementary A4).15 Leveraging this approach allowed us to utilize an automatic contouring method and capitalize on the relative stability of liver contour and position to consistently obtain the same ROI for each contouring.
Image feature extraction and selection
To extract radiomics signature, the Pyradiomics package16 was utilized. We customized the deep residual network in a three-dimensional fashion (3D-ResNet) to devise our Fine-tuned 3D-ResNet50 aimed at effectuating profound learning feature extraction (Supplementary A5).17 The process of feature normalization was conducted using the z-score method to standardize the value range, with the aim of identifying the most significant signatures associated with HCC. Mann–Whitney U test was performed for each feature to select the signatures that were significantly associated with the outcome and had a P-value below the threshold of 0.05, and Spearman's rank correlation coefficient was used to identify highly correlated features. Lastly, the Lasso regression model with 10-fold cross-validation was utilized to remove features with zero weight, ensuring the feature selection process was robust and reducing the risk of overfitting (Supplementary A6).
Model construction and validation
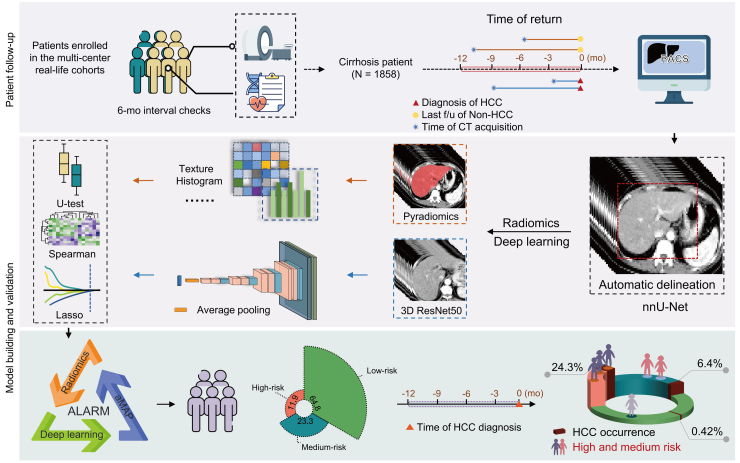
The Lasso regression model was utilized to identify pertinent features, with their coefficients being assigned weights to produce the corresponding radiomics score and deep learning score. This methodology enables the calculation of scores that assess the significance and quality of radiomics and deep learning within the framework of imaging analysis. In our study, univariate and multivariate analyses were implemented to identify independent markers for differentiating liver which would transit to malignance or not, and these markers were subsequently integrated into a combine model. The discovery cohort, which constituted 80% of the primary cohort (Nanfang Hospital) was used to develop the model. To rectify the imbalance between positive and negative samples in the discovery cohort, we employed the Borderline-1 SMOTE method18 (Figure S1). After oversampling, we obtained a new discovery cohort to develop the model. An internal validation cohort representing 20% of the primary cohort, and an external validation cohort composed of patients from the other 10 centers in the PreCar cohort, were utilized to evaluate the detection performance and robustness of the ALARM model. The study flowchart is depicted in Figure S2. The modeling pipeline is illustrated in Fig. 1.
Fig. 1.
Workflow of the current study. f/u, Follow-up; mo, Month; HCC, Hepatocellular carcinoma; CT, Computed Tomography; PACS, Picture Archiving and Communication System; U-test, Mann–Whitney U test; Spearman, Spearman's rank correlation coefficient; Lasso, Lasso regression.
Statistics
Categorical variables were expressed as counts and percentages and analyzed using the Chi-square test or Fisher exact test, as appropriate. Continuous variables were expressed as median (inter-quartile range [IQR]) or mean ± standard deviation and compared using the Mann–Whitney U test and the Kruskal–Wallis test, as appropriate. We consider a two-tailed P-value below 0.05 to indicate statistical significance. The performance of the model was evaluated using receiver operating characteristic (ROC) curves, and the area under the curve (AUC) was calculated to compare its efficacy with that of single-signature models across all cohorts. Delong test, Net Reclassification Index (NRI), and Integrated Discrimination Improvement (IDI) were used to compare the performance of various models. Furthermore, decision curve analysis was conducted to evaluate the clinical usefulness of the model by quantifying the net benefit at various threshold probabilities. Calibration curves were used to evaluate the model's accuracy and reliability. To address potential confounding factors and ensure the robustness of the findings, we conducted subgroup analysis based on age, sex, and alpha-fetoprotein (AFP) levels. In the discovery cohort, X-tile plots were used to generate two optimal cut-off values with the highest χ2 value to separate patients into three deterioration trends, corresponding to the high-risk, medium-risk, and low-risk.19 The above process was carried out using R software (version 4.2) and Python (version 3.7 and 3.9).
Role of the funding source
The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Results
Patient characteristics
A total of 1858 eligible patients from 11 centers were included in the current study, among which 1836 (98.82%) patients with 67 cases of HCC were from PreCar cohort, and the other 22 patients (1.18%, all HCC cases) from the outpatients and Search-B cohort in Nanfang Hospital (Figure S3). Among these patients, 924 were allocated to the discovery cohort, 231 to the internal validation cohort, and 703 to the external validation cohort. 45 participants (4.9%) in the discovery cohort, 13 participants (5.6%) in the internal validation cohort and 31 participants (4.4%) in the external validation cohort were diagnosed with HCC after 3–12 months after CT scans. The average time intervals from CT examination to the diagnosis of HCC for stage 0, A, B, C, and D were 6.0, 6.8, 7.2, 11.0, and 9.7 months, respectively. Characteristics of the patients are shown in Table 1.
Table 1.
Clinical characteristics of enrolled patients.
| All (n = 1858) |
Discovery cohort (n = 924) |
Internal validation cohort (n = 231) |
External validation cohort (n = 703) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Non-HCC |
HCC |
P-value | Non-HCC |
HCC |
P-value | Non-HCC |
HCC |
P value | Non-HCC |
HCC |
P-value | |
| (n = 1769) | (n = 89) | (n = 879) | (n = 45) | (n = 218) | (n = 13) | (n = 672) | (n = 31) | |||||
| Age, years | 49.9 (43.2, 56.4) | 58.2 (50.9, 64.0) | <0.001 | 48.3 (42.0, 55.2) | 57.8 (52.3, 62.9) | <0.001 | 48.6 (41.5, 54.2) | 58.6 (49.5, 64.4) | 0.012 | 52.3 (45.5, 59.8) | 58.2 (51.9, 66.9) | 0.002 |
| Sex (Male), n (%) | 1411 (79.8) | 78 (87.6) | 0.093 | 742 (84.4) | 41 (91.1) | 0.314 | 191 (87.6) | 0 (0.0) | 0.365 | 478 (71.1) | 24 (77.4) | 0.579 |
| aMAP HCC score | 58.3 (53.9, 63.1) | 64.3 (59.4, 69.0) | <0.001 | 57.4 (52.8, 62.0) | 64.2 (59.5, 68.3) | <0.001 | 56.7 (53.0, 62.8) | 65.0 (61.9, 68.6) | 0.003 | 59.4 (55.0, 64.4) | 64.7 (57.8, 69.1) | 0.004 |
| AST, U/L | 29.0 (23.0, 40.2) | 38.0 (26.0, 54.0) | <0.001 | 28.0 (22.0, 38.0) | 35.0 (26.0, 54.0) | 0.001 | 28.0 (23.0, 36.0) | 36.0 (21.0, 44.0) | 0.689 | 32.4 (24.5, 45.1) | 39.0 (29.0, 55.0) | 0.034 |
| ALT, IU/L | 29.0 (21.0, 41.0) | 33.0 (24.0, 46.0) | 0.096 | 28.0 (21.0, 40.0) | 34.0 (24.0, 46.0) | 0.157 | 28.0 (21.0, 39.0) | 30.0 (23.0, 33.0) | 0.871 | 29.0 (21.0, 43.4) | 34.6 (24.5, 56.5) | 0.105 |
| Total bilirubin, μmol/L | 16.5 (12.1, 24.5) | 19.1 (13.4, 32.9) | 0.003 | 15.6 (11.2, 22.1) | 20.7 (14.1, 34.9) | 0.001 | 16.1 (11.9, 24.8) | 17.9 (11.6, 32.9) | 0.431 | 17.8 (13.1, 27.1) | 18.7 (13.8, 30.6) | 0.568 |
| Albumin, g/L | 43.0 (38.8, 46.0) | 40.0 (33.3, 45.2) | <0.001 | 43.2 (40.0, 46.1) | 40.6 (33.3, 44.8) | 0.001 | 44.0 (40.3, 46.7) | 34.5 (34.2, 41.9) | 0.004 | 42.2 (37.0, 45.4) | 42.1 (33.1, 45.8) | 0.688 |
| Platelet, × 10³/mm³ | 114.0 (76.6, 157.0) | 95.0 (68.0, 134.0) | 0.007 | 122.0 (81.0, 163.5) | 95.0 (68.0, 132.0) | 0.008 | 118.5 (76.0, 164.8) | 95.0 (66.0, 138.0) | 0.321 | 104.0 (75.0, 142.0) | 95.0 (71.0, 135.5) | 0.374 |
| Creatinine, μmol/L | 71.0 (60.0, 82.0) | 74.0 (64.0, 83.7) | 0.048 | 74.0 (63.5, 84.0) | 76.0 (64.0, 84.0) | 0.683 | 74.0 (65.2, 84.0) | 72.0 (63.0, 94.0) | 0.760 | 65.0 (54.0, 76.0) | 69.0 (57.0, 79.5) | 0.149 |
| Bun, mmol/L | 4.6 (3.8, 5.5) | 4.8 (4.1, 5.8) | 0.061 | 4.5 (3.8, 5.4) | 4.9 (4.2, 6.4) | 0.005 | 4.5 (3.8, 5.4) | 4.6 (4.5, 5.5) | 0.270 | 4.7 (3.9, 5.7) | 4.8 (4.1, 6.0) | 0.488 |
| Glucose, mmol/L | 5.3 (4.9, 5.9) | 5.4 (5.0, 5.8) | 0.539 | 5.3 (4.9, 5.8) | 5.4 (5.1, 6.0) | 0.075 | 4.5 (3.8, 5.4) | 5.4 (4.9, 5.8) | 0.998 | 5.4 (4.9, 6.0) | 5.4 (4.9, 5.6) | 0.269 |
| AFP, μg/L | 2.9 (1.8, 5.3) | 7.3 (3.0, 14.7) | <0.001 | 2.7 (1.6, 4.8) | 6.2 (2.9, 13.2) | <0.001 | 2.6 (1.6, 4.4) | 8.1 (2.7, 14.7) | 0.005 | 3.4 (2.1, 6.5) | 8.0 (4.2, 15.9) | <0.001 |
| LSM, kPa | 13.7 (9.4, 21.1) | 17.4 (11.8, 26.4) | <0.001 | 13.5 (9.3, 21.8) | 21.8 (14.7, 38.5) | <0.001 | 13.4 (8.5, 22.4) | 16.7 (10.5, 24.9) | 0.218 | 14.0 (9.9, 20.7) | 13.2 (11.2, 20.0) | 0.775 |
| ALBI score | −2.6 (−3.0, −2.2) | −2.3 (−2.8, −1.5) | <0.001 | −2.9 (−3.2, −2.6) | −2.5 (−3.0, 11.9) | <0.001 | −3.0 (−3.2, −2.6) | −2.1 (−2.7, −1.8) | 0.006 | −2.8 (−3.1, −2.2) | −2.9 (−3.1, −2.5) | 0.004 |
| Cirrhosis etiology, n (%) | ||||||||||||
| HBV | 1563 (88.4) | 82 (92.1) | 814 (92.6) | 42 (93.3) | 208 (95.4) | 13 (100.0) | 541 (80.8) | 27 (87.1) | ||||
| HCV | 59 (3.3) | 3 (3.4) | 29 (3.3) | 1 (2.2) | 4 (1.8) | 0 (0.0) | 26 (3.9) | 2 (6.5) | ||||
| NASH | 9 (0.5) | 0 (0) | 1 (0.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 8 (1.2) | 0 (0.0) | ||||
| Alcohol | 49 (2.8) | 2 (2.2) | 20 (2.3) | 1 (2.2) | 3 (1.4) | 0 (0.0) | 26 (3.9) | 1 (3.2) | ||||
| Other | 89 (5.0) | 2 (2.2) | 15 (1.7) | 1 (2.2) | 3 (1.4) | 0 (0.0) | 71 (10.6) | 1 (3.2) | ||||
| HCC BCLC stage, n (%) | ||||||||||||
| 0 | / | 32 (36.0) | / | 13 (28.9) | / | 8 (61.5) | / | 11 (35.5) | ||||
| A | / | 20 (22.5) | / | 14 (31.1) | / | 2 (15.4) | / | 4 (12.9) | ||||
| B | / | 14 (15.7) | / | 3 (6.7) | / | 0 (0.0) | / | 11 (35.5) | ||||
| C | / | 16 (18.0) | / | 11 (24.4) | / | 1 (7.7) | / | 4 (12.9) | ||||
| D | / | 7 (7.9) | / | 4 (8.9) | / | 2 (15.4) | / | 1 (3.2) | ||||
Data are reported as median and IQR, unless otherwise specified. AST, aspartate aminotransferase; ALT, alanine aminotransferase; ALB, serum albumin; AFP, serum alpha-fetoprotein; Bun, blood urea nitrogen.
HBV, hepatitis B virus; HCV, hepatitis C virus; NASH, non-alcoholic steatohepatitis; HCC, Hepatocellular Carcinoma; LSM, Liver stiffness measurement; ALBI, albumin-bilirubin.
aMAP HCC score = (0.06 × Age + 0.89 × Sex (Male: 1; Female: 0) + 0.48 × (((log10 Total bilirubin) × 0.66) + (Albumin × −0.085))–0.01 × PLT+ 7.4)/14.77 × 100.
Image signature analysis
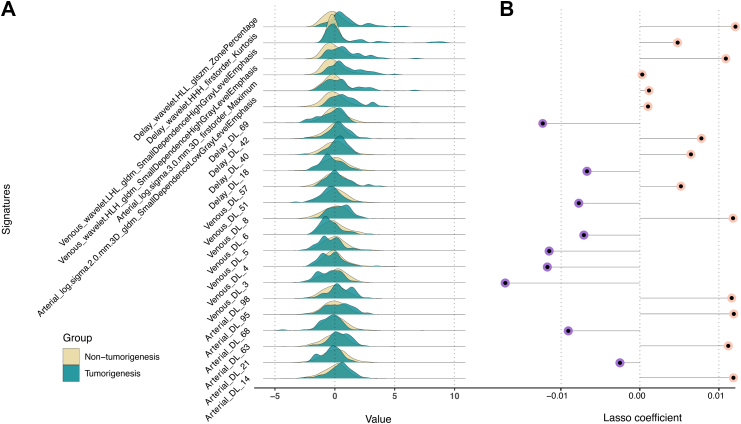
From a single liver phase per patient, we extracted a total of 1223 signatures, resulting in 3669 signatures across three phases (Figure S4, Table S2, and Supplementary A7). Additionally, we obtained 100 × 3 deep learning signatures from the average pooling layer of the Fine-tuned 3D ResNet50 for each patient. After feature selection process, we identified 6 radiomics signatures and 17 deep learning signatures for building single-signature models and calculating radiomics and deep learning scores, respectively (Supplementary A5, Fig. 2 and Figures S5–S11). The formula and the distribution of radiomics score and deep learning score are presented in Supplementary A8. The Mann–Whitney U test indicated that there were significant differences in the radiomics score, deep learning score and aMAP score between patients who would develop HCC or not (Figure S12). Additionally, multivariable linear regression analysis revealed that these signature scores all served as independent markers to distinguish malignant progression in patients with cirrhosis (Figure S13).
Fig. 2.
(A) Comparison of patterns of signatures between subgroups. The y-axis of Ridgeline Plot represents the frequency or density, while the x-axis represents the range of signatures. Distinct peaks and differences in spread between the groups suggest potential variations in the signature patterns. (B) Feature importance in Lasso regression model. The length of the lollipop represents the importance of signatures in the Lasso model. A longer lollipop indicates a greater impact on the outcome, while shorter or nonexistent lollipops suggest a smaller or negligible effect on the target variable. HLL, High Level Features; glszm, Gray Level Size Zone Matrix; HHH, High-Order Histogram of Homogeneous; gldm, Gray Level Dependence Matrix; HLH, Histogram of Local Homogeneity; DL, Deep learning.
Construction and validation of ALARM model
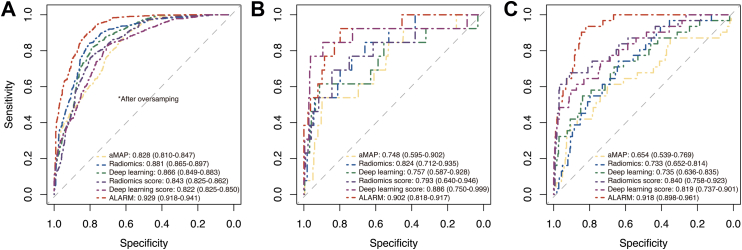
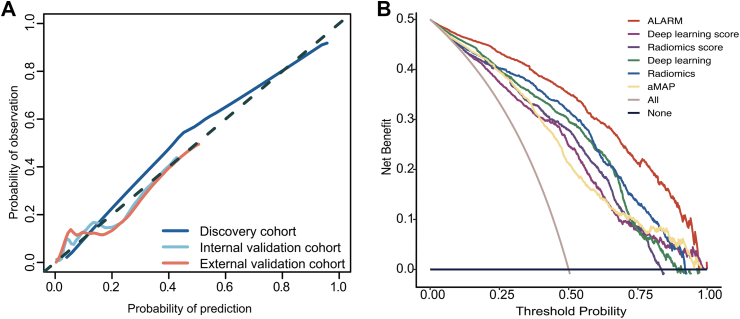
The ALARM model was developed by fitting a logistic regression model using the clinical score, radiomics score, and deep learning score as three individual covariates. For clinical score, we attempted various combinations of clinical variables with radiomics score and deep learning score, including aMAP, age, sex, total bilirubin, albumin, platelet counts, and AFP. The results indicate that the combination of radiomics score, deep learning score and aMAP score performed significantly better than the other combinations in terms of AUC. Even with AFP included in these variables, according to the Delong test, their performance did not significantly improve (Table S3). Hence, aMAP score was selected as the variable included in the clinical score. Then, it was encouragingly found that ALARM exhibited great discriminatory performance, with an AUC of 0.929 (95% confidence interval [CI]: 0.918–0.941) in the discovery cohort, 0.902 (95% CI: 0.818–0.987) in the internal validation cohort, and 0.918 (95% CI: 0.898–0.961) in the external validation cohort, as confirmed by bootstrapping validation (Fig. 3). We also performed cross-validation in each cohort and demonstrated the stable performance of the model under different folds, further confirming the reliability of our approach (Figure S14). The DeLong test revealed a statistically significant disparity (P < 0.05) between ALARM and single-signature models, signifying that ALARM exhibited superior performance in predicting short-term HCC development among cirrhotic patients (Table S4). The performance of the ALARM was also compared with that of single-signature models using the NRI and IDI analyses, which demonstrated that the ALARM had superior performance (Table S5 and S6). The calibration curve generated from our study revealed a strong agreement between the predicted probabilities generated by the ALARM and the actual outcomes (Fig. 4A). In addition, our decision curve analysis results, presented in Fig. 4B, demonstrated that the ALARM offers superior benefits in clinical decision-making when compared to single-signature model. Furthermore, the subgroup analysis showed that the performance of ALARM was consistent regardless of age, sex, and AFP levels (Supplementary A9 and Figure S15).
Fig. 3.
Prediction performance of ALARM vs single-signature models across all cohorts. The receiver operating characteristic curves for early prediction performance of HCC is compared among the three single-modality prediction models and ALARM in the discovery cohort (A), the internal validation cohort (B), and the external validation cohort (C).
Fig. 4.
(A) Calibration curves of ALARM on discriminating HCC and non-HCC. (B) Decision curve analysis based on ALARM, radiomics score, deep learning score and aMAP score to guide clinical practice.
Risk stratification in patients with cirrhosis
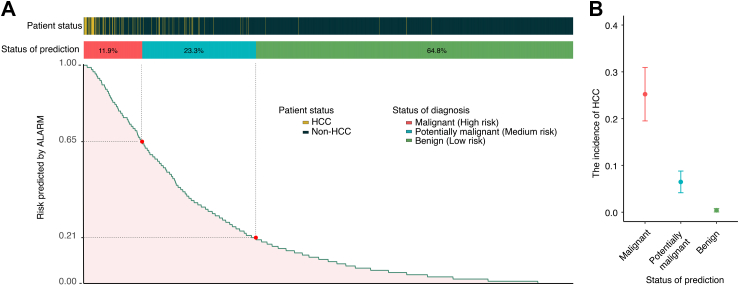
After determining the optimal thresholds of 0.21 and 0.65 using the x-tile software (Figure S16) in the newly generated discovery cohort after SMOTE oversampling, the patients were divided into three groups: high-risk, medium-risk, low-risk groups. The results revealed that the high-risk (n = 221, 11.9%) and medium risk (n = 433, 23.3%) groups, which covered 94.4% (84/89) of the patients who developed HCC, had significantly higher rates of HCC occurrence in comparison to low-risk group (n = 1204, 64.8%) (24.3% vs 6.4% vs 0.42%, P < 0.001) (Fig. 5). Moreover, ALARM achieved an average lead time of 7.2 months for early warning of HCC development by employing a threshold of 0.21, with 33.3% of patients being warned less than 6 months prior to clinical diagnosis, 32.1% between 6 and 9 months, and 34.5% between 9 and 12 months. At the upper threshold, the specificity values for discovery cohort, internal validation cohort, and external validation cohort were 0.899 (95% CI: 0.879–0.918), 0.889 (95% CI: 0.850–0.930), and 0.926 (95% CI: 0.906–0.945), respectively. At the lower threshold, the sensitivity values for discovery cohort, internal validation cohort, and external validation cohort were 0.933 (95% CI: 0.917–0.949), 0.923 (95% CI: 0.889–0.957), and 0.944 (95% CI: 0.955–0.981), respectively (Table 2).
Fig. 5.
(A) Risk stratification and threshold analysis. Composite chart depicting the relationship between ALARM prediction outcomes and actual results, along with a risk stratification system based on optimal threshold values. (B) Comparing of the incidence of HCC. Error bar chart revealing significantly different incidence between each patient status classification, highlighting variations in risk levels. This error bar chart illustrates the incidence of HCC and their corresponding 95% confidence intervals.
Table 2.
Diagnostic performance of ALARM in different cohorts at cutoff values of 0.21 and 0.65.
| Cut-off value: 0.21 |
Cut-off value: 0.65 |
|||
|---|---|---|---|---|
| Value | 95% CI | Value | 95% CI | |
| ALARM | Discovery cohorta | |||
| Sensitivity | 0.933 | (0.917–0.949) | 0.600 | (0.568–0.632) |
| Specificity | 0.671 | (0.641–0.702) | 0.899 | (0.879–0.918) |
| PPV | 0.127 | (0.105–0.148) | 0.233 | (0.206–0.260) |
| NPV | 0.995 | (0.990–0.999) | 0.978 | (0.968–0.987) |
| Internal validation cohort | ||||
| Sensitivity | 0.923 | (0.889–0.957) | 0.692 | (0.633–0.752) |
| Specificity | 0.693 | (0.633–0.752) | 0.889 | (0.850–0.930) |
| PPV | 0.152 | (0.106–0.198) | 0.273 | (0.215–0.330) |
| NPV | 0.993 | (0.983–0.999) | 0.979 | (0.962–0.998) |
| External validation cohort | ||||
| Sensitivity | 0.944 | (0.955–0.981) | 0.645 | (0.610–0.681) |
| Specificity | 0.684 | (0.662–0.730) | 0.926 | (0.906–0.945) |
| PPV | 0.130 | (0.103–0.153) | 0.286 | (0.252–0.319) |
| NPV | 0.996 | (0.994–0.999) | 0.983 | (0.973–0.992) |
Data are mean (95% CI). NPV, negative predictive value; PPV, positive predictive value.
The discovery cohort is the original discovery cohort before SMOTE oversampling.
Discussion
In this multicenter study, we developed and validated a multi-omics model called ALARM, which integrated triple-phase CT deep learning radiomics with common clinical variables, enabling enhance the accuracy and confidence in early warning HCC development for patients with cirrhosis 3–12 months before the HCC clinical diagnosis. The ALARM model exhibited a notable AUC, ranging from 0.902 to 0.929, and the calibration and decision curve analyses results further reinforced the model's robustness and clinical applicability. To our knowledge, this is the first model that can accurately predict the risk of carcinogenesis in patients with cirrhosis 3–12 months prior to the traditional clinical diagnosis of HCC.
Artificial intelligence has revolutionized medical practice through faster, precise data analysis, transforming diagnostics and treatment. This advancement has also prompted a reevaluation of conventional screening methods. Traditionally, HCC surveillance has been recommended to be conducted semiannually, approximately every 6 months, utilizing ultrasound and AFP tests, as per guidelines.20 Previous studies have indicated that ultrasound exhibits a sensitivity of 84% for detecting HCC in patients with cirrhosis, but this sensitivity drops to 47% for early-stage detection. Additionally, combining ultrasound with AFP testing enhances its limited sensitivity (88%), especially for early-stage HCC.21 Currently, HCC risk assessment is shifting towards personalized approaches, focusing on individual-level risk using patient-specific characteristics. Several risk scores have been created for patients, using clinical and lab data to stratify risk.12,22, 23, 24, 25 The aMAP score, derived from five commonly used clinical parameters, reliably predicts HCC development across diverse populations, despite originating from tertiary hospital patients.12 Its widespread validation underscores its effectiveness.26,27 Hence, in this study, we harnessed the synergy between the strengths of deep learning and radiomics analysis with the aMAP score information to construct a model that outperformed traditional methods without significantly increasing the economic burden.
We selected a 3–12 months' timeframe for short-term prediction of HCC in cirrhotic patients is based on the need for early detection and timely intervention. This period is long enough to capture early HCC signals yet short enough to ensure model accuracy, thereby bolstering physician confidence in implementing interventions. We found that ALARM could identify high-risk and medium-risk groups, accounting for around 35.2% (654/1858) of the whole cirrhotic population but covered as high as 94.4% (84/89) of the future HCC cases. Based on the model's prediction of HCC incidence in different strata, we may be able to make the following recommendations to prevent HCC progression. For high-risk individuals, immediate magnetic resonance imaging (MRI) examination or biopsy may be recommended to determine the nature of the lesion. Those at medium-risk are advised to undergo repeat short-interval CT scans approximately every 3–6 months, while individuals at low-risk are advised to have screenings spaced at intervals of 6 months. We acknowledge that these proposed strategies may not be exhaustive or optimal. The proposal of the optimal management strategy would need additional clinical investigation and cost-effective analysis.
Within HCC surveillance, LIRADS-3 lesions (20–40% chance of HCC) are frequently identified on initial CT.20 However, the delineation of these lesions is often impeded by economic constraints and nonspecific clinical signs, leading to diagnostic ambiguity that can result in deferred clinical intervention. In our study, among the 89 patients with HCC, we encountered 3 cases where the CT scans exhibited features indicative of LIRADS-3 (without encountering LIRADS-4 or LIRADS-5). Of these 3 cases, 2 of them were classified as high-risk, and the remaining one was categorized as medium-risk by the model. The predictive insight of models may increase the confidence of clinicians to further confirm the diagnosis by means of advanced imaging modalities such as MRI or to perform biopsies. Our Nomograph (Figure S17) and dynamic web pages (https://dlrasn.shinyapps.io/dynnomapp/) could facilitate the more effective application of ALARM model in clinical practice.
Despite the significant findings, the present study has certain limitations. Firstly, the included population mainly consisted of patients with HBV-related cirrhosis, which is more prevalent among Asian populations, potentially limiting the generalizability of our model to other etiologies. Histological variations can significantly influence the appearance of lesions on CT scans. For instance, non-alcoholic steatohepatitis (NASH)-related cirrhosis commonly exhibits widespread hepatic steatosis,28 whereas HBV-induced cirrhosis may result in irregularities in liver structure. These histopathological differences not only affect the visual characteristics of lesions but also impact the overall texture and architecture of the liver as observed through imaging modalities like CT scans. These factors can impact lesion detection accuracy and subsequently affect the performance of prediction models. Our future research strategy will involve expanding the sample range to include a more extensive representation of patients with cirrhosis caused by different etiologies and from different geographical areas. Secondly, one drawback of the aMAP score is that patients were recruited from tertiary hospitals, where they were particularly prone to having active disease before treatment. In future research, we will further explore the model's performance in primary care settings and integrate other risk scores to provide a more comprehensive risk assessment. Thirdly, we avoided end-to-end techniques,29 focusing on integrating clinical scores to enhance the model's interpretability. This option helps medical professionals better understand the predictions, but it can lead us to fail to take full advantage of complex patterns in the data, which can reduce the model's ability to capture potential predictive signals, which in turn affects the model's accuracy. Future research is needed to further explore how to improve the accuracy of the model while maintaining its usefulness and simplicity in clinical practice.
In summary, the novel ALARM model, based on deep learning radiomics with clinical variables, provides reliable estimates of short-term HCC development for cirrhotic patients, and may have the potential for identifying the early changes transitioning from cirrhosis to HCC. Integrating this tool into clinical decision-making processes is expected to yield substantial progress in the precision clinical decision-making for HCC and individualized preemptive treatments.
Contributors
FR and HJL contributed to the conception and design of the study. HJL, WHY, JS, FR and CL coordinated the study. QYS, WCY, LXL, FXT, JGQ, ZD, GPJ, BHL, WCX, YYL, DWC, HX, GYH, LXE, LJF, and SJ acquired the data. FR, HJL, GLX, and HX did the statistical analysis, interpreted the data and verified the underlying data. FR and GLX wrote the drafting of the manuscript. FR, CL, TJ, WHY and HJL critically reviewed the manuscript. All authors read and approved the final version of the manuscript.
Data sharing statement
All text, tables and figures in this article are available to other researchers. If requested, deidentified data collected for the cohort study and informed consent form can be made available. Please contact the corresponding author.
Declaration of interests
Jinlin Hou has received consulting fee from AbbVie, Arbutus, Bristol Myers Squibb, Gilead Sciences, Johnson &Johnson, Roche and received grants from Bristol Myers Squibb, Berry, and Johnson & Johnson. The other authors declare no conflicts of interest that pertain to this work.
Acknowledgements
We gratefully acknowledge the support provided by the National Key Research and Development Program of China (grant numbers 2022YFC2303600 and 2022YFC2304800), the National Natural Science Foundation of China (grant number 82170610), and the Guangdong Basic and Applied Basic Research Foundation (grant number 2023A1515011211).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2024.102718.
Contributor Information
Hongyang Wang, Email: hywangk@vip.sina.com.
Jinlin Hou, Email: jlhousmu@163.com.
Rong Fan, Email: rongfansmu@163.com.
Appendix A. Supplementary data
References
- 1.Rumgay H., Shield K., Charvat H., et al. Global burden of cancer in 2020 attributable to alcohol consumption: a population-based study. Lancet Oncol. 2021;22:1071–1080. doi: 10.1016/S1470-2045(21)00279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Singal A.G., Kanwal F., Llovet J.M. Global trends in hepatocellular carcinoma epidemiology: implications for screening, prevention and therapy. Nat Rev Clin Oncol. 2023;20:864–884. doi: 10.1038/s41571-023-00825-3. [DOI] [PubMed] [Google Scholar]
- 4.Sartoris R., Gregory J., Dioguardi Burgio M., Ronot M., Vilgrain V. HCC advances in diagnosis and prognosis: digital and Imaging. Liver Int. 2021;41(Suppl 1):73–77. doi: 10.1111/liv.14865. [DOI] [PubMed] [Google Scholar]
- 5.Harding-Theobald E., Louissaint J., Maraj B., et al. Systematic review: radiomics for the diagnosis and prognosis of hepatocellular carcinoma. Aliment Pharmacol Ther. 2021;54:890–901. doi: 10.1111/apt.16563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambin P., Leijenaar R.T.H., Deist T.M., et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 7.Wei J., Jiang H., Gu D., et al. Radiomics in liver diseases: current progress and future opportunities. Liver Int. 2020;40:2050–2063. doi: 10.1111/liv.14555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayerhoefer M.E., Materka A., Langs G., et al. Introduction to radiomics. J Nucl Med. 2020;61:488–495. doi: 10.2967/jnumed.118.222893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park H.J., Park B., Lee S.S. Radiomics and deep learning: hepatic applications. Korean J Radiol. 2020;21:387–401. doi: 10.3348/kjr.2019.0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K., Lu X., Zhou H., et al. Deep learning radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: a prospective multicentre study. Gut. 2019;68:729–741. doi: 10.1136/gutjnl-2018-316204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu F., Liu D., Wang K., et al. Deep learning radiomics based on contrast-enhanced ultrasound might optimize curative treatments for very-early or early-stage hepatocellular carcinoma patients. Liver Cancer. 2020;9:397–413. doi: 10.1159/000505694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan R., Papatheodoridis G., Sun J., et al. aMAP risk score predicts hepatocellular carcinoma development in patients with chronic hepatitis. J Hepatol. 2020;73:1368–1378. doi: 10.1016/j.jhep.2020.07.025. [DOI] [PubMed] [Google Scholar]
- 13.Fan R., Chen L., Zhao S., et al. Novel, high accuracy models for hepatocellular carcinoma prediction based on longitudinal data and cell-free DNA signatures. J Hepatol. 2023;79:933–944. doi: 10.1016/j.jhep.2023.05.039. [DOI] [PubMed] [Google Scholar]
- 14.Yamashita Y., Joshita S., Sugiura A., et al. aMAP score prediction of hepatocellular carcinoma occurrence and incidence-free rate after a sustained virologic response in chronic hepatitis C. Hepatol Res. 2021;51:933–942. doi: 10.1111/hepr.13689. [DOI] [PubMed] [Google Scholar]
- 15.Isensee F., Jaeger P.F., Kohl S.A.A., Petersen J., Maier-Hein K.H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. 2021;18:203–211. doi: 10.1038/s41592-020-01008-z. [DOI] [PubMed] [Google Scholar]
- 16.van Griethuysen J.J.M., Fedorov A., Parmar C., et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He K., Zhang X., Ren S., Sun J. 2016 IEEE conference on computer vision and pattern recognition (CVPR) IEEE; Las Vegas, NV, USA: 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 18.Han H., Wang W.-Y., Mao B.-H. In: Advances in intelligent computing. Huang D.-S., Zhang X.-P., Huang G.-B., editors. Springer Berlin Heidelberg; Berlin, Heidelberg: 2005. Borderline-SMOTE: a new over-sampling method in imbalanced data sets learning; pp. 878–887. [Google Scholar]
- 19.Camp R.L., Dolled-Filhart M., Rimm D.L. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 20.Singal A.G., Llovet J.M., Yarchoan M., et al. AASLD practice guidance on prevention, diagnosis, and treatment of hepatocellular carcinoma. Hepatology. 2023;78:1922–1965. doi: 10.1097/HEP.0000000000000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tzartzeva K., Obi J., Rich N.E., et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta-analysis. Gastroenterology. 2018;154:1706–1718.e1. doi: 10.1053/j.gastro.2018.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujiwara N., Kobayashi M., Fobar A.J., et al. A blood-based prognostic liver secretome signature and long-term hepatocellular carcinoma risk in advanced liver fibrosis. Med. 2021;2:836–850.e10. doi: 10.1016/j.medj.2021.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ioannou G.N., Green P.K., Beste L.A., Mun E.J., Kerr K.F., Berry K. Development of models estimating the risk of hepatocellular carcinoma after antiviral treatment for hepatitis C. J Hepatol. 2018;69:1088–1098. doi: 10.1016/j.jhep.2018.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ioannou G.N., Green P., Kerr K.F., Berry K. Models estimating risk of hepatocellular carcinoma in patients with alcohol or NAFLD-related cirrhosis for risk stratification. J Hepatol. 2019;71:523–533. doi: 10.1016/j.jhep.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papatheodoridis G., Dalekos G., Sypsa V., et al. PAGE-B predicts the risk of developing hepatocellular carcinoma in Caucasians with chronic hepatitis B on 5-year antiviral therapy. J Hepatol. 2016;64:800–806. doi: 10.1016/j.jhep.2015.11.035. [DOI] [PubMed] [Google Scholar]
- 26.El-Serag H., Kanwal F., Ning J., et al. Serum biomarker signature is predictive of the risk of hepatocellular cancer in patients with cirrhosis. Gut. 2024 doi: 10.1136/gutjnl-2024-332034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson P.J., Innes H., Hughes D.M., Kalyuzhnyy A., Kumada T., Toyoda H. Evaluation of the aMAP score for hepatocellular carcinoma surveillance: a realistic opportunity to risk stratify. Br J Cancer. 2022;127:1263–1269. doi: 10.1038/s41416-022-01851-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman S.L., Neuschwander-Tetri B.A., Rinella M., Sanyal A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24:908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L., Wu M., Li R., Xu X., Zhu C., Feng X. MVI-mind: a novel deep-learning strategy using computed tomography (CT)-Based radiomics for end-to-end high efficiency prediction of microvascular invasion in hepatocellular carcinoma. Cancers. 2022;14:2956. doi: 10.3390/cancers14122956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.