Abstract
The internal structural protein of retroviruses, Gag, comprises most of the mass of the virion, and Gag itself can give rise to virus-like particles when expressed in appropriate cells. Previously the stoichiometry of Gag in virions was inferred from indirect measurements carried out 2 decades ago. We now have directly determined the masses of individual particles of the prototypic avian retrovirus, Rous sarcoma virus (RSV), by using scanning transmission electron microscopy. In this technique, the number of scattered electrons in the dark-field image integrated over an individual freeze-dried virus particle on a grid is directly proportional to its mass. The RSV virions had a mean mass of 2.5 × 108 Da, corresponding to about 1,500 Gag molecules per virion. The population of virions was not homogeneous, with about one-third to two-thirds of the virions deviating from the mean by more than 10% of the mass in two respective preparations. The mean masses for virions carrying genomes of 7.4 or 9.3 kb were indistinguishable, suggesting that mass variability is not due to differences in RNA incorporation.
The assembly of retroviruses appears deceptively simple. A single protein, the product of the gag gene, organizes the internal structure of the virion. Expression of the Gag protein, even in the absence of other viral proteins such as Pol and Env, leads to budding of virus-like particles from the plasma membrane of the cell. In a process called maturation, the viral protease proteolytically processes Gag late in the budding process to yield the several internal structural proteins that have been known for many years to comprise the bulk of the protein mass of the virion (7, 8, 12, 35, 38). The remaining protein mass is accounted for by products of the env gene and the pol gene, as well as by minor amounts of host cell proteins. Several particular host proteins have been documented in retroviruses (2, 13, 19, 20, 27, 36), but each comprises no more than a few percent of the total protein mass. The nature of the protein-protein interactions, protein-lipid interactions, and protein-RNA interactions that occur during assembly remains incompletely understood, and the underlying principles that determine the stoichiometry of Gag in the virion are unknown. In human immunodeficiency virus type 1 (HIV-1) (14) and in murine leukemia virus (MLV) (40), and thus probably in all retroviruses, mature and immature virions are somewhat heterogeneous in size as visualized by cryo-electron microscopy (cryo-EM). Neither mature nor immature virions show evidence of icosahedral symmetry, in contrast to common pictoral representations of retroviruses. Small deletions in Rous sarcoma virus (RSV) capsid protein (CA) (18) or HIV p6 (15, 16) can lead to virus-like particles that are grossly heterogeneous in size but that nevertheless appear to bud normally from the plasma membrane.
An understanding of retrovirus structure requires knowledge of the number of Gag molecules in a virion. The most straightforward method to estimate stoichiometry is based on the mass of the particle and the percentage of the mass that is due to Gag protein. The dry mass of retrovirus particles has often been said to be composed of about two-thirds protein, one-third lipid, and a few percent RNA. The viral lipid, compared with total cellular lipids, is enriched for cholesterol and sphingomyelin (1, 28). Perhaps the first attempt to quantify retroviral mass was reported 40 years ago for avian myeloblastosis virus (AMV) (4), a favorite system in early studies because of the vast numbers of particles shed into the blood of infected chicks. The ratio of particles, as determined by EM, to the combined masses of protein, lipid, and RNA led to the retroviral mass estimate of about 4.5 × 108 Da per particle. Perhaps the most careful estimate for the number of Gag molecules in an avian retrovirus particle was derived some 25 years ago (35) and was based on the measured value of 0.038 for the ratio of total RNA to protein in virions, on the measurement of the percentage of protein comprising each Gag protein, and on the assumption that each virion carries one genomic RNA molecule of 1.0 × 107 Da plus 0.4 × 107 Da of tRNA. The authors concluded that an AMV virion contains 4,100 molecules of CA (then called p27), 3,600 molecules of matrix protein (MA) (p19), 5,800 molecules of nucleocapsid protein (NC) (p12), and 5,500 molecules of protease (PR) (p15). A somewhat earlier estimate for the very closely related avian retrovirus MC29 was 3,000 molecules of CA (then called gs1), 1,800 molecules of MA (gs2), 2,000 molecules of NC (gs4), and 3,000 molecules of PR (gs3) (12). It is important to note that both of these studies predated the discoveries that retroviruses have diploid genomes and that the internal structural proteins are derived by proteolytic processing of a single protein, later called Gag. From what is now known, the ratio of RNA to protein in virions used in the study published in reference 35 was too high and the amount of total RNA was too low.
We decided to reinvestigate the mass of RSV virions by using the scanning transmission electron microscope (STEM) at Brookhaven National Laboratory. The STEM is capable of examining individual unstained, unshadowed freeze-dried particles and determining their masses. Virus particles in solution are adsorbed to a thin carbon support film on an EM grid. They are then extensively washed, quick-frozen, freeze-dried overnight, and transferred to the microscope for viewing. Tobacco mosaic virus (TMV) is used as an internal control and calibrator on each grid. The STEM operates in a dark-field mode, and a 40-kV electron probe focussed at 0.25 nm scans an image of 512 by 512 pixels. At each point, the number of scattered electrons, collected by two annular detectors, is recorded digitally. The number of scattered electrons is directly proportional to the mass thickness at that point. The mass of a particle is determined by summing the number of scattered electrons over an entire particle after subtracting the support film contribution. The mass determined for large particles such as viruses should be accurate to within a few percent. STEM analysis, which has been reviewed recently (39), has been used frequently to characterize the masses of nonenveloped viruses but until now has been applied to only one enveloped virus, vesicular stomatitis virus (VSV) (37). The data obtained in that classic study led to the accepted stoichiometry of virion proteins in VSV.
Preparation and biochemical analysis of virus stocks.
Two molecular clones that differ in size by 1.9 kb were used to derive the stocks of virus used in this study. The plasmid pRCASBP is a DNA clone of the Schmidt-Ruppin A strain of RSV with the original pol gene replaced by that of the closely related Bryan strain of RSV and with the original src gene deleted and replaced by a short synthetic linker with a ClaI restriction site (25, 26). The plasmid pRCASBP-AP is identical except for the insertion of the human placental alkaline phosphatase gene in the ClaI site (10a, 11). Virus infections were initiated by transfection of turkey embryo fibroblasts, and after the infection had spread to all of the cells, medium containing virus was collected at 24-h intervals. Parallel plates of turkey embryo fibroblasts were continuously labeled with [14C]glycine (6 μCi/ml, 60 μCi/μmol), [14C]leucine (2.5 μCi/ml, 30 μCi/μmol), or [3H]mannose (100 μCi/ml) or were pulse-labeled for 30 min with [35S]methionine (500 μCi/ml) and then grown in normal, unlabeled medium for 20 h. The media for amino acid labeling were prepared without the respective radioactive amino acids. For each set of plates, the collected medium was centrifuged at 5,000 × g to remove debris and filtered through a 0.45-μm-pore-size filter, and then the unlabeled or labeled virus was collected by centrifugation for 2.5 h at 25,000 rpm (SW28 rotor) through a 5-ml cushion of 15% sucrose in virus suspension buffer (VSB; 10 mM Tris HCl [pH 7.5], 50 mM NaCl, 1 mM EDTA). The supernatant was removed by aspiration, the pellet was resuspended in VSB plus 5% sucrose, and then the virus was purified further by isopycnic centrifugation for 2 h at 50,000 rpm (TLS55 swinging-bucket rotor) in a 2-ml gradient of 15 to 50% (wt/wt) sucrose in VSB. The visible band of virus was collected and diluted fivefold in VSB, and the virus particles were collected again by centrifugation. The yield of RCASBP was about twice that of RCASBP-AP at all stages of purification, for the unlabeled as well as all of the labeled preparations. Parallel samples of labeled and unlabeled virus were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie staining, fluorography, and PhosphorImager analysis.
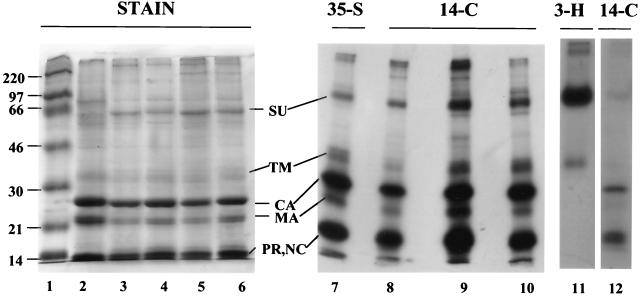
The purified virus showed a pattern of stained polypeptides like that commonly reported for the avian sarcoma and leukosis viruses (24, 34, 38) (Fig. 1). The profiles for RCASBP (lanes 4 and 6) and RCASBP-AP (lanes 3 and 5) were identical, as expected, with virus samples that had been purified by pelleting from the medium (lanes 5 and 6) being only slightly less pure than samples that in addition had been centrifuged to equilibrium in sucrose gradients (lanes 3 and 4). The display of polypeptides was nearly identical to that for AMV used as a standard (lane 2). The Gag proteins CA, MA, NC, PR, and p10 accounted for the bulk of the stained proteins, with NC, PR, and p10 comigrating on the minigels used in these experiments. Proteins labeled with radioactive amino acids and visualized by fluorography yielded the same pattern of bands, as shown in Fig. 1 for [35S]methionine (lane 7), [14C]glycine (lanes 8 and 9), and [14C]leucine (lane 10). The next most abundant proteins after Gag were the Env surface and transmembrane proteins (SU and TM, respectively) (8, 30), as determined by their labeling with [3H]mannose (lane 11). SU migrated faster than implied by the standard nomenclature (gp85) for avian SU proteins, as was reported already many years ago for this strain of RSV (8), presumably due to the presence of fewer glycosylation sites than in the SU proteins of other strains. Barely evident bands corresponding to the α (65-kDa) and β (93-kDa) polypeptides of reverse transcriptase bracketed the SU band, and a band corresponding to integrase (IN) (32 kDa) was visible in the same region as TM. These pol gene products are expected to be present at a stoichiometry of about 1:20 compared with Gag (17). A minor band corresponding to a molecular mass of 40 kDa is likely to be cellular actin, which is known to be incorporated into some retroviruses in small amounts (20) and to interact with Gag (29).
FIG. 1.
Gel analysis of viral proteins. Samples of purified unlabeled and radioactively labeled RCASBP and RCASBP-AP virus were analyzed by SDS-PAGE on three separate minigels. The gel on the left (15% polyacrylamide) (lanes 1 to 6) was stained with Coomassie blue (approximately 20 μg of protein per lane). The gel in the middle (17.5% polyacrylamide) (lanes 7 to 10) and the gel on the right (17.5% polyacrylamide) (lanes 11 and 12) were submitted to fluorography at −70°C after being impregnated with 1 M sodium salicylate and dried (4,000 to 13,000 cpm per lane for 35S and 14C; 50,000 cpm per lane for 3H). The virus preparations shown in lanes 5, 6, and 11 were obtained by pelleting virus particles from the medium, while the remainder of the preparations were further purified by isopycnic banding of the pelleted virus particles. The sample in lane 12 was used as a parallel standard on the same gel as lane 11 to mark the positions of the nonglycosylated proteins. All gels were purposely overloaded to better visualize minor proteins other than Gag. Lane 1, Rainbow marker proteins (Amersham); lane 2, AMV from leukemic chick plasma; lane 3, RCASBP-AP; lane 4, RCASBP; lane 5, RCASBP-AP; lane 6, RCASBP; lane 7, [35S]methionine-labeled RCASBP; lane 8, [14C]glycine-labeled RCASBP-AP; lane 9, [14C]glycine-labeled RCASBP; lane 10, [14C]leucine-labeled RCASBP; lane 11, [3H]mannose-labeled RCASBP; lane 12, [14C]glycine-labeled RCASBP.
Quantitation of the 14C-labeled proteins by PhosphorImager analysis showed that the sum of all Gag proteins represented 78% of the total protein in the gel lane, for both [14C]glycine- and [14C]leucine-labeled RCASBP. For this purpose, Gag proteins were operationally defined as the sum of all radioactive material that migrated with a mobility equal to or faster than that of CA, including material at the buffer front. This value thus should include not only CA, MA, NC, PR, and p10 but also the peptides p2a, p2b (22, 24), and SP (23), which also are cleaved from Gag. This definition of Gag leaves out the variable amount of incompletely processed MA molecules with the attached downstream p2 and p10 domains, which migrate above the CA band. On the other hand, this definition of Gag proteins is an overestimate to the degree that low-molecular weight cellular proteins—for example, ubiquitin (27)—are included. The Env protein SU represented 4.4 and 3.8% of total protein for glycine-labeled and leucine-labeled RCASBP, respectively. SU was operationally defined as the total amino acid-labeled material that migrated with the [3H]mannose marker band run on parallel lanes. The Env protein TM could not be quantitated reliably, since IN and the partially processed MA form migrate in the same part of the gel. A smear of cellular polypeptides extended throughout the lane. We assume that much of this material is incorporated into virions, since preliminary experiments with subtilisin treatment of the virus preparation (20) did not result in a significant reduction of the smear.
Analysis by STEM.
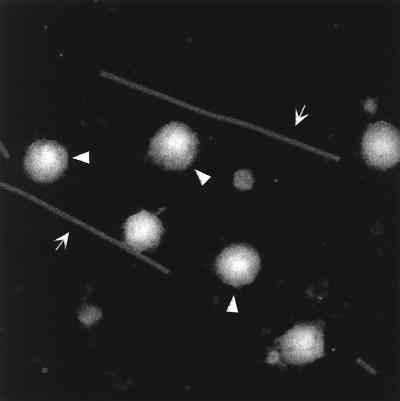
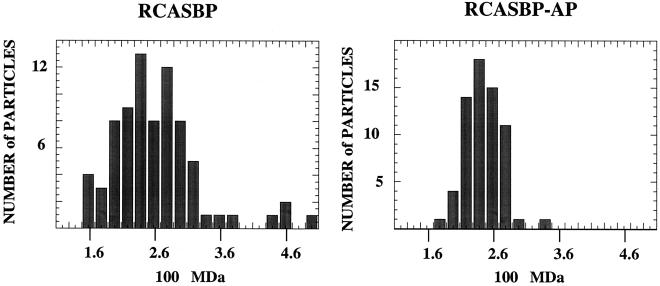
Portions of the sucrose gradient-banded RCASBP and RCASBP-AP virus preps in VSB were used to prepare grids for STEM. A 3-μl aliquot was injected into a drop of buffer and allowed to adsorb for 1 min to the 2- to 3-nm-thick carbon support film (to which TMV had previously been adsorbed). The grids were washed extensively, blotted to a thin layer of liquid, quick frozen, freeze-dried overnight, and then transferred under vacuum to the STEM for viewing. An image of 512 by 512 pixels was acquired by recording the number of scattered electrons at each point. A representative dark-field electron micrograph is shown in Fig. 2. Six round RSV virions can be seen along with two long, thin TMV particles. Some smaller contaminants are visible in the background. The mass of each particle was determined by summing the number of scattered electrons over an entire particle, subtracting the background from the support film (determined in clear areas between particles), and using TMV on the same grid as a microscope calibrator (39). The results yielded a wide distribution of particle masses, with means of 2.62 × 108 Da (standard deviation = 0.65; n = 77) for RCASBP and 2.44 × 108 Da (standard deviation = 0.34; n = 66) for RCASBP-AP (Fig. 3). We consider these two mean masses to be statistically indistinguishable, although for unknown reasons the distribution of masses was wider for the virus carrying the smaller genome. Since the accuracy and precision of the STEM measurements are expected to be better than 5%, the RSV virions analyzed here are distinctly heterogeneous. From recent cryo-EM studies it is known that HIV-1 (14) and MLV (40) also are heterogeneous in size. The mean RSV mass is very similar to that measured by STEM for VSV, 2.65 × 108 Da, in the classic study by Thomas et al. (37).
FIG. 2.
Visualization of virus particles by STEM. This dark-field electron micrograph shows a field with six RCASBP-AP particles. The three indicated by arrowheads were used for mass measurements as described in the text. (The other three could not be used because one is not fully visible, one is too close to a TMV, and one is too close to a contaminating mass.) The arrows point to TMV particles (used as an internal control). Some smaller contaminants can be seen in the background; it is possible that these are similar to microvesicles described by Ott et al. (20). The full scale of the micrograph is 1.024 μm.
FIG. 3.
Distribution of virus particle mass. The masses of RCASBP and RCASBP-AP virions were determined as described in the text. The number of particles in each window of 0.2 × 108 Da is shown on the ordinate, and the measured masses of particles are on the abcissa.
Given the assumption that all virions contain identical ratios of their components—i.e., Gag, Pol, Env, host proteins, RNA, and lipid—it is possible to calculate the stoichiometry of Gag protein. These calculations are based on several measurements carried out previously or on the following assumptions. First, we assume that 95% of radioactive proteins (Fig. 1) and also of lipids are in bona fide virions, with the remaining 5% representing contaminating microvesicles. This assumption is obviously somewhat arbitrary and is based in part on the STEM pictures themselves, which show a scattering of smaller particulate matter that might be microvesicles. Second, we take the lipid content for RSV to be 31%, which is the average of several determinations (range ± 4%) of the percentage of dry mass reported by Quigley et al. (28). In this and other, similar studies, lipid was defined as the percentage of dry mass of the preparation that could be extracted into chloroform-methanol. Virus purity is not a very critical factor in interpreting these data if any contaminating material has the same protein/lipid ratio, as might be expected from the purification by equilibrium sedimentation. It is known that even extensively purified retroviruses can be contaminated with microvesicles that are associated with cellular proteins (20). Third, we take the RNA content to be 2.3% for RCASBP and 3% for RCASBP-AP. These values are based on the assumption that each particle of RCASBP or RCASBP-AP contains two copies of the 7.4- or 9.3-kB genomic RNA, respectively, plus tRNA and other small RNAs comprising an additional 30% of the genomic RNA by mass (9, 10, 31, 32), equaling about 3.5 kb of tRNA. Fourth, using the average PhosphorImager values for SU, i.e., 4% of total radioactive protein, and assuming an equimolar amount of TM, we estimate that the Env polypeptides account for 6% of the radioactive virion protein. Given the more than a dozen Env N-linked glycosylation sites, carbohydrate then should comprise about 4% of the virion protein mass, or about 3% of the total mass.
Finally, we take the sum of Gag proteins to represent 73% of the total protein in bona fide virions, slightly less than the total amount of label migrating with CA or more rapidly. We chose to use a method of calculating this value that is based on PhosphorImager quantitation of the CA band alone, since this is the most cleanly resolved Gag protein and since choosing a single, narrowly defined band obviates the need to make any assumptions about cellular proteins that may migrate in the lower part of the gel. [14C]glycine-labeled CA and [14C]leucine-labeled CA comprised 16.6 and 26.7% of the total radioactivity in the lane of the gel, respectively, i.e., 17.5 and 28.1% of the label in bona fide virions according to the assumption about virus purity. Given that CA has 17 of the 74 Gag glycine residues and 24 of the 60 Gag leucine residues, and assuming that Gag proteins are equimolar, all of the Gag proteins together thus must represent 76% of the glycine-labeled virus and 70% of the leucine-labeled virus, giving an average value of 73%. This calculated percentage of protein that is Gag is consistent with previous analyses of avian retrovirus polypeptides by SDS-PAGE and by gel filtration in guanidine hydrochloride, although in some of these earlier experiments the amount of Gag was as low as 65 to 70% (12, 35, 38). On the other hand, some published SDS-PAGE profiles of [35S]methionine-labeled RSV as well as of other retroviruses appear to indicate that Gag constitutes more than 80% of the total radioactive protein. However, visual estimates of this type are misleading. Furthermore, labeling with this isotope is rarely done under steady-state conditions, and hence the results generally reflect an overestimation of abundance for proteins that are turned over rapidly, like Gag itself.
Based on these several assumptions and measurements, the mean mass of 2.5 × 108 Da for the two RSV viruses then corresponds to about 1,500 Gag molecules per virion. According to the distributions shown in Fig. 3, 40% of the virions in RCASBP and 60% of the virions in RCASBP-AP have 1,300 to 1,700 Gag molecules; the remainder of the virions fall outside of those limits.
Discussion.
The STEM data presented here provide the first direct measurement of the mass of a retrovirus, which leads in turn to an estimate for the average number of each Gag protein in a virion. The accuracy of each virion mass measurement does not significantly affect this number, since with the internal calibrations that are used routinely the STEM data are within ±5%. Rather, the major factors affecting accuracy are the uncertainties in purity, in lipid content, and in content of proteins other than Gag. The purity estimate of 95% is a nominal value. Unlike HIV (20), RSV has never been submitted to a direct analysis for microvesicle contamination. However, clearly the STEM pictures show evidence of particulate material smaller than virus particles. Given the agreement in the lipid data for RSV (28) and the closely related AMV (3, 4), the correct lipid content is likely to be within 5% of the given value (i.e., between 26 and 36% of the total mass). As an example, if RSV contained 36% instead of 31% lipid, the calculated average number of each of the Gag proteins would drop from 1,500 to 1,400. The percentage of protein that is Gag also is probably within 5% of being correct, given the abundance of values in the literature that cluster around this number. If Gag proteins accounted for 78% of virion protein mass instead of 73%, the calculated average number of each Gag protein would rise to about 1,600.
In interpreting the observed heterogeneity in RSV particle mass, we make the simplifying assumption that in particles differing in mass, the ratios of Gag, lipid, and other proteins are identical. This assumption is difficult to test, and while it is likely to hold for Gag/lipid ratios, it may not hold for Gag/Env ratios. Since retroviruses do not need Env protein to bud, virions budding from one part of the plasma membrane plausibly could pick up more Env protein than those budding from another. Also, SU could be lost differentially from some particles by shedding. However, the observed mass heterogeneity is much too large to be explained by variations in Env levels. If the SU that comprised about 4% of the total radioactive protein in the lane on the gel was unevenly distributed, so that the number was 2% for some virions and 6% for others, the virion mass would differ by only about 4%, far less than the standard deviation in observed masses. Furthermore, and more importantly, virion diameters measured by cryo-EM for the same RCASBP preparation used in our analyses showed a standard deviation of ±10% (17a), consistent with the size heterogeneity described for MLV (40). Assuming that the amount of Gag protein and the amount of lipid in a virion increase as the square of the virion diameter (since both form two-dimensional shells), the 10% standard deviation of diameters implies a ca. 20% standard deviation in the mass of Gag plus lipid, roughly consistent with the standard deviation of 25% that we observed.
Since proteolytic cleavage occurs late in budding, it is widely believed that the mature Gag proteins are equimolar in virions, although this assumption has never been tested critically. Because of the comigration of some Gag proteins upon SDS-PAGE as well as upon subjection to the previously used method of high-resolution gel filtration in guanidine, it is difficult to exactly quantify the several Gag proteins independently. This problem is exacerbated by the propensity of the p10 protein to leach out of gels even after fixation (21) and by the nonuniform or incomplete cleavage of MA. Depending on the strain of virus and on the virus preparation, MA may carry C-terminal extensions including p2a, p2b, or p10 or may be cleaved by a host cell protease to yield a polypeptide shorter than MA (22, 24, 33). Thus, it is possible that the apparent modest underrepresentation of MA in virions (Fig. 1), which was already observed many years ago (12, 35, 38), is simply due to alternatively processed forms of this polypeptide that migrate differently on SDS-polyacrylamide gels. However, the possibility that late in budding, after some or all of the mature MA has been generated by proteolysis, some of the MA molecules are resorbed back into the cell has not been rigorously ruled out. In that case, the concept of the number of Gag proteins per virion would have to be reinterpreted.
One of the motivations of our experiments was to examine the possibility that size variation in retroviruses is due to differences in RNA incorporation. That RNA may play a structural role in retroviruses is suggested by in vitro assembly studies; formation of spherical virus-like particles in vitro, from purified HIV-1 and RSV Gag proteins expressed in Escherichia coli, is dependent on the presence of nucleic acid (4a, 5, 6, 16a). Furthermore, the mass ratio of RNA to Gag protein in such particles is constant, regardless of the molecular size of the RNA, implying that the RNA is fully covered by the NC domain of Gag (41). Thus, we expected RCASBP-AP virus to incorporate about 20% more Gag molecules than RCASBP, because of the larger genome size of the former. In fact, this expectation was not fulfilled. However, there appeared to be a distinctly greater heterogeneity of mass in the virus with the smaller genome. We have no convincing hypothesis to explain this difference. Two models could account for the similarity in the masses of the two viruses. In one, the smaller genomic mass in RCASBP is compensated by the incorporation of relatively more tRNA. In the other, there is no obligatory stoichiometric relationship in vivo between the RNA and amount of Gag protein incorporated into a virion. These possibilities could be addressed directly by a careful analysis of RNA content in retroviruses that have different genome sizes, including viruses that have no packageable genome.
Acknowledgments
We thank Steve Hughes and Mark Federspiel for the molecular clones of RSV and Richard Kingston and Michael Rossmann for communicating results on the cryo-EM of RSV particles.
This work was supported by U.S. Public Health Service grant CA20081 to V.M.V. The Brookhaven National Laboratory STEM facility is an NIH-supported resource center (NIH P41-RR01777), with additional support provided by the Department of Energy and the Office of Biological and Environmental Research.
REFERENCES
- 1.Aloia R C, Tian H, Jensen F C. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc Natl Acad Sci USA. 1993;90:5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur L O, Bess J W, Jr, Sowder II R C, Benveniste R E, Mann D L, Chermann J C, Henderson L E. Cellular proteins bound to immunodeficiency viruses: implications for pathogenesis and vaccines. Science. 1992;258:1935–1938. doi: 10.1126/science.1470916. [DOI] [PubMed] [Google Scholar]
- 3.Beard J W. Avian virus growth and their etiologic agents. Adv Cancer Res. 1963;7:1–126. doi: 10.1016/s0065-230x(08)60982-3. [DOI] [PubMed] [Google Scholar]
- 4.Bonar R A, Beard J W. Virus of avian myeloblastosis. XII. Chemical constitution. J Natl Cancer Inst. 1959;23:183–195. [PubMed] [Google Scholar]
- 4a.Campbell, S., and A. Rein. Personal communication.
- 5.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell S, Vogt V M. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duesberg P H, Robinson H L, Robinson W S, Huebner R J, Turner H C. Proteins of Rous sarcoma virus. Virology. 1968;36:73–86. doi: 10.1016/0042-6822(68)90118-9. [DOI] [PubMed] [Google Scholar]
- 8.Duesberg P H, Martin G S, Vogt P K. Glycoprotein components of avian and murine RNA tumor viruses. Virology. 1970;41:631–646. doi: 10.1016/0042-6822(70)90428-9. [DOI] [PubMed] [Google Scholar]
- 9.Erikson R L. Studies on the RNA from avian myeloblastosis virus. Virology. 1969;37:124–131. doi: 10.1016/0042-6822(69)90313-4. [DOI] [PubMed] [Google Scholar]
- 10.Faras A J, Garapin A C, Levinson W E, Bishop J M, Goodman H M. Characterization of the low-molecular-weight RNAs associated with the 70S RNA of Rous sarcoma virus. J Virol. 1973;12:334–342. doi: 10.1128/jvi.12.2.334-342.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Federspiel, M. Personal communication.
- 11.Fekete D M, Cepko C L. Replication-competent retroviral vectors encoding alkaline phosphatase reveal spatial restriction of viral gene expression/transduction in the chick embryo. Mol Cell Biol. 1993;13:2604–2613. doi: 10.1128/mcb.13.4.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleissner E. Chromatographic separation and antigenic analysis of proteins of the oncornaviruses. I. Avian leukemia-sarcoma viruses. J Virol. 1971;8:778–785. doi: 10.1128/jvi.8.5.778-785.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franke E K, Yuan H E H, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 14.Fuller S D, Wilk T, Gowen B E, Kräusslich H-G, Vogt V M. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 15.Garnier L, Ratner L, Rovinski B, Cao S-X, Wills J W. Particle size determinants in the human immunodeficiency virus type 1 Gag protein. J Virol. 1998;72:4667–4677. doi: 10.1128/jvi.72.6.4667-4677.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garnier L, Parent L J, Rovinski B, Cao S-X, Wills J W. Identification of retroviral late domains as determinants of particle size. J Virol. 1999;73:2309–2320. doi: 10.1128/jvi.73.3.2309-2320.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Gross, I., and H.-G. Kräuslich. Personal communication.
- 17.Jacks T, Varmus H E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985;230:1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- 17a.Kingston, R., and M. Rossmann. Personal communication.
- 18.Krishna N K, Campbell S, Vogt V M, Wills J W. Genetic determinants of Rous sarcoma virus particle size. J Virol. 1998;72:564–577. doi: 10.1128/jvi.72.1.564-577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ott D E, Coren L V, Johnson D G, Sowder R C, Arthur L O, Henderson L E. Analysis and localization of cyclophilin A found in the virions of human immunodeficiency virus type 1 MN strain. AIDS Res Hum Retroviruses. 1995;11:1003–1006. doi: 10.1089/aid.1995.11.1003. [DOI] [PubMed] [Google Scholar]
- 20.Ott D E, Coren L V, Kane B P, Busch L K, Johnson D G, Sowder II R C, Chertova E N, Arthur L O, Henderson L E. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J Virol. 1996;70:7734–7743. doi: 10.1128/jvi.70.11.7734-7743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pepinsky R B, Vogt V M. Purification and properties of a fifth major viral gag protein from avian sarcoma and leukemia viruses. J Virol. 1983;45:648–658. doi: 10.1128/jvi.45.2.648-658.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pepinsky R B, Mattaliano R J, Vogt V M. Structure and processing of the p2 region of avian sarcoma and leukemia virus gag precursor polyproteins. J Virol. 1986;58:50–58. doi: 10.1128/jvi.58.1.50-58.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pepinsky R B, Papayannopoulos I A, Chow E P, Krishna N K, Craven R C, Vogt V M. Differential proteolytic processing leads to multiple forms of the CA protein in avian sarcoma and leukemia viruses. J Virol. 1995;69:6430–6438. doi: 10.1128/jvi.69.10.6430-6438.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pepinsky R B, Papayannopoulos I A, Campbell S, Vogt V M. Analysis of Rous sarcoma virus Gag proteins by mass spectrometry indicates trimming by host exopeptidase. J Virol. 1996;70:3313–3318. doi: 10.1128/jvi.70.5.3313-3318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petropoulos C J, Hughes S H. Replication-competent retrovirus vectors for the transfer and expression of gene cassettes in avian cells. J Virol. 1991;65:3728–3737. doi: 10.1128/jvi.65.7.3728-3737.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petropoulos C J, Payne W, Salter D W, Hughes S H. Using avian retroviral vectors for gene transfer. J Virol. 1992;66:3391–3397. doi: 10.1128/jvi.66.6.3391-3397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Putterman D, Pepinsky R B, Vogt V M. Ubiquitin in avian sarcoma and leukemia virus particles. Virology. 1990;176:633–637. doi: 10.1016/0042-6822(90)90035-p. [DOI] [PubMed] [Google Scholar]
- 28.Quigley J P, Rifkin D B, Reich E. Phospholipid composition of Rous sarcoma virus, host cell membranes, and other enveloped RNA viruses. Virology. 1971;46:106–116. doi: 10.1016/0042-6822(71)90010-9. [DOI] [PubMed] [Google Scholar]
- 29.Rey O, Canon J, Krogstad P. HIV-1 gag protein associates with F-actin present in microfilaments. Virology. 1996;220:530–534. doi: 10.1006/viro.1996.0343. [DOI] [PubMed] [Google Scholar]
- 30.Rifkin D B, Compans R W. Identification of the spike proteins of Rous sarcoma virus. Virology. 1971;46:485–489. doi: 10.1016/0042-6822(71)90049-3. [DOI] [PubMed] [Google Scholar]
- 31.Robinson W S, Robinson H L, Duesberg P H. Tumor virus RNAs. Proc Natl Acad Sci USA. 1967;58:825–834. doi: 10.1073/pnas.58.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawyer R C, Dahlberg J E. Small RNAs of Rous sarcoma virus: characterization by two-dimensional polyacrylamide gel electrophoresis and fingerprint analysis. J Virol. 1973;12:1226–1237. doi: 10.1128/jvi.12.6.1226-1237.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shealy D J, Mosser A G, Rueckert R R. Novel p19-related protein in Rous-associated virus type 61: implications for avian gag gene order. J Virol. 1980;34:431–437. doi: 10.1128/jvi.34.2.431-437.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart L, Schatz G, Vogt V M. Properties of avian retrovirus particles defective in viral protease. J Virol. 1990;64:5076–5092. doi: 10.1128/jvi.64.10.5076-5092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stromberg K, Hurley N E, Davis N L, Rueckert R R, Fleissner E. Structural studies of avian myeloblastosis virus: comparison of polypeptides in virion and core component by dodecyl sulfate-polyacrylamide gel electrophoresis. J Virol. 1974;13:513–528. doi: 10.1128/jvi.13.2.513-528.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh C T, Sodroski J, Göttlinger H G. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 37.Thomas D, Newcomb W W, Brown J C, Wall J S, Hainfeld J F, Trus B L, Steven A C. Mass and molecular composition of vesicular stomatitis virus: a scanning transmission electron microscopy analysis. J Virol. 1985;54:598–607. doi: 10.1128/jvi.54.2.598-607.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogt V M, Eisenman R, Diggelmann H. Generation of avian myeloblastosis virus structural proteins by proteolytic cleavage of a precursor polypeptide. J Mol Biol. 1975;96:471–493. doi: 10.1016/0022-2836(75)90174-6. [DOI] [PubMed] [Google Scholar]
- 39.Wall J S, Hainfeld J F, Simon M N. Scanning transmission electron microscopy (STEM) of nuclear structures. Methods Cell Biol. 1998;53:139–166. doi: 10.1016/s0091-679x(08)60878-x. [DOI] [PubMed] [Google Scholar]
- 40.Yeager M, Wilson-Kubalek E M, Weiner S G, Brown P O, Rein A. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc Natl Acad Sci USA. 1998;95:7299–7304. doi: 10.1073/pnas.95.13.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu, F., and V. M. Vogt. Unpublished data.