Abstract
Virus replication relies on complex interactions between viral proteins. In the case of African swine fever virus (ASFV), only a few such interactions have been identified so far. In this study, we demonstrate that ASFV protein p72 interacts with p11.5 using co-immunoprecipitation and liquid chromatography-mass spectrometry (LC-MS). It was found that protein p72 interacts specifically with p11.5 at sites amino acids (aa) 1–216 of p72 and aa 1–68 of p11.5. To assess the importance of p11.5 in ASFV infection, we developed a recombinant virus (ASFVGZΔA137R) by deleting the A137R gene from the ASFVGZ genome. Compared with ASFVGZ, the infectious progeny virus titers of ASFVGZΔA137R were reduced by approximately 1.0 logs. In addition, we demonstrated that the growth defect was partially attributable to a higher genome copies-to-infectious virus titer ratios produced in ASFVGZΔA137R-infected MA104 cells than in those infected with ASFVGZ. This finding suggests that MA104 cells infected with ASFVGZΔA137R may generate larger quantities of noninfectious particles. Importantly, we found that p11.5 did not affect virus-cell binding or endocytosis. Collectively, we show for the first time the interaction between ASFV p72 and p11.5. Our results effectively provide the relevant information of the p11.5 protein. These results extend our understanding of complex interactions between viral proteins, paving the way for further studies of the potential mechanisms and pathogenesis of ASFV infection.
Keywords: African swine fever virus, p72 protein, p11.5 protein, Protein interactions
Highlights
-
•
P72 interacts with p11.5.
-
•
The absence of A137R leads to the production of abundant noninfectious viruses.
-
•
A137R is not required for ASFV attachment to and entry into MA104 cells.
1. Introduction
African swine fever (ASF), caused by the African swine fever virus (ASFV), is an acute and highly infectious disease that affects both domestic pigs and wild boars, resulting in an alarming mortality rate of nearly 100%. ASF has spread in Europe, China, South-East Asia, and more recently, in the Dominican Republic and Haiti, causing unprecedented losses to the pig farming industry (Pujols et al., 2023). Currently, there is no effective commercial vaccine available to fight the ASF pandemic at a global level. The first commercial vaccine for ASFV recently marketed in Vietnam was based on the recombinant deletion mutant lacking the I177L gene from the Georgia 2007 ASFV isolate (Borca et al., 2020). However, its long-term efficacy, safety, and cross-protective ability need to be further evaluated (Liu et al., 2023). Therefore, the global pig industry still faces profound challenges.
ASFV is the only DNA arbovirus harboring a double-stranded DNA (dsDNA) genome of approximately 180–190 kb which encodes more than 150 open reading frames (ORFs) (Dixon et al., 2013; Galindo and Alonso, 2017). Despite the emerging information about the ASFV structural components, little is known about the molecular mechanism of ASFV replication and pathogenesis. The interactions between ASFV proteins play a crucial role in its life cycle. Through the interactions between proteins, ASFV completes the complex assembly process. Thus, it is necessary to investigate the function of ASFV proteins and their interactions, as a means by which to understand the infectious mechanisms of ASFV. The p72 is the most dominant structural component of ASFV virions and it accounts for 31–33% of the total mass (Revilla et al., 2018). As the main component of capsid protein, p72 interacts with H240R, M1249L, p17 and p49 proteins to form a complicated network that stabilizes the entire capsid structure (Alejo et al., 2018; Wang et al., 2019; Andres et al., 2020). The p72 also has a molecular chaperone, pB602L, which interacts with p72 to promote the correct folding of p72 (Liu Q. et al., 2019). Further, repression of pB602L synthesis reduces the expression level of p72 and affects the replication of ASFV (Epifano et al., 2006). Protein pE120R, also called p14.5, is another capsid component that associates with the major capsid protein p72 (Martinez-Pomares et al., 1997). pE120R was found to be essential for virus dissemination and is required for virus egress. Repression of pE120R synthesis drastically inhibited virus release from the host cell, as deduced from one-step virus growth curves (Andres et al., 2001). In addition, Zhou et al. identified pH240R as a capsid protein that interacts with the p72 through pulldown assays (Zhou et al., 2022). Compared with the parental strain, the recombinant ASFV with the deletion of the H240R gene showed a decrease of approximately 2.0 logs in infectious progeny virus titers (Zhou et al., 2022).
Understanding the interactions between proteins and recognizing the pivotal role of viral proteins in viral replication are imperative for developing novel disease control strategies. Therefore, in order to further understand the interactions between p72 and other proteins, explore the pathogenesis and immune escape mechanism of ASFV, we identified ASFV proteins that might interact with p72 from ASFV-infected cells using co-immunoprecipitation (CO-IP) and liquid chromatography-mass spectrometry (LC-MS). Deleting specific genes by genetic manipulation of the virus genome is an extraordinarily powerful approach to study the function of specific genes during protein interactions. To enhance our understanding of the interplay between p11.5 and p72 proteins in virus replication, we employed genetic manipulation to delete the A137R gene from the virus genome. The absence of A137R resulted in an increase in the noninfectious to infectious virus ratio following replication. Our results will further clarify the molecular mechanism of ASFV replication, offering a new idea for the development of anti-ASFV drugs and a novel ASFV vaccine.
2. Materials and methods
2.1. Cells, virus and antibodies
African green monkey cells MA104, human embryonic kidney 293T (HEK293T) cells and porcine kidney cells PK-15 stored in our laboratory were maintained in Dulbecco's modified Eagle medium (DMEM) (Thermo Fisher Scientific, Massachusetts, USA) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Massachusetts, USA). IPAM cells (porcine macrophage-derived cell lines, 3D4/21) were kindly provided by professor Jianzhong Zhu (Yangzhou University, Jiangsu, China) and grown in RPMI 1640 medium (Hyclone Laboratories, Logan, UT, USA) containing 10% FBS with penicillin/streptomycin. All cells were maintained in a humidified incubator with 5% CO2 at 37 °C. The ASFV strain GZ2018 (GenBank accession number: MT496893.1, the open reading frame of MGF100-1R was replaced by an enhanced green fluorescent protein expression cassette, hereafter ASFVGZ) was prepared in the previous study (Liu et al., 2021). The mouse monoclonal antibody (mAb) against the major capsid protein p72 has been described previously (Yin et al., 2022). A homemade mouse anti-p11.5 mAb was raised against the recombinant p11.5 protein of ASFV (GenBank accession number MT496893.1). A rabbit anti-Flag polyclonal antibody (pAb) was purchased from proteintech (Wuhan, China). Fluorescein isothiocyanate-conjugated (FITC-conjugated) goat anti-rabbit IgG (Jackson, USA) and Alexa Fluor 594-conjugated goat anti-mouse IgG (Jackson, USA) antibodies were purchased from Jackson. All operations involving ASFV in the present study were carried out in a biosafety level-3 (BSL-3) laboratory at Yangzhou University (Yangzhou, China).
2.2. Co-immunoprecipitation
For infection experiments, cell monolayers were inoculated with ASFVGZ stock dilutions at 0.1 multiplicity of infections (MOI). Subsequently, cells were collected at various time points post-infection for DNA extraction and growth curve analysis. Employing the identical infection protocol, cells were lysed 48 h post-infection using NP-40 Lysis Buffer (GenStar, Beijing, China) with protease inhibitor (TransGen Biotech, Beijing, China) for CO-IP. After centrifugation, the supernatants were pre-cleared with 20 μL protein G agarose beads (Beyotime, Shanghai, China) coupled with normal mouse IgG for over 2 h. The cell lysates were then centrifuged and incubated overnight with the indicated antibodies or control IgG at 4 °C. Subsequently, samples were mixed with protein G agarose beads for over 3 h. The beads were washed 4 times with 1 mL phosphate-buffered saline (PBS) for 10 min, and boiled in a 5 × Protein SDS PAGE Loading Buffer (Novoprotein, Shanghai, China) for silver staining or Western blot analyses using One step PAGE Gel Fast Preparation Kit (MeilunBio, Dalian, China).
2.3. Protein identification by LC-MS
After silver staining, the differential bands were cut and sent to Shanghai Applied Protein Technology Co. Ltd. (Shanghai, China) and the assays were conducted on a Q Exactive mass spectrometer coupled to Easy nLC 1000 (Thermo Fisher Scientific, Waltham, MA, USA) using a routine method (Huang et al., 2018). Mass spectrometry raw files were processed using Proteome Discoverer 1.4. A peptide search was performed against an UniProt African swine fever virus database. For protein identification, the following options were used: peptide mass tolerance = 20 ppm, enzyme = trypsin, max missed cleavage = 2, fixed modifications = carbamidomethyl (C), dynamical modifications = oxidation (M), and FDR <0.01 at both peptide and protein levels.
2.4. Construction of plasmids and transfection
The B646L gene and the truncated B646L gene fragments were amplified from the genomic DNA of ASFVGZ and cloned into the EcoR I/Xho I sites of the pCAGGS vector, creating pCAGGS-p72, pCAGGS-p72-1-216-Flag, pCAGGS-p72-1-324-Flag, pCAGGS-p72-1-432-Flag, and pCAGGS-p72-1-540-Flag, respectively. The A137R gene and the truncated A137R gene fragments were cloned into the Not I/Xho I sites of the pCAGGS vector, creating pCAGGS-p11.5, pCAGGS-p11.5-Flag, pCAGGS-p11.5-1-100-Flag, and pCAGGS-p11.5-68-138-Flag respectively. The primers for amplification of plasmids are listed in Supplementary Table S1. To transfect the related plasmids, HEK293T cells or MA104 cells were inoculated onto the designated plate at the appropriate density according to the experimental scheme and grew to 80% confluence. The constructed plasmids were transfected into the cells using lipofectamine 3000 transfection reagent (Invitrogen, Carlsbad, CA, USA).
2.5. Laser confocal microscopy
For the colocalization assay of p72 and p11.5, MA104 cells were co-transfected with pCAGGS-p72 and pCAGGS-p11.5-Flag for 24 h. The cells were fixed with 4% paraformaldehyde at room temperature for 30 min, and permeabilized with 0.5% Triton X-100 for 20 min. After washing with PBS, the cells were incubated with the relevant antibodies and examined using laser-scanning confocal microscope (LSCM, Leica SP8, Solms, Germany).
2.6. Generation and identification of ASFVGZΔA137R
ASFVGZΔA137R mutant was generated by homologous recombination between parental strain ASFVGZ and the recombination vector by transfection and infection procedures (Zsak et al., 1996; Kleiboeker et al., 1998). The recombination transfer vectors contain a marker gene mCherry, the upstream and downstream homologous arms flanking the A137R gene. Briefly, A137R gene, a left arm with a size of 1100 bp from 53460 nucleotides (nt) to 54559 nt, and a right arm with a size of 1122 bp from 54948 nt to 56070 nt were amplified using L + A137R + R primers and then connected to the PGEM-T Easy vector, which was named PGEM-L-A137R–R. PGEM-L-A137R–R vector was linearized using PGEM + L + R primers and linked to an expression box containing mCherry gene controlled by the late promoter of ASFV p72. The recombinant transfer vector PGEMΔA137R was obtained using PGEM + L + R and p72+mCherry primers by OK Clon DNA Ligation Kit [ACCURATE BIOTECHNOLOGY (HUNAN) CO., LTD, ChangSha, China]. The primers used in the study are shown in Supplementary Table S1.
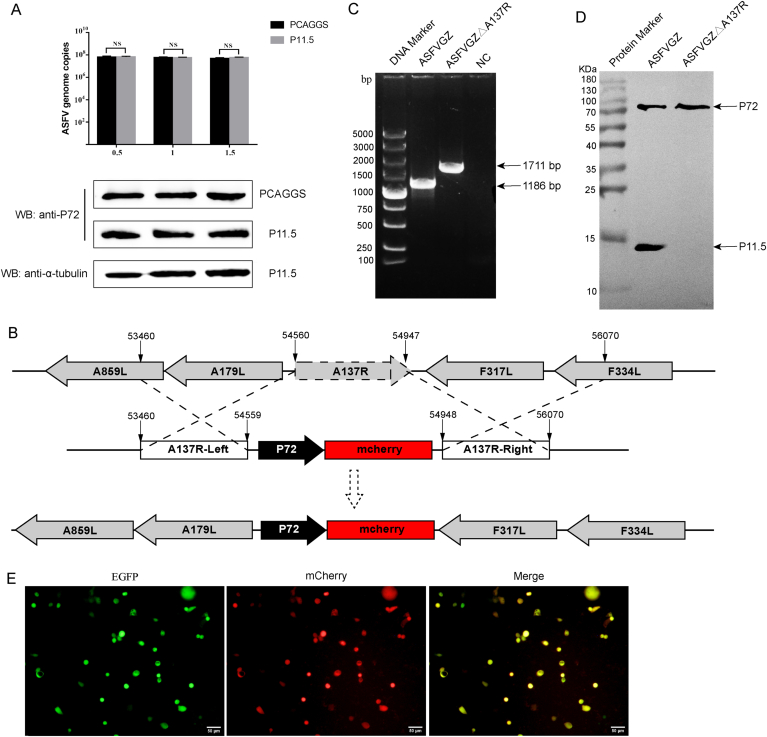
The ASFVGZΔA137R mutant was generated by homologous recombination as depicted in Fig. 3B. MA104 cells were transfected with 2.5 μg of PGEMΔA137R using lipofectamine 3000 transfection reagent (Invitrogen, Carlsbad, CA, USA) and then infected with ASFVGZ at a dose of 0.1 MOI at 12 h post-transfection. After 24 h post-infection, cells expressing both enhanced green fluorescent protein (EGFP) and mCherry were selected from the MA104 monolayers under the fluorescence microscope and rinsed to blind passages in MA104 cells. After 8−10 rounds of screened and further limiting dilution purification processes, the A137R gene-deleted virus was purified to homogeneity. Eventually, we generated A137R gene-deleted ASFV, designated as ASFVGZΔA137R. To ensure the absence of the desired deletion in each recombinant genome, viral DNA was extracted from ASFVGZΔA137R-infected MA104 cells and identified by PCR using specific primers ΔA137R (Supplementary Table S1) and sequencing analysis.
Fig. 3.
Construction of an ASFVGZΔA137R deletion mutant. A MA104 cells were transfected with pCAGGS-p11.5 for 24 h and inoculated with ASFVGZ at an MOI of 0.1. The gene and protein expression levels of p72 were quantified by qPCR and Western blot. B Schematic diagram for the construction of ASFVGZΔA137R. The open reading frame of A137R was replaced by an mCherry expression cassette as shown. C PCR analysis of ASFVGZ and ASFVGZΔA137R. NC, negative control. D Detection of p11.5 expression by Western blot. E MA104 cells were infected with the recombinant ASFVGZΔA137R deletion mutant showing EGFP and mCherry expression.
2.7. Determination of ASFVGZΔA137R replication kinetics
Comparative growth curves between ASFVGZΔA137R and parental ASFVGZ were performed in MA104 cells cultures in 24-well plates and were infected at an MOI of 0.1. The initial inoculum was removed after adsorption for 2 h at 37 °C under 5% CO2. The monolayers were then incubated with medium for 1, 2, 3, 4, 5 and 6 d. At different time points, the cells were frozen at −80 °C, and the thawed lysates were used to determine titers by fifty percent tissue culture infective dose (TCID50) in MA104 cells. The sample was diluted 10 times to 10−1–10−10 and inserted into a 96-well plate containing MA104 cells. Eight repeat wells were set for each dilution. Fluorescence expression was observed under fluorescence microscope for 5 d.
2.8. qPCR assay
ASFV genomic DNA was extracted by the FastPure Blood/Cell/Tissue/Bacteria DNA Isolation Mini Kit (Vazyme Biotech Co., Ltd., Nanjing, China) according to the manufacturer's protocols. ASFV genomic DNA copies were quantified by qPCR on the LightCycler (Roche Diagnostics, Mannheim, Germany) based on a previously described method (King et al., 2003).
2.9. Determination of infectious progeny viruses
The extracellular ASFV genome copies and titers were measured in 24-well plates (NEST Biotechnology). MA104 cells in 24-well plates were infected with 108 or 109 genome copies of the viruses. After 2 h of virus adsorption, the inoculum was removed, and MA104 cells were rinsed twice with PBS. The cells were then incubated with medium for 24 h at 37 °C with 5% CO2. The extracellular ASFV genome copies and titers were measured as described above. All samples were examined in triplicate.
2.10. Virus adsorption and entry assays
To evaluate virus adsorption efficiency, MA104 cells in 24-well cell culture plates were incubated with equal numbers of genome copies (107, 108, and 109) or equal titers (104 and 105) of virions for 2 h at 4 °C to allow virus binding but prevent virus internalization. The virus solution was then discarded, and the cells were washed three times with pre-cooled PBS. ASFV genome copies in the cells were then detected by qPCR.
In order to assess virus internalization, MA104 cells were seeded and exposed to equal amounts of genome copies (107, 108, and 109) or equal titers (104 and 105) of virions for 2 h at 37 °C. After three rounds of washing to eliminate any virus bound to the cell surface, the cells were treated with 0.05% trypsin for 2 min. Subsequently, after three additional washes, ASFV genome copies in the cells were detected using qPCR.
3. Results
3.1. Identification of p72-interacting protein by LC-MS
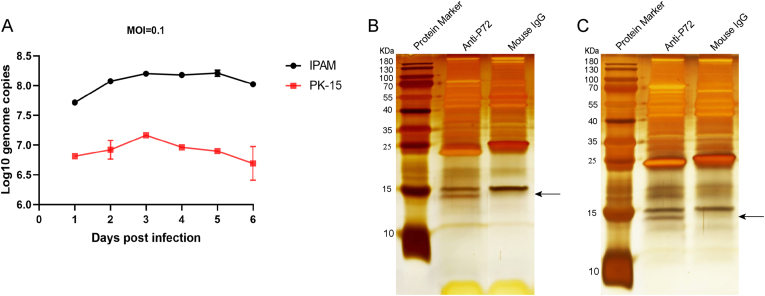
The growth curve data presented in Fig. 1A demonstrate the susceptibility of ASFVGZ strain in IPAM and PK-15 cells. To identify the potential ASFV protein binding to p72, IPAM and PK-15 cells were infected with ASFVGZ for CO-IP and LC-MS/MS analysis. The cell lysates were immunoprecipitated using anti-p72 mAb and stained with silver to visualize p72 and its binding proteins. The normal mouse IgG was used as a negative control to eliminate non-specific interactions. Silver staining of the SDS-PAGE gel of the immunoprecipitation revealed a differential band of approximately 14 KDa that was immunoprecipitated with anti-p72 mAb and not with the control antibody (Fig. 1B and C).
Fig. 1.
Silver staining was used to detect the enrichment of p72 interacting proteins. A Dynamic growth of ASFVGZ in IPAM and PK-15 cells in vitro. IPAM or PK-15 cells were infected (MOI = 0.1) with ASFVGZ and viral genome copies were calculated at the indicated times post-infection. PK-15 cells (B) and IPAM cells (C) were infected with ASFVGZ at 0.1 MOI. Anti-p72 mAb were used for CO-IP 48 h after cell infection. p72-interacting proteins were eluted with protein G sepharose and analyzed on SDS-PAGE followed by silver staining. The normal mouse IgG as a negative control. The data were tested three times independently. Arrows indicate differential bands.
LC-MS identified the interaction between p72 and ASFV protein (Supplementary Table S2). The final p72 binding protein data eliminated the nonspecific interaction through the control antibody. Finally, the p11.5 protein encoded by the ASFV A137R gene that interacts with p72 was identified.
3.2. Validating the interactions between p72 protein and p11.5 protein
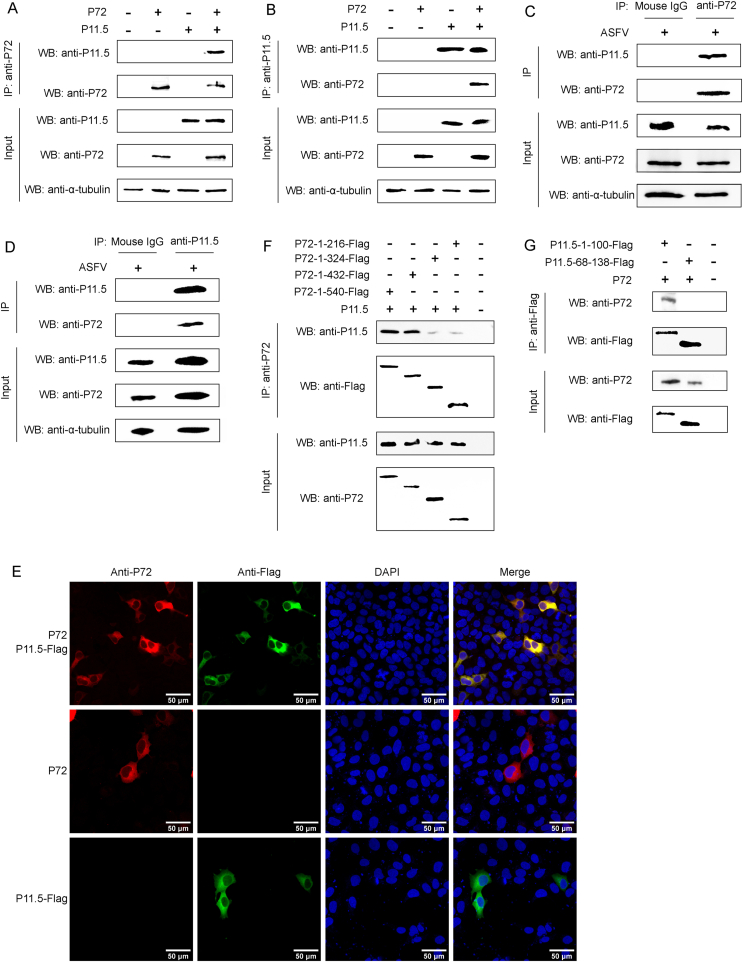
To verify the protein interactions obtained from LC-MS, CO-IP experiments were performed in vitro. The full-length B646L gene encoding p72 and the full-length A137R gene encoding p11.5 were cloned into pCAGGS vectors, named pCAGGS-p72 and pCAGGS-p11.5, respectively. HEK293T cells were transfected with pCAGGS-p72 along with pCAGGS-p11.5, and a CO-IP assay was performed with anti-p72 mAb or anti-p11.5 mAb. We found that the p11.5 protein was immunoprecipitated by p72 protein, and reciprocally, p72 protein could also be immunoprecipitated by p11.5 protein (Fig. 2A and B). Furthermore, endogenous CO-IP experiments indicated that p72 was associated with p11.5 in MA104 cells following ASFVGZ infection (Fig. 2C and D). The interaction between p72 and p11.5 was additionally confirmed by their subcellular colocalization (Fig. 2E). All these data clearly demonstrate that p11.5 is identified as a novel interacting protein with ASFV p72.
Fig. 2.
Protein p11.5 interacts with p72. A-B HEK293T cells were co-transfected with the pCAGGS-p72 and pCAGGS-p11.5 expression plasmids for 48 h, followed by a CO-IP assay for p11.5 protein and p72 protein using anti-p72 (A) or anti-p11.5 (B) antibody. C-D MA104 cells were infected with ASFVGZ at an MOI of 0.1, and cell extracts were analyzed by Co-IP at 48 h post infection using anti-p72 antibody (C) or anti-p11.5 (D) antibody. (E) MA104 cells were transfected with pCAGGS-p72 and/or pCAGGS-p11.5-Flag expression plasmids for 24 h and then fixed and processed for dual labeling. p11.5 (green) and p72 (red) proteins were visualized by immunostaining with rabbit anti-Flag and mouse anti-p72 antibodies. Cell nuclei were counterstained with DAPI (blue). The areas of colocalization in merged images are shown in yellow. (F) The truncated p72 and full-length p11.5 were co-transfected into HEK293T cells. After 48 h, the cell lysates were immunoprecipitated with anti-p72 and analyzed by WB. (G) The truncated p11.5 and full-length p72 were co-transfected into HEK293T cells. After 48 h, the cell lysates were immunoprecipitated with anti-Flag and analyzed by WB.
In order to further verify the domain of the interaction between p72 and p11.5, we constructed a series of truncation mutants. As shown in Fig. 2G, p11.5 (aa 1 to 100) coimmunoprecipitated with p72, whereas p11.5 (aa 68 to 138) did not, suggesting that p11.5 (aa 1 to 68) is essential for the association of p11.5 with p72. Similarly, we also found that the p72 (aa 1 to 216) domain can mediate the interaction between p72 and p11.5 (Fig. 2F).
3.3. Construction of ASFVGZ△A137R deletion mutant
p72 is the major composition of viral icosahedrons, which is closely related to the assembly and replication of the virus (Jia et al., 2017). Since p11.5 interacts with p72, the effect of p11.5 on ASFV replication was studied. pCAGGS-A137R was transfected into MA104 cells. The results showed that the ASFV genome copies and protein expression of p72 remained unchanged (Fig. 3A), indicating that over-expression of p11.5 has no significant effect on ASFV replication.
To determine the function of the p11.5 protein encoded by A137R gene during in vitro virus replication, ASFVGZΔA137R mutant was developed using the ASFVGZ as a template. Deletion of A137R was achieved by replacing the ORF with mCherry gene controlled by the late promoter of ASFV p72, following standard methodologies based on homologous recombination to generate recombinant virus. The ASFVGZΔA137R was purified through successive rounds of plaque purification and limiting dilution, followed by PCR with primers located outside of the deletion site to verify the expected deletion. In Fig. 3C, only the mCherry cassette band (about 1711 bp) was obtained in ASFVGZΔA137R, with no DNA band of parental strain ASFVGZ (about 1186 bp). The expression of the p72 and p11.5 in the MA104 infected with ASFVGZΔA137R was carried out by Western blot analysis using the Super ECL Detection Reagent (Cat No. 36208; Yeasen, Shanghai, China). Universal antibody diluent was purchased from New Cell & Molecular Biotech (NCM, China). In Fig. 3D, the p11.5 band (approx. 14 KDa) from ASFVGZ could be seen while no p11.5 band was detected from the ASFVGZΔA137R infected cell lysate. The p72 band (approx. 72 KDa) can be detected in both viruses. Western blot analysis demonstrated that ASFVGZΔA137R was not contaminated with ASFVGZ. Parental ASFVGZ-infected MA104 cells showed EGFP (green) expression. Building upon this framework, we substituted the A137R gene with the mCherry gene. Consequently, ASFVGZΔA137R-infected MA104 cells showing both EGFP (green) and mCherry (red) expressions (Fig. 3E). The above results indicate that the efficient deletion of the A137R gene.
3.4. Replication of recombinant virus in cell cultures
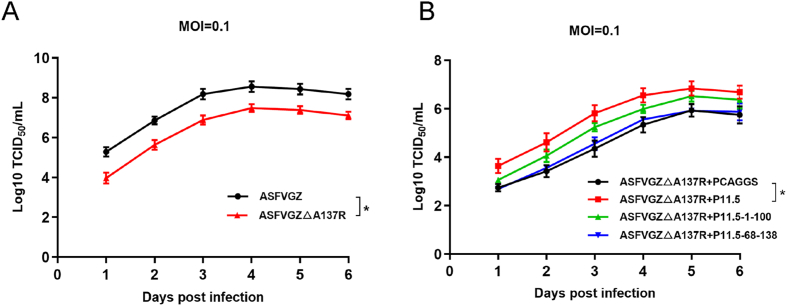
To evaluate the potential role of A137R in virus replication, the in vitro growth characteristics of ASFVGZΔA137R were studied in MA104 cells and compared with that of the parental ASFVGZ in growth curves. Infected cells were used to determine the titers by TCID50/mL in MA104 cell cultures. Results demonstrated that ASFVGZΔA137R displayed lower titers compared to parental ASFVGZ (Fig. 4A). ASFVGZΔA137R yields were nearly 10-fold lower than those of ASFVGZ. To further verify the effect of A137R on virus replication, MA104 cells were transfected with pCAGGS-A137R, pCAGGS-A137R-1–100, pCAGGS-A137R-68–138 or pCAGGS empty vector respectively and then inoculated with ASFVGZΔA137R at 0.1 MOI. The in vitro growth kinetics of ASFVGZΔA137R were compared in MA104 cell cultures transfected with different plasmids. Results demonstrated that ASFVGZΔA137R exhibited higher titers after overexpression of pCAGGS-p11.5 compared with transfection of the empty vector (Fig. 4B). Moreover, overexpression of pCAGGS-p11.5-1-100 induced slightly higher ASFVGZΔA137R yields than overexpression of pCAGGS-p11.5-68-138 (Fig. 4B). These findings suggested that the A137R gene is not necessary for virus replication, but its deletion significantly decreased viral titers in MA104 cells.
Fig. 4.
Kinetics of growth of ASFVGZΔA137R and parental ASFVGZ in MA104 cells in vitro. A MA104 cells were infected (MOI = 0.1) with each of the viruses, and virus yields were titrated with MA104 cells at the indicated times post-infection. B MA104 cells were transfected with pCAGGS-p11.5, pCAGGS-p11.5-1-100, pCAGGS-p11.5-68-138 or pCAGGS empty vector respectively and then infected with ASFVGZΔA137R at 0.1 MOI. The virus yields were titrated with MA104 cells at the indicated times post-infection. Data represent means and SD from three independent experiments. The significance of differences between groups was determined using Student's t-test (∗P < 0.05).
3.5. Deletion of A137R results in decreased infectious progeny virus production
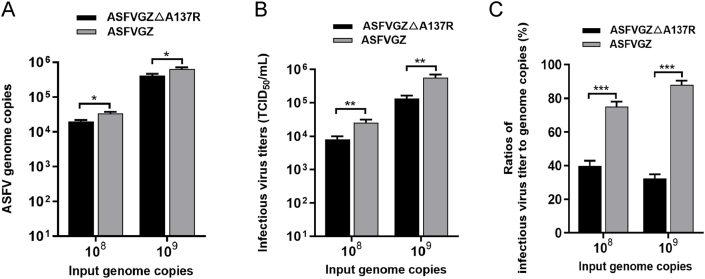
The growth curves indicated that the absence of A137R leaded to the partial production of replication-competent virions but with 1.0-log-fewer infectious progeny virions than parental ASFVGZ. To investigate the reasons why A137R deficiency exhibits lower viral titers, the cell supernatants from the MA104 cells infected with ASFVGZΔA137R or ASFVGZ at equal numbers of genome copies (108 and 109) were harvested at 24 h post infection (hpi), and the ASFV genome copies and titers were measured in the cellular supernatant (Fig. 5A). Upon attachment and entry of mature virus particles to sprouting, the entire cycle of ASFV infection culminates within a concise 24-h timeframe (Gaudreault et al., 2020). Our analysis of ASFV genome copies and titers excreted into the extracellular milieu following a 24-hour infection with ASFV suggests that non-infectious viruses can be classified as viral particles. The results showed that in the cell-free particles released by MA104 cells infected with both viruses, the genome copies and titers of ASFVGZΔA137R were lower than those of ASFVGZ (Fig. 5 B). The infectious virus titer-to-genome copies ratios for ASFVGZΔA137R were significantly lower than those for parental ASFVGZ. That is, approximately 40% progeny of ASFVGZΔA137R had the ability to successfully infect MA104 cells, while approximately 80% progeny of ASFVGZ had the ability to infect MA104 cells at 24 hpi (Fig. 5C), indicating that MA104 cells infected with ASFVGZΔA137R produced a larger proportion of noninfectious virus.
Fig. 5.
ASFVGZΔA137R produces a reduced ratio of infectious virus titer-to-genome copies in MA104 cells. A Genome copies of progeny ASFVGZΔA137R or ASFVGZ. MA104 cells in 24-well plates were infected with either ASFVGZΔA137R or ASFVGZ at genome copies of 108 or 109. The genome copies in supernatant were determined by qPCR at 24 hpi. B Infectious progeny virus production of ASFVGZΔA137R or ASFVGZ. The titers of infectious progeny virions from group A were detected according to the Spearman–Kärber method. C ASFVGZΔA137R or ASFVGZ infectious virus titer-to-genome copies ratios. The infectious virus titer-to-genome copies ratios were calculated based on the ratio of (B) to the (A) at 24 hpi (n = 3). The significance of differences between groups was determined using Student's t-test (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001).
3.6. A137R is not required for virus attachment to and entry into MA104 cells
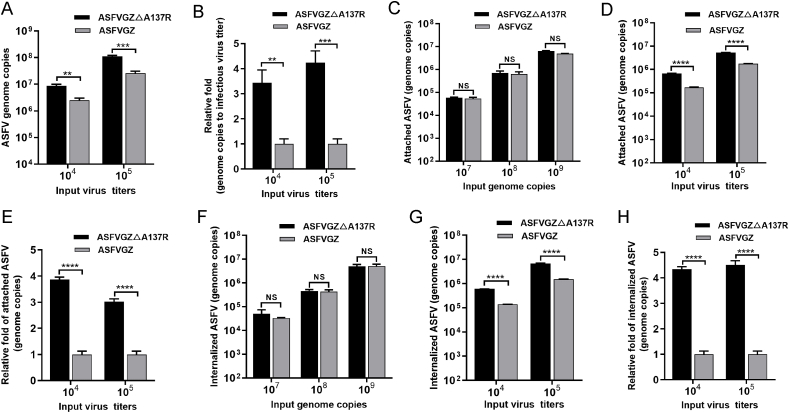
To investigate the involvement of A137R in ASFV entry, we compared the binding and entry abilities of two viruses. Firstly, the genome copies of two viruses with the same titers were detected. As depicted in Fig. 6A, ASFVGZΔA137R exhibited 8.62 × 106 and 1.1 × 108 genome copies per 104 and 105 TCID50, respectively. In contrast, ASFVGZ displayed 2.49 × 106 and 2.61 × 107 genome copies per 104 and 105 TCID50, respectively. These findings indicate that the numbers of genome copies of ASFVGZΔA137R were 3.4- or 4.2-fold higher than those of ASFVGZ at equal titers (Fig. 6B). Consequently, the results suggest that the A137R-deleted virus can generate more viral genomes with lower infectivity.
Fig. 6.
The p11.5 protein is not required for ASFV binding to or entry into MA104 cells. A Genome copies of ASFVGZΔA137R or ASFVGZ per 104 and 105 TCID50. B ASFVGZΔA137R or ASFVGZ genome copies-to-infectious virus titer ratios based on samples from panel A. C-D The attachment levels of ASFVGZΔA137R or ASFVGZ were similar. Equal numbers of genome copies (107, 108, and 109) (C) or equal titers (104 and 105) (D) of ASFVGZΔA137R or ASFVGZ were added to MA104 cells at 4 °C and allowed to attach for 2 h. The numbers of genome copies of attached ASFVs were quantified by qPCR. E The fold change in attached ASFVGZΔA137R was nearly 4 times higher than that of ASFVGZ with equal titers (D). F-G The internalization levels of ASFVGZΔA137R or ASFVGZ were similar. Equal numbers of genome copies (107, 108, and 109) (F) or equal titers (104 and 105) (G) of ASFVGZΔA137R or ASFVGZ were added to MA104 cells at 37 °C and allowed to internalize for 2 h. The genome copies of internalized ASFVs were quantified by qPCR. H The fold change in internalized ASFVGZΔA137R was approximately 4 times higher than that of ASFVGZ with equal titers (G). The data shown are from three independent experiments. The significance of differences between groups was determined using Student's t-test (∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001; NS, not significant).
To investigate the potential involvement of A137R in virus attachment and internalization, equivalent quantities of ASFVGZ and ASFVGZΔA137R virions were subjected to incubation with MA104 cells for 2 h at 4 °C or 37 °C. The quantification of attached and endocytosed virions was accomplished by detecting the genome copies of B646L within the MA104 cells. When the input genome copies of the inoculum are equal, a considerable number of ASFVGZΔA137R and ASFVGZ attach or internalize into MA104 cells (Fig. 6C and F). On the contrary, a noteworthy distinction in the genome copy number of B646L was observed between virions attached to or internalized into MA104 cells when the inoculum contained equivalent titers of ASFVGZΔA137R and ASFVGZ (Fig. 6D and G). Specifically, the genome copies of ASFVGZΔA137R were nearly four-fold higher than those of ASFVGZ (Fig. 6E and H). The results showed that both infectious and non-infectious forms of the A137R-deleted virus bind to and are internalized into MA104 cells, resulting in a higher uptake of the virus genome than parental ASFVGZ.
Taken together, these findings suggest that infection-defective phenotype of virions generated by ASFVGZΔA137R is not attributed to the attachment and entry defects of virions.
4. Discussion
ASFV has a complex structure composed of many proteins. Of more than 150 ORFs encoded by the ASFV genome, only a few protein structures are known and the functions of most of these proteins remain unknown (Dixon et al., 2019; Yang et al., 2023). Analyzing the interaction among proteins as well as the functional roles of viral proteins in ASFV is crucial to understand the viral replication process, which will accelerate the development of novel ASF vaccines and antiviral drugs. The ASFV capsid structure is built from 17,280 proteins, including one major (p72) and four minor (M1249L, p17, p49, and H240R) capsid proteins organized into pentasymmetrons and trisymmetrons (Liu S. et al., 2019; Wang et al., 2019). As the main component of ASFV capsid protein, p72 plays an important role in the process of virus assembly and replication (Salas and Andres, 2013). Previous studies have found that p72 protein interacts with two structural proteins, pH240R and pE120R, and a non-structural protein, pB602L, respectively (Andres et al., 2001; Epifano et al., 2006; Zhou et al., 2022). Inhibiting the expression of any of the above proteins will affect virus replication and reduce virus titer (Andres et al., 2001; Epifano et al., 2006; Zhou et al., 2022). In this study, CO-IP and mass spectrometry analysis were used to discover another important structural protein interacting with p72, that is, the p11.5 protein. Proteome and transcriptome analysis showed that the p11.5 protein encoded by the A137R gene was expressed at high abundance (Kessler et al., 2018; Cackett et al., 2020). Considering the high expression level of p11.5, it is likely important throughout infection, which makes it an interesting candidate as potential drug or vaccine targets.
p11.5 is a late-expressed viral structural protein and associated with ASFV virulence, but the localization of p11.5 in a viral particle remains to be elucidated (Alcami et al., 1993; Gladue et al., 2021). Deletion of the A137R gene from the highly virulent ASFV-Georgia2010 (ASFV-G) isolate significantly reduced virus virulence in swine. Moreover all ASFV-G-ΔA137R-inoculated animals were protected when challenged with the virulent parental strain ASFV-G, but the underlying mechanism remains unknown (Gladue et al., 2021). To further elucidate the mechanism of the A137R gene regulating ASFV virulence, Sun et al. found that p11.5 negatively regulates the cGAS-STING-mediated IFN-β signaling pathway via the autophagy-mediated lysosomal degradation of TBK1, which highlights the involvement of p11.5 regulating ASFV virulence (Sun et al., 2022). Although part of the pathogenic mechanism of the A137R gene has been understood, large gaps still remain to be filled in our knowledge of A137R gene.
This study proves, for the first time, the interaction between p72 and p11.5. We found that aa 1 to 216 of p72 specifically binds to aa 1 to 68 of p11.5. Notably, the immunoprecipitation of p11.5 protein levels by p72 (aa 1 to 216) and p72 (aa 1 to 324) in Fig. 2F reveals a comparatively subdued signal in comparison to the adjacent lanes. This observation prompts speculation on the potential significance of aa 324 to 432 within the p72 protein, suggesting a plausible pivotal role in the interaction with p11.5. Next, the replication of ASFV was unaffected in MA104 cells with p11.5-overexpressed (Fig. 3A), which prompted us to construct the ASFVGZΔA137R deletion strain. Inferred from one-step virus growth curves, the titers of ASFVGZΔA137R are about 1.0 log lower than that of parental strain ASFVGZ, which is consistent with previous studies of Gladue et al. (2021). The above results show that although the A137R gene is not essential for virus replication, its deletion significantly decreased the viral titers in MA104 cell cultures. After supplementing with A137R, the virus titers significantly increased compared to the control group. Meanwhile, the virus titers after supplementing with aa 1–100 encoded by the A137R gene were slightly higher than that of supplementing with aa 68–138, suggesting that the aa 1–68 of the A137R gene may be a key region affecting viral titers.
Based on our data, we speculate that the 1.0-log growth defect in ASFVGZΔA137R-infected MA104 cells is partially attributable to an increase in the actual production of non-infectious virus (Fig. 5). In the realm of genome copies-to-infectious virus titer ratios, ASFVGZΔA137R exhibits an approximately fourfold increase compared to its ASFVGZ counterpart (Fig. 6B). This discrepancy implies that the virus lacking A137R can produce a greater quantity of viral genomes while maintaining lower infectivity levels. The adsorption and internalization experiments of the ASFV indicate that A137R is not required for virus attachment to and entry into MA104 cells. It is hypothesized that A137R may affect virus replication by affecting the assembly and release of the virus. Remarkably, we show that p11.5 interacts with the p72 (Fig. 2), indicating that p11.5 may play an important role in virus assembly. p72, as the main component of the capsid protein, is critical in determining the structure and stability of the viral particle, and interactions with p11.5 are likely to be involved in these processes. In addition, viruses often hijack host cellular machinery to support their replication. The interaction between p72 and p11.5 may contribute to the modulation of cellular processes, creating a favorable environment for viral replication within the host cell. In the annals of ASFV research, a noteworthy revelation emerges from the pages of prior literature: the absence of the A137R gene within the genome of the highly virulent ASFV-Georgia2010 (ASFV-G) isolate markedly diminishes the virulence of the virus in swine (Gladue et al., 2021). Contemplating this intriguing premise, it is conceivable that the interaction between the p72 and p11.5 assumes a pivotal role in dictating the extent of ASFV infection severity and its consequential repercussions on host organisms. This speculation strengthens our understanding of the complex molecular mechanism of ASFV, underscoring the potential significance of the p72-p11.5 interaction in modulating virulence dynamics. Since the ASFV life cycle is regulated by multiple proteins, we cannot rule out the possibility that p11.5 interacts with other viral proteins at different stages of ASFV replication. The aforementioned hypothesis warrants meticulous scrutiny and comprehensive validation in future studies.
5. Conclusion
Analysis of above results led to three important conclusions: (i) p72 interacts with p11.5, (ii) A137R is not essential for virus replication but loss of A137R results in the production of abundant noninfectious viruses, and (iii) A137R is not required for ASFV attachment to and entry into MA104 cells. These findings not only enrich the understanding of ASFV but also provide novel potential targets for the design of drugs or vaccines to prevent and control ASFV.
Data availability
All the data generated during the current study are included in the manuscript.
Ethics statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author contributions
Dan Yin: data curation, writing-original draft. Bin Shi, Renhao Geng, Yingnan Liu, Lang Gong, Hongxia Shao, Kun Qian, Hongjun Chen: data curation. Aijian Qin: methodology, project administration, writing-review & editing.
Conflict of interest
All authors declare that there are no competing interests.
Acknowledgements
This work was supported by the National Key Research and Development Program (Grant No. 2021YFD1800100), the National Natural Science Foundation of China (Grant No. 31941016), the Priority Academic Program Development of Jiangsu Higher Education Institutions, and the Jiangsu Co-innovation Center for the Prevention and Control of Important Animal Infectious Diseases and Zoonoses. The funding bodies did not play direct roles in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2024.05.007
Contributor Information
Kun Qian, Email: qiankun@yzu.edu.cn.
Hongjun Chen, Email: vetchj@shvri.ac.cn.
Aijian Qin, Email: aijian@yzu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Alcami A., Angulo A., Vinuela E. Mapping and sequence of the gene encoding the African swine fever virion protein of M(r) 11500. J. Gen. Virol. 1993;74:2317–2324. doi: 10.1099/0022-1317-74-11-2317. [DOI] [PubMed] [Google Scholar]
- Alejo A., Matamoros T., Guerra M., Andres G. A proteomic atlas of the African swine fever virus particle. J. Virol. 2018;92 doi: 10.1128/JVI.01293-18. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres G., Charro D., Matamoros T., Dillard R.S., Abrescia N.G.A. The cryo-EM structure of African swine fever virus unravels a unique architecture comprising two icosahedral protein capsids and two lipoprotein membranes. J. Biol. Chem. 2020;295:1–12. doi: 10.1074/jbc.AC119.011196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres G., Garcia-Escudero R., Vinuela E., Salas M.L., Rodriguez J.M. African swine fever virus structural protein pE120R is essential for virus transport from assembly sites to plasma membrane but not for infectivity. J. Virol. 2001;75:6758–6768. doi: 10.1128/JVI.75.15.6758-6768.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borca M.V., Ramirez-Medina E., Silva E., Vuono E., Rai A., Pruitt S., Holinka L.G., Velazquez-Salinas L., Zhu J., Gladue D.P. Development of a highly effective African swine fever virus vaccine by deletion of the I177L gene results in sterile immunity against the current epidemic Eurasia strain. J. Virol. 2020;94 doi: 10.1128/JVI.02017-19. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cackett G., Matelska D., Sykora M., Portugal R., Malecki M., Bahler J., Dixon L., Werner F. The African swine fever virus transcriptome. J. Virol. 2020;94 doi: 10.1128/JVI.00119-20. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon L.K., Chapman D.A., Netherton C.L., Upton C. African swine fever virus replication and genomics. Virus Res. 2013;173:3–14. doi: 10.1016/j.virusres.2012.10.020. [DOI] [PubMed] [Google Scholar]
- Dixon L.K., Sun H., Roberts H. African swine fever. Antivir. Res. 2019;165:34–41. doi: 10.1016/j.antiviral.2019.02.018. [DOI] [PubMed] [Google Scholar]
- Epifano C., Krijnse-Locker J., Salas M.L., Rodriguez J.M., Salas J. The African swine fever virus nonstructural protein pB602L is required for formation of the icosahedral capsid of the virus particle. J. Virol. 2006;80:12260–12270. doi: 10.1128/JVI.01323-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo I., Alonso C. African swine fever virus: a review. Viruses. 2017;9:103. doi: 10.3390/v9050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudreault N.N., Madden D.W., Wilson W.C., Trujillo J.D., Richt J.A. African swine fever virus: an emerging DNA arbovirus. Front. Vet. Sci. 2020;7:215. doi: 10.3389/fvets.2020.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladue D.P., Ramirez-Medina E., Vuono E., Silva E., Rai A., Pruitt S., Espinoza N., Velazquez-Salinas L., Borca M.V. Deletion of the A137R gene from the pandemic strain of African swine fever virus attenuates the strain and offers protection against the virulent pandemic virus. J. Virol. 2021;95 doi: 10.1128/JVI.01139-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Liu T., Li K., Song X., Yan R., Xu L., Li X. Proteomic analysis of protein interactions between Eimeria maxima sporozoites and chicken jejunal epithelial cells by shotgun LC-MS/MS. Parasites Vectors. 2018;11:226. doi: 10.1186/s13071-018-2818-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N., Ou Y., Pejsak Z., Zhang Y., Zhang J. Roles of African swine fever virus structural proteins in viral infection. J Vet Res. 2017;61:135–143. doi: 10.1515/jvetres-2017-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler C., Forth J.H., Keil G.M., Mettenleiter T.C., Blome S., Karger A. The intracellular proteome of African swine fever virus. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-32985-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D.P., Reid S.M., Hutchings G.H., Grierson S.S., Wilkinson P.J., Dixon L.K., Bastos A.D., Drew T.W. Development of a TaqMan PCR assay with internal amplification control for the detection of African swine fever virus. J Virol Methods. 2003;107:53–61. doi: 10.1016/s0166-0934(02)00189-1. [DOI] [PubMed] [Google Scholar]
- Kleiboeker S.B., Kutish G.F., Neilan J.G., Lu Z., Zsak L., Rock D.L. A conserved African swine fever virus right variable region gene, l11L, is non-essential for growth in vitro and virulence in domestic swine. J. Gen. Virol. 1998;79:1189–1195. doi: 10.1099/0022-1317-79-5-1189. [DOI] [PubMed] [Google Scholar]
- Liu Q., Ma B., Qian N., Zhang F., Tan X., Lei J., Xiang Y. Structure of the African swine fever virus major capsid protein p72. Cell Res. 2019;29:953–955. doi: 10.1038/s41422-019-0232-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Luo Y., Wang Y., Li S., Zhao Z., Bi Y., Sun J., Peng R., Song H., Zhu D., Sun Y., Li S., Zhang L., Wang W., Sun Y., Qi J., Yan J., Shi Y., Zhang X., Wang P., Qiu H.J., Gao G.F. Cryo-EM structure of the African swine fever virus. Cell Host Microbe. 2019;26:836–843.e3. doi: 10.1016/j.chom.2019.11.004. [DOI] [PubMed] [Google Scholar]
- Liu Y., Li Y., Xie Z., Ao Q., Di D., Yu W., Lv L., Zhong Q., Song Y., Liao X., Song Q., Wang H., Chen H. Development and in vivo evaluation of MGF100-1R deletion mutant in an African swine fever virus Chinese strain. Vet. Microbiol. 2021;261 doi: 10.1016/j.vetmic.2021.109208. [DOI] [PubMed] [Google Scholar]
- Liu Y., Xie Z., Li Y., Song Y., Di D., Liu J., Gong L., Chen Z., Wu J., Ye Z., Liu J., Yu W., Lv L., Zhong Q., Tian C., Song Q., Wang H., Chen H. Evaluation of an I177L gene-based five-gene-deleted African swine fever virus as a live attenuated vaccine in pigs. Emerg. Microb. Infect. 2023;12 doi: 10.1080/22221751.2022.2148560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pomares L., Simon-Mateo C., Lopez-Otin C., Vinuela E. Characterization of the African swine fever virus structural protein p14.5: a DNA binding protein. Virology. 1997;229:201–211. doi: 10.1006/viro.1996.8434. [DOI] [PubMed] [Google Scholar]
- Pujols J., Blazquez E., Segales J., Rodriguez F., Chang C.Y., Argilaguet J., Bosch-Camos L., Rosell R., Pailler-Garcia L., Gavrilov B., Campbell J., Polo J. Feeding spray-dried porcine plasma to pigs improves the protection afforded by the African swine fever virus (ASFV) BA71ΔCD2 vaccine prototype against experimental challenge with the pandemic ASFV-Study 2. Vaccines (Basel) 2023;11:825. doi: 10.3390/vaccines11040825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla Y., Perez-Nunez D., Richt J.A. African swine fever virus biology and vaccine approaches. Adv. Virus Res. 2018;100:41–74. doi: 10.1016/bs.aivir.2017.10.002. [DOI] [PubMed] [Google Scholar]
- Salas M.L., Andres G. African swine fever virus morphogenesis. Virus Res. 2013;173:29–41. doi: 10.1016/j.virusres.2012.09.016. [DOI] [PubMed] [Google Scholar]
- Sun M., Yu S., Ge H., Wang T., Li Y., Zhou P., Pan L., Han Y., Yang Y., Sun Y., Li S., Li L.F., Qiu H.J. The A137R protein of African swine fever virus inhibits type I interferon production via the autophagy-mediated lysosomal degradation of TBK1. J. Virol. 2022;96 doi: 10.1128/jvi.01957-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Zhao D., Wang J., Zhang Y., Wang M., Gao Y., Li F., Wang J., Bu Z., Rao Z., Wang X. Architecture of African swine fever virus and implications for viral assembly. Science. 2019;366:640–644. doi: 10.1126/science.aaz1439. [DOI] [PubMed] [Google Scholar]
- Yang S., Miao C., Liu W., Zhang G., Shao J., Chang H. Structure and function of African swine fever virus proteins: current understanding. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1043129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin D., Geng R., Shao H., Ye J., Qian K., Chen H., Qin A. Identification of novel linear epitopes in P72 protein of African swine fever virus recognized by monoclonal antibodies. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.1055820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Li L.F., Zhang K., Wang B., Tang L., Li M., Wang T., Sun Y., Li S., Qiu H.J. Deletion of the H240R gene of African swine fever virus decreases infectious progeny virus production due to aberrant virion morphogenesis and enhances inflammatory cytokine expression in porcine macrophages. J. Virol. 2022;96 doi: 10.1128/jvi.01667-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsak L., Lu Z., Kutish G.F., Neilan J.G., Rock D.L. An African swine fever virus virulence-associated gene NL-S with similarity to the herpes simplex virus ICP34.5 gene. J. Virol. 1996;70:8865–8871. doi: 10.1128/jvi.70.12.8865-8871.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data generated during the current study are included in the manuscript.