Abstract
As of December 2022, 2603 laboratory-identified Middle East respiratory syndrome coronavirus (MERS-CoV) infections and 935 associated deaths, with a mortality rate of 36%, had been reported to the World Health Organization (WHO). However, there are still no vaccines for MERS-CoV, which makes the prevention and control of MERS-CoV difficult. In this study, we generated two DNA vaccine candidates by integrating MERS-CoV Spike (S) gene into a replicating Vaccinia Tian Tan (VTT) vector. Compared to homologous immunization with either vaccine, mice immunized with DNA vaccine prime and VTT vaccine boost exhibited much stronger and durable humoral and cellular immune responses. The immunized mice produced robust binding antibodies and broad neutralizing antibodies against the EMC2012, England1 and KNIH strains of MERS-CoV. Prime-Boost immunization also induced strong MERS-S specific T cells responses, with high memory and poly-functional (CD107a-IFN-γ-TNF-α) effector CD8+ T cells. In conclusion, the research demonstrated that DNA-Prime/VTT-Boost strategy could elicit robust and balanced humoral and cellular immune responses against MERS-CoV-S. This study not only provides a promising set of MERS-CoV vaccine candidates, but also proposes a heterologous sequential immunization strategy worthy of further development.
Keywords: MERS-CoV, VTT vaccine, DNA vaccine, Humoral and cellular immune responses, Prime/boost strategy
Highlights
-
•
DNA-Prime/VTT-Boost vaccines stimulated strong humoral responses with broader neutralizing antibodies against multiple MERS-CoV strains in mice.
-
•
The DNA/VTT vaccines sequential immunization elicits high percentages of polyfunctional MERS-CoV-S specific CD8+ T cell compared to DNA immunization alone.
-
•
The heterologous vaccines sequential immunization could induce balanced and durable humoral and cellular immune responses with high memory B, memory T and polyfunctional effector CD8+ T cells against MERS-CoV.
1. Introduction
The Middle East Respiratory Syndrome (MERS) was first reported in 2012, and a novel coronavirus (MERS-CoV) was confirmed as its causative pathogen (Zaki et al., 2012; Racaniello et al., 2013). MERS-CoV is one of seven reported human coronaviruses (HCoVs) belonging to the betacoronavirus genus of Coronavirinae in the Coronaviridae family (Sampson et al., 2021). It differs from HCoV-OC43, HCoV-HKU1, HCoV-NL63 and HCoV-229E, which cause “common cold” symptoms in the population, MERS-CoV causes severe respiratory symptoms and can even be fatal in infected individuals (Zaki et al., 2012; Paules et al., 2020). The prominent clinical symptoms of critically ill patients include acute and rapidly progressive viral pneumonia, pneumonitis, hypoxic lung injury, and acute respiratory distress syndrome. Up to half of the adult symptomatic patients require treatment in the intensive care unit (ICU), with 40–70% requiring mechanical ventilation (Senga et al., 2017). The MERS-CoV epidemic has spread to 27 countries across the Middle East, Europe, Asia, and the Americas. Globally, the total number of laboratory-identified MERS-CoV infected individuals reported to the WHO reached 2603, including 935 associated deaths (mortality rate was 36%) as of December 2022 (WHO, 2023). Effective treatments and approved vaccines to control and prevent the MERS-CoV epidemic remain lacking. To cope with the epidemic of highly pathogenic MERS-CoV, the development of MERS-specific vaccines is an effective strategy for preventing infections in individuals. Various vaccine platforms have developed MERS-CoV vaccine candidates with demonstrated protective effects including inactivated vaccines, recombinant protein vaccines, DNA vector vaccines, and viral vector vaccines (Volz et al., 2015; Chi et al., 2017; Hashem et al., 2019; Rodon et al., 2019; Kim et al., 2022). However, MERS-CoV vaccines have yet to receive approval for human use, although three types of MERS-CoV vaccine (GLS-5300, MVA-MERS-S and MERS002) have been evaluated for safety and immunogenicity in clinical phase1 or 1b trials (Modjarrad et al., 2019; Koch et al., 2020; Bosaeed et al., 2022).
The immunogen of MERS-CoV vaccine candidates primarily targets the spike (S) gene, echoing the approach taken in the development of other coronavirus vaccines. The coronavirus S monomer comprises the S1 and S2 subunits. The receptor binding domain (RBD) of MERS-CoV that is located in the S1 subunit is a vital region for binding to the human dipeptidyl peptidase 4 of target cells (hDPP4, known as human CD26) (Wang et al., 2014; Xiong et al., 2022). The homotrimeric S glycoprotein on the viral envelope binds to the target cellular receptor and triggers serial changes in the protein structure, leading to the fusion of target cells and the viral membrane (Pallesen et al., 2017; Lan et al., 2020). The S protein plays a substantial role in the process of virus infection. Over 35 years of monitoring in healthy individuals shows that reinfection with the same seasonal coronavirus often occurs 12 months after recovery from coronavirus infection and may occur six months after reinfection (Edridge et al., 2020), which implied a short duration of humoral immune responses in the human body. Coronavirus-specific T cell immune responses have demonstrated longevity (such as in convalescent SARS-CoV-1) with the specific memory T cell immune responses against the S protein being detected six years post-infection (Tang et al., 2011). Cohort studies show that CD4+ T cells in unexposed individuals develop affinity with SARS-CoV-2 and the common cold human coronavirus (HCoV-OC43, HCoV-229E, HCoV-NL63, and HCoV-HKU1) in human samples (Lipsitch et al., 2020). This suggests that T cell epitopes are predominantly located in the conserved region of the S protein and exhibit cross reactivity to different coronaviruses. Given these aspects, considering a vaccine that induces a cellular immune response is necessary in designing coronavirus vaccines. The preceding researches on coronavirus vaccines offer crucial references for the investigation and development of MERS-CoV vaccines.
While inactivated and recombinant protein vaccines generally induce weak cellular immune responses, the viral vector vaccines, especially the replication-competent ones, have demonstrated the capacity to induce strong cellular immune responses. Our study utilizes the replicating vaccinia TianTan strain (VTT)-based vector vaccine platform to develop novel MERS-CoV vaccines. We constructed two types of DNA vaccines based on MERS-S and MERS-S1 and used them as priming immunization vaccines, with the VTT vaccine serving as the boost vaccine. The heterologous sequential Prime-Boost immunization strategy (DNA-Prime/VTT-Boost) will be tested in mice for humoral and cellular immune responses against MERS-CoV. The quantity and quality of the immune responses, including binding antibodies and neutralizing antibodies, CD8+ T cell responses and their poly-functionality, as well as central and effector memory cells will be evaluated comprehensively. Our study aims to provide scientific data and strategies for the development of an effective MERS-CoV vaccine.
2. Materials and methods
2.1. Cells, viruses, and vectors
Chicken embryo fibroblasts (CEFs) were obtained from 9-day-old chicken embryos. The Chinese Small Pox vaccine strains (Vaccinia TianTan, VTT) were provided by Beijing Institute of Biological Products. We purchased MERS-CoV spike gene (pCMV3-spike plasmid, the catalog number is VG40069-CH) from Sino Biological (China) to construct DNA vector vaccine and VTT vector vaccine, and the translated amino acid sequence is identical with AFS88936.1 in NCBI. The vectors of pSC65gfp and pDRVISV3.0 (p3.0) are preserved in our department, and a pcDNA3.1 vector (Invitrogen, USA) was used to construct pseudoviruses of MERS-EMC/2012, KNIH, and England1.
2.2. Construction of the recombinant replicating VTT-MERS-S vaccine candidate
The MERS-S gene was recombinantly inserted into the pSC-65gfp vector. The correct clone strain was subsequently verified through gene sequencing by Sangon Biotech (China), and designated as pSC-65gfp-MERS-S. The pSC-65gfp-MERS-S was linearized by restriction enzyme PvuⅠ (TaKaRa, Japan). Following the infection of CEFs with the VTT for 2 h, the cells were transfected with 5 μg of the linearized the pSC-65gfp-MERS-S. Subsequently, after a 24 h incubation period, first-generation recombinant virus was obtained. The MERS-CoV full-length spike gene (1–4059 bp, 1353 amino acid) was inserted into the VTT vector. First-generation virus was purified by five consecutive passages in CEFs, then we successfully obtained pure recombinant VTT-MERS-S virus using high-speed centrifugation through 36% sucrose cushions (Garcia-Arriaza et al., 2021). The recombinant VTT-MERS-S vaccine was termed as VTT-S. The amino acid sequence of VTT-S vaccine is in Supplementary Material.
2.3. Construction of the recombinant DNA-MERS-S vaccine candidates
MERS spike gene lacking transmembrane domain (TD) (termed S△TD, 1–3867 bp, 1289 aa) was amplified using PCR with pCMV3-spike plasmid serving as a template, subsequently, the amplification product (3867 bp) was recombined into the pDRVISV3.0 vector (p3.0-MERS-S△TD). The p3.0-MERS-S△TD was duplicated by Escherichia coli JM109 (TaKaRa, Otsu, Shiga, Japan) and extracted with the EndoFree Plasmid Mega kit (QIAGEN GmbH, Shanghai, China) following the manufacturer's instructions. Another p3.0-MERS-S1 vaccine with a histidine tag was provided by our department. The S1 region (1–1503 bp, 501 aa) of MERS spike gene was recombined into the pDRVISV3.0 vector to construct p3.0-MERS-S1 vaccine with a histidine tag. The recombinant vaccines of p3.0-MERS-S△TD and p3.0-MERS-S1 were termed as DNA-S△TD and DNA-S1, respectively. The sequences of both vaccines were codon-optimized, and the amino acid sequences of DNA-S△TD and DNA-S1 vaccines are in Supplementary Material.
2.4. Western blot
DNA-S△TD, DNA-S1 and DNA (p3.0 empty vector) transfected 293T cell, whereas VTT-S and VTT infected CEFs. After a 48-h incubation period, 293T cells and the CEFs were harvested and lysed using sonication. Western blot was used to identify the expression of MERS-S1, MERS-S△TD and MERS-S protein (Sino Biological, Beijing, China). The assay referred to previous references (Hnasko and Hnasko, 2015).
2.5. Polypeptides
MERS-CoV peptides were used to evaluate the cellular immune response elicited by vaccines. Peptides (Supplementary Table S1) were synthesized in GenScript (Nanjing, China). Eight peptide sequences referred from literatures (Alharbi et al., 2017; Choi et al., 2020), and four peptide sequences were predicted from the Immune Epitope Database (IEDB) (https://www.iedb.org/). These twelve peptide sequences are distributed in S1 and S2 regions.
2.6. Animal and immunization
Specific pathogen-free female Balb/c mice (6–8 weeks old) were purchased from Beijing Vital River Laboratory (Beijing, China) and raised at the Chinese Center for Disease Control and Prevention Laboratory Animal Center. Mice were randomly assigned to groups and immunized intramuscularly (i.m.) in the tibialis anterior with electroporation method (electroporation parameters: 100 V, 60 ms, interval 999 ms, 10 times; WJ-2005-Intelligent live cell gene delivery instrument, NINGBO SCIENTZ BIOTECHNOLOGY CO.LTD). The doses of recombinant DNA and VTT-S vaccines were 50 μg/time/each mouse and 5 × 106 pfu/time/mouse, respectively. The blank and control groups received three injections of phosphate buffer saline (PBS) and pDRVISV3.0 vector instead of the vaccine.
2.7. Enzyme-linked immunosorbent assay
The levels of MERS-CoV Receptor binding domain (MERS-RBD) protein-specific IgG, IgG1, IgG2a and IgG2b in sera were measured by an enzyme-linked immunosorbent assay (ELISA). Ninety-six-well plates (Corning, USA) were coated with MERS-RBD protein 100 μL/well (1.5 μg/mL) at 4 °C overnight. The plates were then blocked with 5% milk-2% bovine serum albumin. HRP-conjugated goat anti-mouse IgG, IgG1, Ig2a and IgG2b (1:2500) (Southern Biotech, USA) were used as secondary antibodies. Chromogen solution A and B (Beijing kinghawk pharmaceutical, China) were added to each well, and the reaction was stopped after 10 min. The absorbance was read at 450 and 630 nm. Values 2-fold higher than the control group were considered positive (Chi et al., 2017).
2.8. Pseudovirus neutralization assays
The MERS-CoV pseudovirus was constructed using a vesicular stomatitis virus-based system following a previously-published method (Nie et al., 2020), and the pseudoviruses of EMC2012 (YP_009047204.1), England1 (YP_007188579.1) and KNIH (AKL59401.1) variants were constructed. Sera were inactivated at 56 °C for 1 h. The immune sera were diluted serially for detecting neutralizing antibody titers. Ultra-High Sensitivity luciferase reagents (PerkinElmer, MA) were used to detect the luciferase activity by a chemiluminescence detector (VICTOR3, PerkinElmer), and neutralizing titres at 50% were determined by the Reed-Muench method.
2.9. Enzyme-linked immunosorbent spot assay
The enzyme-linked immunosorbent spot (ELISpot) assay was used to measure MERS-CoV S-specific IFN-γ-secreting cells. We detected specific IFN-γ+ T cells according to the instruction manual. The 96-well nitrocellulose-bottomed plates (BD Biosciences) were coated with purified anti-mouse IFN-γ (5 μg/mL) and then the plates were blocked. Fresh splenocytes were used in the assay for detecting specific IFN-γ+ T cell. The color development of spots was terminated by washing the wells with deionized water, and spots were calculated by using an ELISpot detector.
2.10. Intracellular cytokine staining (ICS) assay
The magnitude, polyfunctionality, and phenotypes of the MERS-CoV-S specific CD8+ T-cell adaptive immune responses were tested by ICS. Splenocytes were seeded in 96-well U-bottom plates (corning) and stimulated for 6 h in complete RPMI 1640 medium (Corning, USA) with GolgiStop™ containing monensin (0.06 μL/sample) (BD Biosciences, NJ, USA), brefeldin A (3 μg/sample) (Sigma-Aldrich, USA), PE anti-mouse CD107a (LAMP-1) antibody (0.16 μL/sample) (Biolegend, CA, USA) and 2 μg/mL MERS-CoV-S peptide pools. The dead cells were distinguished using the violet LIVE/DEAD stain kit (Thermo Fisher Scientific). The cocktail of fluorochrome-conjugated antibodies used for surface markers were Brilliant Violet 711™ anti-mouse CD3ε (Biolegend), CD4 (BD Biosciences), CD8 (BD Biosciences), FITC anti-mouse/human CD44 (Biolegend) and PerCP/Cyanine5.5 anti-mouse CD62L antibody (Biolegend). Intracellular antibodies included APC Rat anti-mouse IFN-γ (BD Biosciences), PE-Cy7™ Rat anti-mouse TNF-α (BD Biosciences), and APC-Cy7™ Rat anti-mouse IL-2(BD Biosciences). Lastly, the cells were fixed with 2% polyformaldehyde (PFA) and tested using a flow cytometer (LSRFortessa, BD Biosciences). Results were analyzed by FlowJo software 9.9.4 (FlowJo LLC, Ashland, OR). Boolean combinations of single functional gates were created to analyze polyfunctional specific T cells. The frequencies of all possible combinations of cytokine expression were calculated.
2.11. Memory B cells studies
The biotinylated recombinant MERS-RBD protein was respectively labeled with streptavidin, R-phycoerythrin (PE) conjugate (Thermo Fisher Scientific) and streptavidin, allophycocyanin (APC) conjugate (Thermo Fisher Scientific) as antigen-specific MBC probes. For immunophenotyping of antigen-specific memory B cells (MBC), splenocytes were incubated with anti-mouse CD3ε BV711 (Biolegend), anti-mouse CD19 BV510 (Biolegend), anti-mouse CD38 PE/Dazzle 594 (Biolegend) (Laczkó et al., 2020; Cancro and Tomayko, 2021), anti-mouse CD45R/B220 PE/Cy7 (Biolegend), anti-mouse IgG FITC (Biolegend) antibodies and two fluorescent labelling recombinant RBD protein probes on ice for 1 h. Then we washed off excess antibodies and fixed splenocytes with PFA. Antigen-specific memory B cells were detected and analyzed using a flow cytometer (LSRFortessa, BD Biosciences) and FlowJo software 9.9.4 (FlowJo LLC, Ashland, OR).
2.12. Statistical analyses
Graphpad Prism 9.5.1 (San Diego, CA, USA) was used for plotting and statistical analysis. Binding antibody titers and neutralizing titer values were based on a geometric mean and geometric mean standard deviation (SD), the results of T and B cells were described as the mean ± SD. Multiple Comparisons were calculated using the Kruskal-Wallis's test (Dunn's multiple comparisons), and comparisons between two groups were calculated using the Mann-Whitney test. A P value < 0.05 was considered significant (∗P < 0.05, ∗∗P < 0.01, or ∗∗∗P < 0.001).
3. Results
3.1. Construction of MERS-CoV-S VTT and DNA vector vaccines and immunization scheme
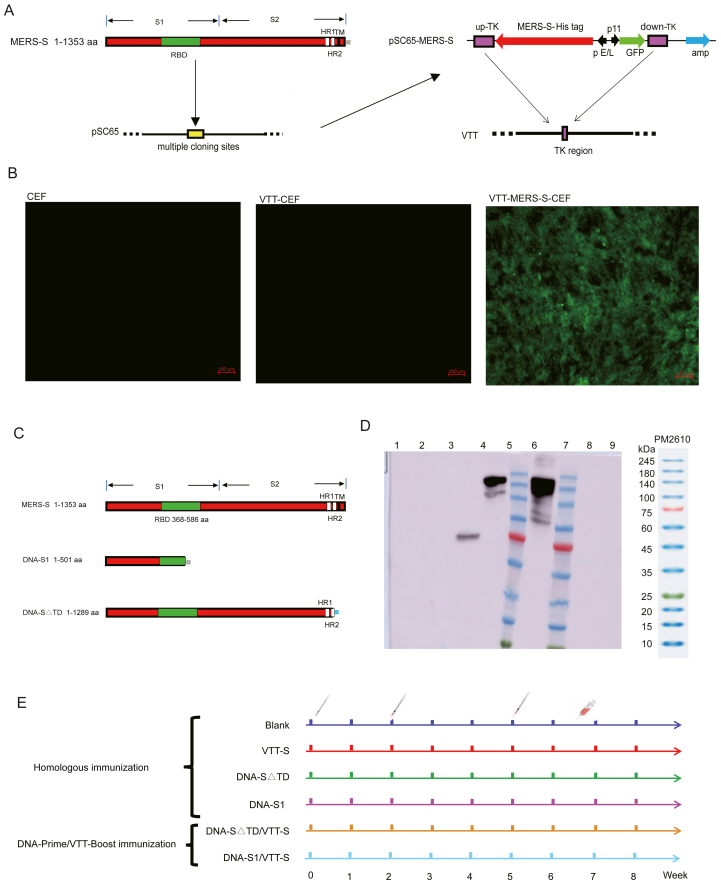
The recombinant plasmid pSC-65gfp-MERS-S was recombined with VTT in CEFs (Fig. 1A). After five generations of purification, a purified VTT-S vaccine was obtained. The VTT-S vaccine in CEFs expressed green fluorescent protein (GFP) and MERS-S protein (Fig. 1B, D). The recombinant DNA vaccines were constructed and confirmed by gene sequencing (Fig. 1C). Additionally, the western blot assay showed that two recombinant plasmids expressed MERS-S△TD or MERS-S1 protein in 293T cells (Fig. 1D).
Fig. 1.
Design and identification of vaccine candidates based on DNA and VTT vectors. A Schematic representation of the recombinant VTT vaccine expressing the MERS-S gene (the vaccine termed VTT-S); the white areas on the MERS-S gene represent HR1 and HR2 respectively, the green and black areas represent RBD and transmembrane, and the grey represents the histidine tag. B VTT-S vaccine was indirectly identified by fluorescence microscopy. Scale bar: 100 μm. C Schematic representation of the recombinant DNA vaccines with expressing the MERS-S1 gene or MERS-S△TD (the vaccines termed DNA-S1 and DNA-S△TD, respectively); the grey area at the end of S1 signifies the histidine tag, while the blue area at the end of S△TD represents the flag tag. D Vaccine candidates based on DNA and VTT vectors were identified by Western blot. Lane 1 represented 293F cell, line 2 represented 293F which transfected DNA-p3.0, line 3 represented 293F which transfected DNA-S1, line 4 represented 293F which transfected DNA-S△TD, line 5 and 7 represented marker, line 6 represented CEFs which infected VTT-S vaccine, line 8 represented CEFs and line 9 represented CEF which infected VTT (752-1). E Immunization strategies of vaccine candidates based on DNA and VTT vectors.
The mice in each group were immunized three times on days 0, 14 and 35. Blood and spleen samples were collected from mice on day 45. Homologous immunization was performed in mice to evaluate the immunogenicity of VTT-S, DNA-S△TD and DNA-S1. Additionally, the DNA-Prime/VTT-Boost immunization strategy involving immunizing mice with two doses of DNA vaccines, followed by booster immunization with the VTT vaccine, was performed as previously described (Liu et al., 2012) to enhance the immune responses of MERS-CoV vaccines. The blank group was injected three times with phosphate buffer saline (PBS) instead of the vaccine. Detailed immunization strategies are shown in Fig. 1E.
3.2. Mice immunized with DNA/VTT vaccines produced strong specific binding and neutralizing antibodies
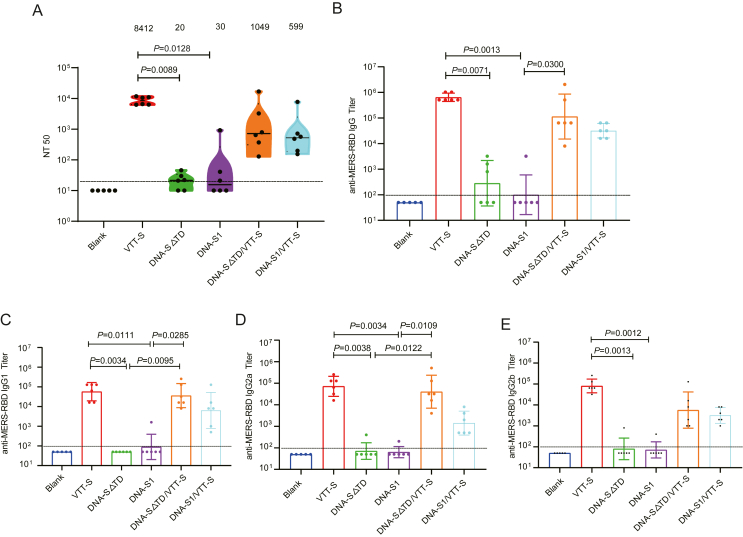
Subsequently, we analyzed the humoral immune responses elicited by MERS-CoV vector vaccines in groups of Babl/c mice immunized using heterologous or homologous immunization strategies. Immunized sera were obtained on day 45, and the neutralizing activity was measured using pseudovirions based on the MERS-CoV EMC/2012 strain. The VTT-S group showed stronger neutralizing responses against EMC/2012 than that of the DNA-S△TD and DNA-S1 groups (P = 0.0089 and P = 0.0128, respectively). Although homologous immunization with DNA vaccines induced a weak neutralizing activity against EMC/2012, boosting with VTT-S enhanced the neutralizing antibody titer against EMC/2012. The neutralizing antibody of the geometric mean titers (GMTs) against EMC/2012 in DNA-S△TD/VTT-S and DNA-S1/VTT-S groups were 1049 and 599, respectively (Fig. 2A). These results revealed that the DNA-Prime/VTT-Boost was an effective vaccination strategy for improving potent humoral immune response in mice.
Fig. 2.
Specific humoral immune responses were elicited by various immunization strategies in mice. A Serum neutralizing activity against MERS-CoV detected by pseudovirus neutralization assay. B Specific MERS-RBD IgG titer was measured by ELISA. C-E Specific MERS-RBD IgG subtype titers were measured by ELISA. Bars represented the GMT ± geometric SD; results were analyzed by Kruskal-Wallis test. Blank group n = 5; VTT, vaccinated groups n = 6.
The specific binding antibody levels against MERS-RBD IgG and IgG subtypes (IgG1, IgG2a and IgG2b) in the immune sera were determined using indirect ELISA. The VTT-S group also exhibited strong binding antibody responses against the MERS-RBD IgG and IgG subtypes. The GTMs against MERS-RBD IgG in the VTT group were significantly higher than those in the DNA-S△TD and DNA-S1 groups (P = 0.0071 and P = 0.0013). A similar antibody response trend was observed in IgG subtypes of the VTT group. The immunized sera of DNA-S△TD and DNA-S1 groups generated weak binding antibodies responses against MERS-RBD IgG, IgG1, IgG2a and IgG2b. The GMTs against MERS-RBD IgG and IgG subtypes were not statistically significant between DNA-S△TD and DNA-S1 groups (Fig. 2B–E). The immunized sera of mice showed high levels of specific binding antibodies against MERS-RBD IgG, IgG1, IgG2a and IgG2b in response to the DNA-Prime/VTT-Boost strategy. The results demonstrated that the specific MERS-RBD IgG and IgG subtype GMTs of DNA-Prime/VTT-Boost groups were improved compared with those of the DNA-S△TD and DNA-S1 groups (Fig. 2B–E).
3.3. Mice immunized with DNA/VTT vaccines generated strong specific T cell immune responses
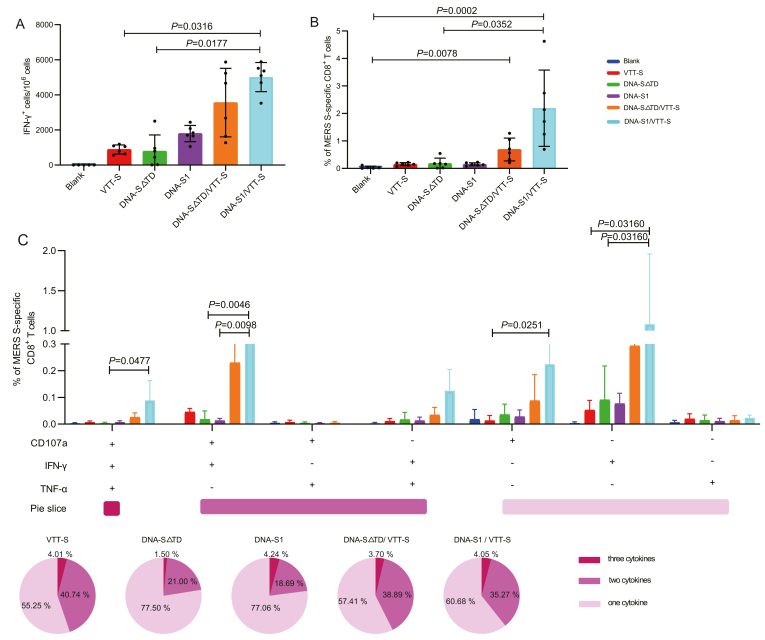
The ELISpot and intracellular cytokine staining (ICS) assays were used to evaluate specific cellular immune responses to MERS-S vector vaccines. The heterologous immunization groups induced a potent cellular immune response compared with the homologous immunization groups. The ELISpot assay showed that all vaccinated groups induced specific interferon (IFN)-γ+ T cells against the MERS-CoV S peptide pool compared to the blank group; moreover, the DNA-S1/VTT-S group elicited higher levels of specific IFN-γ+ T cells against MERS-CoV S than VTT-S (P = 0.0316) and DNA-S△TD (P = 0.0177) groups (Fig. 3A). The VTT-S, DNA-S1, and DNA-S△TD groups induced MERS-S specific CD8+ T cells at relatively low levels. However, the DNA-S1/VTT-S and DNA-S△TD/VTT-S groups elicited high level of MERS-S specific CD8+ T cells. In particular, the frequencies of MERS-S specific CD8+ T cells induced by the DNA-S1/VTT-S groups were statistically higher compared to the blank (P = 0.0002) and DNA-S△TD (P = 0.0352) groups; the frequency of MERS-S specific CD8+ T cells elicited by the DNA-S△TD/VTT-S group was significantly higher than that of the blank group (P = 0.0078) (Fig. 3B). These results demonstrated that the DNA-Prime/VTT-Boost strategy significantly improved the quantity of MERS-S specific IFN-γ+ and the frequencies of MERS-S specific CD8+ T cells compared with those of the homologous immunization strategy.
Fig. 3.
Specific cellular immune responses were elicited by various immunization strategies in mice. A MERS-S specific T cell responses were detected by IFN-γ ELISpot assay. B MERS-S specific CD8+ T cell responses were detected by ICS, the percentage of MERS-S specific CD8+ T cells was sum of CD8+ expressing CD107a and/or producing IFN-γ and/or TNF-a against MERS-S peptide pool in immunized mice. C MERS-S specific polyfunctional CD8+ T cell responses were detected by ICS. Polyfunctional profiles (based on expression of selected markers CD107a, IFN-γ and TNF-a) of total MERS-CoV specific CD8+ T-cell immune responses directed against MERS-S peptide pool. The polyfunctional T cell profiles are shown on the x axis, and the percentages of specific CD8+ T cells for various immunization schemes were shown on the y axis. The pie charts illustrated the percentages of specific CD8+ T cells exhibiting one, two or three markers, which are shown different color. Bars represented the mean ± SD; results were analyzed by Kruskal-Wallis test. Blank group n = 5; VTT, vaccinated groups n = 6.
The quality of the MERS-S specific CD8+ T cell reaction was further characterized by analyzing the types of cytokine production (IFN-γ, and/or TNF-a) and its cytotoxic potential (CD107a) (Fig. 3C). CD8+ T cells expressing CD107a, IFN-γ and TNF-α (CD107a-IFN-γ-TNF-α), CD107a-IFN-γ and IFN-γ-TNF-α were the most induced populations, showing differences among the vaccinated groups. The frequencies of MERS-S specific polyfunctional CD8+ T cell were 44.75%, 22.50% and 22.93% in VTT-S, DNA-S△TD and DNA-S1 groups, respectively, whereas the proportion of polyfunctional CD8+ T cell in DNA-Prime/VTT-Boost groups were 42.59% and 39.32% respectively. The percentage of polyfunctional CD8+ T cells was increased in the DNA-Prime/VTT-Boost groups compared to DNA vaccine groups. The populations of specific polyfunctional CD8+ T cells (CD107a-IFN-γ-TNF-α) in the DNA-S1/VTT-S groups were significantly higher than those in the DNA-S△TD (P = 0.0477) groups. The frequencies of specific polyfunctional CD8+ T cell (CD107a-IFN-γ) in the DNA-S1/VTT-S groups was significantly higher than those in DNA-S△TD (P = 0.0046) and DNA-S1 (P = 0.0098) groups. Additionally, the main specific single cytokine populations in vaccinated groups were those of CD8+ CD107a+ T and CD8+ IFN-γ+ T cells. The frequency of a single cytokine population (CD107a+) in the DNA-S1/VTT-S group was statistically higher than that in the VTT-S group (P = 0.0251). The frequencies of single cytokine population (IFN-γ+) in DNA-S1/VTT-S group was higher than those in VTT-S (P = 0.0316) and DNA-S△TD (P = 0.0316) groups.
3.4. Mice immunized with DNA/VTT vaccines generated specific memory B cells, memory CD8+ T cells and poly-functional CD8+ T effector memory cells
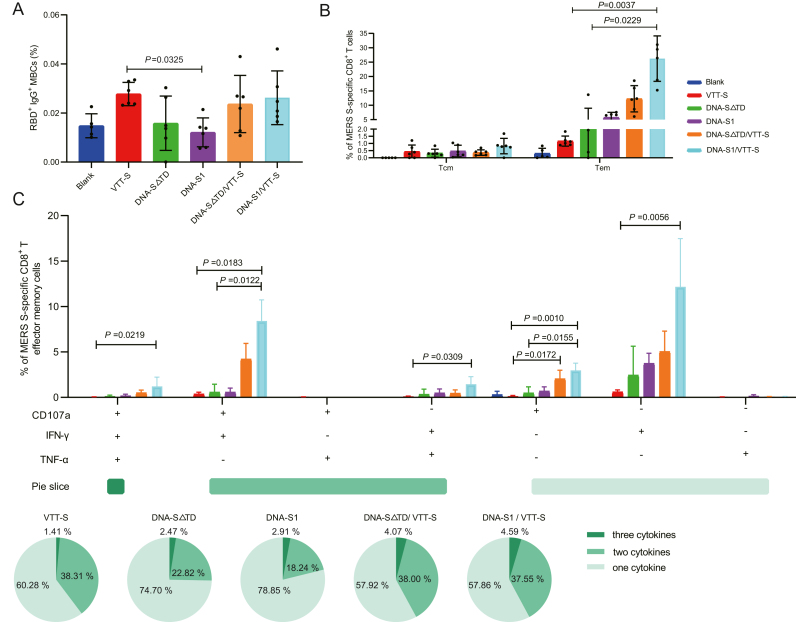
Immunological memory is a significant parameter for successful vaccines. An effective vaccine should induce immunological memory and defend against pathogen reinfections (Quast and Tarlinton, 2021). In our study, both the VTT-S and DNA-Prime/VTT-Boost groups prevailed in inducing memory B cells (MBCs). Particularly, the frequency of MERS-RBD MBCs in the VTT-S group was significantly higher than that of DNA-S1 group (P = 0.0352). The frequency of MBCs was high in DNA-Prime/VTT-Boost groups compared with DNA groups, but there were no significant differences between the DNA-Prime/VTT-Boost groups and DNA groups (Fig. 4A).
Fig. 4.
The characteristic of specific memory B and T cell were elicited by various immunization strategies in mice. A Frequencies of specific memory B cells in mice were analyzed by flow cytometry in mice immunized with various immunization strategies. B Memory phenotypic profiles of the specific MERS-CoV-S CD8+ T cells elicited in mice immunized with various immunization strategies. Percentages of central memory (Tcm) (CD44high and CD62Lhigh) and T effector memory (Tem) (CD44high and CD62Llow) CD8+ T cells specific for MERS-S peptide pool, and expressing any of the markers, CD107a, IFN-g, TNF-a. C MERS-S specific polyfunctional CD8+ Tem cell responses were detected by ICS. Result analysis referenced Fig. 3C. Bars represented the mean ± SD; results were analyzed by Kruskal-Wallis test. Blank group n = 5; VTT, vaccinated groups n = 6.
Subsequently, we distinguished the memory phenotype of MERS-S specific CD8+ T cells via detecting the expression of CD44 and CD62L surface markers. The memory T cell subpopulations included T central memory (Tcm) (CD44highCD62Lhigh) and T effector memory (Tem) (CD44highCD62Llow) (Sahu et al., 2020). The Tem phenotype was higher in the MERS-S specific CD8+ T cell population than in the Tcm in all the vaccinated groups (Fig. 4B). Moreover, the frequencies of specific CD8+ Tem phenotype cells were higher in the DNA-Prime/VTT-Boost groups than those in the homologous immunization group. In particular, the frequencies of Tem in the DNA-S1/VTT-S group were significantly higher than those in the VTT-S (P = 0.0037) and DNA-S△TD (P = 0.0229) groups.
To further broaden the research on T cells immune responses, we measured MERS-S specific CD8+ Tem cells elicited by vaccinated groups stimulated ex vivo with the MERS-S peptide pool. The main specific polyfunctional MERS-S CD8+ Tem were the cell populations expressing CD107a-IFN-γ-TNF-α, CD107a-IFN-γ and IFN-γ-TNF-α. The frequencies of polyfunctional MERS-S specific CD8+ Tem were similar in the DNA-S△TD/VTT-S and DNA-S1/VTT-S groups. The frequency of polyfunctional MERS-S specific CD8+ Tem cells was 39.72% in VTT-S group, which was higher than DNA-S△TD and DNA-S1 groups. Similarly, compared with the DNA vaccine groups, the DNA-Prime/VTT-Boost groups also induced a higher proportion of polyfunctional MERS-S specific CD8+ Tem cells. DNA-Prime/VTT-Boost immunization was a more effective strategy for inducing MERS-S specific CD8+ Tem and polyfunctional CD8+ Tem compared with homologous immunization (Fig. 4C).
3.5. Mice immunized with DNA/VTT vaccines generated strong and durable cellular immune responses against MERS-CoV
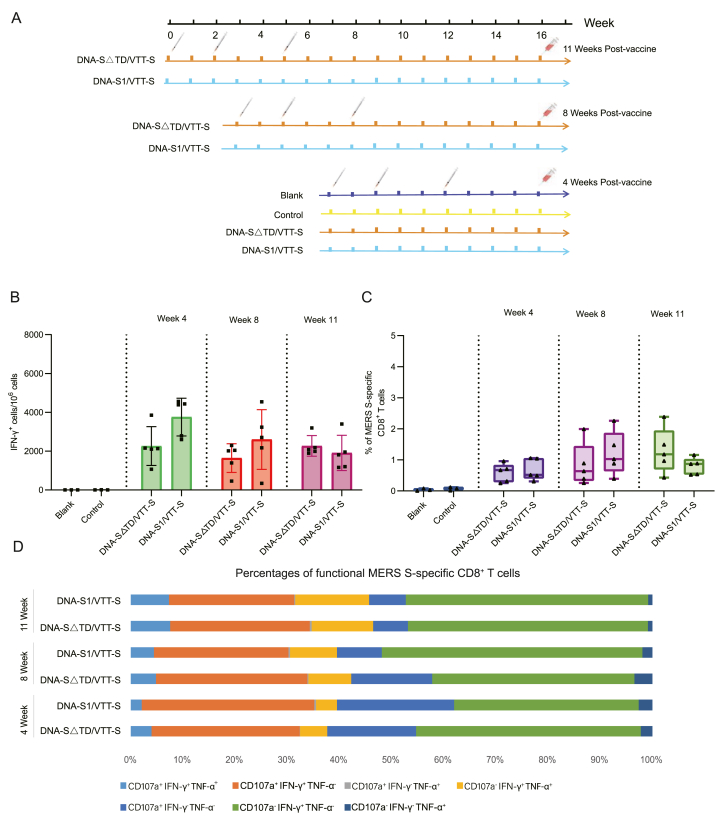
Both the DNA-S△TD/VTT-S and DNA-S1/VTT-S groups presented durable cellular immune responses from 4 to 11 weeks post-vaccination (Fig. 5A and B). No significant differences were observed between the two vaccinated groups at any of the three time points. The specific CD8+ T-cell population was an important parameter for assessing vaccine protection. We observed that the DNA-S△TD/VTT-S and DNA-S1/VTT-S groups could elicit MERS-S specific CD8+ T-cell 4-, 8-, and 11-weeks post-vaccination (Fig. 5C). Furthermore, we evaluated the specific CD8+ T-cell functionality using the intracellular cytokine staining (ICS) assay; the frequencies and populations of specific polyfunctional CD8+ T-cells were similar in the DNA-S△TD/VTT-S and DNA-S1/VTT-S groups (Fig. 5D). Cellular immune responses induced by the DNA-S△TD/VTT-S and DNA-S1/VTT-S groups were strong and durable for almost three months.
Fig. 5.
MERS-S specific cellular immune responses in mice with DNA-Prime/VTT-Boost strategy at different time points. A The scheme of DNA-Prime/VTT-Boost strategy. B MERS-S specific IFN-γ+ T cells were measured by ELISpot at 4-, 8-, and 11-weeks post-vaccination. C MERS-S specific CD8+ T cells were measured by ICS at 4-, 8-, and 11-weeks post-vaccination. Result analysis referenced Fig. 3B. D The percentages of MERS-S specific functional CD8+ T cells were detected by ICS. Result analysis referenced Fig. 3C. Bars represented the mean ± SD.
3.6. Mice immunized with DNA/VTT vaccines elicited strong and durable neutralizing activity against MERS-CoV
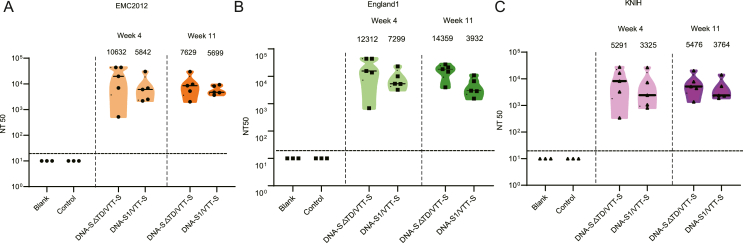
In the DNA-S△TD/VTT-S and DNA-S1/VTT-S groups, the GMTs of neutralizing antibodies against hCoV-EMC2012 reached 10632 and 5842, respectively four weeks post-vaccination; however, the GMTs of the two groups showed no statistical differences. The results showed that both DNA-S△TD/VTT-S and DNA-S1/VTT-S groups elicited robust neutralizing activity and persistence against EMC2012, England1 and KNIH pseudoviruses; the neutralizing antibody GMTs in the DNA-S△TD/VTT-S and DNA-S1/VTT-S groups of the three pseudoviruses showed no significant differences 4-, and 11-weeks post-vaccination (Fig. 6A–C).
Fig. 6.
The sera of neutralizing antibody titers in immunized mice at different time points. A Neutralizing antibody titers against EMC2012 at 4-, and 11-weeks after the last immunization. B Neutralizing antibody titers against England1 at 4-, and 11-weeks after the last immunization. C Neutralizing antibody titers against KNIH at 4 and 11 weeks after the last immunization. Bars represented the GMT ± geometric SD.
4. Discussion
The development of safe and effective vaccines to control the spread of epidemics is an important strategy to fight emerging infectious diseases. Various MERS-CoV vaccine candidates have been developed for camels and human applications (Li et al., 2020; Chi et al., 2022). Effective camelid vaccines could decrease the MERS-CoV epidemic among camel families, thereby reducing the risk of infection in humans who have close contact with these animals. The development of human vaccines requires extremely stringent constraints and standards compared to veterinary vaccines. An effective vaccine against MERS-CoV could provide direct protection to humans. In this study, we successfully developed two promising vaccine candidates against MERS-CoV, utilizing both DNA and viral vector technological platforms, with the ultimate objective of facilitating their application in humans.
Viral vectors (including vaccinia vector) have been used in several vaccines (Ebola, human immunodeficiency virus (HIV) and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and others). Mice immunized with viral vector vaccine showed strong neutralization activity, as well as CD8+ T cell responses (Xie et al., 2019; Routhu et al., 2021; Mei et al., 2022). Here, we reported that three doses of VTT-S vaccine immunization alone could induce a robust humoral immune response but a moderate cellular immune response to MERS-CoV. Similar predominantly humoral responses over cellular responses have also been seen in several clinical trials with repeated immunization with viral vector vaccines, including adenovirus and vaccinia vector. Although the antibodies rise constantly with each viral vector vaccine boosts in volunteers, their cellular immune responses stagnated without further increase (Pantaleo et al., 2019; Flaxman et al., 2021; Stieh et al., 2023). There might be different dynamics in the humoral and cellular responses of the anti-vector immunity that will compromise the designated vaccine induced humoral or cellular responses at different levels. A stronger cellular than humoral anti-vector immune response may result in a weaker vaccine-induced cellular response than vaccine-induced humoral response.
DNA vaccines have the advantages of quick construction, easy-to-manufacture and temperature stability. One of the disadvantages of DNA vaccines is inefficient delivery and expression of target cells, which results in low immunogenicity when used alone (Li and Petrovsky, 2016; Porter and Raviprakash, 2017). In our study, we removed the TD of S gene to increase its soluble expression and released it extracellularly. Meanwhile, the DNA vaccines were immunized using electroporation to improve the delivery efficiency of plasmids. Nonetheless, three doses of DNA-based MERS-CoV S or S1 vaccines elicited a slight humoral and moderate cellular immune response. Genetic modification can be used in vaccine design to enhance the immunogenicity of vaccines; for example, C-terminal fusion with the foldon induces stable rabies virus glycoprotein (RVG), and the fibritin foldon maintains the native antigenic structure of the carboxy part of RVG ectodomain (Sissoëff et al., 2005). Additionally, optimizing the vector and adding adjuvants can enhance the immune response of immunogens (Smith et al., 2020; Hayashi et al., 2022; Song et al., 2023), all of which could be incorporated into future research.
DNA vaccines used as priming vaccine could significantly boost the immune responses of the viral vector vaccines afterwards. A DNA-Prime/VTT-Boost immunization strategy was used in research on HIV-1 vaccines and resulted in a robust potency in inducing balanced humoral and cellular immune responses (Liu et al., 2013, 2015). In this study, regardless of whether MERS DNA-S△TD or DNA-S1 was used as the prime vaccine, both groups induce strong humoral and cellular immune responses after boosted by MERS VTT-S vaccine. Additionally, the heterologous immunization strategy elicited higher levels of Tem than that from homologous immunization with DNA or VTT vaccines. This reconfirmed that sequential DNA-Prime/VTT-Boost was a superior immunization strategy.
DNA vaccine priming with S△TD generated a trend of stronger neutralizing antibody than that of DNA-S1 vaccine, while DNA-S1 priming elicited trend of stronger cellular immune responses. But these differences are not statistically significant. There are multiple factors affecting the immune responses induced by DNA vaccines. DNA-S△TD retains more immune epitopes than S1, especially the ability to form protein trimer, which is beneficial to neutralizing antibodies induction. This may explain why DNA-S△TD generated stronger neutralizing antibodies than DNA-S1. Cellular immunity does not rely on conformational epitopes, which do not compromise the ability for DNA-S1 to generate stronger T cell responses. The study by Cho et al. (2021) showed that DNA-MERS-S1 vaccine elicited higher antibody and cellular immune responses than that by MERS-S, which is partially consistent with our study. In comparison with S and S1 based COVID-19 DNA vaccines, Cho et al. showed that DNA-S vaccine elicited stronger neutralizing antibodies, while DNA-S1 vaccine induced a higher cellular immune response. These results are similar with our results for MERS DNA vaccine constructs. The actual mechanism of humoral and cellular immune responses induced by coronavirus DNA vaccines requires detailed studies in the future.
The immune response will wane to varying degrees with prolonged vaccine immunization. A low level of specific neutralizing antibody titer correlates with a breakthrough infection of betacoronavirus SARS-CoV-2 (Bergwerk et al., 2021). The maintenance of immune responses after immunization is an important indicator. Therefore, we immunized mice with the DNA-Prime/VTT-Boost scheme and monitored the humoral and cellular immune responses for almost three months. Both immunized groups maintained persistent and robust immune responses. The DNA-Prime/VTT-Boost groups produced strong, long lasting and broader neutralizing antibodies against the MERS-CoV variants.
Our research has several limitations. We analyzed the characteristics of specific CD8+ T cell immune responses without measuring the population of regulatory T cells. Thus, the cellular immune response is not comprehensively understood in this study. Moreover, the immune sera were not conducted using the plaque reduction neutralization test (PRNT) with authentic virus against MERS-CoV because limited volume of sera was collected in experimental mice. Lastly, the results were based on a small sample size and necessitate further investigation.
5. Conclusion
The DNA-Prime/replicating VTT-Boost is a good immunization strategy to simulate strong and balanced humoral and cellular specific immune responses against MERS-CoV. Such a heterogeneous Prime/Boost immunization strategy could avoid the anti-vector immunity in repeated immunization with viral vector vaccines. More importantly, this immunization strategy could induce a broader spectrum of neutralizing antibodies and poly-functional T cells responses, advantageous for controlling various MERS-CoV variants. By using the replicating viral vector boost, the induced specific immune responses are durable. In summary, the study demonstrated that the DNA and VTT vector vaccines, alongside a heterologous Prime-Boost immunization strategy, are promising vaccine candidates warranting further development for MERS-CoV vaccines.
Data availability
All the data generated during the current study are included in the manuscript.
Ethics statement
All the animal experimental programs were carried out in accordance with the guidelines of animal welfare and the research were approved by the Institutional Animal Care and Use Committee (IACUC) of China CDC (Approval number: 2022-CCDC-Appl-007).
Author contributions
Xiuli Shen: conceptualization, data curation, formal analysis, investigation, methodology, visualization, writing-original draft, writing review & editing. Shuhui Wang: conceptualization, data curation, investigation, methodology, visualization, writing-original draft. Yanling Hao: investigation, methodology, visualization. Yuyu Fu: data curation, investigation, methodology. Li Ren, Dan Li, Wenqi Tang, Jing Li, Meiling Zhu, Shuo Wang and Ran Chen: methodology, visualization. Ying Liu: conceptualization, funding acquisition, project administration, supervision, visualization, writing-review & editing. Yiming Shao: study design and conceptualization, data curation, formal analysis, funding acquisition, project administration, supervision, visualization, writing-review & editing.
Conflict of interest
Prof. Yiming Shao is an editorial board member for Virologica Sinica and was not involved in the editorial review or the decision to publish this article. All authors declare that there are no competing interests.
Acknowledgements
We thank the Chinese Center for Disease Control and Prevention Laboratory Animal Center for their assistance in animal experiments, especially, we are grateful for Prof. Xuancheng Lu's assistance in animal experiments. The work was financially supported by National Nature Science Foundation of China (U20A20362) and the Subject of SKLID (2020SKLID102). This research is partially supported by Changping Laboratory.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2024.05.005.
Contributor Information
Ying Liu, Email: yingliu@chinaaids.cn.
Yiming Shao, Email: yshao@bjmu.edu.cn, shaoyiming@cpl.ac.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Alharbi N.K., Padron-Regalado E., Thompson C.P., Kupke A., Wells D., Sloan M.A., Grehan K., Temperton N., Lambe T., Warimwe G., Becker S., Hill A.V.S., Gilbert S.C. ChAdOx1 and MVA based vaccine candidates against MERS-CoV elicit neutralising antibodies and cellular immune responses in mice. Vaccine. 2017;35:3780–3788. doi: 10.1016/j.vaccine.2017.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergwerk M., Gonen T., Lustig Y., Amit S., Lipsitch M., Cohen C., Mandelboim M., Levin E.G., Rubin C., Indenbaum V., Tal I., Zavitan M., Zuckerman N., Bar-Chaim A., Kreiss Y., Regev-Yochay G. Covid-19 breakthrough infections in vaccinated health care workers. N. Engl. J. Med. 2021;385:1474–1484. doi: 10.1056/NEJMoa2109072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosaeed M., Balkhy H.H., Almaziad S., Aljami H.A., Alhatmi H., Alanazi H., Alahmadi M., Jawhary A., Alenazi M.W., Almasoud A., Alanazi R., Bittaye M., Aboagye J., Albaalharith N., Batawi S., Folegatti P., Ramos Lopez F., Ewer K., Almoaikel K., Aljeraisy M., Alothman A., Gilbert S.C., Khalaf Alharbi N. Safety and immunogenicity of ChAdOx1 MERS vaccine candidate in healthy Middle Eastern adults (MERS002): an open-label, non-randomised, dose-escalation, phase 1b trial. Lancet Microbe. 2022;3:e11–e20. doi: 10.1016/S2666-5247(21)00193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancro M.P., Tomayko M.M. Memory B cells and plasma cells: the differentiative continuum of humoral immunity. Immunol. Rev. 2021;303:72–82. doi: 10.1111/imr.13016. [DOI] [PubMed] [Google Scholar]
- Chi H., Zheng X., Wang X., Wang C., Wang H., Gai W., Perlman S., Yang S., Zhao J., Xia X. DNA vaccine encoding Middle East respiratory syndrome coronavirus S1 protein induces protective immune responses in mice. Vaccine. 2017;35:2069–2075. doi: 10.1016/j.vaccine.2017.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H., Wang Y., Li E., Wang X., Wang H., Jin H., Han Q., Wang Z., Wang X., Zhu A., Sun J., Zhuang Z., Zhang L., Ye J., Wang H., Feng N., Hu M., Gao Y., Zhao J., Zhao Y., Yang S., Xia X. Inactivated rabies virus vectored MERS-Coronavirus vaccine induces protective immunity in mice, camels, and alpacas. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.823949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Jang Y., Park K.-H., Choi H., Nowakowska A., Lee H.-J., Kim M., Kang M.-H., Kim J.-H., Shin H.Y., Oh Y.-K., Kim Y.B. Human endogenous retrovirus-enveloped baculoviral DNA vaccines against MERS-CoV and SARS-CoV2. NPJ Vaccines. 2021;6:37. doi: 10.1038/s41541-021-00303-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.A., Goo J., Yang E., Jung D.I., Lee S., Rho S., Jeong Y., Park Y.S., Park H., Moon Y.H., Park U., Seo S.H., Lee H., Lee J.M., Cho N.H., Song M., Kim J.O. Cross-protection against MERS-CoV by prime-boost vaccination using viral spike DNA and protein. J. Virol. 2020;94 doi: 10.1128/JVI.01176-20. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edridge A.W.D., Kaczorowska J., Hoste A.C.R., Bakker M., Klein M., Loens K., Jebbink M.F., Matser A., Kinsella C.M., Rueda P., Ieven M., Goossens H., Prins M., Sastre P., Deijs M., van der Hoek L. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- Flaxman A., Marchevsky N.G., Jenkin D., Aboagye J., Aley P.K., Angus B., Belij-Rammerstorfer S., Bibi S., Bittaye M., Cappuccini F., et al. Reactogenicity and immunogenicity after a late second dose or a third dose of ChAdOx1 nCoV-19 in the UK: a substudy of two randomised controlled trials (COV001 and COV002) Lancet. 2021;398:981–990. doi: 10.1016/S0140-6736(21)01699-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Arriaza J., Garaigorta U., Perez P., Lazaro-Frias A., Zamora C., Gastaminza P., Del Fresno C., Casasnovas J.M., Sorzano C.O.S., Sancho D., Esteban M. COVID-19 vaccine candidates based on modified vaccinia virus Ankara expressing the SARS-CoV-2 spike protein induce robust T- and B-cell immune responses and full efficacy in mice. J. Virol. 2021;95 doi: 10.1128/JVI.02260-20. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem A.M., Algaissi A., Agrawal A.S., Al-Amri S.S., Alhabbab R.Y., Sohrab S.S., A S.A., Alharbi N.K., Peng B.H., Russell M., Li X., Tseng C.K. A highly immunogenic, protective, and safe adenovirus-based vaccine expressing Middle East respiratory syndrome coronavirus S1-CD40L fusion protein in a transgenic human dipeptidyl peptidase 4 mouse model. J. Infect. Dis. 2019;220:1558–1567. doi: 10.1093/infdis/jiz137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H., Sun J., Yanagida Y., Otera T., Kubota-Koketsu R., Shioda T., Ono C., Matsuura Y., Arase H., Yoshida S., Nakamaru R., Ju N., Ide R., Tenma A., Kawabata S., Ehara T., Sakaguchi M., Tomioka H., Shimamura M., Okamoto S., Amaishi Y., Chono H., Mineno J., Komatsuno T., Saito Y., Rakugi H., Morishita R., Nakagami H. Preclinical study of a DNA vaccine targeting SARS-CoV-2. Current Research in Translational Medicine. 2022;70 doi: 10.1016/j.retram.2022.103348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnasko T.S., Hnasko R.M. The western blot. Methods Mol. Biol. 2015;1318:87–96. doi: 10.1007/978-1-4939-2742-5_9. [DOI] [PubMed] [Google Scholar]
- Kim N., Lee T.Y., Lee H., Yang J.S., Kim K.C., Lee J.Y., Kim H.J. Comparing the immunogenicity and protective effects of three MERS-CoV inactivation methods in mice. Vaccines. 2022;10:1843. doi: 10.3390/vaccines10111843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch T., Dahlke C., Fathi A., Kupke A., Krahling V., Okba N.M.A., Halwe S., Rohde C., Eickmann M., Volz A., Hesterkamp T., Jambrecina A., Borregaard S., Ly M.L., Zinser M.E., Bartels E., Poetsch J.S.H., Neumann R., Fux R., Schmiedel S., Lohse A.W., Haagmans B.L., Sutter G., Becker S., Addo M.M. Safety and immunogenicity of a modified vaccinia virus Ankara vector vaccine candidate for Middle East respiratory syndrome: an open-label, phase 1 trial. Lancet Infect. Dis. 2020;20:827–838. doi: 10.1016/S1473-3099(20)30248-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laczkó D., Hogan M.J., Toulmin S.A., Hicks P., Lederer K., Gaudette B.T., Castaño D., Amanat F., Muramatsu H., Oguin T.H., et al. A single immunization with nucleoside-modified mRNA vaccines elicits strong cellular and humoral immune responses against SARS-CoV-2 in mice. Immunity. 2020;53:724–732.e7. doi: 10.1016/j.immuni.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Li L., Petrovsky N. Molecular mechanisms for enhanced DNA vaccine immunogenicity. Expert Rev. Vaccines. 2016;15:313–329. doi: 10.1586/14760584.2016.1124762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.D., Chi W.Y., Su J.H., Ferrall L., Hung C.F., Wu T.C. Coronavirus vaccine development: from SARS and MERS to COVID-19. J. Biomed. Sci. 2020;27:104. doi: 10.1186/s12929-020-00695-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M., Grad Y.H., Sette A., Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat. Rev. Immunol. 2020;20:709–713. doi: 10.1038/s41577-020-00460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Qiu C., Huang Y., Xu J., Shao Y. Potent T cell responses induced by single DNA vaccine boosted with recombinant vaccinia vaccine. Virol. Sin. 2013;28:109–115. doi: 10.1007/s12250-013-3303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Hao Y., Luo Z., Huang Y., Hu X., Liu Y., Shao Y. Broad HIV-1 neutralizing antibody response induced by heterologous gp140/gp145 DNA prime-vaccinia boost immunization. Vaccine. 2012;30:4135–4143. doi: 10.1016/j.vaccine.2012.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Li Y., Luo Z., Yang G., Liu Y., Liu Y., Sun M., Dai J., Li Q., Qin C., Shao Y. HIV-1 vaccines based on replication-competent Tiantan vaccinia protected Chinese rhesus macaques from simian HIV infection. AIDS. 2015;29:649–658. doi: 10.1097/QAD.0000000000000595. [DOI] [PubMed] [Google Scholar]
- Mei S., Fan Z., Liu X., Zhao F., Huang Y., Wei L., Hu Y., Xie Y., Wang L., Ai B., Liang C., Xu F., Guo F. Immunogenicity of a vaccinia virus-based severe acute respiratory syndrome coronavirus 2 vaccine candidate. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.911164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modjarrad K., Roberts C.C., Mills K.T., Castellano A.R., Paolino K., Muthumani K., Reuschel E.L., Robb M.L., Racine T., Oh M-d, et al. Safety and immunogenicity of an anti-Middle East respiratory syndrome coronavirus DNA vaccine: a phase 1, open-label, single-arm, dose-escalation trial. Lancet Infect. Dis. 2019;19:1013–1022. doi: 10.1016/S1473-3099(19)30266-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Fan C., Huang W., Xu M., Wang Y. Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay. Nat. Protoc. 2020;15:3699–3715. doi: 10.1038/s41596-020-0394-5. [DOI] [PubMed] [Google Scholar]
- Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., Kong W.P., Andres E.L., Kettenbach A.N., Denison M.R., Chappell J.D., Graham B.S., Ward A.B., McLellan J.S. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaleo G., Janes H., Karuna S., Grant S., Ouedraogo G.L., Allen M., Tomaras G.D., Frahm N., Montefiori D.C., Ferrari G., Ding S., Lee C., Robb M.L., Esteban M., Wagner R., Bart P.-A., Rettby N., McElrath M.J., Gilbert P.B., Kublin J.G., Corey L. Safety and immunogenicity of a multivalent HIV vaccine comprising envelope protein with either DNA or NYVAC vectors (HVTN 096): a phase 1b, double-blind, placebo-controlled trial. The Lancet HIV. 2019;6:e737–e749. doi: 10.1016/S2352-3018(19)30262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paules C.I., Marston H.D., Fauci A.S. Coronavirus infections-more than just the common cold. JAMA. 2020;323:707–708. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- Porter K.R., Raviprakash K. DNA vaccine delivery and improved immunogenicity. Curr. Issues Mol. Biol. 2017;22:129–138. doi: 10.21775/cimb.022.129. [DOI] [PubMed] [Google Scholar]
- Quast I., Tarlinton D. B cell memory: understanding COVID-19. Immunity. 2021;54:205–210. doi: 10.1016/j.immuni.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello V., Coleman C.M., Frieman M.B. Emergence of the Middle East respiratory syndrome coronavirus. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodon J., Okba N.M.A., Te N., van Dieren B., Bosch B.J., Bensaid A., Segales J., Haagmans B.L., Vergara-Alert J. Blocking transmission of Middle East respiratory syndrome coronavirus (MERS-CoV) in llamas by vaccination with a recombinant spike protein. Emerg. Microb. Infect. 2019;8:1593–1603. doi: 10.1080/22221751.2019.1685912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routhu N.K., Cheedarla N., Gangadhara S., Bollimpelli V.S., Boddapati A.K., Shiferaw A., Rahman S.A., Sahoo A., Edara V.V., Lai L., et al. A modified vaccinia Ankara vector-based vaccine protects macaques from SARS-CoV-2 infection, immune pathology, and dysfunction in the lungs. Immunity. 2021;54:542–556.e9. doi: 10.1016/j.immuni.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu R., Dixit S., Verma R., Duncan S.A., Coats M.T., Giambartolomei G.H., Singh S.R., Dennis V.A. A nanovaccine formulation of Chlamydia recombinant MOMP encapsulated in PLGA 85:15 nanoparticles augments CD4+ effector (CD44high CD62Llow) and memory (CD44high CD62Lhigh) T-cells in immunized mice. Nanomedicine. 2020;29 doi: 10.1016/j.nano.2020.102257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson A.T., Heeney J., Cantoni D., Ferrari M., Sans M.S., George C., Di Genova C., Mayora Neto M., Einhauser S., Asbach B., Wagner R., Baxendale H., Temperton N., Carnell G. Coronavirus pseudotypes for all circulating human coronaviruses for quantification of cross-neutralizing antibody responses. Viruses. 2021;13:1579. doi: 10.3390/v13081579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senga M., Arabi Y.M., Fowler R.A. Clinical spectrum of the Middle East respiratory syndrome coronavirus (MERS-CoV) J Infect Public Health. 2017;10:191–194. doi: 10.1016/j.jiph.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sissoëff L., Mousli M., England P., Tuffereau C. Stable trimerization of recombinant rabies virus glycoprotein ectodomain is required for interaction with the p75NTR receptor. J. Gen. Virol. 2005;86:2543–2552. doi: 10.1099/vir.0.81063-0. [DOI] [PubMed] [Google Scholar]
- Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., Gary E.N., Walker S.N., Schultheis K., Purwar M., et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Wang Q., Wen Y., Tan R., Cui Y., Xiong D., Jiao X., Pan Z., Rajao D.S. Enhanced immunogenicity elicited by a novel DNA vaccine encoding the SARS-CoV-2 S1 protein fused to the optimized flagellin of Salmonella typhimurium in mice. Microbiol. Spectr. 2023;11 doi: 10.1128/spectrum.02549-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieh D.J., Barouch D.H., Comeaux C., Sarnecki M., Stephenson K.E., Walsh S.R., Sawant S., Heptinstall J., Tomaras G.D., Kublin J.G., et al. Safety and immunogenicity of Ad26-vectored HIV vaccine with mosaic immunogens and a novel mosaic envelope protein in HIV-uninfected adults: a phase 1/2a study. J. Infect. Dis. 2023;227:939–950. doi: 10.1093/infdis/jiac445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Quan Y., Xin Z.T., Wrammert J., Ma M.J., Lv H., Wang T.B., Yang H., Richardus J.H., Liu W., Cao W.C. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- Volz A., Kupke A., Song F., Jany S., Fux R., Shams-Eldin H., Schmidt J., Becker C., Eickmann M., Becker S., Sutter G. Protective efficacy of recombinant modified vaccinia virus Ankara delivering Middle East respiratory syndrome coronavirus spike glycoprotein. J. Virol. 2015;89:8651–8656. doi: 10.1128/JVI.00614-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Qi J., Yuan Y., Xuan Y., Han P., Wan Y., Ji W., Li Y., Wu Y., Wang J., Iwamoto A., Woo P.C., Yuen K.Y., Yan J., Lu G., Gao G.F. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Middle East respiratory syndrome coronavirus – Oman. 2023. https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON436
- Xie L., Zai J., Yi K., Li Y. Intranasal immunization with recombinant Vaccinia virus Tiantan harboring Zaire Ebola virus gp elicited systemic and mucosal neutralizing antibody in mice. Vaccine. 2019;37:3335–3342. doi: 10.1016/j.vaccine.2019.04.070. [DOI] [PubMed] [Google Scholar]
- Xiong Q., Cao L., Ma C., Tortorici M.A., Liu C., Si J., Liu P., Gu M., Walls A.C., Wang C., Shi L., Tong F., Huang M., Li J., Zhao C., Shen C., Chen Y., Zhao H., Lan K., Corti D., Veesler D., Wang X., Yan H. Close relatives of MERS-CoV in bats use ACE2 as their functional receptors. Nature. 2022;612:748–757. doi: 10.1038/s41586-022-05513-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data generated during the current study are included in the manuscript.