Abstract
Background:
CGD is caused by defects in any of the 6 subunits forming the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase complex 2 (NOX2), leading to severely reduced or absent phagocyte-derived ROS production. Almost 50% of patients with chronic granulomatous disease (CGD) have IBD (CGD-IBD). While conventional IBD therapies can treat CGD-IBD, their benefits must be weighed against the risk of infection. Understanding the impact of NOX2 defects on the intestinal microbiota may lead to the identification of novel CGD-IBD treatments.
Objective:
To identify microbiome and metabolome signatures that can distinguish patients with CGD and CGD-IBD.
Methods:
We conducted a cross-sectional, observational study of 79 patients with CGD, 8 pathogenic variant carriers and 19 healthy controls followed at the National Institutes of Health Clinical Center (NIH CC). We profiled the intestinal microbiome (amplicon sequencing) and stool metabolome, and validated our findings in a second cohort of 36 CGD patients recruited through the Primary Immune Deficiency Treatment Consortium (PIDTC).
Results:
We identified distinct intestinal microbiome and metabolome profiles in patients with CGD compared to healthy individuals. We observed enrichment for Erysipelatoclostridium spp., Sellimonas spp. and Lachnoclostridium spp. in CGD stool samples. Despite differences in bacterial alpha and beta diversity between the NIH CC and PIDTC cohorts, several taxa correlated significantly between both cohorts. We further demonstrated that patients with CGD-IBD have a distinct microbiome and metabolome profile compared to patients without CGD-IBD.
Conclusion:
Intestinal microbiome and metabolome signatures distinguished patients with CGD and CGD-IBD, and identified potential biomarkers and therapeutic targets.
Keywords: Chronic granulomatous disease, CGD, inflammatory bowel disease, IBD, dysbiosis, microbiome, metabolome, intestinal inflammation, NADPH oxidase, inborn errors of immunity, primary immune deficiency
CAPSULE SUMMARY
Clinical markers of conventional IBD disease activity may be applicable to patients with CGD-associated IBD. The intestinal microbiome and metabolome distinguish patients with CGD and CGD-associated IBD leading to the identification of potential therapeutic targets for CGD-associated IBD.
INTRODUCTION
Reactive oxygen species (ROS) are involved in inflammatory responses 1. Genetic variants reducing ROS production are associated with inflammatory bowel disease (IBD), including very early onset IBD 2. Notably, 40–50% of patients with chronic granulomatous disease (CGD) have IBD 3, 4. CGD is caused by inherited defects in any of the 6 subunits (gp91phox, p47phox, p22phox, p67phox, p40phox) forming the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase complex 2 (NOX2) or supporting its assembly (essential for ROS - EROS). Defects in NOX2 lead to reduced or absent phagocyte-derived ROS production 5.
Clinically, CGD is characterized by recurrent infections and inflammation notably in the gut 6. CGD-associated IBD (CGD-IBD) can appear at any age, and its prevalence and severity are independent of genotype or residual ROS 7. Although the majority have colitis, Crohn-like manifestations are also present 8, 9. Besides allogeneic hematopoietic cell transplantation (HCT) 10, treatment of CGD-IBD is limited to standard therapies including 5-aminosalicylates, corticosteroids, immunomodulators (azathioprine, thalidomide) 11, 12 and biologics such as anti-tumor necrosis factor agents (infliximab) 13, anti-interleukin (IL)-12/23 antibodies (ustekinumab) 14 IL-1R antagonists (anakinra) 15 or anti-alpha-4-beta-7 integrin antibodies (vedolizumab) 16. Long-term use of these agents is associated with side-effects, and their benefits must be weighed against the risk of infectious complications in this immunocompromised population 3, 13. The identification of alternative treatment options, including the modulation of the intestinal microbiota, is hence imperative.
Limited treatment options for CGD-IBD are due to incomplete understanding of the role of ROS at the intestinal mucosal barrier 17. Mucosal barrier compromise leads to the recruitment of intestinal phagocytes, where NOX2-derived ROS contribute to antimicrobial defense, and hypoxic environment that supports tissue repair 18. However, tightly controlled ROS production by non-hematopoietic cells highlight the significance of low physiological ROS levels in signaling pathways and in modulating the microbiota for mucosal barrier repair 19. The intestinal microbiota also contributes to mucosal ROS levels 20. Several Lactobacillus strains secrete H2O2 and protect the intestinal barrier 21. Thus, the interplay between the microbiota, ROS and immunological homeostasis at the mucosal barrier is a key consideration in CGD-IBD.
We have previously demonstrated that the microbiota at birth plays an important role in mediating colitis susceptibility in CGD mice 22. Studies examining the microbiota in patients with NOX2 defects are limited and include small cohorts (n≤10) 23, 24. Based on these data, and the well-accepted role of the microbiota in modulating immune responses in conventional IBD 25, we hypothesize that patients with NOX2 defects, even without intestinal inflammation, have qualitatively and functionally distinct intestinal microbiome and metabolome signatures resulting from the impact of the immune defect on the mucosal milieu. We present the largest study published to date profiling the intestinal microbiome and metabolome in patients with NOX2 defects, including patients with CGD from 2 distinct cohorts and heterozygous carriers of NOX2 defects.
METHODS
Study design and population
In this cross-sectional observational study, patients were recruited from the National Institutes of Health Clinical Center (NIH CC; n=106; primary cohort) and Primary Immune Deficiency Treatment Consortium (PIDTC; n=36; validation cohort) between August 2012 and November 2018. All patients providing consent for their stool sample were included and each cohort was analysed separately.
NIH CC cohort
Healthy individuals (Healthy; n=17), healthy individuals on trimethoprim-sulfamethoxazole (TMP-SMX) prophylaxis for recurrent urinary tract infections (Healthy PPX; n=2) and patients with NOX2 defects including CGD (n=79), heterozygous carriers of gp91phox (CYBB) defects (Carriers; n=6), and digenic heterozygous carriers of DUOX2 (a NOX2 homologue associated with IBD 26) and p67phox (NCF2) defects (DUOX2-NCF2; n=2), were enrolled into the National Institute of Allergy and Infectious Diseases (NIAID) institutional review board (IRB)-approved protocol 93-I-0119 – Detection and characterization of host defense defects (Clinicaltrials.gov, NCT00001355), a prospective longitudinal observational cohort study.
PIDTC cohort
We included a validation cohort (n=36) of CGD patients recruited from 11 centers across the USA and Canada into the PIDTC Protocol 6903 (Clinicaltrials.gov, NCT02082353), a retrospective, cross-sectional and prospective observational study evaluating patients before, during and after HCT 27. The protocol was IRB-approved at each participating center. The PIDTC cohort was included to validate the findings of the NIH CC cohort and identify microbial biomarkers that are consistent among CGD patients. Only samples and clinical metadata collected during the pre-HCT (baseline) visit were included.
Study procedures
Stool samples were self-collected in sterile dry tubes and immediately stored at −80 °C 28. PIDTC stools were also immediately frozen upon collection and shipped overnight on dry ice. For NIH CC patients, complete blood count (CBC), C-reactive protein (CRP), erythrocyte sedimentation rate (ESR) and albumin measurements were performed. All participants completed a self-administered questionnaire which included the Numeric Rating Scale (NRS) 29, the Short Quality of Life IBD Questionnaire (SIBDQ) 30 and the Patient-modified Simple Clinical Colitis Activity Index (P-SCCAI) 31. A Clinical Activity Index (CAI) was assigned based on the following criteria: CAI 1= 0–2 bowel movements (BM)/day, CAI 2= 2–4 BMs/day, CAI 3= > 4 BMs/day and/or presence of blood or mucus in stool and/or fistulae and/or perianal disease. All fecal samples were evaluated for the presence of occult blood using the Hemoccult II Sensa Single Slides System (Beckman Coulter). Fecal calprotectin was measured using a Calprotectin ELISA Assay Kit (Eagle Biosciences).
Amplicon sequencing and analysis
Details on fecal DNA extraction and amplicon sequencing are provided in the Online Repository Text. Raw sequence reads were analyzed using QIIME2 (Quantitative Insights Into Microbial Ecology version 2). Sequence quality analysis was performed using FastQC and DADA2 v1.10 32 was used for quality-score based filtering. The alpha rarefaction maximum depth was 127,928 sequences. The sequence variants were identified and classified against the SILVA v132 database. For all analyses, feature tables were filtered requiring a microbial feature to have at least 0.01% relative abundance. The following R packages were used to perform the indicated analyses: ggplot2 for diversity trends and community composition visualizations; Phyloseq for Alpha diversity 33 with comparisons using the Kruskal-Wallis test; EdgeR 34 for differential abundance analyses; Metacoder 35 for heat tree analysis; FactoMineR to identify the metadata variables contributing to microbiome diversity and to generate Factor Analysis of Mixed Data plots with principal component method for quantitative variables and multiple correspondence analysis for the qualitative variables 36; lefser version 1.8.0 37 for Linear Discriminant Analysis Effect Size (LEfSe) analysis to identify microbiome biomarkers. 38. For further accurate prediction of biomarkers, the machine learning algorithm was performed by averaging 1000 decision trees using the randomForest package in R 39. Beta diversity was evaluated by principal coordinate analyses (PCoA) based on the weighted UniFrac distance method and significance determined by Permutational MANOVA (PERMANOVA, 9999 permutations). Predictive functional profiling of the microbial communities was performed using PICRUSt2 40. The heat tree analysis depicting taxonomic differences utilized the quantitative median abundance value and the non-parametric Wilcoxon Rank Sum test to leverage the hierarchical structure of taxonomic classifications. The Spearman Rank correlation analysis between the NIH CC and PIDTC cohorts and network generation was performed at the genus level, setting p<0.05 and r>0.3 by incorporation of the SparCC method and 100 iterations All sequences, metadata and supplementary files associated with data analysis are available in NCBI Gene Expression Omnibus (GEO; accession number: GSE220260).
Fecal metabolomics
Samples were subjected to liquid chromatography-mass spectrometry (LC-MS) analysis and metabolite extraction was performed as described 41. Full details on the hybrid metabolomics methods, data processing and relative quantification of metabolites are provided in the Online Repository Text.
Statistical analyses
Statistical analyses of clinical data were performed using GraphPad Prism Version 9.0. Continuous variables were expressed as means with standard deviations (SD) and categorical variables as absolute values with percentages. Unpaired t-test with Welch correction was used for two-group comparisons, and one-way ANOVA with Welch correction for multiple comparisons. All tests were two-sided and p<0.05 was considered significant. The correlogram of clinical and laboratory variables was constructed with corrplot package in R. The R versions 4.1.3 to 4.2.2 were used for the data analysis and visualization. The figures were assembled using Adobe Illustrator or Photoshop.
RESULTS
NIH CC and PIDTC cohorts
The NIH CC cohort included 19 healthy participants, 79 CGD patients and 8 pathogenic variant carriers including 2 highly lyonized carriers (HL Carriers) and 2 DUOX2-NCF2 (Table 1). Although the age distribution was comparable, there were more females in the control group compared to the CGD group, presumably due to the fact that 65% of CGD patients have gp91phox (CYBB) defects, which are inherited in an X-linked manner and therefore predominantly affect males. The PIDTC validation cohort included 36 pre-HCT patients with CGD (see Table E1 in online repository). Patients in the PIDTC cohort were younger (median age: 2.1 years), had a lower prevalence of CGD-IBD (19.4%) and were less likely to be on non-prophylactic antimicrobials (16.7%).
Table 1.
Demographic and clinical characteristics of participants from the NIH CC cohort.
| Healthy n=19 | CGD n=79 | Carriers n=8 | |
|---|---|---|---|
| Age, median (IQR), years | 29 (12) | 23 (20.5) | 38 (13.5) |
|
| |||
| Female, n (%) | 14 (73.7) | 17 (21.5) | 7 (87.5) |
|
| |||
| Race / Ethnicity, n (%) | |||
| White | 10 (52.5) | 55 (69.6) | 4 (50) |
| Black | 1 (5.3) | 10 (12.7) | 0 (0) |
| Latino(a) or Hispanic | 3 (15.8) | 6 (7.6) | 1(12.5) |
| Asian | 2 (10.5) | 3 (3.8) | 1 (12.5) |
| Mixed | 0 (0) | 2 (2.5) | 0 (0) |
| Unknown | 3 (15.8) | 3 (3.8) | 2 (25) |
|
| |||
| CGD genotype [affected protein], n (%) | |||
| CYBB0/− [gp91phox] | NA | 48 (60.8) | NA |
| CYBA−/− [p22phox] | NA | 5 (6.3) | NA |
| NCF1−/− [p47phox] | NA | 23 (29.1) | NA |
| NCF2−/− [p67phox] | NA | 2 (2.5) | NA |
| NCF4−/− [p40phox] | NA | 0 (0) | NA |
| CYBB+/− carriers | |||
| All carriers, n | NA | NA | 6 |
| Highly lyonized carriers, n | NA | NA | 2 |
| DUOX2+/− and NCF2+/− digenic heterozygous carriers | NA | NA | 2 |
|
| |||
| History of CGD-IBD, n (%) | 0 (0) | 54 (68.4) | 3 (37.5) |
|
| |||
| Active GI symptoms at visit a, n (%) | 0 (0) | 35 (44.3) | 4 (50) |
|
| |||
| On prophylactic antibiotics, n (%) | 2 (10.5) | 73 (92.4) | 3 (37.5)b |
| TMP-SMX | 2 (10.5) | 64 (81) | 3 (37.5) |
| Other prophylactic antibiotic | NA | 8 (10) | 0 (0) |
| Azole | NA | 71 (89.9) | 3 (37.5) |
|
| |||
| On non-prophylactic antibiotics, n (%) | |||
| Any | 0 (0) | 29 (36.7) | 2 (25) |
| Carbapenem | NA | 6 (7.6) | 0 (0) |
| Metronidazole | NA | 4 (5.1) | 0 (0) |
| Cephalosporin | NA | 4 (5.1) | 2 (25) |
| Quinolone | NA | 10 (12.6) | 0 (0) |
| Macrolide | NA | 3 (3.7) | 0 (0) |
| Tetracycline | NA | (5.1) | 0 (0) |
|
| |||
| On antifungal other than azole, n (%) | 0 (0) | 6 (7.6) | 0 (0) |
|
| |||
| On any antiviral, n (%) | 0 (0) | 1 (1.3) | 0 (0) |
|
| |||
| On steroids, n (%) | 0 (0) | 17 (21.5) | 1 (16.7) |
| On another immune modulator, n (%) | |||
| Any | 0 (0) | 29 (36.7) | 1 (16.7) |
| IFN-γ | NA | 12 (15.2) | 0 (0) |
| 5-Aminosalicylates | NA | 21 (26.6) | 1 (16.7) |
| Azathioprine | NA | 4 (5.1) | 0 (0) |
| Biologic | NA | 5 (6.3) | 0 (0) |
| Other | NA | 11 (13.9) | 1 (16.7) |
NIH CC: National Institutes of Health Clinical Center, CGD: chronic granulomatous disease, IQR: interquartile range, NA: not applicable, TMP-SMX: trimethoprim-sulfamethoxazole.
Active GI symptoms: watery bowel movements (BM) or more than 2 BM per day or blood/mucus in stool or active fistulizing or perianal disease.
The 3 patients on prophylactic antibiotics include the 2 HL carriers and 1 carrier (non-HL and non-DUOX2-NCF2).
Correlation between clinical symptoms and biomarkers associated with IBD in patients with CGD
We evaluated GI symptoms, quality of life scores, and biomarkers of local (fecal occult blood and calprotectin) and systemic (CBC, CRP, ESR, and albumin) inflammation in Healthy and CGD patients with or without IBD (NIH CC cohort only; see Table E2 in online repository). Fecal calprotectin levels and both the NRS and P-SCCAI distinguished patients with CGD-IBD. Positive correlations were observed between clinical parameters and biomarkers in CGD patients (see Figure E1 in online repository). There was also agreement between both quality of life measures (NRS and SIBDQ) and between both clinical activity index scores (P-SCCAI and CAI). These data suggest that a quality of life measure as simple as a numeric rating scale (NRS) combined with fecal calprotectin measurements may be helpful clinical markers when assessing patients with CGD-IBD.
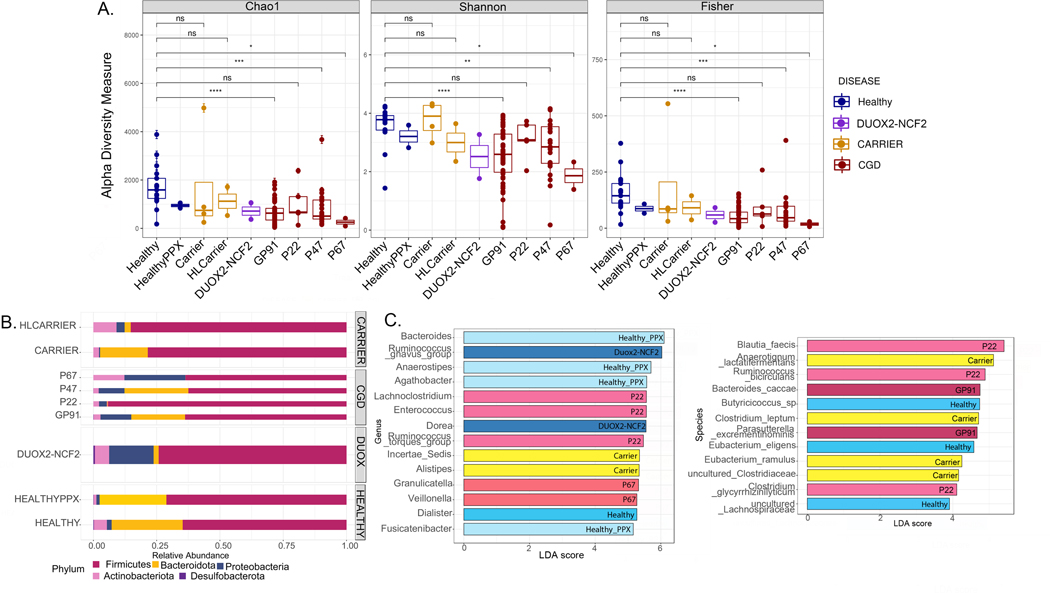
Impact of genotype on microbiome signature of patients with CGD
We examined whether different NOX2 defects were associated with changes in microbiome alpha diversity (Figure 1A) and composition (Figure 1B). The richness analysis giving weight to rare species (Chao1), and diversity indices for richness and evenness (Shannon and Fisher) are presented as alpha diversity metrics. Fecal samples from CGD patients (all genotypes except p22phox [CYBA−/−]) had significantly lower bacterial alpha diversity compared to Healthy. The relative abundance of bacterial phyla was similar between Healthy and Carriers (i.e. the 4 out of 8 carriers that were not HL carriers and not DUOX2-NCF2), whereas samples from DUOX2-NCF2, HL Carriers and CGD patients were enriched for Proteobacteria (Figure 1B). Of note, the 2 HL carriers and 1 Carrier were on antimicrobial prophylaxis (Table 1). Patterns of bacterial composition at the genus level were similar among samples from patients with gp91phox, p47phox and p22phox defects (see Figure E2A in online repository). The LEfSe graph shows bacterial taxa identified as markers for certain genotypes (Figure 1C). In particular, samples from Healthy were distinguished by uncultured Lachnospiraceae and Dialister spp., which ferment plant polysaccharides to short chain fatty acids (SCFA) 42, 43, Butyricicoccus spp. (producers of the SCFA butyrate 44) and Eubacterium eligens sp. (IL-10 producer 45). These bacterial taxa may therefore be contributing to intestinal health by decreasing inflammation. In contrast, samples from patients with gp91phox defects were distinguished by Bacteroides caccae and Parasutterella excrementi hominis sp., while samples from patients with p22phox defects were distinguished by Lachnoclostridium spp., Enterococcus spp., Ruminococcus torques group and others. The genera and species with high discriminatory power between the genotypes identified by random forest classification are presented in Figure E2B and the differential abundance as heat trees (Figure E2C in online repository).
Figure 1. Association of CGD genotype with microbiome signature.
(A) Alpha diversity (within-group variations) analyses (Chao1, Shannon, Fisher) comparing different genotypes (Healthy [n= 17]; Healthy PPX [i.e. healthy patients on infection prophylaxis with TMP-SMX for recurrent cystitis, n=2]; Carriers [CYBB−/+, n=4]; HL [highly lyonized, n=2] carriers; DUOX2-NCF2 [DUOX2−/+ and NCF2−/+ digenic heterozygous carriers, n=2]; GP91 [CYBB0/−, n=48], P22 [CYBA−/−, n=5], P47 [NCF1−/−, n=23], and P67 [NCF2−/−, n=2] CGD patients. * = p<0.05, **=p<0.01, ***=p<0.001, ****=p<0.0001, ns = non-significant. (B) Bar graphs showing relative abundance of bacterial phyla. (C) Linear discriminant analysis (LDA) score determined by the LEfSe analysis for identification of biomarkers, showing significantly enriched taxa in specific genotypes (p<0.05).
Variables contributing to the microbial diversity in CGD patients
One of the challenges with identifying microbiome signatures in patients with inborn errors of immunity (IEI) is the potential for confounders such as age, sex, antibiotics, immunomodulators, and inflammation, which can impact the intestinal microbiota. Controlling for these variables must be weighed against smaller samples sizes when studying rare IEIs. We therefore performed a factor analysis of mixed data on samples from the NIH CC cohort to identify variables most impacting the microbiome composition in CGD (Figure E3A in online repository). Besides CGD-IBD and certain CGD genotypes, non-prophylactic antibiotics (acute antibiotic use, carbapenems, cephalosporins and metronidazole), steroids and azathioprine use were identified as principal confounders (Figure E3A). The top 20 amplicon sequence variants contributing to the observed beta diversity are shown in Figure E3B. For each contributing factor, the defining genera as identified by the LEfSe analysis are presented (Figure E3C in online repository).
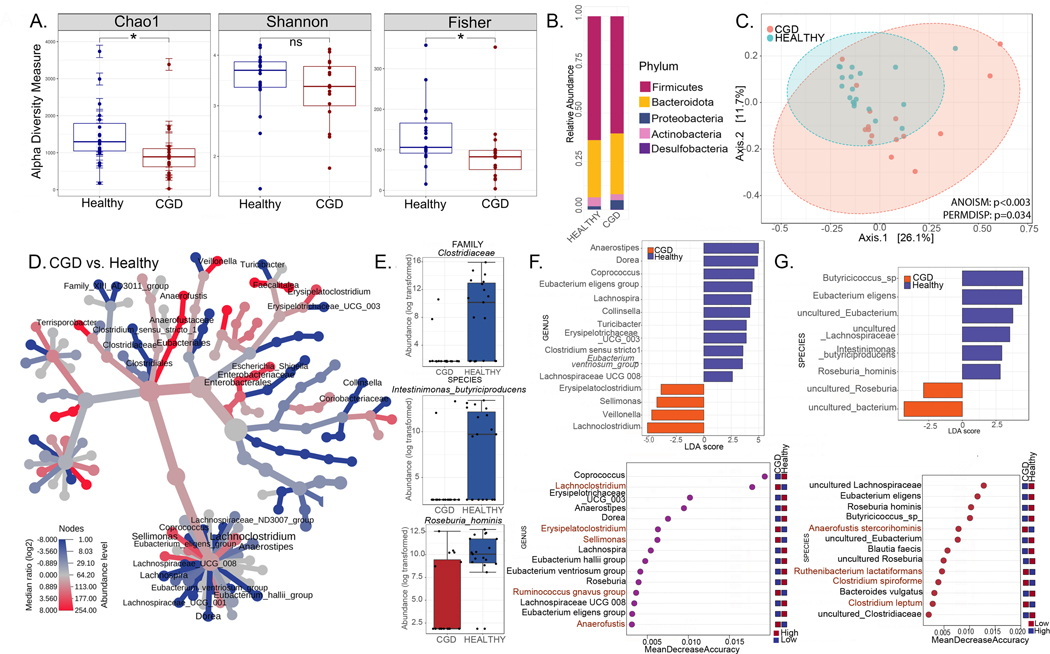
Intestinal microbiome signatures distinguish patients with CGD from healthy individuals
Given that the factor analysis of mixed data highlighted IBD, antibiotics and treatment with steroids or azathioprine as variables having the most impact on the microbiome, our next objective was to capture authentic CGD-associated changes on the microbiome by sub-setting CGD patient samples from the NIH CC cohort unaffected by these confounding variables. We therefore compared fecal samples from Healthy (n=19) with those from CGD patients who were not on any medications other than prophylactic antimicrobials (i.e., TMP-SMX and azole antifungal) and with no history of IBD or GI symptoms at the time of sample collection (n=16) (Figure 2). As expected, alpha diversity was significantly decreased in CGD patients (Figure 2A) and the relative abundance of Proteobacteria was increased (Figure 2B). Beta diversity analysis showed that samples from patients with CGD clustered separately from Healthy (p<0.003; Figure 2C). The taxa Clostridiaceae, Coriobacteriaceae, Turicibacter, Lachnospiraceae, Intestinimonas butyriciproducens and Roseburia hominis were significantly diminished in CGD patients (Figure 2D and 2E). LEfSe and random forest analyses identified Lachnoclostridium, Erysipelatoclostridium, and Sellimonas as being enriched in CGD (Figure 2F and 2G). These bacterial genera may represent intestinal biomarkers of CGD to be further investigated.
Figure 2. Intestinal microbiome signatures distinguish patients with CGD from healthy individuals.
(A-G) Comparisons for the CGD group (without a history of IBD and only on prophylactic antimicrobials, n=16) with the Healthy group (n=17). (A) Alpha diversity (within group variations) analyses (Chao1, Shannon, Fisher; * = p<0.05, **=p<0.01, ***=p<0.001, ****=p<0.0001, ns = non-significant) for CGD compared to Healthy. (B) Relative abundance of amplicon sequence variants in both groups presented at phylum level. (C) PCoA plot of beta diversity (between-group variations) based on weighted Unifrac distances for the two comparison groups with p values determined by Analysis of similarities (ANOSIM; p<0.003) and PERMDISP test for significant differences in dispersion (p =0.034). (D) Heat tree depicting the phylogenetic relationship and significant differential abundance (p<0.05) of bacterial genera between CGD and Healthy groups (red = higher abundance; blue = lower abundance). (E) Top differentially abundant family and species as identified by edgeR analysis (p values for all comparisons are <0.0001). (F) Linear discriminant analysis (LDA) score determined by the LEfSe analysis showing biomarkers at genus and species levels that are present or absent in the experimental groups shown. (G) Random forest analysis where genera and species with highest discriminatory power between CGD and Healthy groups are listed (red = high abundance in the experimental group, blue = low abundance in the experimental group).
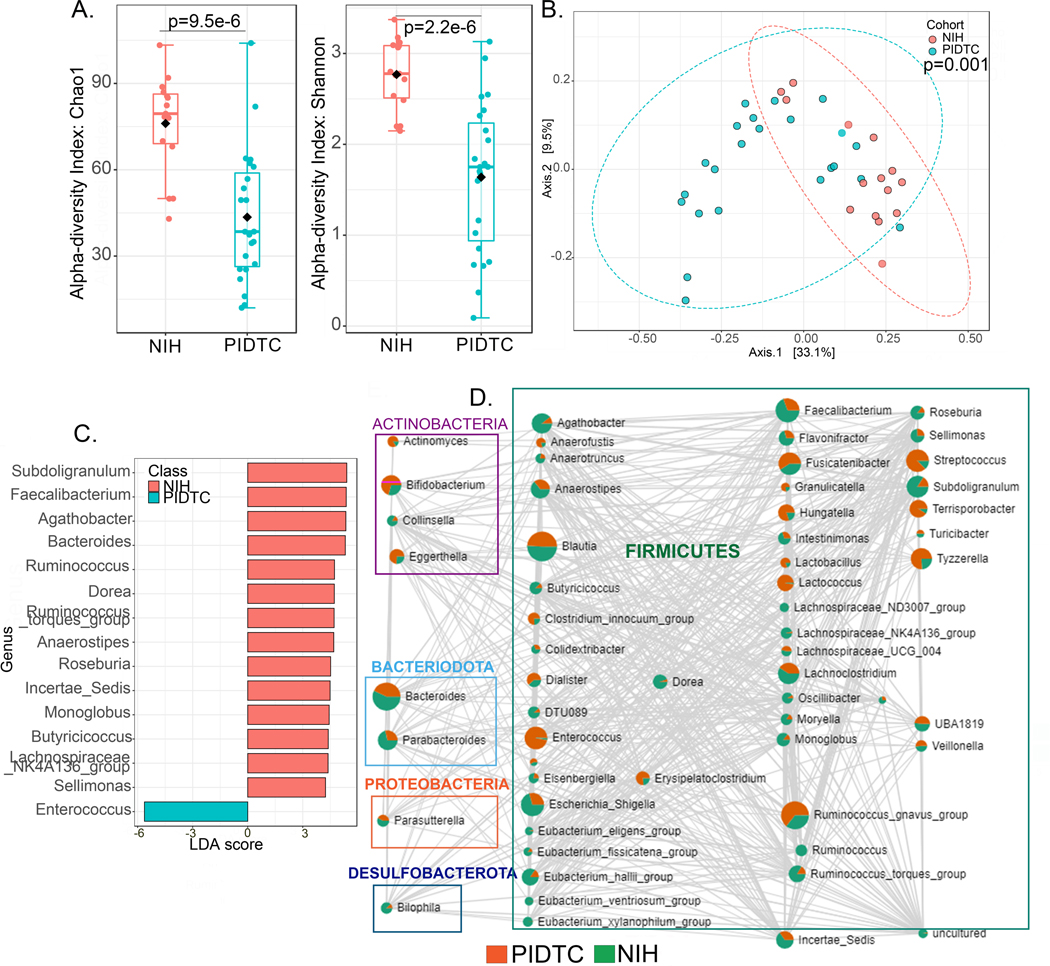
Comparison of microbiome signatures between NIH CC and PIDTC cohorts
To evaluate the generalizability of our findings, we compared the intestinal microbiome profiles of patients with CGD from the NIH CC cohort with those from the PIDTC cohort. Comparisons are provided for patients with CGD without a history of IBD or active IBD and who are not on any medications other than prophylactic antimicrobials (Figure 3) and all patients with CGD regardless of their IBD status or antimicrobial use (see Figure E4 in online repository). In both comparisons, there were significant differences in alpha and beta diversity, albeit the differences were more pronounced when the contribution of IBD and antimicrobials was removed (Figure 3A and 3B; Figure E4A and E4B). More genera and species were depleted in patients from the PIDTC cohort resulting in more positively defining genera and species in the NIH CC cohort, as identified by LEfSe analysis (Figure 3C and Figure E4C). Despite these differences, there were several bacterial taxa that correlated significantly between both cohorts (p<0.05), in positive or negative directions, as shown in Figures 3D and E4D where each node represents a taxon and the size of the node corresponds to the number of connections. Two taxa are connected by an edge when p<0.05 and the correlation threshold was >0.3. The edge size also reflects the magnitude of the correlation. Notable taxa included Bacteroides, Blautia, Faecalibacterium, Enterococcus, Ruminococcus gnavus group, Streptococcus, Lachnoclostridium, Fusicatenibacter, Subdoligranulum and Tyzzerella.
Figure 3. Comparison of microbiome signatures between patients with CGD from NIH CC and PIDTC cohorts.
(A-D) Comparisons for the CGD group (without a history of IBD and not on any medications other than prophylactic antimicrobials) between the NIH CC (n=16) and PIDTC cohorts (n=23). (A) Alpha diversity (within group variations) analyses (Chao1 and Shannon) at genus level. (B) PCoA plot of beta diversity (between group variations) based on weighted Unifrac distances for the two comparison groups with p values determined by permutational MANOVA (PERMANOVA, p=0.001). (C) Linear discriminant analysis (LDA) score determined by the LEfSe analysis for biomarker identification, showing the defining genera of the cohorts. (D) Correlation at genus level of the two cohorts is depicted as the network. Each node represents a taxon and the size of the node corresponds to the number of connections. Two taxa connected by an edge when p<0.05 and the correlation threshold >0.3. Within each taxon circle, green represents distribution in NIH CC cohort and red in PIDTC cohort.
The median age of participants in the NIH CC and PIDTC cohorts was 23 and 2.1 years, respectively as the PIDTC cohort included mostly pediatric patients. To investigate the effect of age on the CGD microbiome in the absence of confounding factors, we selected samples from both cohorts, from subjects without a history of IBD and not on medications other than prophylactic antimicrobials (n=38; 15 from NIH CC cohort and 23 from PIDTC cohort). For the analysis, two groups were generated: one with patients ≤ 12 years of age and the other with patients over 12 years of age. The alpha diversity measure showed significantly decreased microbial diversity in participants ≤ 12 years of age, without significant beta diversity (between-group comparison) differences when compared with samples from participants with ages above 12 years (Figure E5A, B in online repository). At the phylum level, Desulfobacterota were present only in participants over 12 years of age (Figure E5C in online repository). The LEfSe analysis identified 7 genera as markers for age over 12 years (Figure E5D in online repository).
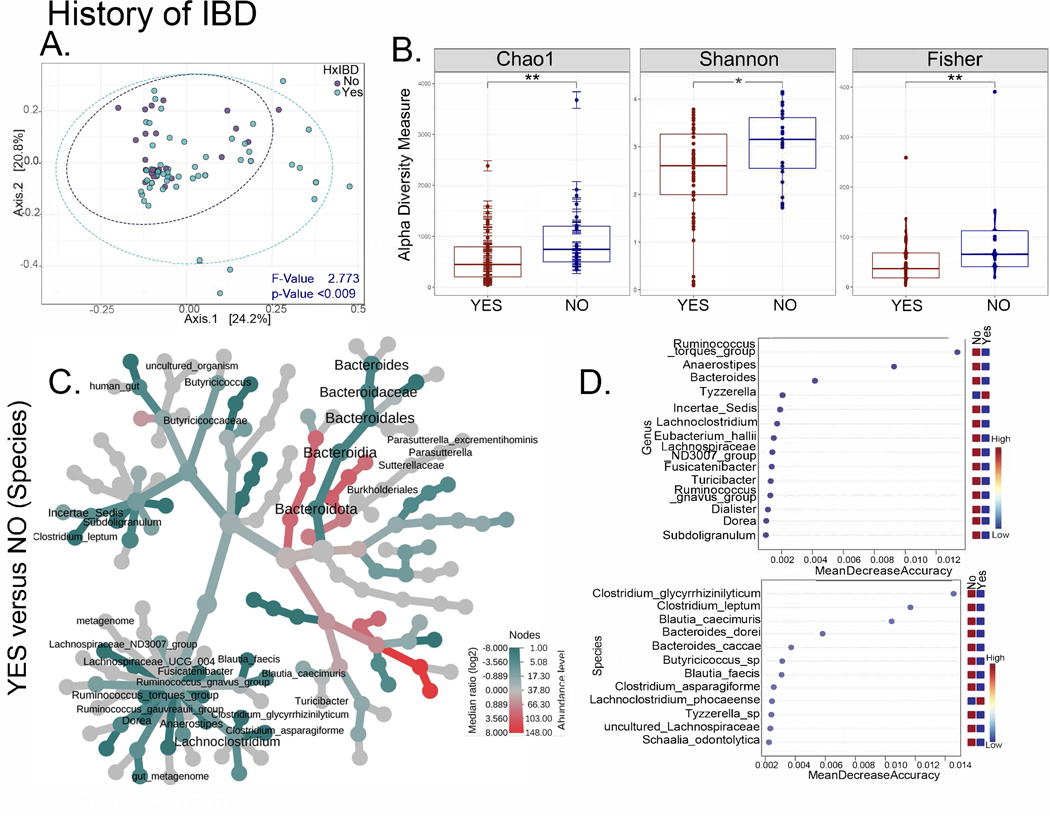
The intestinal microbiome distinguishes patients with CGD-IBD
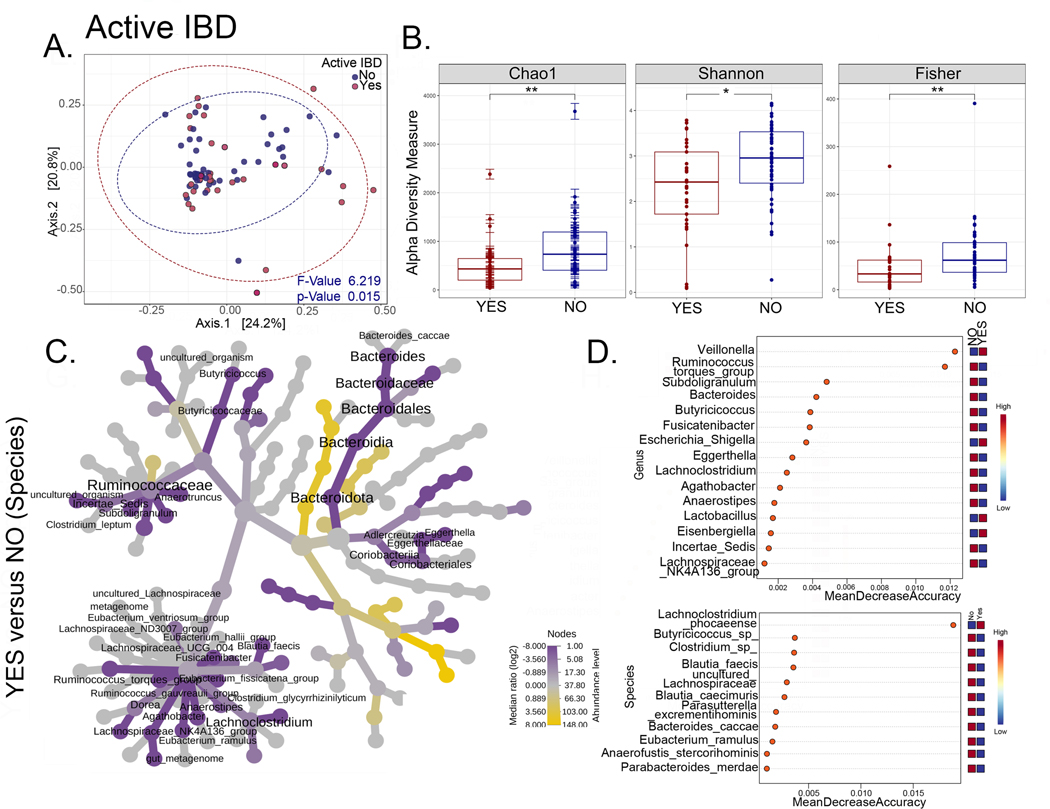
To identify microbial biomarkers of CGD-IBD and/or novel therapeutic targets, we compared microbiome profiles between CGD patients from the NIH CC cohort with (n=54) or without (n=25) a history of IBD, and with (n=35) or without (n=44) active IBD at the time of stool collection (Figures 4, 1; Figure E6 in online repository). Beta diversity analysis showed that samples from patients with a history of IBD or active IBD clustered separately from patients without (p<0.009 and p=0.015 respectively) (Figure 4A and 5A). A history of IBD or active IBD was associated with decreased alpha diversity (Figure 4B and 5B). Bacterial taxa enriched in patients with either a history of IBD or active IBD are shown in Figure 4C and 5C, which mostly include Bacteroidota and Ruminococcus group. Distinguishing markers identified by random forest analysis (Figure 4D and 5D) and bacterial genera and species that were unique, as well as common to patients with a history of IBD and active IBD are shown in Figure E6 in online repository. Species that were enriched and common to both patients with a history of IBD and active IBD include Eubacterium eligens, Butyricicoccus, Clostridium leptum, Blautia faecis, Clostridium spiroforme, and Eubacterium ramulus. Species that were enriched only in patients with active IBD include Lachnoclostridium phocaeense, Bacteroides dorei, Schaalia odontolytica and Bacteroides caccae. These bacterial species may therefore represent biomarkers of active IBD in patients with CGD.
Figure 4. The intestinal microbiome distinguishes patients with CGD and a history of IBD.
(A-D) Comparisons for the NIH CC CGD group with or without the history of IBD (Yes n=54, No n=25). (A) PCoA plots of beta diversity based on weighted Unifrac distances for the two comparison groups with p values determined by permutational MANOVA (PERMANOVA, p<0.009). (B) Alpha diversity plots (Chao1, Shannon, Fisher; * = p<0.05, **=p<0.01) comparing patients with CGD, with and without a history of IBD. (C) Heat tree depicting phylogenetic relationship and significant differential abundance (p<0.05) of bacterial genera for the comparison groups. (D) Random forest plots where genera and species with highest discriminatory power between patients with CGD with vs. without a history of IBD are shown (red = high abundance, blue = low abundance).
Figure 5. The intestinal microbiome distinguishes patients with CGD and active IBD.
(A-D) Comparisons for the NIH CC CGD group with or without active IBD (Yes n=33 and No n=46). (A) PCoA plots of beta diversity based on weighted Unifrac distances for the two comparison groups with p values determined by permutational MANOVA (PERMANOVA, p=0.015). (B) Alpha diversity plots (Chao1, Shannon, Fisher; * = p<0.05, **=p<0.01). (C) Heat tree depicting phylogenetic relationship and significant differential abundance (p<0.05) of bacterial genera for the comparison groups. (D) Random forest plots where genera and species with highest discriminatory power between patients with CGD with vs. without a active IBD are shown (red = high abundance, blue = low abundance).
Predictive functional profiling of the intestinal microbiome in CGD patients with or without IBD
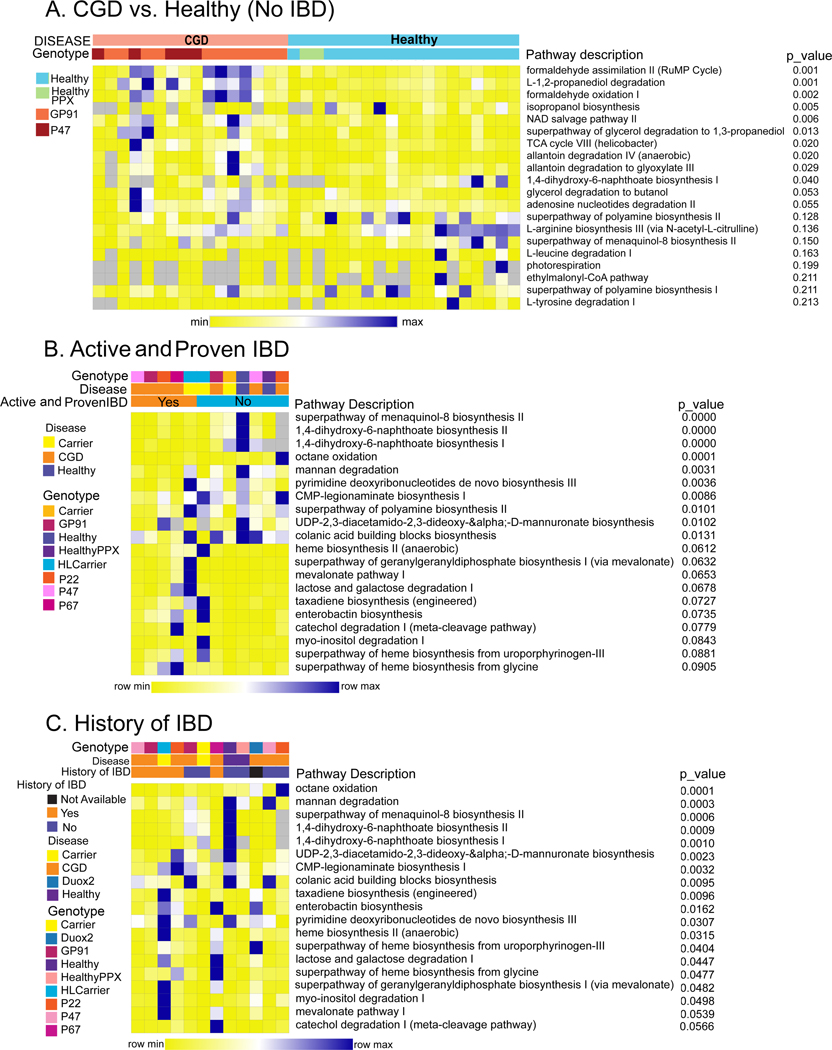
Predictive functional profiling by PICRUSt2 analysis of the NIH CC cohort microbiome comparing stool samples from CGD patients (without active or a history of IBD and not on antimicrobials) to Healthy showed differential regulation of 10 pathways (p<0.05; Figure 6A), mostly pertaining to TCA cycle, formate and allantoin metabolism (end-product of purines). The presence of active IBD or a history of IBD in CGD was associated with severe reductions in 1,4-Dihydroxy-2-naphthoic acid pathways (Figure 6B and 6C). 1,4-DHNA is a bacterial-derived metabolite that binds the aryl hydrocarbon receptor (AhR) and exhibits anti-inflammatory activity in the gut 46, 47. Activation of AhR pathways also attenuates colitis in animal models 48, 49. Other pathways affected by past or current CGD-IBD include octane oxidation, mannan degradation and heme biosynthesis.
Figure 6. Distinct functional profiles of the intestinal microbiome in patients with CGD from the NIH CC cohort.
Heatmap of significantly different functional profiles inferred by PICRUSt2 analysis performed to identify the pathways associated with changes in amplicon sequence variants (blue represents higher abundance and yellow represents lower abundance). The relative abundance was normalized to a Z-score and utilized to generate the heatmaps. (A) Pathway comparisons of the NIH CC CGD group (no history of IBD and only on prophylactic antimicrobials, n=16) with Healthy (n=17). (B) Pathway comparisons of patients with vs. without active and endoscopically proven CGD-IBD (Yes n=33 and No n=46). (C) Pathway comparisons of patients with vs. without a history of CGD-IBD (Yes n=54 and No n=25). The predicted p-values values are shown alongside the heatmap.
Intestinal metabolomic profiles in patients with CGD, with and without IBD.
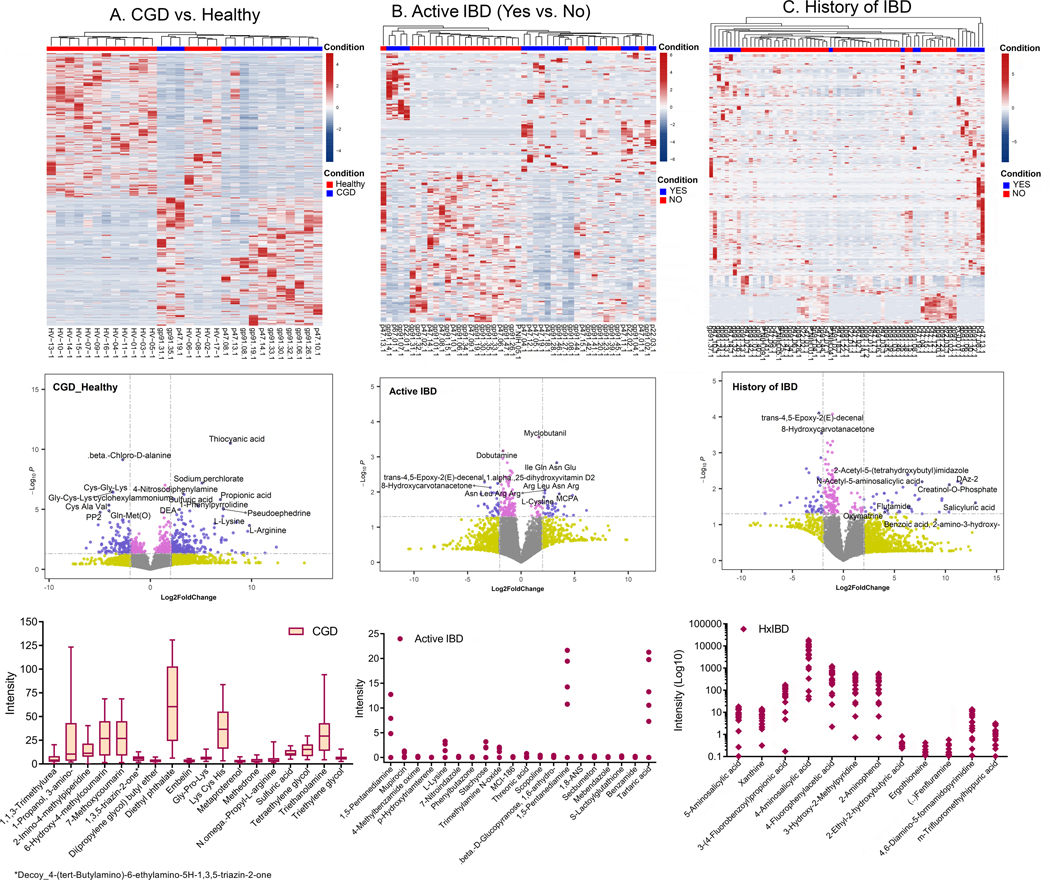
To validate some of the microbial functional pathways predicted to be relevant in patients with CGD or CGD-IBD based on the PICRUSt2 analyses, we also profiled the fecal metabolome in the NIH CC cohort. Patients with CGD (without active/history of IBD and only on prophylactic antimicrobials) had a distinct intestinal metabolomic profile and their samples clustered separately from Healthy (Figure 7A). The top 10% of metabolites with the highest impact scores (low p-value and high fold change) in CGD patients compared to Healthy are shown in Figure 7A (see Table E3 in online repository). Patients with active IBD (Figure 7B) or a history of IBD (Figure 7C) also had distinct metabolomic profiles (top 10% in Table E4 in online repository). The potential metabolites that could serve as the markers for CGD alone, active CGD-IBD or CGD with a history of IBD are shown as box plots (Figure 7).
Figure 7. Distinct metabolome profiles in patients with CGD from the NIH CC cohort.
Top to Bottom: Heatmap of the metabolomic profile, volcano plot displaying high metabolic diversity between the comparison groups, and the plots for the intensity measurements of the metabolites that are expressed only in the indicated groups, which are potential biomarkers. (A) Comparison of CGD group (without a history of IBD and only on prophylactic antimicrobials, n=14) to Healthy (n=16), (B) comparison within the CGD group of those with (Yes n=30) vs. without active IBD (No n=36), and (C) comparison within the CGD group of those with (Yes n=50) vs. without a history of IBD (No n=19). In the heatmaps, the rows display the metabolites and the columns represent the samples (blue = decreased, red = increased). The brightness of each color corresponds to the magnitude of the difference when compared with the average value. The lines in the volcano plots indicate the significance cut-off for the p-value (-log10 P value of 1.3013 corresponding to p<0.05) and fold change (log2 Fold Change >2, log2 Fold Change <−2). All metabolites that are significant and over an absolute log2 Fold Change of 2 are shown in violet and those that are significant but have an expression change less than an absolute log2 Fold Change of 2 are shown in pink. The bar plot indicates metabolites identified only in the CGD group, the dot plot indicates metabolites identified only in CGD patients with active IBD, and the diamond plot indicates metabolites identified only in patients with CGD and a history of IBD.
DISCUSSION
To our knowledge, this is the largest study to date evaluating the intestinal microbiome and metabolome in a cohort of patients with CGD. Our study is also novel insofar as it includes the analysis of both the fecal microbiome and metabolome in patients with NOX2 defects, including pathogenic variant carriers and a validation cohort recruited from 11 centers across North America. Moreover, we identified recent treatment with acute antibiotics, steroids, azathioprine use and having CGD-IBD as the most important variables influencing the microbiome in our CGD cohort, and therefore controlled for these variables in our analysis to uncover the true impact of NOX2 defects on the intestinal microbiome.
We demonstrated that patients with CGD, but not pathogenic variant carriers, had a less diverse and distinct intestinal microbiome composition compared to healthy individuals. While different CGD genotypes did not have major impacts, samples from patients with p22phox defects were specifically enriched with 6 bacterial taxa. This may be explained by the involvement of p22phox in NOX1 and NOX4 complexes, which are NOX2 homologs in the intestinal epithelium 50. p22phox defects may lead to decreased ROS production at the intestinal barrier that can impact the microbiome as described in mouse models 20.
After controlling for major confounders, we identified several bacterial species enriched in healthy individuals compared to CGD patients (Butyricicoccus spp., Eubacterium eligens, Roseburia hominis, Intestimonas butyriciproducens). All of these species produce butyrate, an anti-inflammatory SCFA that helps maintain intestinal barrier homeostasis 51–53. While we do not have the full annotation for the Butyricicoccus species identified, Butyricicoccus pullicaecorum, a butyrate-producer 44 and a probiotic that is safe in humans 54 was shown to strengthen the epithelial barrier in rat colitis models 55 and is depleted in patients with conventional IBD. Moreover, Eubacterium eligens promotes IL-10 production 45, while Roseburia hominis is associated with intestinal health and its depletion defines dysbiosis in patients with ulcerative colitis 56.
We also identified bacterial taxa that were enriched in patients with CGD (without IBD) Erysipelatoclostridium spp., Sellimonas spp. and Lachnoclostridium spp., thereby highlighting their potential relevance as biomarkers or therapeutic targets. Both Erysipelatoclostridium spp. and Sellimonas spp. have been previously associated with conventional IBD 57, 58. Further, Lachnoclostridium has been identified as a marker of non-invasive colorectal cancer 59 and Erysipelatoclostridium is enriched in patients with gout associated with altered uric acid 60. Similar to a previous study comparing CGD patients to healthy (n=10), we identified Ruminococcus gnavus as a species of interest 23. Our changes at the phylum-level, however, differed with an expansion of Proteobacteria and Bacteroidota, and reduction in Firmicutes.
We next focused on the comparison of CGD patients with or without IBD to identify microbial markers of disease activity and targets to treat CGD-IBD. Several markers of clinical disease activity used in conventional IBD also applied to CGD-IBD. In particular, fecal calprotectin, the NRS and the SIBDQ all significantly correlated with each other and with patient reports of GI symptoms. Patients with either an IBD history or active IBD had decreased alpha diversity and distinct microbiome profiles compared to their counterparts. Of the 24 bacterial genera and 8 bacterial species that were found to be significantly altered, relevant bacterial species that could be considered as potential therapeutic targets include Eubacterium eligens, Butyricicoccus spp., Clostridium leptum, Blautia faecis, Anaerofustis stercorihominis, Clostridium spiroforme and Eubacterium ramulus. Clostridium leptum, a carbohydrate fermenter with anti-inflammatory potential, has also been reported to be decreased in patients with conventional IBD 61, 62. Similar to Eubacterium eligens and Butyricicoccus spp., Blautia faecis is a butyrate producer that is decreased in Crohn disease 63. Clostridium spiroforme is a toxin-producing pathobiont 64, while Eubacterium ramulus is a butyrate-producing and flavonoid-degrading bacterium 65, 66. Flavonoids found in fruits, vegetables and grains are well-known for their health benefits 67. We also identified taxa that were specific to patients with active CGD-IBD. Namely, Lachnoclostridium phocaeense and Veillonella spp. were significantly increased, thereby highlighting their potential as biomarkers of active GI disease.
Consistent with our intestinal microbiome findings, CGD patients have distinct fecal metabolomic profiles. Since the intestinal metabolome represents the intersection of metabolic by-products of the environment (medications, diet), the host and the microbiota, observed metabolomic profiles reflected in part medication use, while bringing forward metabolites that corresponded to upregulated microbial metabolic pathways highlighted by the PICRUSt2 analysis. For instance, L-1,2-propanediol degradation was upregulated in CGD compared to healthy individuals and intestinal metabolomic profiles showed a 6.9-fold increase in propionic acid in stool from CGD patients. Meanwhile, active or a history of CGD-IBD was associated with severe reductions in 1,4-DHNA, a bacteria-derived vitamin K precursor that binds AhR 47. AhR activation has been shown to have anti-inflammatory effects through mechanisms including apoptosis, regulatory T cell induction, cytokine suppression and epigenetic modifications 68. Interestingly, a decrease in microbiota-derived AhR agonists was observed in patients with IBD and mutations in CARD9 (caspase recruitment domain-containing protein 9), an IBD susceptibility gene 69 and studies suggest that AhR activation may represent a therapeutic avenue for IBD 70, 71. While Lactobacilli have been shown to produce high levels of AhR ligands 72, 73, samples from patients with CGD without IBD were not significantly enriched for Lactobacilli. Thus, future studies will include isolation of differentially represented bacterial between patients with and without CGD-IBD for evaluation of AhR agonist production. These findings raise the possibility that colonizing patients with bacterial strains that produce AhR ligands may be a potential treatment for patients with CGD and IBD.
Our study has uncovered several candidate bacteria and metabolic pathways that should be further explored as potential biomarkers and therapies for CGD-IBD. However, our study has some limitations: 1) more females in the Healthy compared to CGD cohort, 2) the PIDTC cohort included more pediatric patients compared to the NIH CC cohort, and 3) lack of a healthy cohort on antimicrobial prophylaxis with TMP-SMX and an azole (not feasible). Nevertheless, we included 2 healthy individuals on TMP-SMX and their microbiome signature did not differ significantly from the rest of the Healthy cohort. Similarly, a previous study evaluating the impact of continuous prophylaxis with low-dose TMP-SMX on the intestinal microbiome in a cohort of pediatric patients with vesicoureteral reflux showed that the effect of TMP-SMX on the intestinal microbiome was minimal 74.
Our network analysis highlighted several bacterial taxa that correlated between the NIH CC and PIDTC cohorts. The significant differences in alpha and beta diversity between both cohorts were likely due to the younger age of the PIDTC cohort, which largely included pediatric patients, in comparison to largely adult population of the NIH CC cohort. In fact, one of the objectives of the PIDTC study is to evaluate the CGD microbiome before and after HCT, which is increasingly performed in younger patients 75. Several studies have demonstrated that age can have an important impact on the composition of the microbiome, especially under 3 years of age, a critical period for microbiota stabilization and early immune education 76. It is therefore plausible that younger CGD patients who have likely been exposed to antimicrobials early in life have a less diverse intestinal microbiota. Given the discrepancy in age distribution between the 2 study cohorts, we investigated the effect of age, and found that the alpha diversity was decreased in the ≤12 years group compared to the >12 years group, which agrees with previous studies 77, 78. Beta diversity differences between both cohorts did not reach statistical significance, suggesting that while some differences between the cohorts were driven by age, other factors such as geography and diet likely also played a role. Even though there were some age-driven differences in microbiome signatures, our results clearly demonstrated the microbial changes that were specifically driven by the CGD genotype in our combined cohort correlation analysis (Figure 3D).
Although 16S rRNA gene sequencing analysis combined with fecal metabolomics uncovered some interesting new findings, future studies should include metagenomic sequencing analysis of CGD stool and intestinal mucosal samples to identify bacterial strains distinguishing patients with CGD and CGD-IBD. These analyses would lead to the identification of more specific biomarkers and novel therapeutic targets for CGD-IBD, while providing additional insight into the relationship between NOX2 defects, microbial metabolism and pathogenicity.
Supplementary Material
CLINICAL IMPLICATION.
Our study has identified candidate microbes and metabolites to be further investigated as biomarkers of gastrointestinal disease activity in chronic granulomatous disease (CGD) and/or as novel therapeutic targets for CGD-associated inflammatory bowel disease (IBD).
FUNDING STATEMENT
This project was supported by the Division of Intramural Research (DIR), the National Institute of Allergy and Infectious Diseases (NIAID), the National Human Genomic Research Institute (NHGRI), and the Bioinformatics and Computational Biosciences Branch (BCBB) Service Contract HHSN3162013000006W of the National Institutes of Health (NIH). The work involving the Primary Immune Deficiency Treatment Consortium (PIDTC) was supported by the Division of Allergy, Immunology and Transplantation, NIAID, the Office of Rare Diseases Research (ORDR), National Center for Advancing Translational Sciences (NCATS), NIH (grant U54AI082973, MPI: J.M.P., C.C.D., E.H.; grants U54NS064808 and U01TR001263). The PIDTC is a part of the Rare Diseases Clinical Research Network (RDCRN) of ORDR, NCATS and received support from the Data Management and Coordinating Center (DMCC), RDCRN, at the University of South Florida (Jeffrey Krischer, Ph.D.). The collaborative work of the PIDTC with the Pediatric Transplantation and Cellular Therapy Consortium was supported by the U54 grants listed, along with support of the PBMTC Operations Center by the St. Baldrick’s Foundation and grant U10HL069254. This work utilized the computational resources of the National Institutes of Health (NIH) High-Performance Computation (HPC) Biowulf cluster (http://hpc.nih.gov) and the Nephele platform from the NIAID Office of Cyber Infrastructure and Computational Biology (OCICB; http://nephele.niaid.nih.gov). E.L.F is supported by a Canada Research Chair (Tier 2) in Role of the Microbiome in Primary Immune Deficiencies, Canadian Institutes of Health Research (CIHR), Fonds de Recherche du Québec-Santé (FRQ-S) and J-Louis Lévesque Foundation Research Chair. M.A.P. is supported by 1U01AI126612-01A1, P30CA040214, and 2UG1HL069254. E.H. is supported by the Bank of Montreal Chair of Pediatric Immunology. L.D.N. is supported by the Division of Intramural Research, NIAID, NIH (grant 1 ZIA Al001222-02, Pl: L.D.N). S.-Y.P. is supported by funding from the Intramural Research Program, NIH, National Cancer Institute, Center for Cancer Research.
COMPETING INTEREST STATEMENT
M.A.P has done advisory activities for Novartis, Gentibio, Bluebird, Vertex, and Equillium, has received honoraria for educational talks from Novartis, and is receiving study support from Miltenyi and Adaptive. J.W.L. is currently employed by Bluebird. E.H. has received consultancy fees from Jaspers, Takeda, CSL-Behring, Octapharma and Leadiant Biosciences. All other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The content and opinions expressed are solely the responsibility of the authors and do not represent the official policy or position of the NIAID, ORDR, NCATS, NIH, or any other agency of the US Government. The sponsors had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
ABBREVIATIONS
- ROS
reactive oxygen species
- IBD
inflammatory bowel disease
- CGD
chronic granulomatous disease
- NADPH
nicotinamide adenine dinucleotide phosphate
- EROS
essential for reactive oxygen species
- NOX2
NADPH oxidase complex 2
- CGD
associated IBD: CGD-IBD
- HCT
hematopoietic cell transplantation
- IL
interleukin
- NIH-CC
National Institutes of Health Clinical Center
- PIDTC
Primary Immune Deficiency Treatment Consortium
- NIAID
National Institute of Allergy and Infectious Diseases
- IRB
institutional review board
- CBC
complete blood count
- CRP
C-reactive protein
- ESR
erythrocyte sedimentation rate
- NRS
Numeric Rating Scale
- SIBDQ
Short Quality of Life IBD Questionnaire
- P-SCCAI
Patient-modified Simple Clinical Colitis Activity Index
- CAI
Clinical Activity Index
- BM
bowel movements
- TMP-SMX
trimethoprim-sulfamethoxazole
- QIIME2
Quantitative Insights Into Microbial Ecology version 2
- LDA
Linear Discriminant Analysis
- LEfSe
Linear Discriminant Analysis Effect Size
- PCoA
principal coordinate analyses
- LC-MS
liquid chromatography-mass spectrometry
- IEI
inborn errors of immunity
- CARD9
caspase recruitment domain-containing protein 9
- AhR
aryl hydrocarbon receptor
Footnotes
CONTRIBUTORSHIP STATEMENT
E.L.F. conceived the project, designed the study and experiments, collected and analyzed data, performed experiments, wrote the manuscript. P.C. analyzed data, produced figures and wrote the manuscript. Y.H., Y.S, D.J, C.D. performed experiments, analyzed data and edited the manuscript. C.Z., T.H, S.A.K., B.E.M., S.S.D., R.S.A., M.C.S., A.M.T., S.S., cared for patients, participated in recruitment and data/sample collection, and edited the manuscript. C.G., V.C., P.S., M.Q., S.C., B.L. performed bioinformatics and/or statistical analyses and edited the manuscript. L.M.G., J.A.S., C.E.B., S.M.H. contributed to study and experimental design, provided valuable feedback and support, and edited the manuscript. The following authors participated in study design, patient recruitment, data extraction and analysis, and editing of the manuscript: J.W.L., D.E.A., H.L.M., E.M.K., R.A.M., C.D., E.H., T.T., M.J.C., D.B.K., L.D.N., S.Y.P., J.M.P., M.A.P., F.T., S.P., K.M., D.C., L.M.F., S.S.L., E.G., C.L.D., L.M.B., T.M., N.K., J.R.H., J.L.B., M.K., N.W., A.R., S.C., L.A.B.
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Mittal M, Siddiqui MR, Tran K, Reddy SP, Malik AB. Reactive oxygen species in inflammation and tissue injury. Antioxid Redox Signal 2014; 20:1126–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dhillon SS, Fattouh R, Elkadri A, Xu W, Murchie R, Walters T, et al. Variants in nicotinamide adenine dinucleotide phosphate oxidase complex components determine susceptibility to very early onset inflammatory bowel disease. Gastroenterology 2014; 147:680–9 e2. [DOI] [PubMed] [Google Scholar]
- 3.Falcone EL, Holland SM. Gastrointestinal Complications in Chronic Granulomatous Disease. Methods Mol Biol 2019; 1982:573–86. [DOI] [PubMed] [Google Scholar]
- 4.Magnani A, Brosselin P, Beaute J, de Vergnes N, Mouy R, Debre M, et al. Inflammatory manifestations in a single-center cohort of patients with chronic granulomatous disease. J Allergy Clin Immunol 2014; 134:655–62 e8. [DOI] [PubMed] [Google Scholar]
- 5.Segal BH, Leto TL, Gallin JI, Malech HL, Holland SM. Genetic, biochemical, and clinical features of chronic granulomatous disease. Medicine (Baltimore) 2000; 79:170–200. [DOI] [PubMed] [Google Scholar]
- 6.Arnold DE, Heimall JR. A Review of Chronic Granulomatous Disease. Adv Ther 2017; 34:2543–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhns DB, Alvord WG, Heller T, Feld JJ, Pike KM, Marciano BE, et al. Residual NADPH oxidase and survival in chronic granulomatous disease. The New England journal of medicine 2010; 363:2600–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alimchandani M, Lai JP, Aung PP, Khangura S, Kamal N, Gallin JI, et al. Gastrointestinal histopathology in chronic granulomatous disease: a study of 87 patients. The American journal of surgical pathology 2013; 37:1365–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khangura SK, Kamal N, Ho N, Quezado M, Zhao X, Marciano B, et al. Gastrointestinal Features of Chronic Granulomatous Disease Found During Endoscopy. Clin Gastroenterol Hepatol 2016; 14:395–402 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marsh RA, Leiding JW, Logan BR, Griffith LM, Arnold DE, Haddad E, et al. Chronic Granulomatous Disease-Associated IBD Resolves and Does Not Adversely Impact Survival Following Allogeneic HCT. J Clin Immunol 2019; 39:653–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marciano BE, Rosenzweig SD, Kleiner DE, Anderson VL, Darnell DN, Anaya-O’Brien S, et al. Gastrointestinal involvement in chronic granulomatous disease. Pediatrics 2004; 114:462–8. [DOI] [PubMed] [Google Scholar]
- 12.Noel N, Mahlaoui N, Blanche S, Suarez F, Coignard-Biehler H, Durieu I, et al. Efficacy and safety of thalidomide in patients with inflammatory manifestations of chronic granulomatous disease: a retrospective case series. J Allergy Clin Immunol 2013; 132:997–1000 e1–4. [DOI] [PubMed] [Google Scholar]
- 13.Uzel G, Orange JS, Poliak N, Marciano BE, Heller T, Holland SM. Complications of tumor necrosis factor-alpha blockade in chronic granulomatous disease-related colitis. Clin Infect Dis 2010; 51:1429–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butte MJ, Park KT, Lewis DB. Treatment of CGD-associated Colitis with the IL-23 Blocker Ustekinumab. J Clin Immunol 2016; 36:619–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Luca A, Smeekens SP, Casagrande A, Iannitti R, Conway KL, Gresnigt MS, et al. IL-1 receptor blockade restores autophagy and reduces inflammation in chronic granulomatous disease in mice and in humans. Proc Natl Acad Sci U S A 2014; 111:3526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamal N, Marciano B, Curtin B, Strongin A, DeRavin SS, Bousvaros A, et al. The response to vedolizumab in chronic granulomatous disease-related inflammatory bowel disease. Gastroenterol Rep (Oxf) 2020; 8:404–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dumas A, Knaus UG. Raising the ‘Good’ Oxidants for Immune Protection. Front Immunol 2021; 12:698042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell EL, Bruyninckx WJ, Kelly CJ, Glover LE, McNamee EN, Bowers BE, et al. Transmigrating neutrophils shape the mucosal microenvironment through localized oxygen depletion to influence resolution of inflammation. Immunity 2014; 40:66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leoni G, Alam A, Neumann PA, Lambeth JD, Cheng G, McCoy J, et al. Annexin A1, formyl peptide receptor, and NOX1 orchestrate epithelial repair. J Clin Invest 2013; 123:443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pircalabioru G, Aviello G, Kubica M, Zhdanov A, Paclet MH, Brennan L, et al. Defensive Mutualism Rescues NADPH Oxidase Inactivation in Gut Infection. Cell Host Microbe 2016; 19:651–63. [DOI] [PubMed] [Google Scholar]
- 21.Singh AK, Hertzberger RY, Knaus UG. Hydrogen peroxide production by lactobacilli promotes epithelial restitution during colitis. Redox Biol 2018; 16:11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falcone EL, Abusleme L, Swamydas M, Lionakis MS, Ding L, Hsu AP, et al. Colitis susceptibility in p47(phox−/−) mice is mediated by the microbiome. Microbiome 2016; 4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sokol H, Mahlaoui N, Aguilar C, Bach P, Join-Lambert O, Garraffo A, et al. Intestinal dysbiosis in inflammatory bowel disease associated with primary immunodeficiency. J Allergy Clin Immunol 2019; 143:775–8 e6. [DOI] [PubMed] [Google Scholar]
- 24.Davrandi M, Harris S, Smith PJ, Murray CD, Lowe DM. The Relationship Between Mucosal Microbiota, Colitis, and Systemic Inflammation in Chronic Granulomatous Disorder. J Clin Immunol 2022; 42:312–24. [DOI] [PubMed] [Google Scholar]
- 25.Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019; 569:655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grasberger H, Magis AT, Sheng E, Conomos MP, Zhang M, Garzotto LS, et al. DUOX2 variants associate with preclinical disturbances in microbiota-immune homeostasis and increased inflammatory bowel disease risk. J Clin Invest 2021; 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffith LM, Cowan MJ, Notarangelo LD, Kohn DB, Puck JM, Shearer WT, et al. Primary Immune Deficiency Treatment Consortium (PIDTC) update. J Allergy Clin Immunol 2016; 138:375–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang Y, Dong T, Chen M, He L, Wang T, Liu X, et al. Systematic Analysis of Impact of Sampling Regions and Storage Methods on Fecal Gut Microbiome and Metabolome Profiles. mSphere 2020; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Surti B, Spiegel B, Ippoliti A, Vasiliauskas EA, Simpson P, Shih DQ, et al. Assessing health status in inflammatory bowel disease using a novel single-item numeric rating scale. Dig Dis Sci 2013; 58:1313–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irvine EJ, Zhou Q, Thompson AK. The Short Inflammatory Bowel Disease Questionnaire: a quality of life instrument for community physicians managing inflammatory bowel disease. CCRPT Investigators. Canadian Crohn’s Relapse Prevention Trial. Am J Gastroenterol 1996; 91:1571–8. [PubMed] [Google Scholar]
- 31.Bennebroek Evertsz F, Nieuwkerk PT, Stokkers PC, Ponsioen CY, Bockting CL, Sanderman R, et al. The patient simple clinical colitis activity index (P-SCCAI) can detect ulcerative colitis (UC) disease activity in remission: a comparison of the P-SCCAI with clinician-based SCCAI and biological markers. J Crohns Colitis 2013; 7:890–900. [DOI] [PubMed] [Google Scholar]
- 32.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016; 13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMurdie PJ, Holmes S. Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data. Pac Symp Biocomput 2012:235–46. [PMC free article] [PubMed] [Google Scholar]
- 34.Varet H, Brillet-Gueguen L, Coppee JY, Dillies MA. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS One 2016; 11:e0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foster ZS, Sharpton TJ, Grunwald NJ. Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput Biol 2017; 13:e1005404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lê S, Josse J, Husson F. FactoMineR: An R Package for Multivariate Analysis. Journal of Statistical Software 2008; 25:1–18. [Google Scholar]
- 37.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011; 12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chong J, Liu P, Zhou G, Xia J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc 2020; 15:799–821. [DOI] [PubMed] [Google Scholar]
- 39.Breiman L. Random Forests. Machine Learning 2001; 45:5–32. [Google Scholar]
- 40.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013; 31:814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pacold ME, Brimacombe KR, Chan SH, Rohde JM, Lewis CA, Swier LJ, et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat Chem Biol 2016; 12:452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 2009; 294:1–8. [DOI] [PubMed] [Google Scholar]
- 43.Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 2017; 19:29–41. [DOI] [PubMed] [Google Scholar]
- 44.Geirnaert A, Steyaert A, Van den Abbeele P, Eeckhaut V, Van Immerseel F, Boon N, et al. In vitro characterization of gastrointestinal behavior of Butyricicoccus pullicaecorum, a novel butyrate producing isolate with probiotic potential to counterbalance dysbiosis in inflammatory bowel disease. Commun Agric Appl Biol Sci 2013; 78:157–63. [PubMed] [Google Scholar]
- 45.Chung WSF, Meijerink M, Zeuner B, Holck J, Louis P, Meyer AS, et al. Prebiotic potential of pectin and pectic oligosaccharides to promote anti-inflammatory commensal bacteria in the human colon. FEMS Microbiol Ecol 2017; 93. [DOI] [PubMed] [Google Scholar]
- 46.Furumatsu K, Nishiumi S, Kawano Y, Ooi M, Yoshie T, Shiomi Y, et al. A role of the aryl hydrocarbon receptor in attenuation of colitis. Dig Dis Sci 2011; 56:2532–44. [DOI] [PubMed] [Google Scholar]
- 47.Cheng Y, Jin UH, Davidson LA, Chapkin RS, Jayaraman A, Tamamis P, et al. Editor’s Highlight: Microbial-Derived 1,4-Dihydroxy-2-naphthoic Acid and Related Compounds as Aryl Hydrocarbon Receptor Agonists/Antagonists: Structure-Activity Relationships and Receptor Modeling. Toxicol Sci 2017; 155:458–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cui X, Ye Z, Wang D, Yang Y, Jiao C, Ma J, et al. Aryl hydrocarbon receptor activation ameliorates experimental colitis by modulating the tolerogenic dendritic and regulatory T cell formation. Cell Biosci 2022; 12:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goettel JA, Gandhi R, Kenison JE, Yeste A, Murugaiyan G, Sambanthamoorthy S, et al. AHR Activation Is Protective against Colitis Driven by T Cells in Humanized Mice. Cell Rep 2016; 17:1318–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rada B, Leto TL. Oxidative innate immune defenses by Nox/Duox family NADPH oxidases. Contributions to microbiology 2008; 15:164–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen G, Ran X, Li B, Li Y, He D, Huang B, et al. Sodium Butyrate Inhibits Inflammation and Maintains Epithelium Barrier Integrity in a TNBS-induced Inflammatory Bowel Disease Mice Model. EBioMedicine 2018; 30:317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang RX, Lee JS, Campbell EL, Colgan SP. Microbiota-derived butyrate dynamically regulates intestinal homeostasis through regulation of actin-associated protein synaptopodin. Proc Natl Acad Sci U S A 2020; 117:11648–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li G, Lin J, Zhang C, Gao H, Lu H, Gao X, et al. Microbiota metabolite butyrate constrains neutrophil functions and ameliorates mucosal inflammation in inflammatory bowel disease. Gut Microbes 2021; 13:1968257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steppe M, Van Nieuwerburgh F, Vercauteren G, Boyen F, Eeckhaut V, Deforce D, et al. Safety assessment of the butyrate-producing Butyricicoccus pullicaecorum strain 25–3(T), a potential probiotic for patients with inflammatory bowel disease, based on oral toxicity tests and whole genome sequencing. Food Chem Toxicol 2014; 72:129–37. [DOI] [PubMed] [Google Scholar]
- 55.Eeckhaut V, Machiels K, Perrier C, Romero C, Maes S, Flahou B, et al. Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut 2013; 62:1745–52. [DOI] [PubMed] [Google Scholar]
- 56.Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014; 63:1275–83. [DOI] [PubMed] [Google Scholar]
- 57.Nagayama M, Yano T, Atarashi K, Tanoue T, Sekiya M, Kobayashi Y, et al. TH1 cell-inducing Escherichia coli strain identified from the small intestinal mucosa of patients with Crohn’s disease. Gut Microbes 2020; 12:1788898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yuan X, Chen B, Duan Z, Xia Z, Ding Y, Chen T, et al. Depression and anxiety in patients with active ulcerative colitis: crosstalk of gut microbiota, metabolomics and proteomics. Gut Microbes 2021; 13:1987779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang JQ, Li T, Nakatsu G, Chen YX, Yau TO, Chu E, et al. A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer. Gut 2020; 69:1248–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shao T, Shao L, Li H, Xie Z, He Z, Wen C. Combined Signature of the Fecal Microbiome and Metabolome in Patients with Gout. Front Microbiol 2017; 8:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kabeerdoss J, Sankaran V, Pugazhendhi S, Ramakrishna BS. Clostridium leptum group bacteria abundance and diversity in the fecal microbiota of patients with inflammatory bowel disease: a case-control study in India. BMC Gastroenterol 2013; 13:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang F, Zhang Y, Bai XQ, Wang CX, Li YN. Clostridium leptum induces the generation of interleukin-10(+) regulatory B cells to alleviate airway inflammation in asthma. Mol Immunol 2022; 145:124–38. [DOI] [PubMed] [Google Scholar]
- 63.Takahashi K, Nishida A, Fujimoto T, Fujii M, Shioya M, Imaeda H, et al. Reduced Abundance of Butyrate-Producing Bacteria Species in the Fecal Microbial Community in Crohn’s Disease. Digestion 2016; 93:59–65. [DOI] [PubMed] [Google Scholar]
- 64.Uzal FA, Navarro MA, Li J, Freedman JC, Shrestha A, McClane BA. Comparative pathogenesis of enteric clostridial infections in humans and animals. Anaerobe 2018; 53:11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rodriguez-Castano GP, Dorris MR, Liu X, Bolling BW, Acosta-Gonzalez A, Rey FE. Bacteroides thetaiotaomicron Starch Utilization Promotes Quercetin Degradation and Butyrate Production by Eubacterium ramulus. Front Microbiol 2019; 10:1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schneider H, Blaut M. Anaerobic degradation of flavonoids by Eubacterium ramulus. Arch Microbiol 2000; 173:71–5. [DOI] [PubMed] [Google Scholar]
- 67.Panche AN, Diwan AD, Chandra SR. Flavonoids: an overview. J Nutr Sci 2016; 5:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cannon AS, Nagarkatti PS, Nagarkatti M. Targeting AhR as a Novel Therapeutic Modality against Inflammatory Diseases. Int J Mol Sci 2021; 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 2016; 22:598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Busbee PB, Menzel L, Alrafas HR, Dopkins N, Becker W, Miranda K, et al. Indole-3-carbinol prevents colitis and associated microbial dysbiosis in an IL-22-dependent manner. JCI Insight 2020; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen J, Haller CA, Jernigan FE, Koerner SK, Wong DJ, Wang Y, et al. Modulation of lymphocyte-mediated tissue repair by rational design of heterocyclic aryl hydrocarbon receptor agonists. Sci Adv 2020; 6:eaay8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krishnan S, Ding Y, Saedi N, Choi M, Sridharan GV, Sherr DH, et al. Gut Microbiota-Derived Tryptophan Metabolites Modulate Inflammatory Response in Hepatocytes and Macrophages. Cell Rep 2018; 23:1099–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Natividad JM, Agus A, Planchais J, Lamas B, Jarry AC, Martin R, et al. Impaired Aryl Hydrocarbon Receptor Ligand Production by the Gut Microbiota Is a Key Factor in Metabolic Syndrome. Cell Metab 2018; 28:737–49 e4. [DOI] [PubMed] [Google Scholar]
- 74.Akagawa Y, Kimata T, Akagawa S, Yamaguchi T, Kato S, Yamanouchi S, et al. Impact of Long-Term Low Dose Antibiotic Prophylaxis on Gut Microbiota in Children. J Urol 2020; 204:1320–5. [DOI] [PubMed] [Google Scholar]
- 75.Dedieu C, Albert MH, Mahlaoui N, Hauck F, Hedrich C, Baumann U, et al. Outcome of chronic granulomatous disease - Conventional treatment vs stem cell transplantation. Pediatr Allergy Immunol 2021; 32:576–85. [DOI] [PubMed] [Google Scholar]
- 76.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science 2016; 352:539–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature 2012; 486:222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang X, Zhong H, Li Y, Shi Z, Ren H, Zhang Z, et al. Sex- and age-related trajectories of the adult human gut microbiota shared across populations of different ethnicities. Nat Aging 2021; 1:87–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.