Abstract
Fibroblast growth factor receptor 1 (FGFR1) mutations are associated with congenital hypogonadotropic hypogonadism (CHH) through inheritance or spontaneous occurrence. We detected FGFR1 mutations in a Chinese cohort of 210 CHH patients at Peking Union Medical College Hospital (Beijing, China) using next-generation and Sanger sequencing. We assessed missense variant pathogenicity using six bioinformatics tools and compared clinical features and treatment outcomes between inherited and de novo mutation groups. Among 19 patients with FGFR1 mutations, three were recurrent, and 16 were novel variants. Sixteen of the novel mutations were likely pathogenic according to the American College of Medical Genetics and Genomics (ACMG) guidelines, with the prevalent P366L variant. The majority of FGFR1 mutations was inherited (57.9%), with frameshift mutations exclusive to the de novo mutation group. The inherited mutation group had a lower incidence of cryptorchidism, short stature, and skeletal deformities. In the inherited mutation group, luteinizing hormone (LH) levels were 0.5 IU l−1, follicle-stimulating hormone (FSH) levels were 1.0 IU l−1, and testosterone levels were 1.3 nmol l−1. In contrast, the de novo group had LH levels of 0.2 IU l−1, FSH levels of 0.5 IU l−1, and testosterone levels of 0.9 nmol l−1, indicating milder hypothalamus–pituitary–gonadal axis (HPGA) functional deficiency in the inherited group. The inherited mutation group showed a tendency toward higher spermatogenesis rates. In conclusion, this study underscores the predominance of inherited FGFR1 mutations and their association with milder HPGA dysfunction compared to de novo mutations, contributing to our understanding of the genetic and clinical aspects of FGFR1 mutations.
Keywords: congenital hypogonadotropic hypogonadism, de novo, FGFR1, inherited
INTRODUCTION
Congenital hypogonadotropic hypogonadism (CHH) represents a rare disorder capable of disrupting the natural progression of puberty and fertility.1 This condition results from a diminishment in the quantity of gonadotropin-releasing hormone (GnRH) neurons and aberrations in both the secretion and function of GnRH. The prevalence of this disease is approximately 1 in 8000 for males, though it is lower for females.2 When CHH is accompanied by symptoms of hyposmia or anosmia, it is commonly referred to as Kallmann syndrome (KS).3 Those CHH patients who possess an intact sense of smell are characterized as normosmic CHH (nCHH) patients.4
Thus far, over 30 genes have been implicated in the pathogenesis of CHH.5 Gene mutations, including those in Kallmann syndrome 1 (KAL1), fibroblast growth factor 8 (FGF8), fibroblast growth factor 17 (FGF17), NMDA receptor synaptonuclear signaling and neuronal migration factor (NSMF), interleukin 17 receptor D (IL17RD), and prokineticin receptor 2 (PROKR2), have the potential to affect the development, differentiation, and migration of GnRH neurons from the olfactory epithelium to the hypothalamus, leading to the KS phenotype.6 On the other hand, mutations in genes linked to GnRH synthesis and secretion, such as gonadotropin-releasing hormone 1 (GNRH1), KiSS-1 metastasis suppressor (KISS1), KISS1 receptor (KISS1R), tachykinin precursor 3 (TAC3), leptin (LEP), and leptin receptor (LEPR),7 give rise to the nCHH phenotype. Causative gene variants have been identified in nearly 50% of CHH cases,5 with roughly 10% of patients having pathogenic variants in FGFR1 as the underlying cause.8
The FGFR1 gene, located on chromosome 8p11.23, comprises 18 exons and encodes the FGFR1 receptor protein, which belongs to the receptor tyrosine kinase (RTK) superfamily.9 The FGFR1 protein is composed of 822 amino acids and exhibits wide expression throughout various tissues (Uniprot: P16092). It is structurally organized from the amino-terminus to the carboxyl-terminus, consisting of an extracellular domain, a transmembrane domain, a juxta-membrane (JM) region, and an intracellular catalytic TK domain (TKD). The extracellular domain encompasses three extracellular immunoglobulin (Ig)-like domains (D1–3), with an acidic box (AB) positioned between the first and second Ig-like loops10. Importantly, FGFR1 signaling has been demonstrated to play a pivotal role in the development of the olfactory system, as well as in the migration, differentiation, and survival of GnRH neurons.11
FGFR1 mutations were first identified in KS patients in 2003, and it was later found that these mutations can also give rise to nCHH.12,13 In addition to reproductive system-related symptoms, individuals bearing pathogenic FGFR1 variants exhibit a spectrum of other clinical features, including anosmia, hearing impairment, and skeletal anomalies.14 A substantial number of over 140 FGFR1 variants have been documented in CHH patients.6 However, due to the complex pathogenesis of CHH, the clinical manifestations associated with these mutations are quite diverse. Furthermore, FGFR1 mutations can either be inherited from a parent or arise sporadically, potentially resulting in distinct clinical manifestations.12,15 The difference in clinical manifestations and the response to spermatogenesis therapy between inheritance and de novo groups, have not been extensively investigated. This research seeks to explore the inheritance patterns of FGFR1 gene mutations and subsequently compare the clinical manifestations and response to spermatogenic treatment between these two distinct groups.
PATIENTS AND METHODS
Patients
This study enlisted a cohort comprising 210 unrelated Chinese CHH patients, who had been admitted to Peking Union Medical College Hospital (Beijing, China) between May 2013 and May 2021. The diagnostic criteria employed for CHH included the following: (a) the absence of pubertal development by the age of 18 years for males and 16 years for females; (b) in males, a serum testosterone level ≤100 ng dl−1, or estradiol level ≤25 pg ml−1 in females, while concurrently presenting low levels of serum gonadotropin; (c) the normalcy of other anterior pituitary hormones; and (d) the absence of anomalies in hypothalamic–pituitary region magnetic resonance imaging (MRI). In cases where anosmia symptoms were evident, a diagnosis of KS was established.16 Ethical approval for the study was granted by the Ethics Committee of Peking Union Medical College Hospital (Approval No. JS-2111), and comprehensive informed consent was obtained from all participating individuals.
Clinical data
Age, body mass index (BMI), history of cryptorchidism, testicular volume (TV), history of testosterone replacement therapy before spermatogenesis therapy, time required for sperm appearance, as well as luteinizing hormone (LH), follicle-stimulating hormone (FSH), and testosterone levels during treatment in male CHH patients with FGFR1 mutations were retrospectively reported. Testicular size was measured using an orchidometer by clinical specialists, and the mean bilateral testicular volume was used for data analysis. Seminal samples were collected via masturbation, and the successful induction of spermatogenesis in CHH patients was defined as the presence of at least one sperm visible under a microscope following the centrifugation of the seminal sample.17
Molecular genetic analysis
Genomic DNA was extracted from peripheral leukocytes obtained from 210 CHH patients using the QIAGEN Midi Blood kit (QIAGEN, Hilden, Germany) and subsequently subjected to sequencing with the high-throughput sequencing system (Illumina Nextseq 500; Illumina, Inc., San Diego, CA, USA). A next-generation sequencing panel including 97 CHH-related genes (www.hgmd.cf.ac.uk/) was used in this study. The reference genome version employed was GRCh37/HG19. Additionally, 1000g2015aug_all, ESP6500 (NHLBI Exome Sequencing Project), EXAC (The Exome Aggregation Consortium), and ExAC-EAS (EXAC about 4000 East Asians) databases were also referenced. The sequencing results underwent BLAST analysis, and candidates were selected with reference to the following sequences: GenBank NG_007729 (FGFR1, genomic DNA [gDNA]), NM_023110.2 (FGFR1, complementary DNA [cDNA]), NP_075598.2 (FGFR1, protein), and the Human Gene Mutation Database (HGMD). Sanger sequencing was performed on both patients and their parents to validate and trace the origins of FGFR1 mutations.
Pathogenicity analysis
1000g2015aug_all,18 GnomAD,19 and GERP20 were used to evaluate the frequency of mutations and the conservation of mutated amino acids. The pathogenicity of newly discovered rare missense variants was determined using the American College of Medical Genetics and Genomics (ACMG) standards21 in conjunction with the results obtained from six in silico tools, namely SIFT,22 PolyPhen-2,23 Mutation Taster,24 REVEL,25 M-CAP,26 and LRT,27 to evaluate the pathogenicity of FGFR1 mutations. If a variant initially identified was not in NCBI dbSNP, Ensembl, Exome Variant Server, and 500 normal Chinese controls, or if the allele frequency was <0.0003 in the aforementioned databases and three or more in silico tools predicted pathogenicity, the variant was categorized as likely pathogenic (LP). This classification implies a high probability of pathogenicity at the gene function level, potentially leading to clinical manifestations.
For splicing site mutations, pathogenicity was assessed with the aid of two in silico tools, SPIDEX28 and Splice Site Score Calculation.29 If a mutation consistently received a pathogenic judgment from both in silico tools, it was designated as LP.
Statistical analyses
Data analysis was conducted using SPSS version 26.0 (SPSS Inc., Chicago, IL, USA). Normally distributed data were presented as mean ± standard deviation (s.d.), while nonnormally distributed data were expressed as median values (quartiles). Data comparison between the inheritance and de novo groups, in the case of normally distributed data, was executed using the nonpaired t-test. For nonnormally distributed data, differences between the two groups were assessed using nonparametric tests. The comparison of rates between groups was accomplished with the Fisher’s exact test. Statistical significance was defined as a two-sided P < 0.05.
RESULTS
Gene variant and in silico analysis
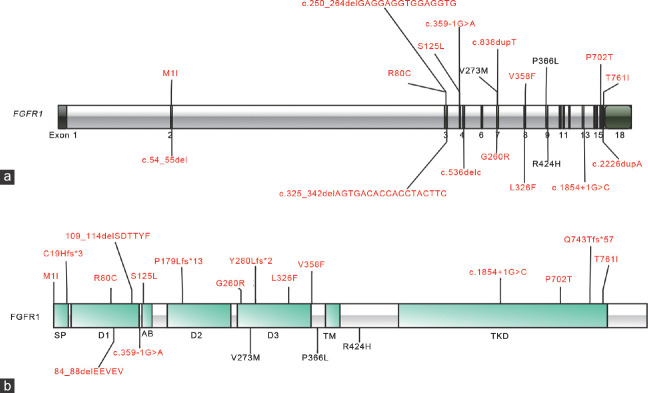
Nineteen CHH patients, with a male-to-female ratio of 15 to 4, presented a spectrum of FGFR1 variants, encompassing 10 KS and 9 nCHH cases. These variants exhibited a broad distribution across the entire FGFR1 gene (Figure 1a) and were distributed across nearly all functional domains of FGFR1 (Figure 1b). Reference sequences for FGFR1 cDNA and protein are NM_023110.2 and NP_075598.2, respectively. Among these variants, three had been previously reported in CHH patients: p.V273M,30 p.P366L,31 and p.R424H.32 The remaining 16 variants were characterized as novel rare variants, comprising eight missense variants (G260R, V358F, T761I, L326F, S125L, M1I, R80C, and P702T), and eight likely truncating variants, which included frameshift, splice site, and nonsense mutations (c.536delC, c.838dupT, c.2226dupA, c.54_55del, c.1854+1G>C, c.359-1G>A, c.250_264delGAGGAGGTGGAGGTG, and c.325_342delAGTGACACCACCTACTTC), as shown in Supplementary Table 1. Notably, one variant (P366L) was observed in two patients, and a single patient harbored two rare FGFR1 variants (L326F and S125L).
Figure 1.
(a) Distribution of rare variants in the FGFR1 gene. (b) Distribution of rare variants across the FGFR1 protein. Recurrent variants are in black. Novel variants are in red. Likely pathogenic variants are represented in regular font. In words with “c.”, G: guanine; C: cytosine; T: thymine; A: adenine. In others, G: glycine; R: arginine; V: valine; F: phenylalanine; T: threonine; I: isoleucine; L: leucine; S: serine; M: methionine: C: cysteine; H: histidine; P: praline;Y: tyrosine; Q: glutamine; E: glutamic acid; D: aspartic acid. FGFR1: fibroblast growth factor receptor 1.
Supplementary Table 1.
Fibroblast growth factor receptor 1 mutants identified in our patient series
| Mutation type | Nucleotide | Amino acid | Novel | ACMG |
|---|---|---|---|---|
| Missense mutations | c.817G>A | V273M | − | LP |
| Missense mutations | c.1097C>T | P366L | − | US LP |
| Missense mutations | c.1271G>A | R424H | − | US |
| Missense mutations | c.778G>C | G260R | + | LP |
| Missense mutations | c.1072G>T | V358F | + | P |
| Missense mutations | c.2282C>T | T761I | + | US |
| Missense mutations | c.976C>T | L326F | + | US |
| Missense mutations | c.374C>T | S125L | + | US |
| Missense mutations | c.3G>A | M1I | + | LP |
| Missense mutations | c.238C>T | R80C | + | US |
| Missense mutations | c.2104C>A | P702T | + | US |
| Splice site mutation | c.1854+1G>C | Splicing | + | LP |
| Splice site mutation | c.359-1G>A | Splicing | + | LP |
| Deletions | c.536delC | P179Lfs*13 | + | P |
| Insertions | c.2226dupA | Q743Tfs*57 | + | P |
| Insertions | c.838dupT | Y280Lfs*2 | + | P |
| Deletions | c.54_55del | C19Hfs*3 | + | P |
| Deletions | c.250_264delGAGGAGGTGGAGGTG | 84_88delEEVEV | + | LP |
| Deletions | c.325_342delAGTGACACCACCTACTTC | 109_114delSDTTYF | + | LP |
Nucleotid - G: guanine; C: cytosine; T: thymine; A: adenine. Amino acid - G: glycine; R: arginine; V: valine; F: phenylalanine; T: threonine; I: isoleucine; L: leucine; S: serine; M: methionine; C: cysteine; H: histidine; P: praline; Y: tyrosine; Q: glutamine; E: glutamic acid; D: aspartic acid. Novel - +: yes; −: no. ACMG - P: pathogenic; LP: likely pathogenic; US: uncertain. ACMG: American College of Medical Genetics and Genomics
The analysis of pathogenicity for the 11 single-nucleotide substitution mutants is summarized in Table 1, while the analysis of pathogenicity for the 2 splice site mutants is presented in Table 2. According to the ACMG classification, five novel missense variants (T761I, L326F, S125L, R80C, and P702T) were categorized as variants of uncertain significance (VUS). Two mutants (G260R and M1I) were identified as LP, and one mutant (V358F) was deemed pathogenic. Notably, all variants garnered pathogenic classifications from more than three out of six in silico tools (Table 1).
Table 1.
Pathogenicity analysis of rare single-nucleotide variants of fibroblast growth factor receptor 1 (FGFR1) in congenital hypogonadotropic hypogonadism patients
| Site | Nucleotide | Amino acid | Hom/het | Novel | GnomeAD | 1000g2015aug_all | ACMG | Clinvar | SIFT | PolyPhen_2 | MutationTaster | REVEL | M-CAP | LRT | GERP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exon 7 | c.778G>C | G260R | Het | Yes | − | − | LP | − | + | + | + | ++ | ++ | + | + |
| Exon 8 | c.1072G>T | V358F | Het | Yes | − | − | P | − | + | + | + | ++ | ++ | + | + |
| Exon 17 | c.2282C>T | T761I | Het | Yes | − | − | UC | − | − | − | + | − | ++ | + | + |
| Exon 8 | c.976C>T | L326F | Het | Yes | − | − | UC | − | + | + | + | ++ | ++ | + | + |
| Exon 8 | c.374C>T | S125L | Het | Yes | 0.00001 | − | UC | − | + | − | + | − | ++ | + | + |
| Exon 2 | c.3G>A | M1I | Het | Yes | − | − | LP | − | + | − | + | − | ++ | − | + |
| Exon 3 | c.238C>T | R80C | Het | Yes | 0.000004 | − | UC | − | − | + | + | − | ++ | + | + |
| Exon 7 | c.817G>A | V273M | Het | No | 0 | − | LP | LP | + | + | + | ++ | ++ | + | + |
| Exon 9 | c.1271G>A | R424H | Het | Yes | 0.00003 | 0.0003 | UC | − | + | − | + | − | ++ | + | + |
| Exon 16 | c.2104C>A | P702T | Het | Yes | − | − | UC | − | + | + | + | + | ++ | + | + |
| Exon 9 | c.1097C>T | P366L | Het | No | − | − | UC | LP | + | − | + | − | ++ | + | + |
| Exon 9 | c.1097C>T | P366L | Het | No | − | − | LP | LP | + | − | + | − | ++ | + | + |
In SIFT column, +: damaging; −: tolerated. In PolyPhen_2 column, +: probably damaging; −: benign. In MutationTaster column, +: disease causing. In REVEL column, ++: pathogenic; +: likely pathogenic; −: uncertain. In M-CAP column, ++: pathogenic. In LRT column, +: deleterious; −: neutral. In GERP column, +: conserved. In nucleotide column, G: guanine; C: cytosine; T: thymine; A: adenine. In amino acid column, G: glycine; R: arginine; V: valine; F: phenylalanine; T: threonine; I: isoleucine; L: leucine; S: serine; M: methionine: C: cysteine; H: histidine; P: praline; Y: tyrosine; Q: glutamine; E: glutamic acid; D: aspartic acid. In Hom/het column, hom: homozygous; het: heterozygous. In ACMG column, P: pathogenic; LP: likely pathogenic; UC: uncertain. In Clinvar column, LP: likely pathogenic. In GnomeAD and 1000g2015aug_all columns, −: unknown. ACMG: American College of Medical Genetics and Genomics
Table 2.
Pathogenicity analysis of splice site mutations of fibroblast growth factor receptor 1 in congenital hypogonadotropic hypogonadism patients
| Site | Nucleotide | Amino acid | Hom/het | Novel | GnomeAD | 1000g2015aug_all | ACMG | Clinvar | SPIDEX | Calculation |
|---|---|---|---|---|---|---|---|---|---|---|
| Exon 13 | c.1854+1G>C | Splicing | Het | Yes | − | − | Pathogenic | − | + | + |
| Exon 4 | c.359-1G>A | Splicing | Het | Yes | − | − | Pathogenic | − | + | + |
In GnomeAD/1000g2015aug_all/Clinvar column, −: none. In SPIDEX column, +: pathogenic. In calculation column, +: pathogenic. In nucleotid column, G: guanine; C: cytosine; T: thymine; A: adenine. In Hom/het column, Hom: homozygous; het: heterozygous. ACMG: American College of Medical Genetics and Genomics
An FGFR1 protein alignment, facilitated by GERP, revealed the strict conservation of all amino acids at the identified loci in this study (Table 1).
In summary, novel LP variants were categorized into three groups: missense variants (n=8), including G260R, V358F, T761I, L326F, S125L, M1I, R80C, and P702T; splice site variants (n=2), namely c.1854+1G>C and c.359-1G>A; and insertions or deletions (n=6), including c.536delC, c.838dupT, c.2226dupA, c.54_55del, c.250_264delGAGGAGGTGGAGGTG, and c.325_342delAGTGACACCACCTACTTC (Supplementary Table 2). In total, our clinical study identified sixteen novel LP variants among 19 CHH patients, including 15 males and 4 females.
Supplementary Table 2.
Types of 16 novel mutations in fibroblast growth factor receptor 1
| Mutation type | Nucleotide | Amino acid | Pathogenic |
|---|---|---|---|
| Missense mutations | c.778G>C | G260R | LP |
| Missense mutations | c.1072G>T | V358F | LP |
| Missense mutations | c.2282C>T | T761I | LP |
| Missense mutations | c.976C>T | L326F | LP |
| Missense mutations | c.374C>T | S125L | LP |
| Missense mutations | c.3G>A | M1I | LP |
| Missense mutations | c.238C>T | R80C | LP |
| Missense mutations | c.2104C>A | P702T | LP |
| Splice site mutation | c.1854+1G>C | Splicing | LP |
| Splice site mutation | c.359-1G>A | Splicing | LP |
| Deletions | c.536delC | P179Lfs*13 | LP |
| Insertions | c.2226dupA | Q743Tfs*57 | LP |
| Insertions | c.838dupT | Y280Lfs*2 | LP |
| Deletions | c.54_55del | C19Hfs*3 | LP |
| Deletions | c.250_264delGAGGAGGTGGAGGTG | 84_88delEEVEV | LP |
| Deletions | c.325_342delAGTGACACCACCTACTTC | 109_114delSDTTYF | LP |
Nucleotid - G: guanine; C: cytosine; T: thymine; A: adenine. Amino acid - G: glycine; R: arginine; V: valine; F: phenylalanine; T: threonine; I: isoleucine; L: leucine; S: serine; M: methionine; C: cysteine; H: histidine; P: praline; Y: tyrosine; Q: glutamine; E: glutamic acid; D: aspartic acid. Pathogenic - LP: likely pathogenic
FGFR1 mutants inheritance mode
Eleven patients displayed mutations of inherited origin, constituting the inheritance group, while 8 patients manifested de novo mutations, forming the de novo group. It is noteworthy that all frameshift mutations exclusively occurred in the de novo group (Table 3).
Table 3.
Inheritance mode of fibroblast growth factor receptor 1 mutations
| Site | Nucleotide | Amino acid | Hom/het | Inheritance |
|---|---|---|---|---|
| Exon 7 | c.778G>C | G260R | Het | De novo |
| Exon 8 | c.1072G>T | V358F | Het | De novo |
| Exon 17 | c.2282C>T | T761I | Het | Paternal |
| Exon 8 | c.976C>T | L326F | Het | Maternal |
| Exon 8 | c.374C>T | S125L | Het | Paternal |
| Exon 2 | c.3G>A | M1I | Het | Paternal |
| Exon 3 | c.238C>T | R80C | Het | Paternal |
| Exon 7 | c.817G>A | V273M | Het | Maternal |
| Exon 9 | c.1271G>A | R424H | Het | Maternal |
| Exon 16 | c.2104C>A | P702T | Het | Paternal |
| Exon 9 | c.1097C>T | P366L | Het | Paternal |
| Exon 9 | c.1097C>T | P366L | Het | Maternal |
| Exon 5 | c.536delC | P179Lfs*13 | Het | De novo |
| Exon 7 | c.838dupT | Y280Lfs*2 | Het | De novo |
| Exon 17 | c.2226dupA | Q743Tfs*57 | Het | De novo |
| Exon 2 | c.54_55del | C19Hfs*3 | Het | De novo |
| Exon 13 | c.1854+1G>C | Splicing | Het | De novo |
| Exon 4 | c.359-1G>A | Splicing | Het | Paternal |
| Exon 3 | c.250_264delGAGGAGGTGGAGGTG | 84_88delEEVEV | Het | De novo |
| Exon 3 | c.325_342delAGTGACACCACCTACTTC | 109_114delSDTTYF | Het | Paternal |
In nucleotide column, G: guanine; C: cytosine; T: thymine; A: adenine. In amino acid column, G: glycine; R: arginine; V: valine; F: phenylalanine; T: threonine; I: isoleucine; L: leucine; S: serine; M: methionine; C: cysteine; H: histidine; P: praline; Y: tyrosine; Q: glutamine; E: glutamic acid; D: aspartic acid. In Hom/het column, Hom: homozygous; het: heterozygous
Clinical characteristics
Eighteen patients carrying mutations in FGFR1 were included in this study to investigate the association between the type of variants and specific symptoms. Notably, hearing loss was exclusively observed in patients harboring frameshift mutations (Supplementary Table 3). Among the 14 male patients, 8 were classified in the inheritance group, while 6 belonged to the de novo group. The incidence of cryptorchidism displayed a somewhat lower trend in the inheritance group compared to the de novo group (25.0% vs 33.3%). Patients with short stature and hearing loss were solely found in the de novo group. The baseline testicular volume was greater in the inheritance group (2.3 ml vs 1.6 ml). Moreover, the inheritance group appeared to exhibit higher levels of LH (mean ± s.d.: 0.5 ± 0.4 IU l−1 vs 0.2 ± 0.1 IU l−1) and testosterone (mean ± s.d.: 1.3 ± 1.0 nmol l−1 vs 0.9 ± 0.3 nmol l−1) than the de novo group (both P>0.05). Furthermore, the FSH levels were notably higher in the inheritance group (mean ± s.d.: 1.0 ± 0.5 IU l−1 vs 0.5 ± 0.2 IU l−1; P = 0.03; Table 4).
Supplementary Table 3.
The relationship between fibroblast growth factor receptor 1 mutation types and symptoms
| Clinical characteristic | Missense mutation (n=10) | Splice site mutation (n=2) | Frameshift mutation (n=4) | Deletion mutation (n=2) |
|---|---|---|---|---|
| Incidence of cryptorchidism, n (%) | 2 (20) | 1 (50) | 1 (25) | 0 |
| Incidence of short stature, n (%) | 0 | 0 | 2 (50) | 0 |
| Incidence of skeletal anomalies, n (%) | 1 (10) | 0 | 2 (50) | 0 |
| Incidence of hearing loss, n (%) | 0 | 0 | 1 (25) | 0 |
| Abnormal olfactory bulb and tract, n (%) | 6 (60) | 1 (50) | 2 (50) | 0 |
n: number of congenital hypogonadotropic hypogonadism patients
Table 4.
Comparison of clinical characteristics of male congenital hypogonadotropic hypogonadism patients in inheritance and de novo groups
| Clinical characteristic | Inheritance group (n=8) | De novo group (n=6) | P |
|---|---|---|---|
| Age at diagnosis (year), mean±s.d. | 17.8±4.5 | 17.0±2.1 | 0.68 |
| BMI (kg m−2), mean±s.d. | 24.2±5.8 | 22.2±4.2 | 0.50 |
| Incidence of cryptorchidism, n (%) | 2 (25.0) | 2 (33.3) | 0.99 |
| Incidence of short stature, n (%) | 0 (0) | 1 (16.7) | 0.42 |
| Incidence of skeletal anomalies, n (%) | 1 (12.5) | 2 (33.3) | 1.00 |
| Incidence of hearing loss, n (%) | 0 (0) | 1 (16.7) | 0.42 |
| Abnormal olfactory bulb and tract, n (%) | 4 (50.0) | 4 (66.7) | 0.62 |
| Baseline testis size (ml), mean±s.d. | 2.3±1.7 | 1.6±1.3 | 0.38 |
| Baseline LH (IU l−1), mean±s.d. | 0.5±0.4 | 0.2±0.1 | 0.07 |
| Baseline FSH (IU l−1), mean±s.d. | 1.0±0.5 | 0.5±0.2 | 0.03 |
| Baseline testosterone (nmol l−1), mean±s.d. | 1.3±1.0 | 0.9±0.3 | 0.22 |
BMI: body mass index; LH: luteinizing hormone; FSH: follicle-stimulating hormone; s.d.: standard deviation
Among the four female patients, two were allocated to the inheritance group, and two to the de novo group. Patients with anosmia and skeletal abnormalities were exclusively identified in the inheritance group, whereas short stature was observed solely in the de novo group (Supplementary Table 4). Analogous to male patients, the inheritance group displayed elevated levels of LH (0.4 IU l−1 vs 0.3 IU l−1), FSH (2.0 IU l−1 vs 0.4 IU l−1), and estradiol (18.5 pg ml−1 vs 17.0 pg ml−1) in comparison to the de novo group.
Supplementary Table 4.
Clinical characteristics of female congenital hypogonadotropic hypogonadism patients in inheritance and de novo groups
| Patient | Age at diagnosis (years) | Diagnosis | Comorbidities | BMI (kg m−2) | LH (IU/l) (baseline) | FSH (IU/l) (baseline) | Estradiol (pg/ml) (baseline) | Inheritance |
|---|---|---|---|---|---|---|---|---|
| Female 1 | 14 | KS | - | 16.9 | 0.65 | 2.40 | 22 | Yes |
| Female 2 | 13 | nCHH | Skeletal abnormalities | 17.3 | 0.20 | 1.54 | 15 | Yes |
| Female 3 | 15 | nCHH | - | 23.1 | 0.20 | 0.24 | 19 | No |
| Female 4 | 15 | nCHH | Short stature | 20.3 | 0.48 | 0.64 | 15 | No |
Comorbidities (-): no. KS: Kallmann syndrome; nCHH: normosmic congenital hypogonadotropic hypogonadism; BMI: body mass index; LH: luteinizing hormone; FSH: follicle-stimulating hormone
Treatment and spermatogenesis outcomes
Among the 14 male patients, four with deleterious mutations in FGFR1 (V273M, T761I, c.1854+1G>C, and Q743Tfs*57) underwent testosterone-only treatment, while ten patients (six in the inheritance group and four in the de novo group) received sperm-inducing therapy due to their initial inability to ejaculate. Following treatment, significant increases were observed in testicular volume (mean ± s.d.) from 2.6 ± 1.3 ml to 6.6 ± 3.9 ml, P < 0.05) and sperm concentration.
For patients receiving human chorionic gonadotropin and human menopausal gonadotropin (hCG/HMG) treatment, successful sperm production was observed in two patients from the inheritance group, while one patient from the de novo group did not achieve this outcome. In contrast, among patients subjected to pulsatile GnRH therapy, successful spermatogenesis was observed in 3 out of 4 patients from the inheritance group and 2 out of 3 patients from the de novo group (Table 5).
Table 5.
Sperm-inducing treatment information for 10 male congenital hypogonadotropic hypogonadism patients carrying mutations in fibroblast growth factor receptor 1 (FGFR1)
| Patient | Inheritance | TRT | TRT time (month) | ST | Before spermatogenesis therapy | Age (year) | Time required for spermatogenesis (month) | Last visit during spermatogenesis therapy | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||||
| LH (IU l−1) | FSH (IU l−1) | Testosterone (ng ml−1) | TV (ml) | Sperm concentration (×106 ml−1) | LH (IU l−1) | FSH (IU l−1) | Testosterone (ng ml−1) | TV (ml) | Sperm concentration (×106 ml−1) | |||||||
| Male 1 | + | + | 24 | hCG/HMG | 0.20 | 1.10 | 3.46 | 2.0 | − | 24 | 36 | 0.65 | 2.36 | 1.7 | 5 | 2.00 |
| Male 2 | + | + | 17 | hCG/HMG | 0.90 | 2.20 | 1.28 | 3.0 | − | 18 | 32 | 0.78 | 2.85 | 7.0 | 6 | 7.79 |
| Male 3 | + | + | 16 | GnRH | 0.20 | 0.32 | 0.49 | 2.0 | − | 29 | 11 | 8.45 | 13.66 | 3.9 | 3 | 9.92 |
| Male 4 | + | + | 36 | GnRH | 0.60 | 1.30 | 5.90 | 3.0 | − | 17 | 5 | 13.00 | 8.00 | 7.4 | 15 | 10.73 |
| Male 5 | + | + | 24 | GnRH | 0.76 | 1.80 | 2.00 | 3.0 | − | 26 | 10 | 3.14 | 3.81 | 2.1 | 6 | 4.00 |
| Male 6 | + | + | 48 | GnRH | 0.20 | 0.26 | 4.42 | 5.0 | − | 17 | − | 11.06 | 8.63 | 3.7 | 5 | 0 |
| Male 7 | − | + | 24 | GnRH | 0.20 | 0.30 | 1.30 | 4.0 | − | 19 | 9 | 4.85 | 11.92 | 2.1 | 5 | 2.00 |
| Male 8 | − | + | 65 | GnRH | 0.20 | 0.20 | 3.14 | 1.0 | − | 27 | 10 | 9.52 | 26.42 | 3.3 | 6 | 7.97 |
| Male 9 | − | + | 36 | hCG/HMG | 0.20 | 0.20 | 3.80 | 2.0 | − | 20 | − | 0.84 | 2.25 | 2.2 | 7 | 0 |
| Male 10 | − | + | 5 | GnRH | 0.20 | 0.30 | 0.90 | 0.5 | − | 17 | − | 5.78 | 2.68 | 1.7 | 8 | 0 |
In inheritance column, +: mutations derived from parents; −: de novo mutants. In sperm concentration column, −: none. In time required for spermatogenesis column, −: none. TRT: testosterone replacement therapy; ST: spermatogenesis therapy; hCG/HMG: human chorionic gonadotropin and human menopausal gonadotropin; GnRH: gonadotropin-releasing hormone; LH: luteinizing hormone; FSH: follicle-stimulating hormone; TV: testis volume
In summary, sperm-inducing therapy proved effective in enhancing ejaculation capability, increasing testicular volume, and potentially improving spermatogenesis. Of note, 5 patients (83.3%) in the inheritance group and 2 patients (50.0%) in the de novo group achieved successful spermatogenesis. The final testicular volumes (mean ± s.d.) for the inheritance and de novo groups were 7.0 ± 4.6 ml and 5.5 ± 0.7 ml, respectively. The average follow-up durations for the inheritance and de novo groups were 18.8 months and 9.5 months, respectively (Supplementary Table 5).
Supplementary Table 5.
Comparison of spermatogenesis outcomes in patients with inheritance and de novo mutations
| Clinical characteristic | Inheritance group (n=6) | De novo group (n=4) | P |
|---|---|---|---|
| Spermatogenesis success rate, n (%) | 5 (83.3) | 2 (50.0) | 0.50 |
| Basal TV (ml), mean±s.d. | 3.0±1.1 | 1.9±1.6 | 0.21 |
| TV (ml) at spermatogenesis, mean±s.d. | 7.0±4.6 | 5.5±0.7 | 0.68 |
| Time required for spermatogenesis (months), mean±s.d. | 18.8±14.2 | 9.5±0.7 | 0.21 |
n: number of congenital hypogonadotropic hypogonadism patients; s.d.: standard deviation; TV: testicular volume
Furthermore, it is worth highlighting that pulsatile GnRH therapy appeared to require less time to yield sperm compared to hCG/HMG therapy in seven male CHH patients with FGFR1 mutations who reported successful spermatogenesis (9.0 months vs 34.0 months; P < 0.05).
DISCUSSION
In a cohort of 210 CHH patients from a single medical center, we found 19 individuals bearing 19 LP variants in the FGFR1 gene. Notably, almost 60% of these FGFR1 mutations were inherited from phenotypically normal parents. Comparative analysis of LH, FSH, T, or estradiol levels between the inheritance and de novo groups unveiled higher hormone levels in the inheritance group, implying a less severe impairment of the gonadal axis function in this group. Additionally, extra-axial manifestations, such as short stature, exhibited milder presentations in the inheritance group. Concerning spermatogenesis, the inheritance group displayed a notable trend toward higher success rates and larger testicular volume during follow-up.
The prevalence of FGFR1 LP variants in this CHH series was 9.0%, a figure consistent with those reported in other studies.13,33 While no in vitro functional studies were conducted, the collective evidence consistently suggests that these 16 novel variants may detrimentally impact gene function.
For eight missense variants, which had not been previously reported in CHH patients, the application of ACMG guidelines and bioinformatics analysis indicated their likely pathogenic nature. First, these variants demonstrated a high degree of conservation, as evidenced by GERP scores. Second, the majority of in silico tools predicted their pathogenicity.
The presence of signal peptide region variations can influence the intracellular trafficking of FGFR1, with the M1I (methionine substituted by isoleucine) substitution occurring within this signal peptide region.12 The interaction between the acid box and the heparan sulfate-binding region on the D2 domain has the potential to facilitate the folding of D1 onto D2 and D3, impacting interactions with FGFs.34 In an independent study by Sanchez-Heras et al.35 a decrease in binding between FGFR1 and neurocalcineurin, a neuronal cell adhesion molecule, was observed in the absence of the AB region, leading to neuronal migration failures. Mutations such as R80C, situated in the D1 region, and S125L, located in the AB region, may perturb the autoinhibition mechanisms of FGFR136 and subsequently contribute to abnormal migration of GnRH neurons and olfactory neurons.
The ligand binding site, formed by D2, D3, and the junction connecting the two,37 appears to be influenced by mutations such as G260R and L326F, positioned in D2 and D3. These mutations may reduce the ligand’s affinity for the FGFR1 protein, thus affecting signal transduction and protein activation.13,38 The P366L mutation, situated at the junction of D3 and the transmembrane region, has been demonstrated to be associated with KS.13 Interestingly, V358F is located in the same domain as P366L, and is suspected to disrupt protein function and consequently influence GnRH neurons’ function.
Mutations like P702T and T761I, located in the TKD domain, are likely to alter the molecular size, electronic charge, and hydrophobicity, potentially disrupting the three-dimensional protein structure and impairing protein function.12 These 16 novel variants were categorized as detrimental mutants, although in vitro functional studies are warranted for further validation.
Our investigation revealed a 28.6% incidence of cryptorchidism in participants with FGFR1 mutations, consistent with the 13.0%–60.0% range reported in other studies.8,30 Notably, the incidence of cryptorchidism was slightly lower in the inheritance group compared to the de novo group (25.0% and 33.3%, respectively). Furthermore, the inheritance group exhibited higher levels of gonadotropins and sex hormones than the de novo group.
FGFR1 mutations may manifest in extra-gonadal features, with short stature occurring in 15.0% of CHH patients bearing FGFR1 mutations,39 skeletal anomalies in 23.0%,40 and hearing loss in one-third (33.3%) of cases.41 In our study, the inheritance group displayed less severe phenotypes compared to the de novo group.
Notably, seminiferous tubules contribute significantly to testicular volume, with larger testicular size correlating with heightened sperm concentration.42 Therefore, the augmentation in testicular size during treatment can serve as a vital indicator of the potential for spermatogenesis.43,44,45,46 Our investigation found that patients exhibited increased testicular size and yielded sperm in the ejaculate following sperm-inducing therapy, consistent with previous findings.47,48,49
In general, the rates of successful spermatogenesis in CHH patients with FGFR1 mutations range from 76.0% to 80.0%.17,50,51 In our study, the two groups demonstrated distinct responses to spermatogenic treatment, with spermatogenesis rates of 83.3% and 50.0% in the inheritance and de novo groups, respectively. FGFR1 signaling plays a pivotal role in spermatogenesis, and FGFR1-inactivating mutations significantly diminish daily sperm output.52 Notably, our study revealed that the inheritance group exhibited a trend toward more favorable spermatogenic responses and larger testicular volume. Additionally, pulsatile GnRH therapy was associated with earlier spermatogenesis when compared to combined gonadotropin therapy,53,54 a finding corroborated in our male CHH patients carrying FGFR1 mutations. These cumulative evidence suggest that mutations in the inheritance group appear to have less detrimental effects on FGFR1 protein function and result in milder clinical manifestations.
Nonetheless, it is imperative to acknowledge several limitations in our study. First, the pathogenicity of FGFR1 variants was assessed primarily based on ACMG guidelines and bioinformatics tools, with a lack of in vitro functional and morphological assays. Second, the limited number of female patients precluded a comprehensive analysis of differences between the two groups. Third, although the observed differences offer valuable insights into individualized therapy, the data did not attain statistical significance. Future studies encompassing larger sample sizes may ameliorate these limitations.
In conclusion, our investigation identified 19 patients harboring LP variants in FGFR1, with 11 out of the 19 variants being inherited from phenotypically normal parents. The inheritance group exhibited less pronounced damage to the pituitary–gonadal axis function, evident through a lower incidence of cryptorchidism, elevated levels of gonadotropins and testosterone, and a more favorable response to sperm induction therapy. This study broadens our knowledge of loss-of-function FGFR1 variants and enhances our comprehension of the pathogenesis of CHH induced by FGFR1 mutations.
AUTHOR CONTRIBUTIONS
JFM and XYW conceived and designed the experiments. JFM, XW, and HLM collected clinical data. YFY analyzed data and drafted the manuscript. JFM, XYW, and MN revised the paper. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declare no competing interests.
ACKNOWLEDGMENTS
This work was supported by Natural Science Foundation of Beijing (grant No. 7212080), National Natural Science Foundation of China (grant No. 81971375), National High Level Hospital Clinical Research Funding (2022-PUMCH-D-002), CAMS Innovation Fund for Medical Sciences (CIFMS 2021-I2M-1-003), and National High Level Hospital Clinical Research Funding (2022-PUMCH-C-028). We would like to thank all patients and their family members for participating in the study.
Supplementary Information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Al Sayed Y, Howard SR. Panel testing for the molecular genetic diagnosis of congenital hypogonadotropic hypogonadism –a clinical perspective. Eur J Hum Genet. 2023;31:387–94. doi: 10.1038/s41431-022-01261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young J, Xu C, Papadakis GE, Acierno JS, Maione L, et al. Clinical management of congenital hypogonadotropic hypogonadism. Endocr Rev. 2019;40:669–710. doi: 10.1210/er.2018-00116. [DOI] [PubMed] [Google Scholar]
- 3.Boehm U, Bouloux PM, Dattani MT, de Roux N, Dodé C, et al. Expert consensus document: European Consensus Statement on congenital hypogonadotropic hypogonadism –pathogenesis, diagnosis and treatment. Nat Rev Endocrinol. 2015;11:547–64. doi: 10.1038/nrendo.2015.112. [DOI] [PubMed] [Google Scholar]
- 4.Federici S, Cangiano B, Goggi G, Messetti D, Munari EV, et al. Genetic and phenotypic differences between sexes in congenital hypogonadotropic hypogonadism (CHH):large cohort analysis from a single tertiary centre. Front Endocrinol (Lausanne) 2022;13:965074. doi: 10.3389/fendo.2022.965074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lima Amato LG, Latronico AC, Gontijo Silveira LF. Molecular and genetic aspects of congenital isolated hypogonadotropic hypogonadism. Endocrinol Metab Clin North Am. 2017;46:283–303. doi: 10.1016/j.ecl.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Maione L, Dwyer AA, Francou B, Guiochon-Mantel A, Binart N, et al. Genetics in endocrinology:genetic counseling for congenital hypogonadotropic hypogonadism and Kallmann syndrome:new challenges in the era of oligogenism and next-generation sequencing. Eur J Endocrinol. 2018;178:R55–80. doi: 10.1530/EJE-17-0749. [DOI] [PubMed] [Google Scholar]
- 7.Turkyilmaz A, Cayir A, Yarali O, Kurnaz E, Kartal Baykan E, et al. Clinical characteristics and molecular genetic analysis of a cohort with idiopathic congenital hypogonadism. J Pediatr Endocrinol Metab. 2021;34:771–80. doi: 10.1515/jpem-2020-0590. [DOI] [PubMed] [Google Scholar]
- 8.Salenave S, Chanson P, Bry H, Pugeat M, Cabrol S, et al. Kallmann's syndrome:a comparison of the reproductive phenotypes in men carrying KAL1 and FGFR1/KAL2 mutations. J Clin Endocrinol Metab. 2008;93:758–63. doi: 10.1210/jc.2007-1168. [DOI] [PubMed] [Google Scholar]
- 9.Xie Y, Su N, Yang J, Tan Q, Huang S, et al. FGF/FGFR signaling in health and disease. Signal Transduct Target Ther. 2020;5:181. doi: 10.1038/s41392-020-00222-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim SH, Hu Y, Cadman S, Bouloux P. Diversity in fibroblast growth factor receptor 1 regulation:learning from the investigation of Kallmann syndrome. J Neuroendocrinol. 2008;20:141–63. doi: 10.1111/j.1365-2826.2007.01627.x. [DOI] [PubMed] [Google Scholar]
- 11.Misrahi M. β-Klotho sustains postnatal GnRH biology and spins the thread of puberty. EMBO Mol Med. 2017;9:1334–7. doi: 10.15252/emmm.201708180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonçalves C, Bastos M, Pignatelli D, Borges T, Aragüés JM, et al. Novel FGFR1 mutations in Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism:evidence for the involvement of an alternatively spliced isoform. Fertil Steril. 2015;104:1261–7.e1. doi: 10.1016/j.fertnstert.2015.07.1142. [DOI] [PubMed] [Google Scholar]
- 13.Trarbach EB, Costa EM, Versiani B, de Castro M, Baptista MT, et al. Novel fibroblast growth factor receptor 1 mutations in patients with congenital hypogonadotropic hypogonadism with and without anosmia. J Clin Endocrinol Metab. 2006;91:4006–12. doi: 10.1210/jc.2005-2793. [DOI] [PubMed] [Google Scholar]
- 14.Ohtaka K, Fujisawa Y, Takada F, Hasegawa Y, Miyoshi T, et al. FGFR1 analyses in four patients with hypogonadotropic hypogonadism with split-hand/foot malformation:implications for the promoter region. Hum Mutat. 2017;38:503–6. doi: 10.1002/humu.23178. [DOI] [PubMed] [Google Scholar]
- 15.Akkuş G, Kotan LD, Durmaz E, Mengen E, Turan İ, et al. Hypogonadotropic hypogonadism due to novel FGFR1 mutations. J Clin Res Pediatr Endocrinol. 2017;9:95–100. doi: 10.4274/jcrpe.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koenigkam-Santos M, Santos AC, Versiani BR, Diniz PR, Junior JE, et al. Quantitative magnetic resonance imaging evaluation of the olfactory system in Kallmann syndrome:correlation with a clinical smell test. Neuroendocrinology. 2011;94:209–17. doi: 10.1159/000328437. [DOI] [PubMed] [Google Scholar]
- 17.Nie M, Yu B, Chen R, Sun B, Mao J, et al. Novel rare variants in FGFR1 and clinical characteristics analysis in a series of congenital hypogonadotropic hypogonadism patients. Clin Endocrinol (Oxf) 2021;95:153–62. doi: 10.1111/cen.14436. [DOI] [PubMed] [Google Scholar]
- 18.Lin Y, Jiang JB, Xia B, Cao J, Yu AZ, et al. [Alveolar capillary dysplasia with misalignment of the pulmonary veins:a case report and literature review. Zhonghua Er Ke Za Zhi. 2020;58:838–42. doi: 10.3760/cma.j.cn112140-20200427-00441. [Ariticle in Chinese] [DOI] [PubMed] [Google Scholar]
- 19.Hanany M, Yang RR, Lam CM, Beryozkin A, Sundaresan Y, et al. An in-depth single-gene worldwide carrier frequency and genetic prevalence analysis of CYP4V2 as the cause of Bietti crystalline dystrophy. Transl Vis Sci Technol. 2023;12:27. doi: 10.1167/tvst.12.2.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huber CD, Kim BY, Lohmueller KE. Population genetic models of GERP scores suggest pervasive turnover of constrained sites across mammalian evolution. PLoS Genet. 2020;16:e1008827. doi: 10.1371/journal.pgen.1008827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richards S, Aziz N, Bale S, Bick D, Das S, et al. Standards and guidelines for the interpretation of sequence variants:a joint consensus recommendation of the American College of Medical Genetics and Genomics and the association for molecular pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng PC, Henikoff S. SIFT:predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013 doi: 10.1002/0471142905.hg0720s76. Chapter 7: Unit7.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–6. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 25.Tian Y, Pesaran T, Chamberlin A, Fenwick RB, Li S, et al. REVEL and BayesDel outperform other in silico meta-predictors for clinical variant classification. Sci Rep. 2019;9:12752. doi: 10.1038/s41598-019-49224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarnovskaya SI, Kostareva AA, Zhorov BS. L-type calcium channel:predicting pathogenic/likely pathogenic status for variants of uncertain clinical significance. Membranes (Basel) 2021;11:599. doi: 10.3390/membranes11080599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–61. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riepe TV, Khan M, Roosing S, Cremers FP, ‘t Hoen PA. Benchmarking deep learning splice prediction tools using functional splice assays. Hum Mutat. 2021;42:799–810. doi: 10.1002/humu.24212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato C, Fujii K, Arai Y, Hatsuse H, Nagao K, et al. Nevoid basal cell carcinoma syndrome caused by splicing mutations in the PTCH1 gene. Fam Cancer. 2017;16:131–8. doi: 10.1007/s10689-016-9924-2. [DOI] [PubMed] [Google Scholar]
- 30.Pitteloud N, Meysing A, Quinton R, Acierno JS, Jr, Dwyer AA, et al. Mutations in fibroblast growth factor receptor 1 cause Kallmann syndrome with a wide spectrum of reproductive phenotypes. Mol Cell Endocrinol. 2006;254-255:60–9. doi: 10.1016/j.mce.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 31.Chen J, Yuan K, He MF, Wang CL, Chen C, et al. [Clinical and genetic features of Kallmann syndrome:an analysis of 5 cases. Zhongguo Dang Dai Er Ke Za Zhi. 2018;20:925–9. doi: 10.7499/j.issn.1008-8830.2018.11.009. [Ariticle in Chinese] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abir R, Garor R, Felz C, Nitke S, Krissi H, et al. Growth hormone and its receptor in human ovaries from fetuses and adults. Fertil Steril. 2008;90:1333–9. doi: 10.1016/j.fertnstert.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 33.Wang D, Niu Y, Tan J, Chen Y, Xu H, et al. Combined in vitro and in silico analyses of FGFR1 variants:genotype-phenotype study in idiopathic hypogonadotropic hypogonadism. Clin Genet. 2020;98:341–52. doi: 10.1111/cge.13814. [DOI] [PubMed] [Google Scholar]
- 34.Wang F, Kan M, Yan G, Xu J, McKeehan WL. Alternately spliced NH2-terminal immunoglobulin-like Loop I in the ectodomain of the fibroblast growth factor (FGF) receptor 1 lowers affinity for both heparin and FGF-1. J Biol Chem. 1995;270:10231–5. doi: 10.1074/jbc.270.17.10231. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez-Heras E, Howell FV, Williams G, Doherty P. The fibroblast growth factor receptor acid box is essential for interactions with N-cadherin and all of the major isoforms of neural cell adhesion molecule. J Biol Chem. 2006;281:35208–16. doi: 10.1074/jbc.M608655200. [DOI] [PubMed] [Google Scholar]
- 36.Opalinski L, Szczepara M, Sokolowska-Wedzina A, Zakrzewska M, Otlewski J. The autoinhibitory function of D1 domain of FGFR1 goes beyond the inhibition of ligand binding. Int J Biochem Cell Biol. 2017;89:193–8. doi: 10.1016/j.biocel.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 37.Groth C, Lardelli M. The structure and function of vertebrate fibroblast growth factor receptor 1. Int J Dev Biol. 2002;46:393–400. [PubMed] [Google Scholar]
- 38.Pitteloud N, Acierno JS, Jr, Meysing A, Eliseenkova AV, Ma J, et al. Mutations in fibroblast growth factor receptor 1 cause both Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci U S A. 2006;103:6281–6. doi: 10.1073/pnas.0600962103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao J, Chen R, Wu X, Liu Z, Zheng J, et al. [Association of the extra-gonadal manifestations with different pathogenic gene mutations in male patients with congenital hypogonadotropic hypogonadism. Zhonghua Yi Xue Za Zhi. 2015;95:3424–7. [Ariticle in Chinese] [PubMed] [Google Scholar]
- 40.Costa-Barbosa FA, Balasubramanian R, Keefe KW, Shaw ND, Al-Tassan N, et al. Prioritizing genetic testing in patients with Kallmann syndrome using clinical phenotypes. J Clin Endocrinol Metab. 2013;98:E943–53. doi: 10.1210/jc.2012-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zenaty D, Bretones P, Lambe C, Guemas I, David M, et al. Paediatric phenotype of Kallmann syndrome due to mutations of fibroblast growth factor receptor 1 (FGFR1) Mol Cell Endocrinol. 2006;254–5:78–83. doi: 10.1016/j.mce.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 42.Fuse H, Akashi T, Kazama T, Katayama T. Gonadotropin therapy in males with hypogonadotropic hypogonadism:factors affecting induction of spermatogenesis after gonadotropin replacement. Int Urol Nephrol. 1996;28:367–74. doi: 10.1007/BF02550500. [DOI] [PubMed] [Google Scholar]
- 43.Warne DW, Decosterd G, Okada H, Yano Y, Koide N, et al. A combined analysis of data to identify predictive factors for spermatogenesis in men with hypogonadotropic hypogonadism treated with recombinant human follicle-stimulating hormone and human chorionic gonadotropin. Fertil Steril. 2009;92:594–604. doi: 10.1016/j.fertnstert.2008.07.1720. [DOI] [PubMed] [Google Scholar]
- 44.Liu PY, Baker HW, Jayadev V, Zacharin M, Conway AJ, et al. Induction of spermatogenesis and fertility during gonadotropin treatment of gonadotropin-deficient infertile men:predictors of fertility outcome. J Clin Endocrinol Metab. 2009;94:801–8. doi: 10.1210/jc.2008-1648. [DOI] [PubMed] [Google Scholar]
- 45.Farhat R, Al-zidjali F, Alzahrani AS. Outcome of gonadotropin therapy for male infertility due to hypogonadotrophic hypogonadism. Pituitary. 2010;13:105–10. doi: 10.1007/s11102-009-0203-1. [DOI] [PubMed] [Google Scholar]
- 46.Miyagawa Y, Tsujimura A, Matsumiya K, Takao T, Tohda A, et al. Outcome of gonadotropin therapy for male hypogonadotropic hypogonadism at university affiliated male infertility centers:a 30-year retrospective study. J Urol. 2005;173:2072–5. doi: 10.1097/01.ju.0000158133.09197.f4. [DOI] [PubMed] [Google Scholar]
- 47.Schopohl J, Mehltretter G, von Zumbusch R, Eversmann T, von Werder K. Comparison of gonadotropin-releasing hormone and gonadotropin therapy in male patients with idiopathic hypothalamic hypogonadism. Fertil Steril. 1991;56:1143–50. [PubMed] [Google Scholar]
- 48.Sahib BO, Hussein IH, Alibrahim NT, Mansour AA. Management outcomes in males with hypogonadotropic hypogonadism treated with gonadotropins. Cureus. 2023;15:e35601. doi: 10.7759/cureus.35601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sachdev K, Agrawal S, Ish P, Gupta N, Raheja K. Neurological manifestations of COVID-19:a brief review. Indian J Med Res. 2020;152:41–7. doi: 10.4103/ijmr.IJMR_1395_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rastrelli G, Corona G, Mannucci E, Maggi M. Factors affecting spermatogenesis upon gonadotropin-replacement therapy:a meta-analytic study. Andrology. 2014;2:794–808. doi: 10.1111/andr.262. [DOI] [PubMed] [Google Scholar]
- 51.Li S, Zhao Y, Nie M, Ma W, Wang X, et al. Clinical characteristics and spermatogenesis in patients with congenital hypogonadotropic hypogonadism caused by FGFR1 mutations. Int J Endocrinol. 2020;2020:8873532. doi: 10.1155/2020/8873532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cotton L, Gibbs GM, Sanchez-Partida LG, Morrison JR, de Kretser DM, et al. FGRF-1 signaling is involved in spermiogenesis and sperm capacitation. J Cell Sci. 2006;119:75–84. doi: 10.1242/jcs.02704. [DOI] [PubMed] [Google Scholar]
- 53.Mao JF, Liu ZX, Nie M, Wang X, Xu HL, et al. Pulsatile gonadotropin-releasing hormone therapy is associated with earlier spermatogenesis compared to combined gonadotropin therapy in patients with congenital hypogonadotropic hypogonadism. Asian J Androl. 2017;19:680–5. doi: 10.4103/1008-682X.193568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei C, Long G, Zhang Y, Wang T, Wang S, et al. Spermatogenesis of male patients with congenital hypogonadotropic hypogonadism receiving pulsatile gonadotropin-releasing hormone therapy versus gonadotropin therapy: a systematic review and meta-analysis. World J Mens Health. 2021;39:654–65. doi: 10.5534/wjmh.200043. [DOI] [PMC free article] [PubMed] [Google Scholar]